Abstract
Rationale: Automated analysis of computed tomographic (CT) lung images for epidemiologic and genetic association studies is increasingly common, but little is known about the utility of visual versus semiautomated emphysema and airway assessments for genetic association studies.
Objectives: Assess the relative utility of visual versus semiautomated emphysema and airway assessments for genetic association studies.
Methods: A standardized inspection protocol was used to visually assess chest CT images for 1,540 non-Hispanic white subjects within the COPDGene Study for the presence and severity of radiographic features representing airway wall thickness and emphysema. A genome-wide association study (GWAS) was performed, and two sets of candidate single-nucleotide polymorphisms with a higher prior likelihood of association were specified a priori for separate analysis. For each visual CT examination feature, a corresponding semiautomated CT feature(s) was identified for comparison in the same subjects.
Measurements and Main Results: GWAS for visual features of chest CT scans identified a genome-wide significant association with visual emphysema at the 15q25 locus (P = 6.3e−9). In the a priori–specified set of 19 previously identified GWAS loci, 7 and 8 loci were associated with airway measures or emphysema measures, respectively. In the a priori–specified candidate gene set, 13 of 196 candidate genes harbored a nearby single-nucleotide polymorphism significantly associated with an emphysema phenotype. Visual CT examination associations were robust to adjustment for semiautomated correlates in many cases.
Conclusions: Standardized visual assessments of emphysema and airway disease are significantly associated with genetic loci previously associated with chronic obstructive pulmonary disease susceptibility or semiautomated CT examination phenotypes in GWAS. Visual CT measures of emphysema and airways disease offer independent information for genetic association studies in relation to standard semiautomated measures.
Keywords: genomics, emphysema, airway remodeling, radiology
In subjects with chronic obstructive pulmonary disease (COPD), lung computed tomographic (CT) imaging enables detailed assessment of emphysema and airway thickness. CT scans can be assessed visually or through the use of semiautomated image processing algorithms. The correlation between semiautomated CT measures with visual assessments and physiologic parameters has been well established (1–4). However, there is increasing recognition that semiautomated evaluation of emphysema and airway disease may not always reflect visual analysis, and that manual, visual evaluation of findings may provide important complementary information (5).
From the perspective of genetic association studies, semiautomated measures are used more commonly than visual assessments, due primarily to the greater availability of semiautomated CT measures in the large research cohorts required for such studies. Genome-wide association studies (GWASs) of emphysema and airway phenotypes, which demonstrate independent familial aggregation in subjects with COPD (6), have identified genetic associations at genome-wide significance (7–12). However, the relative utility of visual versus semiautomated CT measures for genetic association studies has not been explored.
We hypothesized that genetic variants previously associated with COPD or semiautomated measures of emphysema or airway wall thickness would also be associated with visual assessments of COPD using chest CT examinations. We further hypothesized that visual measures may identify novel genetic associations. To test these hypotheses, we performed GWASs for visual emphysema and airway wall thickness in 1,540 subjects from the COPDGene Study. Given our modest sample size, we also focused on two single-nucleotide polymorphism (SNP) sets with a higher prior likelihood of association, specified a priori for targeted investigation. One set consisted of SNPs previously associated with COPD, emphysema, or airway phenotypes at genome-wide significance (prior GWAS SNPs), and the other consisted of SNPs located within 100 kb of a previously published set of COPD candidate genes (candidate gene SNPs) (13).
Methods
Subjects
The COPDGene Study is comprised of 10,192 non-Hispanic white and African American current and former cigarette smokers, with and without COPD. Details of patient selection and data collection have been described previously (14). Subjects provided blood samples for genotyping, underwent spirometry testing, and had inspiratory and expiratory chest CT imaging. Institutional review board approval was obtained at all 21 clinical centers, and written informed consent was obtained from all subjects (see the online supplement).
Image Analysis
At the time of this study, analysis of visual phenotypes was performed for all non-Hispanic white subjects for which complete visual scores were available (n = 1,540). Subjects represented the full spectrum of spirometric abnormalities, including preserved ratio impaired spirometry subjects (see Methods in the online supplement for details) (15).
Visual imaging assessments were performed by two research analysts trained to score scans according to a standardized categorization developed by the Fleischner Society (16). These analysts were blinded to the genotype and case/control status of the subjects. Emphysema was assessed according to six categories of ascending severity: no emphysema; trace centrilobular emphysema; and mild, moderate confluent, or advanced destructive emphysema.
Airway wall thickness was assessed according to three categories of ascending severity: absent, mild, and substantial. For scans in which both reviewers identified some level of abnormality, but the scores for a particular phenotype differed by one level, scores were averaged. For all other instances of disagreement, one of two radiologists (D.A.L. or D.N.) independently reviewed the scans and adjudicated all disagreements. Further details on visual scoring are included in the online supplement.
For each visual phenotype, a semiautomated quantitative measure was used for comparison. The quantitative correlate for visual emphysema was low attenuation area at −950 Hounsfield units (LAA-950), and for visual airway thickness two correlates were used: the square root of wall area for airways standardized at an internal perimeter of 10 mm (Pi10), and wall area percent of airways for segmental (third-generation) bronchi (WAP), both as calculated by VIDA software (VIDA Diagnostics, Coralville, IA).
Measurements were obtained along the centerline of the lumen, in the middle third of the airway segment, for one segmental airway of each lung lobe, including the lingula; the mean value across all lobes was used for analysis. Details of the imaging techniques have been described previously (14).
GWASs
We performed GWASs for of 6,173,964 genotyped and imputed SNPs with minor allele frequencies of 5% or greater across the 22 autosomal chromosomes for visual and semiautomated emphysema and airway measures. Information on genotyping and imputation methods has been previously reported (17) and is included in the online supplement. To ensure equal sample size between the visual measure and its semiautomated correlate, genetic association analysis was limited to subjects with complete data for both the visual and quantitative measure (n = 1,513 and 1,473 for emphysema and airway phenotypes, respectively). In addition, two a priori–specified sets of candidate SNPs were assessed at more lenient significance thresholds based on their higher prior likelihood of association to COPD.
The prior GWAS set was based on 33 lead SNPs across 19 loci previously associated at genome-wide significance with COPD, emphysema, or airway phenotypes (7–12, 17). The candidate gene set consisted of SNPs located within 100 kb of 184 candidate genes from a published review of COPD genetics (13) as well as 12 genes—RIN3, SNRPF, MYO1D, VWA8, HYKK, DLC1, SERPINA10, MIR2054, MAGI2, SERPINE2, NT5C3B, and C10orf90—located near SNPs from the prior GWAS SNP set. Additional details on gene selection are included in the online supplement.
Statistical Analysis
Genetic association tests were conducted separately for visual emphysema and airway scores, using linear regression with visual scores as response and SNP genotype as predictor. In each analysis, we adjusted for sex, age, smoking pack-years, and principal components of genetic ancestry (see details in the supplemental Methods). The same regression model was used for the semiautomated phenotypes as for the visual phenotypes: the semiautomated phenotype served as response variable and the SNP genotype served as predictor, with the aforementioned covariates included as a way to adjust for potential confounders.
For all significant results, we repeated the analysis adjusting for body mass index and current smoking status, and using ordinal logistic regression. The statistical significance threshold for the GWAS analysis was P less than 5.0 × 10−8. For candidate gene analysis, the false discovery rate (FDR) was controlled at 5%, using the method of Storey (18). Linear regression tests were performed in PLINK (19). FDR adjustment was performed using the qvalue package from Bioconductor (20). We generated locus plots and linkage disequilibrium (LD) calculations using LocusZoom (21).
Results
The 1,540 analyzed subjects were non-Hispanic white, 50.1% male, median age of 63.3 years, with median 42.0 pack-year smoking history, and median forced expiratory volume in 1 second (FEV1) of 78.8%. Additional characteristics of the cohort appear in Table 1.
Table 1.
Characteristics of study cohort
| Characteristics | Visual Grade | PRISm | Smoking Controls | GOLD 1 | GOLD 2 | GOLD 3 | GOLD 4 |
|---|---|---|---|---|---|---|---|
| n | 162 | 564 | 176 | 309 | 200 | 128 | |
| Sex (% male) | 41 | 43 | 62 | 55 | 62 | 50 | |
| Age | 62 (8) | 61 (8) | 64 (8) | 65 (8) | 65 (8) | 64 (7) | |
| Pack-years | 40 (30–54) | 35 (23–47) | 42 (31–60) | 47 (35–68) | 52 (40–75) | 49 (38–70) | |
| FEV1/FVC | 0.76 (0.04) | 0.78 (0.05) | 0.64 (0.05) | 0.57 (0.09) | 0.44 (0.10) | 0.31 (0.06) | |
| FEV1 % predicted | 71 (8) | 97 (11) | 92 (10) | 64 (8) | 40 (6) | 23 (5) | |
| LAA-950 | 0.9 (0.4–2.8) | 1.8 (0.7–4.2) | 4.6 (2.0–9.0) | 5.9 (2.5–13.2) | 16.0 (6.7–25.6) | 26.3 (15.5–40.1) | |
| Pi10 | 3.68 (0.11) | 3.64 (0.09) | 3.61 (0.10) | 3.68 (0.13) | 3.72 (0.13) | 3.75 (0.13) | |
| WAP | 61.5 (2.8) | 59.6 (2.6) | 59.9 (2.5) | 62.1 (2.8) | 63.2 (2.8) | 63.6 (2.8) | |
| Emphysema visual grade* | 0 | 98 [6.4] | 389 [25.3] | 53 [3.4] | 79 [5.1] | 16 [1.0] | 4 [0.3] |
| 1 | 36 [2.3] | 100 [6.5] | 30 [1.9] | 51 [3.3] | 18 [1.2] | 3 [0.2] | |
| 2 | 21 [1.4] | 57 [3.7] | 42 [2.7] | 68 [4.4] | 33 [2.1] | 6 [0.4] | |
| 3 | 4 [0.3] | 14 [0.9] | 29 [1.9] | 50 [3.2] | 42 [2.7] | 22 [1.4] | |
| 4 | 3 [0.2] | 4 [0.3] | 16 [1.0] | 37 [2.4] | 43 [2.8] | 29 [1.9] | |
| 5 | 0 [0.0] | 0 [0.0] | 6 [0.4] | 24 [1.6] | 48 [3.1] | 64 [4.2] | |
| Airway wall thickness visual grade† | 0 | 24 [1.6] | 141 [9.2] | 27 [1.8] | 9 [0.6] | 1 [0.1] | 0 [0] |
| 1 | 66 [4.3] | 312 [20.3] | 86 [5.6] | 82 [5.3] | 19 [1.2] | 5 [0.3] | |
| 2 | 72 [4.7] | 111 [7.2] | 63 [4.1] | 218 [14.2] | 180 [11.7] | 123 [8.0] |
Definition of abbreviations: GOLD = Global Initiative for Chronic Obstructive Lung Disease; LAA-950 = percentage of low attenuation areas below −950 Hounsfield units; Pi10 = square root of wall area for airways standardized at an internal perimeter of 10 mm as calculated by VIDA software; PRISm = preserved ratio impaired spirometry (postbronchodilator FEV1/FVC ≥ 0.7 and FEV1 < 80% predicted); smoking controls = smoker subjects with postbronchodilator FEV1 % predicted ≥ 80% and FEV1/FVC ≥ 0.7; WAP = wall area percent of airways for segmental (third-generation) bronchi as calculated by VIDA software.
Values are mean (SD) or median (interquartile range). Numbers shown for visual score categories include fractional scores (i.e., averaged scores of 2.5 are reported under the 3 category).
Data presented in the Emphysema section are visual grade and n [% of total].
Data presented in the Airway wall thickness section are visual grade and n [% of total].
CT scans were scored in duplicate by two research analysts according to a standardized categorization developed by the Fleischner Society (16), with adjudication by a radiologist in instances of disagreement (see Methods). The weighted κ values for agreement between analysts were 0.83 (95% confidence interval = 0.82–0.85) and 0.50 (95% confidence interval = 0.47–0.54) for emphysema and airway wall thickness, respectively. The Spearman correlation coefficients between visual scores, their semiautomated correlates, and postbronchodilator FEV1 (% predicted) appear in Table 2. Visual measures were more highly correlated with FEV1 than were their semiautomated correlates. The correlation between visual emphysema and LAA-950 was 0.61 (Table 2). The correlation between visual airway wall thickness with Pi10 and WAP was 0.28 and 0.55, respectively (Table 2).
Table 2.
Spearman correlation between visual imaging scores and their semiautomated imaging correlates
| Visual Emphysema | Visual Airway Wall Thickness | LAA-950 | Pi10 | WAP | FEV1 % Predicted | |
|---|---|---|---|---|---|---|
| Visual emphysema | 1 | 0.50 (0.45–0.54) | 0.61 (0.57–0.64) | 0.15 (0.09–0.20) | 0.23 (0.17–0.27) | −0.58 (−0.61 to −0.54) |
| Visual airway wall thickness | — | 1 | 0.34 (0.29–0.38) | 0.28 (0.24–0.33) | 0.55 (0.51–0.59) | −0.63 (−0.65 to −0.59) |
| LAA-950 | — | — | 1 | −0.03 (−0.08 to 0.01) | 0.10 (0.04–0.14) | −0.49 (−0.53 to −0.44) |
| Pi10 | — | — | — | 1 | 0.41 (0.36–0.46) | −0.33 (−0.37 to −0.28) |
| WAP | — | — | — | — | 1 | −0.53 (−0.56 to −0.49) |
Definition of abbreviations: LAA-950 = percentage of low attenuation areas below −950 Hounsfield units; Pi10 = square root of wall area for airways standardized at an internal perimeter of 10 mm as calculated by VIDA software; WAP = wall area percent of airways for segmental (third-generation) bronchi as calculated by VIDA software.
Values in parentheses are 95% confidence intervals for Spearman rank correlation coefficients.
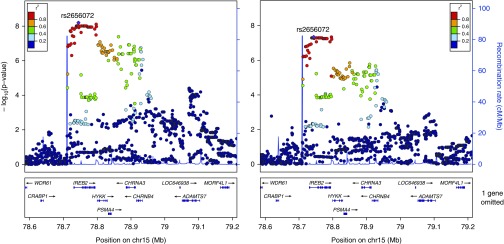
GWAS for visual emphysema and GWAS for LAA-950 identified genome-wide significant associations at the 15q25 locus (lead SNP rs2656072, minor allele frequency = 0.21, P = 6.3 × 10−9 for visual emphysema; P = 4.5 × 10−8 for LAA-950; Figure 1). No genome-wide significant associations were identified for visual airway wall thickness, Pi10, or WAP. The lead visual emphysema SNP at 15q25, rs2656072, was not in LD with any of the six previously identified GWAS lead SNPs at this locus (strongest pairwise LD was for rs12914385 with r2 of 0.14). Conditioning the regression test for rs2656072 on rs12914385 had only a small effect on the rs2656072 association for both visual and semiautomated emphysema (conditioned P = 1.7 × 10−6 for visual emphysema and P = 4.6 × 10−7 for semiautomated emphysema). However, conditioning the regression test for rs12914385 on rs2656072 attenuated the rs12914385 association (conditioned P = 0.19 for visual emphysema and P = 0.92 for semiautomated emphysema).
Figure 1.
Emphysema visual score (left) and low attenuation area (LAA)-950 (right) local association plot at the15q25 locus (n = 1,513): genome-wide significant association signal for both visual emphysema and LAA-950 at the 15q25 locus.
We then tested 33 SNPs in 19 loci from our prior GWAS SNP set previously associated with COPD susceptibility, emphysema, or airway wall thickness based on GWAS. SNPs in eight loci were associated with an emphysema phenotype at P less than 0.05 (see Table E1 in the online supplement), with six loci significantly associated with both the visual score and LAA-950. SNPs in seven loci were associated with an airway phenotype at P less than 0.05 (Table E2), with six loci associated with visual airway thickness and three loci associated with either WAP or Pi10. For all SNPs meeting the P less than 0.05 threshold, the direction of the effect was in agreement with previous reports. In total, 10 of 19 previously identified GWAS loci were nominally associated with visual emphysema or visual airway thickness (Tables E1 and E2).
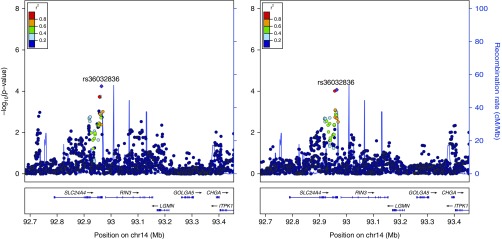
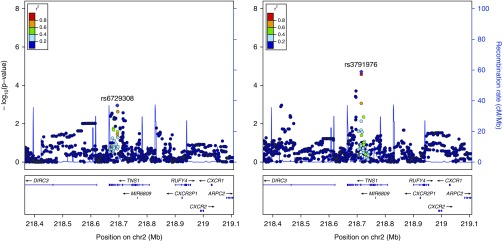
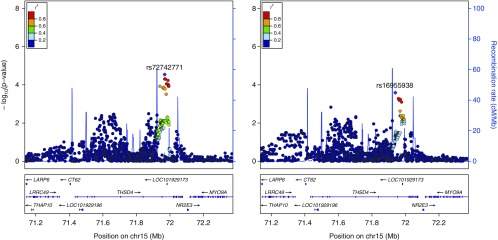
For the candidate gene study, three loci were significantly associated with visual emphysema (Figures 1–3, Table E3) and eight loci were significantly associated with LAA-950 (Table E3, Figures 1 and 4, Figures E1–E6) when using an FDR of 5%. No SNPs from the candidate gene set were significantly associated with visual airway wall thickness or one of the airway semiautomated correlates at 5% FDR.
Figure 3.
Emphysema visual score (left) and low attenuation area (LAA)-950 (right) local association plot at the RIN3 locus (n = 1,513): association signal at the 14q32 locus significant at the false discovery rate less than 0.05 threshold for visual emphysema alongside LAA-950 association signal.
Figure 4.
Emphysema visual score (left) and low attenuation area (LAA)-950 (right) local association plot at the TNS1 locus (n = 1,513): association signal at the 2q35 locus significant at the false discovery rate less than 0.05 threshold for LAA-950 alongside visual emphysema association signal.
Figure 2.
Emphysema visual score (left) and low attenuation area (LAA)-950 (right) local association plot at the THSD4 locus (n = 1,513): association signal at the 15q23 locus significant at the false discovery rate less than 0.05 threshold for visual emphysema alongside LAA-950 association signal.
To determine whether visual measures provided independent information from semiautomated correlates, we repeated genetic association analysis on SNPs with P less than 0.05 (seven visual emphysema loci from Table E1 and six visual airway loci from Table E2), adjusting for semiautomated correlates. After adjustment for LAA-950, three loci remained statistically significant at P less than 0.05 (Table E8). After adjusting the visual airway associations for semiautomated airway measures, three loci remained significant at P less than 0.05 (Table E9). When we performed the opposite analysis (i.e., adjusting the semiautomated associations for the visual correlate), two of the LAA-950 associations remained significant (Table E8), and two of the semiautomated airway associations remained significant (Table E9).
To confirm that our significant results were not due to improper model specification, we performed additional testing for significant results using ordinal logistic regression. Results were similar to those under a linear regression model for all tested lead SNPs (Tables E10 and E11).
Discussion
We performed an analysis of visually assessed emphysema and airway phenotypes in COPDGene, and noted three main findings. First, visual phenotypes are valid for use in genetic association studies based upon reproducibility of previous COPD GWAS findings. Second, visual measures are moderately correlated with corresponding semiautomated phenotypes, indicating correspondence, but incomplete overlap, between these measures. Third, visual and semiautomated measures contain independent information related to genetic association. Some significant genetic associations for both visual and semiautomated measures remained significant after adjustment for the corresponding visual or semiautomated measure.
When threshold-based semiautomated measures for quantifying emphysema from CT scans were introduced, it was demonstrated that semiautomated measures and visual assessment showed similar levels of correlation to pathologic emphysema (22), though other studies have demonstrated better performance for semiautomated measures (23). In what is perhaps the most comprehensive comparison of visual versus semiautomated pulmonary CT assessments, with 58 visual readers, interobserver agreement for presence/absence of emphysema was moderate, with fair but lower levels of agreement for distinct emphysema subtypes (5).
The findings of our study—namely, that visual and semiautomated emphysema assessment both provide useful, partially independent information—support the findings of the previously discussed study, as well as the study by Madani and colleagues (24) that concludes that the correlation with pathologic emphysema is improved by combining multiple different types of measures.
Our study is of interest in part because it relates emphysema measures not to pathology, but to COPD genetic associations, providing a novel metric by which to evaluate visual and semiquantitative measures. Our perspective is that comparisons of currently available emphysema measures are poorly served by overly simplistic questions of which emphysema measure is “better.” It has been extensively demonstrated that various measures of emphysema, and even various Hounsfield unit thresholds for semiautomated measures (22–24), capture different aspects of pathologic emphysema. Although it is a desirable goal to establish a single optimal method for characterizing emphysema from CT scans, our interpretation of our findings supports the perspective that no current emphysema measure is perfect, but many are useful for capturing distinct aspects of emphysema.
The results of this study agree well with previous findings in COPD genetics. The genome-wide significant association at the 15q25 locus is well established, though the lead SNP from the visual emphysema analysis is independent from previously reported associations. This is in line with previous functional investigations of this locus that demonstrate multiple functional variants that regulate different genes in different tissues (25). In addition to the nicotine receptor cluster of genes in this region, DeMeo and coworkers (26) have also implicated IREB2 as a likely COPD candidate gene in this region through an integrative genomics approach, and Cloonan and colleagues (27) used cell-based studies and murine COPD models to demonstrate that IREB2 likely influences COPD pathogenesis by regulating mitochondrial iron. Roughly half of previously identified loci from prior emphysema and airway GWASs were recovered by at least one of the emphysema phenotypes (7/11) and airway phenotypes (3/6). Our study identified significant associations with emphysema for SNPs near 13 out of 196 candidate genes. None of these loci was associated with airway measures, but this may be due to the fact that the majority of candidate gene studies focused on emphysema or COPD affection status rather than airway measures.
In the current study, the quantitative measures of emphysema and airway disease showed only moderate concordance with corresponding visual measures. Reasons for this may include difficulty in defining thresholds for quantitative measures and dependence of quantitative measures on technical parameters (24, 28, 29). Interestingly, the visual scores recovered slightly more signals from prior GWAS and lost fewer of these signals after correlate correction than the semiautomated measures. The RIN3 and MMP3/12 loci were good examples of this phenomenon. RIN3 was strongly associated with both visual emphysema and visual airway thickness, but none of the semiautomated correlates. MMP3/12 was strongly associated with visual airway thickness, but with neither Pi10 nor WAP. These associations remained essentially unchanged after correlate correction.
This finding emerged despite the fact that the prior GWAS signals were mainly discovered using semiautomated measures, and should therefore have biased our replication of previous findings toward the semiautomated correlates rather than the visual phenotypes. Compared with semiautomated CT airway and emphysema phenotypes, these data indicate that visual phenotypes provide independent information. It is not clear whether visual or semiautomated phenotypes are more powerful for genetic discovery, but such statements may be made more definitively with additional analyses of both methods when a larger sample size is available.
Strengths and Limitations
This study has the following strengths. First, CT reviewers were specifically trained to perform visual CT assessment according to consensus standards, with high levels of observer agreement for emphysema. Second, the subjects in our dataset had both visual assessments and semiautomated correlate information, allowing for direct comparison between the visual and the semiautomated correlates. Third, we were able to relate visual and automated radiographic measures to genome-wide assessments of genetic variation, allowing us to assess one aspect of the biologic relevance of these measures.
This study also has important limitations. Although our dataset is similar in size to previous datasets containing visual CT assessment of emphysema and is, to our knowledge, the largest to date in regard to visually assessed airway disease, it is nonetheless small compared with the largest GWAS studies in COPD and other diseases. Thus, our study has limited power to detect modest genetic effects.
Many of our significant associations have previously been identified in COPD GWASs, suggesting that these associations are likely to be true positives, but many other true associations are likely to have gone undetected, and will require future studies with more subjects. We addressed this relative lack of power by specifying in advance two SNP sets with a higher a priori likelihood of harboring genetic associations. Despite the small sample size, our study identified a genome-wide significant GWAS signal at the 15q25 locus for visual emphysema (Figure 1).
Although interobserver agreement for emphysema was high, agreement was more modest for airway measures. It should be noted that, due to the nature of the study, lung tissue was not available for pathologic correlation to emphysema using a histologic approach.
Conclusions
We examined both visual and semiautomated, quantitative COPD measures for association with genetic variants in a dataset of non-Hispanic white subjects. The visual assessments demonstrated independent information, especially in regard to the airway phenotypes, from the quantitative measures relative to genetic association. We anticipate that a larger sample size using visual phenotypes may allow for identification of novel variants at genome-wide significance.
Additional material
Supplementary data supplied by authors.
Footnotes
Supported by a National Institutes of Health (NIH)–Library of Medicine postdoctoral training fellowship, National Library of Medicine grant 5T15LM009451 (E.H.-S.); additional funding came from NIH grant P20 HL113445-01 for the study of chronic obstructive pulmonary disease genomics (S.L.) and NIH grants R01 HL124233 and K08 HL102265 (P.J.C.); the COPDGene Study is supported by grant awards R01 HL089856 and R01 HL089897 from the National Heart, Lung, and Blood Institute; the COPDGene project is also supported by the COPD Foundation through contributions made to an industry advisory board comprised of AstraZeneca, Boehringer Ingelheim, Novartis, Pfizer, Siemens, and Sunovion.
Author Contributions: Study design—E.H.-S., P.J.C., D.A.L., J.D.C., and E.K.S.; data acquisition and quality control—E.H.-S. and D.A.L.; data analysis—E.H.-S., M.H.C., P.J.C., S.L., and D.N.; critical revision of manuscript—all authors.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1513/AnnalsATS.201606-427OC on October 14, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Gould GA, Redpath AT, Ryan M, Warren PM, Best JJ, Flenley DC, MacNee W. Lung CT density correlates with measurements of airflow limitation and the diffusing capacity. Eur Respir J. 1991;4:141–146. [PubMed] [Google Scholar]
- 2.Haruna A, Muro S, Nakano Y, Ohara T, Hoshino Y, Ogawa E, Hirai T, Niimi A, Nishimura K, Chin K, et al. CT scan findings of emphysema predict mortality in COPD. Chest. 2010;138:635–640. doi: 10.1378/chest.09-2836. [DOI] [PubMed] [Google Scholar]
- 3.Grydeland TB, Dirksen A, Coxson HO, Eagan TM, Thorsen E, Pillai SG, Sharma S, Eide GE, Gulsvik A, Bakke PS. Quantitative computed tomography measures of emphysema and airway wall thickness are related to respiratory symptoms. Am J Respir Crit Care Med. 2010;181:353–359. doi: 10.1164/rccm.200907-1008OC. [DOI] [PubMed] [Google Scholar]
- 4.Washko GR, Criner GJ, Mohsenifar Z, Sciurba FC, Sharafkhaneh A, Make BJ, Hoffman EA, Reilly JJ. Computed tomographic-based quantification of emphysema and correlation to pulmonary function and mechanics. COPD. 2008;5:177–186. doi: 10.1080/15412550802093025. [DOI] [PubMed] [Google Scholar]
- 5.COPDGene CT Workshop Group. Barr RG, Berkowitz EA, Bigazzi F, Bode F, Bon J, Bowler R, Chiles C, Crapo JD, Criner GJ, Curtis JL, et al. A combined pulmonary–radiology workshop for visual evaluation of COPD: study design, chest CT findings and concordance with quantitative evaluation. COPD. 2012;9:151–159. doi: 10.3109/15412555.2012.654923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel BD, Coxson HO, Pillai SG, Agustí AG, Calverley PM, Donner CF, Make BJ, Müller NL, Rennard SI, Vestbo J, et al. International COPD Genetics Network. Airway wall thickening and emphysema show independent familial aggregation in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;178:500–505. doi: 10.1164/rccm.200801-059OC. [DOI] [PubMed] [Google Scholar]
- 7.Castaldi PJ, Cho MH, San José Estépar R, McDonald M-LN, Laird N, Beaty TH, Washko G, Crapo JD, Silverman EK COPDGene Investigators. Genome-wide association identifies regulatory Loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190:399–409. doi: 10.1164/rccm.201403-0569OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho MH, Castaldi PJ, Hersh CP, Hobbs BD, Barr RG, Tal-Singer R, Bakke P, Gulsvik A, San José Estépar R, Van Beek EJ, et al. NETT Genetics, ECLIPSE, and COPDGene Investigators. A genome-wide association study of emphysema and airway quantitative imaging phenotypes. Am J Respir Crit Care Med. 2015;192:559–569. doi: 10.1164/rccm.201501-0148OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dijkstra AE, Postma DS, van Ginneken B, Wielpütz MO, Schmidt M, Becker N, Owsijewitsch M, Kauczor H-U, de Koning HJ, Lammers JW, et al. Novel genes for airway wall thickness identified with combined genome-wide association and expression analyses. Am J Respir Crit Care Med. 2015;191:547–556. doi: 10.1164/rccm.201405-0840OC. [DOI] [PubMed] [Google Scholar]
- 10.Kong X, Cho MH, Anderson W, Coxson HO, Muller N, Washko G, Hoffman EA, Bakke P, Gulsvik A, Lomas DA, et al. ECLIPSE Study NETT Investigators. Genome-wide association study identifies BICD1 as a susceptibility gene for emphysema. Am J Respir Crit Care Med. 2011;183:43–49. doi: 10.1164/rccm.201004-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manichaikul A, Hoffman EA, Smolonska J, Gao W, Cho MH, Baumhauer H, Budoff M, Austin JHM, Washko GR, Carr JJ, et al. Genome-wide study of percent emphysema on computed tomography in the general population: the Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilk JB, Shrine NR, Loehr LR, Zhao JH, Manichaikul A, Lopez LM, Smith AV, Heckbert SR, Smolonska J, Tang W, et al. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am J Respir Crit Care Med. 2012;186:622–632. doi: 10.1164/rccm.201202-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bossé Y. Updates on the COPD gene list. Int J Chron Obstruct Pulmon Dis. 2012;7:607–631. doi: 10.2147/COPD.S35294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, Curran-Everett D, Silverman EK, Crapo JD. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan ES, Castaldi PJ, Cho MH, Hokanson JE, Regan EA, Make BJ, Beaty TH, Han MK, Curtis JL, Curran-Everett D, et al. COPDGene Investigators. Epidemiology, genetics, and subtyping of preserved ratio impaired spirometry (PRISm) in COPDGene. Respir Res. 2014;15:89. doi: 10.1186/s12931-014-0089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch DA, Austin JHM, Hogg JC, Grenier PA, Kauczor H-U, Bankier AA, Barr RG, Colby TV, Galvin JR, Gevenois PA, et al. CT-definable subtypes of chronic obstructive pulmonary disease: a statement of the Fleischner Society. Radiology. 2015;277:192–205. doi: 10.1148/radiol.2015141579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho MH, McDonald M-LN, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, DeMeo DL, Sylvia JS, Ziniti J, Laird NM, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storey JD. A direct approach to false discovery rates. J R Stat Soc B. 2002;64:479–498. [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller NL, Staples CA, Miller RR, Abboud RT. “Density mask”: an objective method to quantitate emphysema using computed tomography. Chest. 1988;94:782–787. doi: 10.1378/chest.94.4.782. [DOI] [PubMed] [Google Scholar]
- 23.Bankier AA, De Maertelaer V, Keyzer C, Gevenois PA. Pulmonary emphysema: subjective visual grading versus objective quantification with macroscopic morphometry and thin-section CT densitometry. Radiology. 1999;211:851–858. doi: 10.1148/radiology.211.3.r99jn05851. [DOI] [PubMed] [Google Scholar]
- 24.Madani A, Zanen J, de Maertelaer V, Gevenois PA. Pulmonary emphysema: objective quantification at multi-detector row CT—comparison with macroscopic and microscopic morphometry. Radiology. 2006;238:1036–1043. doi: 10.1148/radiol.2382042196. [DOI] [PubMed] [Google Scholar]
- 25.Castaldi PJ, Cho MH, Zhou X, Qiu W, Mcgeachie M, Celli B, Bakke P, Gulsvik A, Lomas DA, Crapo JD, et al. Genetic control of gene expression at novel and established chronic obstructive pulmonary disease loci. Hum Mol Genet. 2015;24:1200–1210. doi: 10.1093/hmg/ddu525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeMeo DL, Mariani T, Bhattacharya S, Srisuma S, Lange C, Litonjua A, Bueno R, Pillai SG, Lomas DA, Sparrow D, et al. Integration of genomic and genetic approaches implicates IREB2 as a COPD susceptibility gene. Am J Hum Genet. 2009;85:493–502. doi: 10.1016/j.ajhg.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cloonan SM, Glass K, Laucho-Contreras ME, Bhashyam AR, Cervo M, Pabón MA, Konrad C, Polverino F, Siempos II, Perez E, et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat Med. 2016;22:163–174. doi: 10.1038/nm.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madani A, De Maertelaer V, Zanen J, Gevenois PA. Pulmonary emphysema: radiation dose and section thickness at multidetector CT quantification—comparison with macroscopic and microscopic morphometry. Radiology. 2007;243:250–257. doi: 10.1148/radiol.2431060194. [DOI] [PubMed] [Google Scholar]
- 29.Gevenois PA, de Maertelaer V, De Vuyst P, Zanen J, Yernault JC. Comparison of computed density and macroscopic morphometry in pulmonary emphysema. Am J Respir Crit Care Med. 1995;152:653–657. doi: 10.1164/ajrccm.152.2.7633722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.