Abstract
Mycobacterium marinum is closely related to Mycobacterium tuberculosis, the cause of tuberculosis in humans. M. marinum has become an important model system for the study of the molecular mechanisms involved in causing tuberculosis in humans. Through molecular genetic analysis of the differences between pathogenic and nonpathogenic mycobacteria, we identified two loci that affect the ability of M. marinum to infect macrophages, designated mel1 and mel2. In silico analyses of the 11 putative genes in these loci suggest that mel1 encodes secreted proteins that include a putative membrane protein and two putative transglutaminases, whereas mel2 is involved in secondary metabolism or biosynthesis of fatty acids. Interestingly, mel2 is unique to M. marinum and the M. tuberculosis complex and not present in any other sequenced mycobacterial species. M. marinum mutants with mutations in mel1 and mel2, constructed by allelic exchange, are defective in the ability to infect both murine and fish macrophage cell lines. These data suggest that the genes in mel1 and mel2 are important for the ability of M. marinum to infect host cells.
Although mycobacteria were among the first organisms associated with disease in humans (19, 23), they remain possibly the most important cause of death due to a single infectious agent throughout the world. Possible reasons for this are the relative difficulty of manipulating mycobacteria in the laboratory and the relatively low growth rate (∼20-h generation time) of the most important mycobacterial species, Mycobacterium tuberculosis. Investigators have sought appropriate models that are both relevant and easy to manipulate to speed progress in tuberculosis research. Recently, there has been a great deal of interest in Mycobacterium marinum as a model for study of M. tuberculosis pathogenesis because of its ease of genetic manipulation (2, 17, 33, 37), close genetic relationship to M. tuberculosis (36, 42, 46), relatively high growth rate (∼4-h generation time) (10), and the presence of a number of useful laboratory models for in vitro (16) and in vivo (14, 34, 38, 45) virulence studies.
Pathogenic mycobacterial species, such as M. tuberculosis, differ from nonpathogenic species, such as Mycobacterium smegmatis, in that they invade mammalian cells more efficiently (6, 13, 39), block lysosomal fusion (4, 20), and replicate well in eukaryotic cells (29, 40, 47, 48). Investigators have taken advantage of these differences to identify the genes involved in host cell interactions by cloning genomic DNA from pathogenic species into nonpathogenic mycobacterial species (6, 29, 47, 48). Although these studies have resulted in identification of genes that are potentially important, further analyses have been slowed by the fact that they were isolated from slow-growing pathogenic mycobacterial species. As a result, no strains have been constructed with mutations in these genes, and it remains unclear whether mutants would be defective for host cell interactions. Our group recently found that the relatively rapidly growing pathogenic mycobacterium M. marinum also differs from nonpathogenic mycobacteria in its ability to invade, prevent lysosomal fusion, and replicate in macrophages (16). However, no genetic analyses of these differences in host cell interactions have been carried out.
In the present study we further characterized the differences between M. marinum and M. smegmatis at the cellular and molecular level in order to better understand the mechanisms of host cell interaction by pathogenic mycobacteria. We observed significant differences in the abilities of M. marinum to adhere to and invade mammalian monocytic cell lines. Using the ability to infect the human monocytic cell line THP-1 as a selection, we identified two loci that confer enhanced host cell infection and designated them mel1 and mel2 for “mycobacterial enhanced infection loci. ” All of the genes in these loci are present in M. tuberculosis in nearly the same gene order. In addition, mel2 appears to be present only in M. marinum and M. tuberculosis complex. We constructed a specific M. marinum mutation in each of these loci by allelic exchange and demonstrated that the resulting mutants were defective in the ability to infect both fish and murine macrophages. These data indicate that mel1 and mel2 are involved in the ability of M. marinum to infect macrophages and support the use of M. marinum as a model to speed progress in molecular analysis of mycobacterial virulence mechanisms.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. marinum strain M, a clinical isolate obtained from the skin of a patient (32), was used in all studies. M. marinum was grown at 33°C in 7H9 broth (Difco, Detroit, Mich.) supplemented with 0.5% glycerol, 10% albumin-dextrose complex (ADC), and 0.05% Tween 80 (M-ADC-TW broth) for 5 days. M. smegmatis strain mc2155 (41) cultures were grown in the same manner for 3 days at 37°C. The number of viable bacteria was determined for each assay by the LIVE-DEAD assay (Molecular Probes, Eugene, Oreg.) and by plating dilutions for CFU on 7H9 (M-ADC) agar (Difco). All inocula used were >99% viable. Escherichia coli strains were grown in Luria-Bertani (LB; Difco) medium at 37°C. Where appropriate, sucrose was added at 10% weight per volume, kanamycin was added at a concentration of 25 μg/ml, and chloramphenicol was added at 10 μg/ml. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) was used at a concentration of 40 μg/ml in E. coli and 80 μg/ml in M. marinum.
Cell lines and culture conditions.
The murine cell lines J774A.1 (ATCC TIB67) and RAW264.7 (ATCC CRL-2278) were maintained at 37°C and 5% CO2 in high-glucose Dulbecco's modified Eagle medium (Gibco, Bethesda, Md.) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 2 mM l-glutamine. The human monocytic cell line THP-1 (ATCC TIB202) and the human epithelial cell line HEp-2 (ATCC CCL23) were maintained at 37°C and 5% CO2 in RPMI medium (Gibco) supplemented with 10% (THP-1) or 5% (HEp-2) heat-inactivated fetal bovine serum (Gibco) and 2 mM l-glutamine. The adherent carp monocyte/macrophage cell line CLC (European Collection of Cell Cultures, 95070628) was maintained at 28°C and 5% CO2 as described previously (16).
Uptake, cell association, and adherence assays.
Uptake and adherence assays with adherent macrophages and epithelial cells were carried out in 24-well tissue culture plates (Costar) as described previously (8, 9). J774A.1 and RAW264.7 cells were seeded at a density of 1 × 106 cells/well, CLC monocytes were seeded at 2.5 × 105 cells/well, and HEp-2 cells were seeded at 1 × 105 cells/well, 18 to 24 h prior to use. In the case of THP-1 cells, all assays were done in suspension, requiring that the cells be pelleted by centrifugation at 100 × g for 1 min before each change of solution, and 106 cells were used per tube. The medium was replaced before use for all cell types, and they were infected at a multiplicity of infection (MOI) of 10 in each assay. In most uptake assays the cells and bacteria were coincubated for 30 min at 37°C for all cells except fish monocytes, which were incubated at 28°C. In some assays the coincubation period was varied to test the effects of this time on the data obtained. After coincubation, the cells were washed twice with phosphate buffered saline (PBS) and incubated in fresh medium plus 200 μg of amikacin (Sigma Chemicals, St. Louis, Mo.)/ml for 2 h. The cells were then washed once with PBS and lysed using 1 ml of 0.1% Triton X-100 (Sigma) for 10 min. Dilutions were plated to determine intracellular CFU. Cell association assays were carried out in a similar manner, except that, after 30 min of coincubation, the cells were washed five times with PBS prior to lysis. We used two methods to measure adherence of the bacteria to host cells, immediate assays and fixed-cell adherence assays. Immediate assays were carried out in a manner similar to that of cell association assays except that, after the bacteria were added to the cells, they were immediately washed five times with PBS. This assay measures cell association at an early time point before uptake can occur. In order to ensure that uptake was not a significant aspect of the data obtained in these assays, we also utilized adherence assays with fixed cells, carried out as previously described (8). Basically, cells were fixed in 3.7% formaldehyde for 10 min at room temperature and washed three times with PBS prior to addition of bacteria. Bacteria were coincubated with fixed cells for 30 min, and then the cells were washed and lysed and CFU were determined. Triton X-100 (0.1%) had no effect on the viability of M. marinum or M. smegmatis, and all mycobacterial strains used displayed comparable levels of killing by amikacin.
Intracellular survival assays.
Intracellular survival assays were carried out in a manner similar to that of uptake assays, but after amikacin treatment fresh medium containing 30 μg of amikacin/ml was added. The cells were incubated at the appropriate temperature, lysed, and plated as described above at different time points. Survival is expressed as the percentage of CFU present at each time point compared to time zero (30 min), i.e., percent survival = (CFU Tx/CFU T0) × 100.
Construction of M. marinum total DNA contiguous cosmid library in M. smegmatis.
The shuttle cosmid pJDC16, which was used as the vector for this library, was constructed by cloning the mycobacterial origin of replication from pAL5000 (35) into the cosmid pYUB289 (9). The plasmid pMV262 (44) was digested with the restriction enzymes MluI and XbaI and ligated to the purified pYUB289 fragment containing the aminoglycoside transferase gene that confers kanamycin resistance after digestion with MluI and NheI (XbaI and NheI are compatible). M. marinum chromosomal DNA was digested with Sau3AI to produce fragments between 45 and 50 kbp in length. The fragments were dephosphorylated and ligated into the BamHI site of pJDC16. The resulting ligation was in vitro packaged with the Gigapack Gold III system (Stratagene) and used to infect E. coli strain χ2819 (9) for in vivo packaging. More than 10,000 kanamycin-resistant χ2819 colonies were pooled for in vivo packaging (22), producing a lysate that had a titer of 109 cosmid-containing phages/ml. This lysate was used to infect E. coli strain XL1-Blue and plated on LB agar plates with kanamycin. The resulting colonies (∼20,000) were pooled, and plasmid DNA was prepared from them. The DNA was then transformed into M. smegmatis by electroporation. Approximately 5,000 kanamycin-resistant M. smegmatis transformants were stocked as independent pools of approximately 1,000 clones each and enriched for M. marinum fragments that confer enhanced cell association on M. smegmatis.
Bacterial transformation by electroporation.
Electroporation of E. coli was carried out as described previously (15). Electroporation of M. marinum was carried out by first inoculating 500 ml of M-ADC-TW broth with 500 μl of a refrigerated culture of M. marinum previously grown to stationary phase (A600 of >1.00); the culture was incubated at 33°C until A600 was 0.9 to 1.2; the culture was incubated on ice for >30 min; and bacteria were washed with 5 ml of ice-cold 10% glycerol three times, washed with 50 ml of ice-cold 10% glycerol, and suspended in 1 ml of 10% ice-cold glycerol. Approximately 100-μl aliquots of bacteria were electroporated in 0.2-cm cuvettes at 2.5 kV, 25 μF, and 1,000 Ω parallel resistance, and bacteria were transferred into 1 ml of M-ADC-TW and incubated for 3 h at 33°C prior to plating for CFU. The same protocol was used for electroporation of M. smegmatis after growth at 37°C.
Identification of cosmids that confer enhanced infection.
Five independent pools of approximately 1,000 cosmid library clones in M. smegmatis were used to infect THP-1 cells in a manner similar to that used for standard cell association assays. Each of the five pools was separately used to infect 106 THP-1 cells at an MOI of 10 bacteria per cell for 30 min at 37°C. The cells were then washed five times with warm PBS and lysed, and dilutions were plated to determine CFU. Ten individual colonies from each infection were selected at random and screened individually in cell association assays to evaluate their ability to confer enhanced monocyte infection.
Characterization of cosmids that confer enhanced infection.
Plasmid DNA was isolated from M. smegmatis strains displaying an enhanced-infection phenotype, transformed into E. coli strain XL1-Blue (Stratagene), and restriction mapped using NotI and PstI. The DNA was transformed back into M. smegmatis to confirm that the cosmids and not a secondary mutation in the M. smegmatis chromosome were responsible for the enhanced-infection phenotype. To localize the region responsible for the phenotype, each cosmid was mutagenized using the TGS Template Generation System F-700 (Finnzymes), with a Mu-based transposon carrying the chloramphenicol resistance gene (cat) and transformed back into XL1-Blue. E. coli transformants were selected on LB agar plates containing kanamycin and chloramphenicol. More than 200 colonies carrying transposon insertions in each cosmid were picked, and plasmid DNA was isolated. Transposon insertions were then mapped by restriction analysis with NotI. Forty cosmids representing insertions in approximately every 1 kbp were selected and transformed back into M. smegmatis. M. smegmatis clones were then retested in cell association assays to identify cosmids with transposon insertions that failed to confer the enhanced-infection phenotype on M. smegmatis.
Determination of the sequence of mel loci.
Sequencing reactions were carried out as described previously (9). Initially, sequencing was performed outwards from both ends of the Mu transposon with the oligonucleotides SeqA (ATCAGCGGCCGCGATCC) and SeqB (GTTTTTCGTGCGCCGCTTCA). Sequencing was continued by primer walking directly on the cosmid of interest. All regions were sequenced completely on both strands with Big Dye Terminator (Applied Biosystems) cycle sequencing and subsequent analysis on an ABI 310 automated sequencing apparatus (Applied Biosystems). Sequence analysis and assembly were carried out using Gene Construction Kit 2 (Textco) software, and comparison with known sequences was carried out using BLAST (3).
In silico analysis of mel loci.
All putative open reading frames (ORFs) within the mel loci were examined for similarity to other proteins in GenBank by using protein-protein National Center for Biotechnology Information (NCBI) BLAST (3), and putative protein motifs were identified using the NCBI Conserved Domain Search (26). Putative gram-positive bacterial signal sequences were identified within the ORFs by using SignalP 3.0 (30). Genome sequences were analyzed for sequences similar to mel loci by using the most recent updates available in the NCBI database for the completed genomes of Mycobacterium avium subsp. paratuberculosis (www.ncbi.nlm.nih.gov), Mycobacterium bovis (18), Mycobacterium leprae (12), and M. tuberculosis (11) and the nearly complete Mycobacterium avium subsp. avium (www.tigr.org), M. marinum (www.sanger.ac.uk), and M. smegmatis (www.tigr.org) genome sequences at their individual websites.
Construction of M. marinum melE and melF mutants.
The plasmid pJDC55 was constructed by deletion of the chloramphenicol resistance gene (cat) from pJDC15 (9). pJDC15 was digested with SacI and ScaI, to release the cat-containing fragment. The vector was blunt ended using mung bean endonuclease and then self-ligated to produce pJDC55. pJDC55 carries an R6K origin of replication from pJDC15 and thus is a suicide plasmid. This plasmid also carries kanamycin resistance (aph) and the counterselectable sucrose sensitivity (sacB) marker. In order to clone the Mu transposon-mutagenized melE and melF genes into pJDC55, we digested the cosmids with PmlI (melF) or FspI (melE), purified the appropriate fragment, and ligated it to FspI-digested pJDC55. Ligations were transformed into E. coli strain Ψec47 (XL1-Blue::λpir) (9), and clones were selected on LB agar with kanamycin and chloramphenicol. The constructs were purified and linearized by digestion with NheI. The NheI-digested fragments were blunt ended using mung bean endonuclease and ligated to ScaI-linearized pYUB174 (5). pYUB174 contains the β-galactosidase gene driven from the mycobacteriophage L5 promoter, allowing for blue-white selection on X-Gal. The ligations were transformed into E. coli strain XL1-Blue and selected on LB agar with kanamycin, chloramphenicol, and X-Gal. Plasmid DNA was isolated from positive clones and used to transform M. marinum. Plasmid integration events were selected on M-ADC agar with kanamycin and X-Gal and grown in M-ADC-TW broth for 5 days. Clones that carry the appropriate mutation as a result of allelic exchange were then selected on M-ADC agar containing 10% sucrose and X-Gal. Allelic-exchange mutants were confirmed by PCR and Southern analyses (data not shown).
Complementation of M. marinum mel mutants with M. tuberculosis homologues.
Complementing constructs for the melE and melF mutations in M. marinum were constructed by cloning the appropriate M. tuberculosis homologue into pMV262 such that it would be expressed from the pMV262 hsp60 promoter. The M. tuberculosis melE (Rv2569c) and melF (Rv1936) genes were cloned from M. tuberculosis H37Rv total chromosomal DNA by PCR with the oligonucleotide pairs EcomF (TATAAAGCTTGCGGCATGCAACCGCTGTGG) with EcomR (TATACTGCAGGCAGACGTGCCGCCGCTACG) and FcomF (TATACTGCAGCGTGAACGGCTGACCTGTGC) with FcomR (TATAAAGCTTCGGACGGCACG CACAAGACG), respectively. The PCR products were then digested with PstI and HindIII and directionally cloned into PstI-and HindIII-digested pMV262. Constructs were confirmed by restriction analyses and complete sequencing of the M. tuberculosis gene to ensure the absence of PCR-incorporated mutations. The constructs were then transformed into the appropriate M. marinum mutant and selected on kanamycin. The presence of the correct complementing constructs in M. marinum, designated pJDC77 (melE) and pJDC79 (melF), was confirmed by PCR.
Statistical analyses.
All experiments were carried out in triplicate and repeated at least three times. The significance of the results was determined by analysis of variance. P values of <0.05 were considered significant.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in GenBank with accession numbers AY623663 (mel1) and AY623664 (mel2).
RESULTS
M. marinum is taken up by monocytes more efficiently than is M. smegmatis.
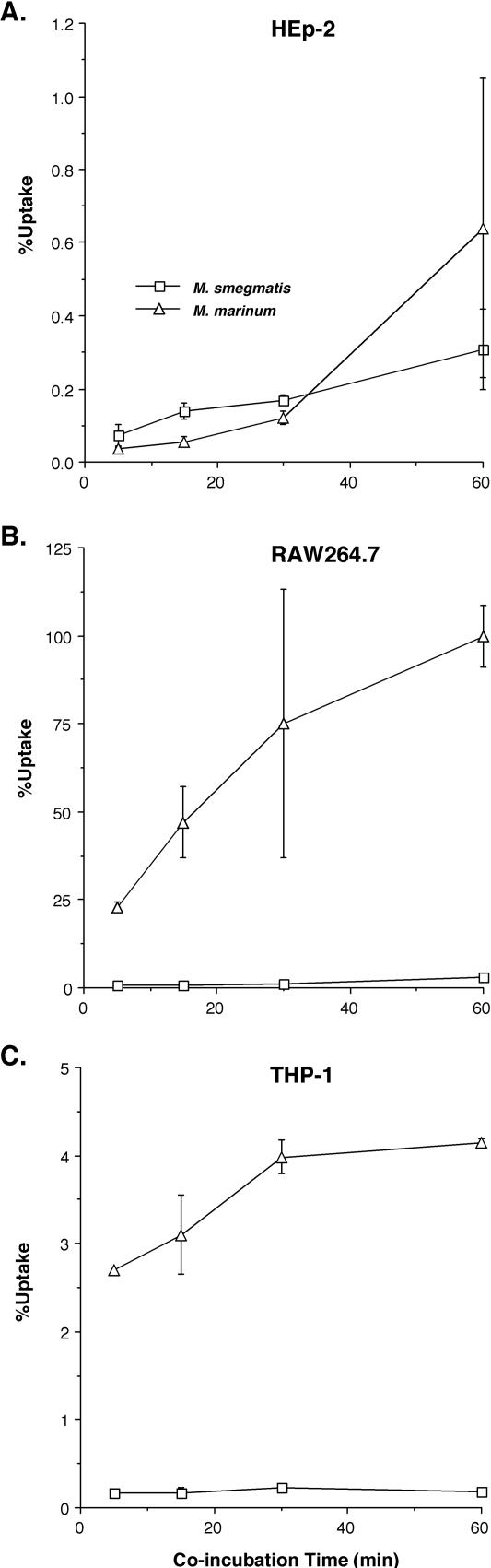
In our previous studies we found a significant difference between the ability of the human and fish pathogen M. marinum and the nonpathogenic mycobacterial species M. smegmatis to enter both murine and fish monocytic cells (16). This difference is not due solely to intracellular killing of bacteria but must be related to different levels of uptake, since microscopic quantitation of cell-associated bacteria, which is not affected by bacterial viability, correlates well with data obtained from CFU. In order to better understand the molecular mechanisms of enhanced uptake and intracellular survival in macrophages by M. marinum, we further characterized uptake into additional cell types (Fig. 1), including the human epithelial cell line HEp-2, the murine macrophage cell line RAW264.7, and the human monocytic cell line THP-1. We found that, similarly to our previous observations in the murine macrophage cell line J774A.1 and the fish monocytic cell line CLC (16), M. marinum displays higher levels of uptake than does M. smegmatis into each of these monocytic cell lines and this difference is mostly independent of the bacterium-host cell coincubation period (Fig. 1). In contrast, there is no significant difference between the level of uptake of M. marinum and that of M. smegmatis by HEp-2 cells after any length of coincubation, suggesting that the mechanism of uptake into monocytic cells is different from that of uptake into epithelial cells. The difference between the percentage of M. marinum taken up by RAW264.7 (25 to 100%) and that take up by THP-1 (2.5 to 4%) is most likely due to the fact that RAW264.7 cells are more fully differentiated than THP-1 cells and thus are more phagocytic. Although the differences between uptake of M. marinum and that of M. smegmatis are somewhat greater in RAW264.7 cells, 37- to 40-fold compared to 20- to 23-fold in THP-1 cells, the differences are more consistent between experiments in THP-1 cells, making these cells the best choice for use in later experiments.
FIG. 1.
Percentage of the bacterial inoculum that is taken up by cells after different periods of coincubation of M. smegmatis or M. marinum with the human epithelial cell line HEp-2 (A), murine macrophage cell line RAW264.7 (B), and human monocytic cell line THP-1(C) as measured by uptake assays. Data represent the means and standard deviations of assays done in triplicate from a representative experiment. Experiments were repeated at least three times independently.
M. marinum adheres to monocytes at higher levels than M. smegmatis does.
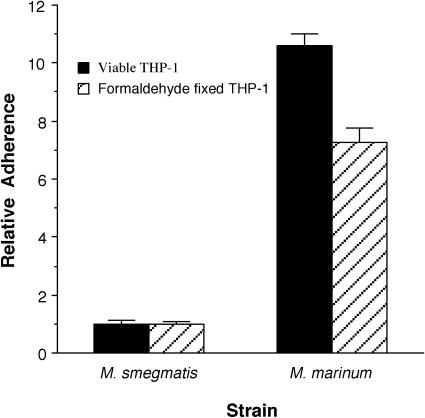
We were curious whether these differences in uptake were the result of very early events during the interactions of M. marinum with monocytic cells, such as adherence. In order to examine this possibility more carefully, we conducted two different types of adherence assays, the first using fixed THP-1 cells for coincubation with the bacteria and the second using addition of the bacteria to the host cells followed by mixing and rapid washing to remove nonadherent bacteria; this assay has also been termed an “immediate assay” (8). Under both assay conditions more M. marinum bacteria than M. smegmatis bacteria associate with THP-1 cells (Fig. 2). Although fixed-cell assays result in between two- and threefold-lower numbers of bacteria associated with cells than do immediate assays, both methods of measuring adherence identify similar differences between the adherence of M. marinum and M. smegmatis. There is a sevenfold difference in adherence between M. marinum and M. smegmatis in fixed cells and a 10-fold difference in the immediate assay. These data confirm that M. marinum adheres more efficiently to monocytic cells than M. smegmatis does, suggesting that there are differences between the initial interactions of M. marinum and M. smegmatis with monocytic cells.
FIG. 2.
Adherence of M. smegmatis and M. marinum to the human monocytic cell line THP-1 assayed by using viable or formaldehyde-fixed host cells. Data represent the means and standard deviations of assays done in triplicate from a representative experiment. Data are expressed relative to the levels of M. smegmatis adherence under the same experimental conditions. Experiments were repeated at least three times independently.
Cell association assays in THP-1 cells allow measurement of differences between M. marinum and M. smegmatis.
Although we did not observe any differences between the levels of susceptibility of M. smegmatis and M. marinum to amikacin, which is used in our uptake assays to kill extracellular bacteria, we wished to develop an assay for host cell infection that would not involve antibiotics to ensure that we would not select for resistant bacteria. Since the differences between M. marinum and M. smegmatis are extremely consistent in THP-1 cells and always at least 20-fold in uptake assays, we chose these cells for use in our assays. We chose a cell association assay that involved a 30-min coincubation of THP-1 cells with M. marinum or M. smegmatis followed by extensive washing to remove extracellular bacteria. This assay resulted in greater than 50-fold-higher levels of cell association with M. marinum than with M. smegmatis. We obtained similar results at MOIs of 1 to 1,000 and when assays were carried out at 37 or 33°C, the normal growth temperature for M. marinum. Thus, although the numbers of bacteria that become cell associated increase linearly with increasing MOI, the relative numbers of M. marinum and of M. smegmatis bacteria that are cell associated are similar under all of these experimental conditions. As a result, all further cell association experiments were conducted at the relatively low MOI of 10 and at the most physiologically relevant condition for the THP-1 cells, 37°C.
Isolation of cosmids that have the ability to confer enhanced infection.
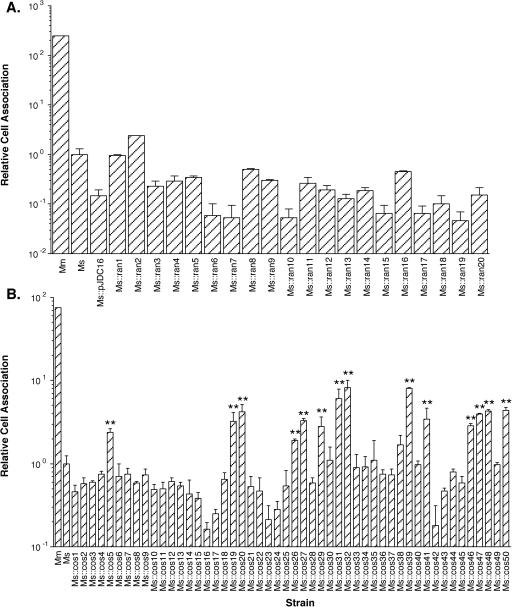
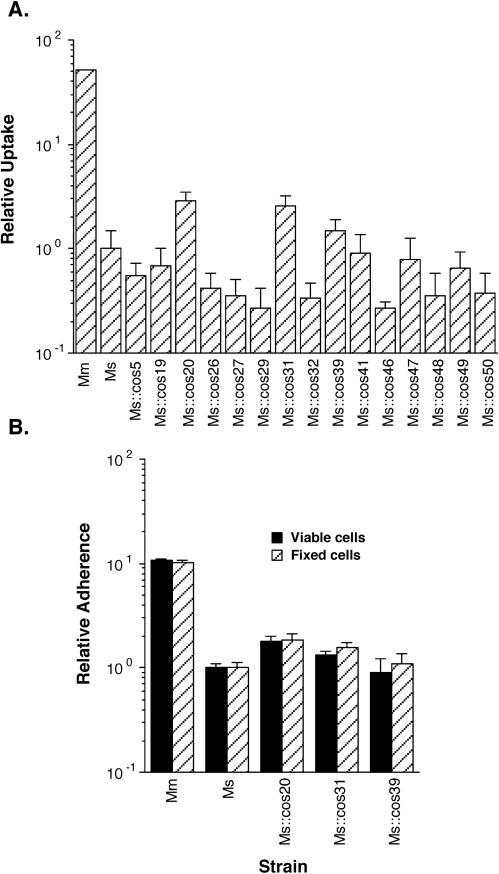
We constructed a cosmid library of contiguous total DNA fragments from M. marinum of between 40 and 50 kbp in length and transformed this library as a pool of ∼20,000 clones from E. coli into M. smegmatis. We examined 20 random cosmid-containing clones in M. smegmatis individually for their ability to confer enhanced infection of THP-1 cells. None of these 20 clones displayed enhanced host cell infection (Fig. 3A). Five pools of ∼1,000 M. smegmatis clones each were enriched for clones that carry enhanced monocytic cell infection genes by their ability to associate with THP-1 cells in a standard cell association assay. After enrichment, 10 clones from each pool (a total of 50) were screened individually in cell association assays. We found that 14 of these clones (28%) displayed enhanced cell association after enrichment (Fig. 3B). Interestingly, only three of these cosmid-containing clones, Ms::cos20, Ms::cos31, and Ms::cos39, displayed enhanced uptake (P < 0.05), and only Ms::cos20 displayed enhanced adherence (P < 0.05) to THP-1 cells, whether the host cells were viable or were fixed with formaldehyde (Fig. 4).
FIG. 3.
Cell association of M. marinum (Mm), M. smegmatis (Ms), and M. smegmatis containing the cosmid vector backbone alone (Ms::pJDC16), random cosmids (Ms::ran1 to Ms::ran20) (A), or individual cosmid clones after one round of cell association assay enrichment (Ms::cos1 to Ms::cos50) (B) with THP-1 cells. Data represent the means and standard deviations of assays done in triplicate from a representative experiment. Data are expressed relative to the level of M. smegmatis cell association. The asterisks indicate statistically significant differences between the individual clone and wild-type M. smegmatis (P < 0.01). Experiments were repeated at least three times independently.
FIG. 4.
Uptake (A) and adherence (B) of M. marinum (Mm), M. smegmatis (Ms), and M. smegmatis containing individual cosmid clones with THP-1 cells. Data represent the means and standard deviations of assays done in triplicate from a representative experiment. Data are expressed relative to M. smegmatis uptake (A) and adherence (B). Experiments were repeated at least three times independently.
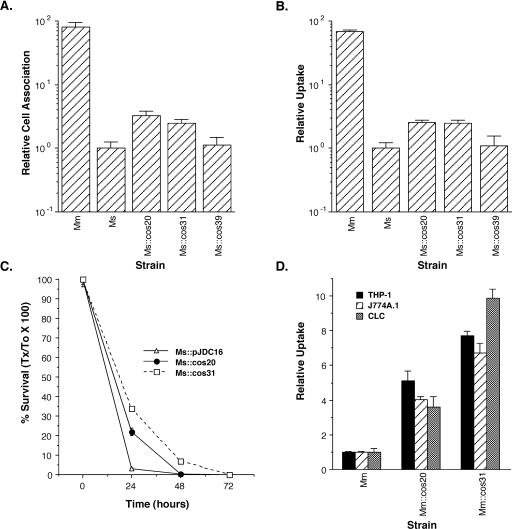
In order to ensure that the cosmid itself, rather than a spontaneous mutation in the M. smegmatis genome, was responsible for the observed phenotype, we purified these cosmids from M. smegmatis, transformed them into E. coli for amplification, and then retransformed them back into M. smegmatis. The resulting retransformed M. smegmatis clones carrying cosmids from the M. marinum genomic library were then screened for cell association and uptake compared to M. smegmatis (Fig. 5). Only cos20 and cos31 had the ability to confer enhanced cell association and uptake (P < 0.01) on M. smegmatis. Since cos20 and cos31 displayed the most consistent ability to confer enhanced cell association and uptake on M. smegmatis, only these two cosmid clones were chosen for further analysis.
FIG. 5.
Individual cosmid clones purified from M. smegmatis, retransformed into M. smegmatis (Ms::cos20, Ms::cos31, and Ms::cos39) or M. marinum (Mm::cos20 and Mm::cos31), and tested for their ability to confer enhanced cell association (A), uptake (B and D), and intracellular survival (C). Assays were carried out in the human monocytic cell line THP-1 (A, B, and D), the murine macrophage cell line J774A.1 (C and D), and the fish monocytic cell line CLC (D). Data represent the means and standard deviations of assays done in triplicate from a representative experiment. Data in panels A, B, and D are expressed relative to M. smegmatis cell association (Ms) (A), M. smegmatis uptake (Ms) (B), or M. marinum uptake (Mm) (D). Experiments were repeated at least three times independently.
Cosmids cos20 and cos31 confer enhanced intracellular survival on M. smegmatis.
It is possible that the enhanced cell association and uptake that cos20 and cos31 confer on M. smegmatis are at least partially due to enhanced survival subsequent to uptake. In order to test this possibility, we examined the abilities of Ms::cos20 and Ms::cos31 to survive in the murine macrophage cell line J774A.1 over time (Fig. 5C). We found that both cosmids conferred an increased ability to survive during the first 24 h after infection (P < 0.001) and that Ms::cos31 could persist out to 72 h, whereas wild-type M. smegmatis was almost completely killed by 24 h and was not detectable by 48 h. Although it is possible that these differences in survival were at least in part the result of an increased bacterial load due to enhanced uptake of the cosmid-containing clones, these observations suggest that either these cosmids contribute to the ability of M. smegmatis to survive inside mammalian macrophages or the mechanism of uptake that they confer on M. smegmatis affects subsequent intracellular events that impact intracellular survival.
Cosmids cos20 and cos31 confer enhanced uptake on M. marinum.
Possibly the functions of the regions that each of these cosmids encodes are affected by genes present in M. smegmatis that are not present in M. marinum. Thus, the genes that they carry may actually play a different role in M. marinum. In order to test this possibility, we transformed cos20 and cos31 into M. marinum and examined the phenotype of the strains containing these cosmids. Although the pAL5000 mycobacterial plasmid origin of replication, which is present on these shuttle cosmids, is thought to be a low-copy-number plasmid (43), it is likely that carrying more than one copy of these genes, i.e., the chromosomal copy and that on the plasmid, will confer a phenotype due to gene dosage effects. When we compared the ability of M. marinum containing these cosmids to the ability of M. marinum without them, we observed between 5- and 10-fold-enhanced uptake (Fig. 5D). This observation suggests that these genes have the ability to confer enhanced uptake on M. marinum and that their ability to confer this phenotype is not dependent on genes present only in M. smegmatis.
Identification of the M. marinum genes that confer enhanced uptake.
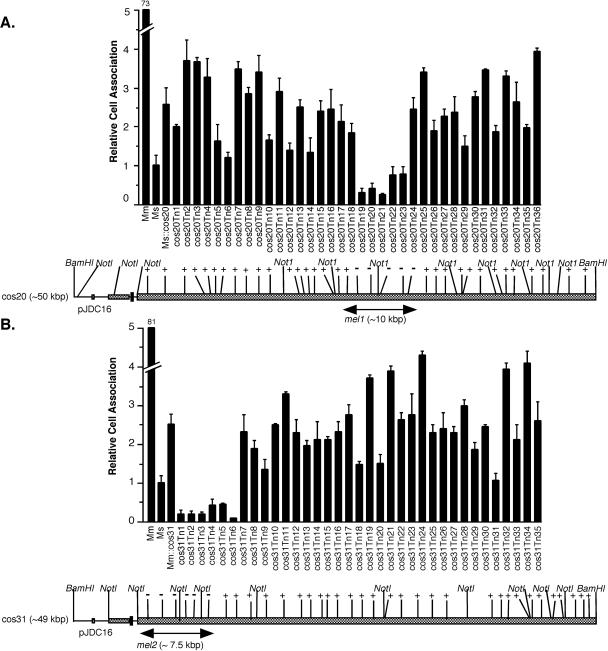
In order to identify the mycobacterial enhanced infection loci (mel) carried on cos20 and cos31, we conducted saturating in vitro transposon mutagenesis on each cosmid followed by transformation back into M. smegmatis to examine their ability to confer enhanced uptake. Over 200 Mu insertions were obtained in both cos20 and cos31. Restriction mapping was used to determine the position of each insertion, and more than 40 insertions in approximately every 1 kbp of both cosmids were selected for transformation back into M. smegmatis. When these mutagenized cosmids were screened in cell association assays, we identified five transposon insertions in cos20 and six in cos31 that could no longer confer enhanced host cell infection (Fig. 6). Sequence analysis of the junctions of each transposon insertion allowed determination of the position for each insertion. The mel locus on cos20, designated mel1, was localized to a region of approximately 10 kbp, and that on cos31, designated mel2, was localized to a region of approximately 7.5 kbp (Fig. 6). The intervening regions between each transposon insertion and at least 2 kbp upstream were then sequenced by primer walking on cosmid DNA. Five putative ORFs were identified in mel1, and six were identified in mel2; they were given the designations melA to K (Fig. 7). All 11 of these ORFs include putative genes that are very similar to genes present in M. tuberculosis, and the majority are in the same gene order as in M. tuberculosis.
FIG. 6.
Cell association of M. smegmatis (Ms), M. marinum (Mm), and M. smegmatis containing cosmid 20 (Ms::cos20) or cosmid 31 (Ms::cos31) as well as cosmid 20, which has a mini-Mu transposon insertion in it (A) (cos20Tn1 to cos20Tn36), or cosmid 31, which has a mini-Mu transposon insertion in it (B) (cos31Tn1 to cos31Tn35), with THP-1 cells. Structure, approximate length and restriction map for each cosmid, and positions of insertions are shown below cell association data. Plus signs indicate Mu insertions that do not affect the ability of the cosmid to confer enhanced cell association, and minus signs indicate insertions that affect the phenotype conferred. The sizes of mel1 and mel2 loci as estimated from saturating transposon mutagenesis data are shown below the cosmids. Data represent the means and standard deviations of assays done in triplicate from a representative experiment. Data are expressed relative to M. smegmatis cell association. Experiments were repeated at least three times independently.
FIG. 7.
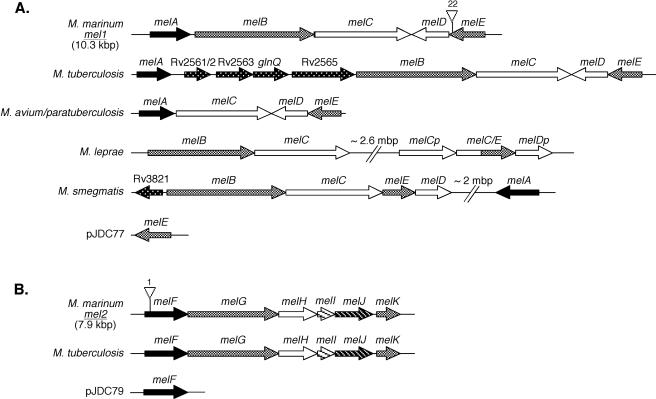
Organization of mel1 (A) and mel2 (B) loci in different mycobacterial species where similar loci could be identified as well as the structure of complementing constructs (pJDC77 and pJDC79) used in the present study. Numbers in parentheses indicate the approximate size of the locus in M. marinum. Broken lines indicate very distant loci for which the distance that the break represents is indicated above the construct. The gene or corresponding Rv number from the M. tuberculosis H37Rv genome sequence is shown above the construct. Arrows on constructs indicate the deduced direction of transcription for each gene. Triangles above mel1 and mel2 indicate the position of insertion into each M. marinum locus and the transposon number that corresponds with that used to construct M. marinum mutant strains by allelic exchange. Gene designations that end in “p” indicate putative pseudogenes, and melC/E indicates an apparent translational fusion between melC and melE in the M. leprae mel1 locus. M. avium/paratuberculosis indicates locus organization that is the same for M. avium subsp. avium and M. avium subsp. paratuberculosis. M. tuberculosis indicates locus organization that is the same for all M. tuberculosis complex mycobacteria.
In silico analysis of melA to melK genes from M. marinum.
We examined the presence of ORFs similar to those in the mel1 and mel2 loci in other mycobacterial species where genome sequences are available. Our analyses indicate that all mycobacterial species, including M. tuberculosis complex, M. avium complex, M. leprae, and M. smegmatis, carry at least a portion of the mel1 locus (Fig. 7). The organization of mel1 genes in M. tuberculosis and M. avium is very similar to that in M. marinum. The greatest differences are in M. leprae, where melA could not be found and both an intact melC and a putative melC pseudogene as well as a single melD pseudogene were identified. In addition, the melE gene appears to be a translational fusion with part of melC in M. leprae. Interestingly, the mel2 locus is present in the same gene order in both M. marinum and M. tuberculosis complex mycobacteria but appears to be absent in other mycobacterial species. We examined each ORF in the mel1 and mel2 loci for similarity to putative ORFs in the M. tuberculosis genome as well as motifs that might provide insight into the functions of the proteins that they encode (Table 1). Highly similar proteins exist in M. tuberculosis for all putative ORFs in both loci. All of the putative ORFs in mel1 carry putative gram-positive bacterial signal sequences except for melD, suggesting that these proteins are secreted. In contrast to the mel1 locus, all of the putative ORFs in the mel2 locus carry significant motifs that suggest biochemical functions of the proteins that they encode. Interestingly, several of the orthologous clusters that display the highest similarity to ORFs in the mel2 locus are eukaryotic clusters (KOG). This observation suggests that this locus is involved in metabolism of complex fatty acids obtained from eukaryotic cells.
TABLE 1.
Characteristics of genes in mel locia
| Locus and gene | M. tuberculosis homologue (% identity, no. of aa)b | E value (score)c | Deduced length (aa) | Other similarity or motif(s)d | Putative activity |
|---|---|---|---|---|---|
| mel1 | |||||
| melA | Rv2560 (63, 249) | 8e-79 (296) | 377 | CO65473, integral membrane protein; proline-rich amino terminus; gram-positive bacterial signal sequence | Membrane protein (unknown) |
| melB | Rv2566 (78, 1,121) | 0.0 (1,730) | 1,106 | CO61305, cysteine protease-like transglutaminases; gram-positive bacterial signal sequence | Transglutaminase or protease |
| melC | Rv2567 (77, 894) | 0.0 (1,311) | 896 | COG2307, COG2308, uncharacterized conserved protein, gram-positive bacterial signal sequence | Unknown |
| melD | Rv2568c (80, 336) | 1e-159 (563) | 350 | COG4307, uncharacterized conserved protein | Unknown |
| melE | Rv2569c (84, 282) | 1e-134 (478) | 336 | COG1305, cysteine protease-like transglutaminases, gram-positive bacterial signal sequence | Transglutaminase or protease |
| mel2 | |||||
| melF | Rv1936 (95, 365) | 0.0 (687) | 379 | COG2141, flavin-dependent oxidoreductases and luciferase-like monooxygenase, luxA, alkanal monooxygenase | Oxidoreductase |
| melG | Rv1937 (83, 837) | 0.0 (1,434) | 839 | Fer2, 2Fe-2S, NqrF, UbiH, dioxygenase, dehydrogenase, luxG | Dehydrogenase |
| melH | Rv1938, ephB (82, 355) | 1e-173 (608) | 352 | KOG4178, epoxide hydrolase, luxH | Lipid transport and metabolism |
| melI | Rv1939 (78, 170) | 5e-71 (267) | 177 | COG1853, oxidoreductase | Flavoprotein oxygenase |
| melJ | Rv1940, ribA (68, 344) | 1e-116 (419) | 365 | KOG1284, bifunctional GTP cyclohydrolase II-DHBP synthase | Cyclohydrolase |
| melK | Rv1941 (78, 221) | 3e-77 (290) | 256 | KOG0725, broad-specificity-range reductase, fabG | Lipid metabolism-synthesis |
Abbreviations: aa, amino acids; COG, cluster of orthologous groups; KOG, cluster of orthologous groups for eukaryotic genome; Fer2, 2Fe-2S, iron-sulfur cluster binding domain; NqrF, Na+-transporting NADH:ubiquinone oxidoreductase; UbiH, 2-polyprenyl-6-methoxyphenol hydroxylase; DHBP, 3,4-dihydroxy-2-butanone-4-phosphate.
M. tuberculosis gene to which the M. marinum gene displays similarity, along with percent identity at amino acid level and number of amino acids that could be aligned to show this level of identity.
Expectation frequency (E value) and score according to NCBI protein-protein BLAST in comparison with M. tuberculosis homologue.
Placement of putative protein product into a cluster of orthologues (COG or KOG) by use of CD-Search at NCBI and SignalP 3.0 at the Center for Biological Sequence Analysis Website.
M. marinum mel mutants are defective in mammalian cell infection.
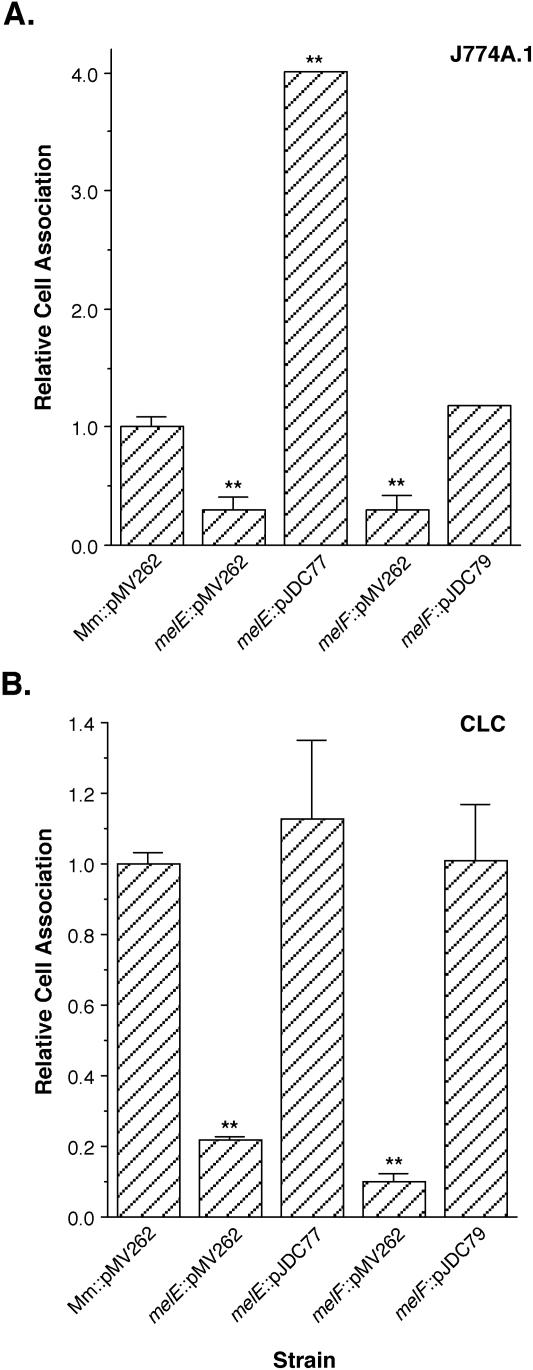
The fact that mel1 and mel2 can confer enhanced host cell infection on both M. smegmatis and M. marinum suggests that these genes are involved in the ability of mycobacteria to infect mammalian cells. However, it remains possible that these findings are the result of an artifact related to their overexpression due to increased gene dosage. In order to confirm that the mel genes are actually involved in host cell infection by M. marinum, we took advantage of the transposon insertions in these loci generated during localization of the mel regions on the cosmids. Two transposon insertions, Tn22 in the melE gene within mel1 and Tn1 in the melF gene within mel2, were cloned into an allelic-exchange vector that carries lacZ and sacB as counterselectable markers. Through the use of a two-step selection procedure, we identified more than 12 white sucrose-resistant colonies for each gene and screened for the appropriate mutation by Southern and PCR analyses (data not shown). Slightly less than 20% of the putative melE mutants and 50% of the putative melF mutants carry the appropriate mutation. To ensure that no secondary mutations were incorporated during genetic manipulation that could be responsible for the observed phenotype and that the M. tuberculosis homologues of the mel genes had the ability to confer similar functions, we complemented the melE and melF mutants. We constructed pJDC77, which carries M. tuberculosis melE (Rv2569c), and pJDC79, which carries M. tuberculosis melF (Rv1936), respectively, expressed from the pMV262 hsp60 promoter (44), and transformed these complementing constructs into each of the mutants. We found that mutants with mutations in both melE and melF display a defect in the ability to infect the murine and fish macrophage cell lines J774A.1 and CLC, respectively, and that their defects can be complemented by the appropriate homologous gene from M. tuberculosis (Fig. 8). Interestingly, melE expressed from pMV262 confers enhanced cell association when J774A.1 cells are used, even with the melE mutant, suggesting that this gene is sufficient to confer enhanced cell association if present in multiple copies and expressed from a strong promoter. However, no significant enhancement was observed in CLC cells, suggesting that the effects of melE overexpression on cell association are somewhat cell type specific. The defect in cell association observed with the melE mutant is consistent in both cell types, though, indicating that the cell type-specific effects are most likely an artifact resulting from overexpression and not an inherent characteristic of melE under normal conditions. Thus, we have confirmed that the mel loci are involved in the ability of M. marinum to infect macrophages and that the homologous gene from M. tuberculosis can complement mutations in the M. marinum mel loci.
FIG. 8.
Cell association of M. marinum containing the vector backbone alone (Mm::pMV262), the M. marinum melE mutant containing the vector alone (melE::pMV262) or the complementing construct containing the M. tuberculosis melE (Rv2569c) gene (melE::pJDC77), and the M. marinum melF mutant containing vector alone (melF::pMV262) or the complementing construct containing the M. tuberculosis melF (Rv1936) gene (melF::pJDC79) in the murine macrophage cell line J774A.1 (A) and the fish monocytic cell line CLC (B). Data represent the means and standard deviations of assays done in triplicate from a representative experiment. The asterisks indicate cell association significantly different from that of wild-type M. marinum carrying pMV262 (P < 0.01). Data are expressed relative to the cell association of M. marinum that carries the vector backbone alone, pMV262 (Mm::pMV262). Experiments were repeated at least three times independently.
DISCUSSION
We identified two mycobacterial loci, mel1 and mel2, that affect the ability of M. marinum to infect macrophages. These loci are composed of 11 genes that are similar to M. tuberculosis genes and when present on a low-copy-number mycobacterial plasmid confer enhanced infection of host cells on both M. smegmatis and M. marinum. These loci were identified as a result of attempts to determine the molecular bases for differences in the abilities of pathogenic and nonpathogenic mycobacteria to infect macrophages. Transfer of genomic DNA to M. smegmatis from M. marinum as large (∼45-kbp) genomic fragments was chosen because of the likelihood that a large number of genes would be involved in and possibly required for the ability of mycobacteria to efficiently infect host cells. Genomic loci involved were identified by enrichment for clones that associate with THP-1 cells. Although 14 clones out of 50 were identified that associate with mammalian cells at higher levels than wild-type M. marinum does after enrichment, none of the clones out of 20 examined had the ability to confer this phenotype beforehand. This observation suggests that cell association assays were very effective at enriching for clones in the library that display enhanced host cell infection.
The majority of the 14 clones that displayed enhanced cell association did not significantly affect uptake into THP-1 cells, but this could be the result of a lesser effect either on adherence or on the ability to survive intracellularly. This is because uptake assays involve both a 30-min coincubation period to allow uptake and a 2-h amikacin treatment to kill extracellular organisms, ensuring that only intracellular bacteria are counted. Although this assay is considered the “gold standard” for evaluating uptake of bacteria into eukaryotic cells, it does not differentiate adherence and intracellular survival if a significant percentage of the bacteria are killed within the 2.5-h assay period. The possibility that mel1 and mel2 can contribute to intracellular survival is supported by our observation that they can confer enhanced intracellular survival on M. smegmatis. Thus, further investigation of the other 12 clones that display enhanced cell association is likely to result in identification of additional mycobacterial factors involved in adherence to monocytic cells.
It is interesting that, although we used a similar approach of transferring genes from pathogenic mycobacteria to nonpathogenic mycobacteria, we did not identify the same genes that previous investigators did using different pathogenic mycobacterial species (6, 29, 47, 48). This is most likely due to differences in the assays used, since the majority of these groups examined solely enhancement of intracellular survival and not earlier events such as adherence and uptake. It is also possible that our own studies have not characterized all of the M. marinum genes, since it is likely that a large number of genes are involved. Continued examination of loci from M. marinum that affect, in particular, intracellular survival of mycobacteria would be likely to identify a set of genes overlapping those from previous studies. Thus, the differences in our enrichment procedure were advantageous, since they allowed identification of a new class of genes that play a role in the ability of mycobacteria to infect mammalian cells.
The genes present in the mel loci are very intriguing, since they may encode potential membrane-localized or secreted components (mel1) and complex lipid-related components involved in host cell infection (mel2). In the mel1 locus, three of the genes are of unknown function, though the suggestion that melA could be a membrane protein fits well with our intuitive understanding of bacterial factors that might be directly involved in adherence and uptake into mammalian cells. In addition, the presence of two putative transglutaminases or cysteine proteases in the mel1 locus is of great interest, since transglutaminases can play a role in signal transduction in eukaryotic cells as well as bacterial attachment. Transglutaminases have been shown to play an important role in pathogenesis by other bacterial species. The Bordetella bronchiseptica dermonecrotic toxin is a transglutaminase that is toxic for mammalian cells because of its ability to modify host cell GTPases (21), Erlichia spp. can recruit transglutaminases from host cells to cross-link proteins in the cell leading to internalization of the bacteria (24), and Staphylococcus spp. appear to use transglutaminases for attachment to host proteins, which is believed to be important for colonization (28). However, insight into the potential functions of these transglutaminases is complicated by the fact that they are also usually proteases and that eukaryotic transglutaminases may have evolved from bacterial cysteine proteases (25). The presence of both putative transglutaminases in M. smegmatis suggests that these genes may not function solely during disease. However, it is equally possible that they are regulated or localized differently in pathogenic and nonpathogenic mycobacterial species. Further studies are necessary to differentiate between these possibilities and to determine whether the biochemical activity conferred by the putative transglutaminases plays a role in host cell infection by mycobacteria.
The fact that all of the genes present in the mel2 locus are found only in M. marinum and M. tuberculosis complex mycobacteria suggests that this locus is important for pathogenesis. Although putative proteins encoded by the genes in the mel2 locus have similarity to other bacterial proteins with known biochemical functions, the exact structure of the molecule that they produce remains unclear. Each of these genes appears to have a function related to fatty acid or polyketide biosynthesis. The fact that M. marinum carries these genes points toward a potentially important parallel in the mechanisms that M. tuberculosis and M. marinum use to cause disease. The absence of these genes in other mycobacterial species supports the great potential of M. marinum as a model for tuberculosis. Recently genomic comparison of M. tuberculosis to M. leprae, M. avium, M. marinum, and M. smegmatis was carried out. At least six additional loci are present in both M. marinum and M. tuberculosis but not in M. avium and M. smegmatis (27). However, unlike mel2, all of these loci were also found in M. leprae, though this could be due to the fact that a “core” set of genes was chosen for these analyses. Because of the differences between pathogenesis of M. leprae and that of M. tuberculosis and the relatively greater similarity between M. tuberculosis and M. marinum, detailed analysis of genes that fit within the same category as the mel2 locus, specific to M. tuberculosis complex and M. marinum, is likely to provide insight into M. tuberculosis pathogenic mechanisms. In addition, these analyses will allow us to better understand what aspects of tuberculosis pathogenesis the M. marinum model can be best applied to.
With the identification of the mel1 and mel2 loci and demonstration of their role in the ability of mycobacteria to infect macrophages, we have made an important step toward understanding the molecular events involved in parasitism of host cells by mycobacteria. Since both of these loci have the ability to confer enhanced host cell infection on M. marinum, it is likely that gene dosage effects could be used to upregulate a number of different mycobacterial genes, thereby allowing their isolation based on functional biological assays similar to our cell association assay. We have previously been successful at using this type of approach for the identification of genes involved in host cell infection by Legionella spp. (9). Since Legionella spp. and mycobacteria are very distantly related, this approach is broadly applicable to many bacterial species. In addition, one could imagine the design of a number of different selection-enrichment strategies that would allow identification of genes with functions other than those involved solely in host cell infection. Indeed, these are not the first studies that have used gene dosage effects to regulate a specific class of genes of interest (1, 7, 31). Application of this and other strategies to the study of mycobacterium-host cell interactions with M. marinum as a facile genetic model system is likely to lead to a more comprehensive understanding of mycobacterial pathogenesis.
Acknowledgments
This work was supported by grant AI47866 from the National Institutes of Health.
Editor: V. J. DiRita
REFERENCES
- 1.Aiba, S., H. Tsunekawa, and T. Imanaka. 1982. New approach to tryptophan production by Escherichia coli: genetic manipulation of composite plasmids in vitro. Appl. Eviron. Microbiol. 43:289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, D. C., J. R. Jones, T. Tan, J. M. Chen, and J. Liu. 2004. PimF, a mannosyltransferase of mycobacteria, is involved in the biosynthesis of phosphatidylinositol mannosides and lipoarabinomannan. J. Biol. Chem. 279:18824-18833. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker, L. P., K. M. George, S. Falkow, and P. L. C. Small. 1997. Differential trafficking of live and dead Mycobacterium marinum organisms in macrophages. Infect. Immun. 65:1497-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barletta, R. G., D. D. Kim, S. B. Snapper, B. R. Bloom, and W. R. Jacobs, Jr. 1992. Identification of expression signals of the mycobacteriophages Bxb1, L1, and TM4 using Escherichia-Mycobacterium shuttle plasmids pYUB75 and pYUB76 designed to create translational fusions to the lacZ gene. J. Gen. Microbiol. 138:23-30. [DOI] [PubMed] [Google Scholar]
- 6.Bermudez, L. E., K. Shelton, and L. S. Young. 1995. Comparison of the ability of Mycobacterium avium, M. smegmatis and M. tuberculosis to invade and replicate within HEp-2 epithelial cells. Tubercle Lung Dis. 76:240-247. [DOI] [PubMed] [Google Scholar]
- 7.Blanc-Potard, A. B., N. Figueroa-Bossi, and L. Bossi. 1999. Histidine operon deattenuation in dnaA mutants of Salmonella typhimurium correlates with a decrease in the gene dosage ratio between tRNAHis and histidine biosynthetic loci. J. Bacteriol. 181:2938-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo, S. L. G., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila uptake gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cirillo, S. L. G., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 10.Clark, H. F., and C. C. Shepard. 1963. Effect of environmental temperatures on infection with Mycobacterium marinum (Balnei) of mice and a number of poikilothermic species. J. Bacteriol. 86:1057-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 12.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 13.Cywes, C., H. C. Hoppe, M. Daffé, and M. R. W. Ehlers. 1997. Nonopsonic binding of Mycobacterium tuberculosis to complement receptor type 3 is mediated by capsular polysaccharides and is strain dependent. Infect. Immun. 65:4258-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, J. M., H. Clay, J. L. Lewis, N. Ghori, P. Herbomel, and L. Ramakrishnan. 2002. Real-time visualization of mycobacterium-macrophage interactions leading to initiation of granuloma formation in zebrafish embryos. Immunity 17:693-702. [DOI] [PubMed] [Google Scholar]
- 15.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Etr, S. H., L. Yan, and J. D. Cirillo. 2001. Fish monocytes as a model for mycobacterial host-pathogen interactions. Infect. Immun. 69:7310-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, L. Y., F. Laval, E. H. Lawson, R. K. Groger, A. Woodruff, J. H. Morisaki, J. S. Cox, M. Daffe, and E. J. Brown. 2003. Requirement for kasB in Mycobacterium mycolic acid biosynthesis, cell wall impermeability and intracellular survival: implications for therapy. Mol. Microbiol. 49:1547-1563. [DOI] [PubMed] [Google Scholar]
- 18.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, G. A. 1874. Undersøgelser Angående Spedalskhedens Årsager. Norsk Magazin Laegevidenskaben 4:1-88. [Google Scholar]
- 20.Hart, P. D. A., and J. A. Armstrong. 1974. Strain virulence and the lysosomal response in macrophages infected with Mycobacterium tuberculosis. Infect. Immun. 10:742-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horiguchi, Y., T. Senda, N. Sugimoto, J. Katahira, and M. Matsuda. 1995. Bordetella bronchiseptica dermonecrotizing toxin stimulates assembly of actin stress fibers and focal adhesions by modifying the small GTP-binding protein rho. J. Cell Sci. 108:3243-3251. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs, W. R., J. F. Barrett, J. E. Clark-Curtiss, and R. Curtiss III. 1986. In vivo repackaging of recombinant cosmid molecules for analyses of Salmonella typhimurium, Streptococcus mutans, and mycobacterial genomic libraries. Infect. Immun. 52:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch, R. 1882. Die Aetiologie der Tuberkulose. Berl. Klin. Wochenschr. 19:221. [Google Scholar]
- 24.Lin, M., M. X. Zhu, and Y. Rikihisa. 2002. Rapid activation of protein tyrosine kinase and phospholipase C-γ2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect. Immun. 70:889-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarova, K. S., L. Aravind, and E. V. Koonin. 1999. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases. Protein Sci. 8:1714-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glaser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core' genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483-496. [DOI] [PubMed] [Google Scholar]
- 28.Matsuka, Y. V., E. T. Anderson, T. Milner-Fish, P. Ooi, and S. Baker. 2003. Staphylococcus aureus fibronectin-binding protein serves as a substrate for coagulation factor XIIIa: evidence for factor XIIIa-catalyzed covalent cross-linking to fibronectin and fibrin. Biochemistry 42:14643-14652. [DOI] [PubMed] [Google Scholar]
- 29.Miller, B. H., and T. M. Shinnick. 2001. Identification of two Mycobacterium tuberculosis H37Rv ORFs involved in resistance to killing by human macrophages. BMC Microbiol. 1:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 31.O'Sullivan, D. J., S. A. Walker, S. G. West, and T. R. Klaenhammer. 1996. Development of an expression strategy using a lytic phage to trigger explosive plasmid amplification and gene expression. Biotechnology 14:82-87. [DOI] [PubMed] [Google Scholar]
- 32.Ramakrishnan, L., and S. Falkow. 1994. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect. Immun. 62:3222-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 34.Ramakrishnan, L., R. H. Valdivia, J. H. McKerrow, and S. Falkow. 1997. Mycobacterium marinum causes both long-term subclinical infection and acute disease in the leopard frog (Rana pipiens). Infect. Immun. 65:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ranes, M. G., J. Rauzier, M. Lagranderie, M. Gheorghiu, and B. Gicquel. 1990. Functional analysis of pAL5000, a plasmid from Mycobacterium fortuitum: construction of a “mini” Mycobacterium-Escherichia coli shuttle vector. J. Bacteriol. 172:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogall, T., J. Wolters, T. Flohr, and E. Böttger. 1990. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 40:323-330. [DOI] [PubMed] [Google Scholar]
- 37.Ruley, K. M., J. H. Ansede, C. L. Pritchett, A. M. Talaat, R. Reimschuessel, and M. Trucksis. 2004. Identification of Mycobacterium marinum virulence genes using signature-tagged mutagenesis and the goldfish model of mycobacterial pathogenesis. FEMS Microbiol. Lett. 232:75-81. [DOI] [PubMed] [Google Scholar]
- 38.Ruley, K. M., R. Reimschuessel, and M. Trucksis. 2002. Goldfish as an animal model system for mycobacterial infection. Methods Enzymol. 358:29-39. [DOI] [PubMed] [Google Scholar]
- 39.Schlesinger, L. S. 1993. Macrophage phagocytosis of virulent but not attenuated strains of M. tuberculosis is mediated by mannoside receptors in addition to complement receptors. J. Immunol. 150:2920-2930. [PubMed] [Google Scholar]
- 40.Shepard, C. C. 1958. A comparison of the growth of selected mycobacteria in HeLa, monkey kidney and human amnion cells in tissue culture. J. Exp. Med. 107:237-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 42.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Bottger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stolt, P., and N. G. Stoker. 1996. Functional definition of regions necessary for replication and incompatibility in the Mycobacterium fortuitum plasmid pAL5000. Microbiology 142:2795-2802. [DOI] [PubMed] [Google Scholar]
- 44.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennet, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R. Jacobs, Jr., and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 45.Talaat, A. M., R. Reimschuessel, S. S. Wasserman, and M. Trucksis. 1998. Goldfish, Carassius auratus, a novel animal model for the study of Mycobacterium marinum pathogenesis. Infect. Immun. 66:2938-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tonjum, T., D. B. Welty, E. Jantzen, and P. L. Small. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 36:918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei, J., J. L. Dahl, J. W. Moulder, E. A. Roberts, P. O'Gaora, D. B. Young, and R. L. Friedman. 2000. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J. Bacteriol. 182:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wieles, B., T. H. Ottenhoff, T. M. Steenwijk, K. L. Franken, R. R. de Vries, and J. A. Langermans. 1997. Increased intracellular survival of Mycobacterium smegmatis containing the Mycobacterium leprae thioredoxin-thioredoxin reductase gene. Infect. Immun. 65:2537-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]