Abstract
Objectives
This meta-analysis investigates the prognostic effect of SET binding protein 1 (SETBP1) mutations in patients with myelodysplastic syndromes (MDS), chronic myelomonocytic leukemia (CMML), or chronic neutrophilic leukemia (CNL).
Methods
Eligible studies from Pubmed, Embase, and Web of Science were searched from database inception through April 2016. Hazard ratios (HRs) and 95% confidence interval (CI) of overall survival (OS) were pooled to calculate the prognostic significance of SETBP1 mutation in patients.
Results
A total of 12 studies with 2321 patients were included in this meta-analysis; 4 studies for MDS, 5 studies for CMML, and 3 studies for CNL. Pooled results suggested that MDS and CMML patients with SETBP1 mutations had a significantly poorer prognosis when compared with patients with wild-type SETBP1 (MDS: HR = 1.808, 95% CI (1.218–2.685), P = 0.001; CMML: HR = 2.223, 95% CI (1.493–3.308), P<0.001). SETBP1 mutations in CNL patients however, showed no significant effect on the overall survival (HR = 1.773, 95% CI (0.877–3.582), P = 0.111). The Begg’s and Egger’s tests did not show significant publication bias in any groups.
Conclusions
Current evidence shows that SETBP1 mutation is associated with a poor prognosis in patients with MDS and CMML, but not in patients with CNL.
Introduction
SETBP1, a gene located on chromosome 18q21.1, encodes SET binding protein 1 that binds with SET nuclear oncoprotein to form a heterodimer complex. Although incompletely characterized, in vitro studies have suggested that this heterodimer inhibits the activity of protein phosphatase type 2A (PP2A), a tumor suppressor, and increases the rate of cell proliferation [1, 2].
Germline mutations in SETBP1 were first identified in children with Schinzel-Giedion syndrome (SGS), where SETBP1 mutations caused congenital disorder characterized by mental retardation, facial deformities and congenital anomalies [3, 4]. More recently, studies have revealed the association of somatic SETBP1 mutations with different types of leukemia. Cristobal et al. demonstrated that the t(12;18)(p13;q12) translocation induced the overexpression of SETBP1, leading to poor prognosis in elderly patients with acute myeloid leukemia (AML) [1]. In 2013, exome-sequencing analysis of patients with atypical chronic myeloid leukemia (aCML) identified the presence of missense SETBP1 mutations [2]. Patients who had SETBP1 mutations had poorer prognosis with a higher white blood cell count when compared with patients who had wild-type SETBP1 [2]. These mutations were clustered from residue 858 to 871 located in the highly conserved SKI-homologous domain [3], suggesting a functional alteration in the mutations.
SETBP1 mutations are also detected in myelodysplastic syndromes (MDS), chronic myelomonocytic leukemia (CMML), and chronic neutrophilic leukemia (CNL). For example, Piazza et al. detected SETBP1 mutations in 3 out of 82 patients with CMML but no SETBP1 mutations were found in 100 patients with MDS [2]. Makishima et al. detected SETBP1 mutations in 22 out of 152 patients with CMML [5] and Damm et al. observed SETBP1 mutations in 12 out of 195 patients with CMML and in 5 out of 222 patients with MDS [6]. Elliott et al. also detected SETBP1 mutations in 5 out of 14 patients with CSF3R-mutated CNL [7]. Recently, through whole exome sequencing, Mason et al. identified SETBP1 mutations in 11 out of 69 patients with CMML [8] and Merlevede et al. reported detection of SETBP1 mutations in one out of 49 patients with CMML [9]. However, the prognostic significance of SETBP1 mutations in MDS, CMML, and CNL remains inconclusive. Reports have suggested that the presence of SETBP1 mutations in these myeloid malignancies reduced survival or are associated with poorer prognosis in patients [10, 11]. However, some studies have also reported that SETBP1 had no significant effect on the overall survival [12, 13]. Therefore, our meta-analysis aims to evaluate the prognostic effect of SETBP1 mutations in patients with MDS, CMML, and CNL. To the best of our knowledge, this is first meta-analysis to evaluate the overall survival (OS) or hazard ratio (HR) for SETBP1 mutations in these patients.
Methods
Literature search
Relevant literatures were systematically searched from electronic databases including PubMed, Embase, and Web of Science. The last search date was April 21, 2016. The following search terms were used: "SET binding protein 1" OR SETBP1 OR SEB AND (leukemia OR leukaemia OR leucocythaemia OR leukemia OR MDS OR "myelodysplastic syndrome" OR "myelodysplasia" OR "preleukemia" OR "myelomonocytic leukemia" OR acml OR “atypical chronic myeloid leukemia” OR “myeloid malignancies” OR “neutrophilic leukemia”) AND (prognosis OR survival OR prognostic OR "Kaplan-Meier analyses").
Study selection
Studies were included when they met the following inclusion criteria: All published articles (including letter to editor) focused on SETBP1 mutations in MDS, CMML, and CNL patients, and provided data on the overall survival (OS) or leukemia-free survival (LFS).
The exclusion criteria were as followed: (1) Articles on the association of SETBP1 expression levels with the OS and LFS of patients with MDS, CMML and CNL; (2) Type of disease was not specified in the article; (3) Diseases other than MDS, CMML, CNL and aCML; (4) Conference proceedings that contained only summaries; (5) Literature published in languages other than Chinese or English; (6) Literature with data that could not be extracted or data that were duplicated.
Data extraction
Two reviewers independently extracted the following information from the included studies: first author, year of publication, source of publication, types of diseases, population studies, total number of patients and number of patients with SETBP1 mutation, diagnostic criteria for disease, mutation detection methods, types of SETBP1 mutations and the corresponding hazard ratios (HRs) with 95% confidence interval (95% CI) for OS. When the HR values were not provided and the only available data were presented in the form of graphical survival curves, the Engauge Digitizer software version 4.1 was used to extract the X- and Y-coordinates of each event from the curve, and converted the coordinates into estimated survival rates at specified time points. SPSS19.0 was then used to reconstruct the Kaplan-Meier curve, and the HR estimates and 95% CIs were obtained using Cox regression analysis. For the remaining data that could not be extracted, contacts with the authors were made to fill in the missing data. Disagreements were resolved through consensus of a third reviewer.
Methodological quality assessment
The methodological quality of the included studies was evaluated using the Newcastle-Ottawa Scale (NOS). The NOS consisted of 3 categories (Selection, Comparability, and Exposure/Outcome). The categories were assigned 4, 2, and 3 stars respectively, with the distribution of the stars as follows: In the Selection and Exposure/Outcome categories, a maximum of one star could be awarded for each numbered item within each category, whereas in the Comparability category, a maximum of two stars could be awarded for a high quality study. Therefore, the quality of an included study could range between 0–9 stars.
Statistical analysis
All meta-analyses were performed using Stata software version 12.0 (College Station, TX, USA). The effects of SETBP1 mutations on OS were assessed using overall HR and 95% CI. Heterogeneity between studies was evaluated using the Q test (where P<0.10 was considered significant) and I2 statistic (I2 = 0–25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2 = 50–75%, large heterogeneity; I2 = 75–100% extreme heterogeneity). When there was no statistically significant heterogeneity (I2<50%) present, fixed-effects model was adopted as the pooling method. Otherwise, the random-effects model was conducted. HR>1 indicated that patients with SETBP1 mutations have a poorer prognosis than patients with wild-type SETBP1. The sensitivity analysis was performed using the sequential omission of individual studies to assess the quality and robustness of the results. Publication bias was assessed using Begg’s and Egger’s tests. A two-sided P value less than 0.05 was considered to be statistically significant.
Results
Study selection
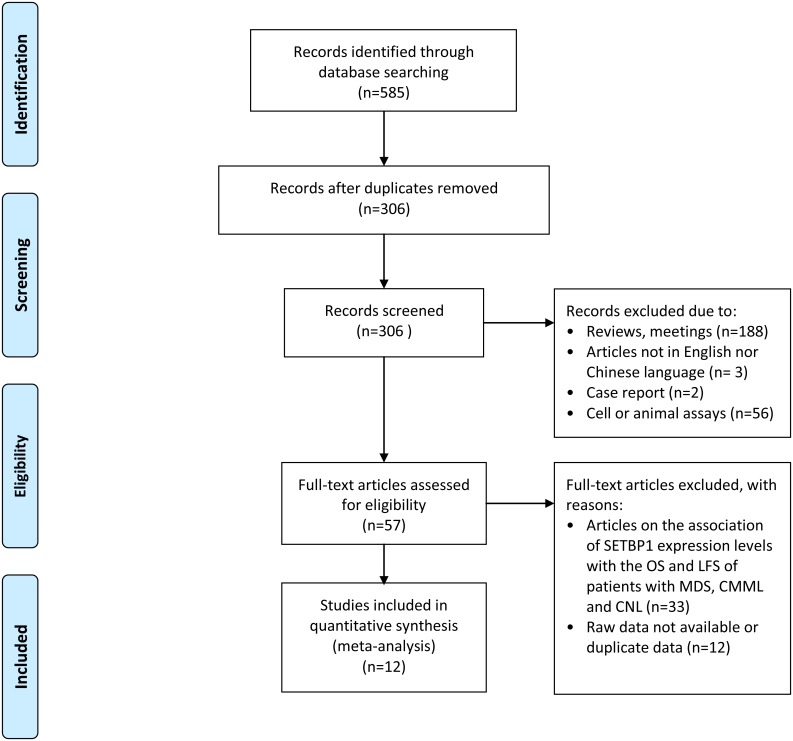
Our systematic literature search identified a total of 585 publications and 279 were removed because they were duplicates. The remaining 306 publications were reviewed by their titles and abstracts, 249 articles were then eliminated due to the following reasons: reviews or meetings abstracts (188), articles neither in English nor Chinese (3), case reports (2), and articles related to cell or animal assays (56). Another 45 articles were excluded after full text review as they were articles on the association of SETBP1 expression levels with the OS and LFS of patients with MDS, CMML and CNL or the raw data was not available or duplicated. This left 12 studies that met the criteria for this meta-analysis (Fig 1).
Fig 1. Flow chart of study selection.
Study characteristics
Twelve studies with a total of 2587 patients were included for meta-analysis; 1167 patients with MDS (4 studies), 1364 patients with CMML (5 studies), and 56 patients with CNL (3 studies). The characteristics and patient demographic data are summarized in Table 1. All studies examined patients with MDS, CMML, or CNL screened for SETBP1 mutations and their corresponding OS. These studies were reported between 2013 and 2015. Of these studies, 4 originated from USA, 4 from Europe, and 4 from Asia. Sample sizes of these studies varied considerably, from as few as 8 patients to as many as 727 patients. Most of the diagnosed diseases were based on the WHO classification except for one study where the disease classification was not mentioned. Ten studies identified SETBP1 mutations using PCR and Sanger sequencing; however, two studies did not mention the methods for detection.
Table 1. Summary of data extracted from 12 studies included in the meta-analysis.
| Study | Year of publication | Journal | Types of diseases | Population studied | Patient (total) | Diagnosis criteria of disease | Rate of SETBP1 mutation case | Method to distinguish mutations | SETBP1 mutations |
|---|---|---|---|---|---|---|---|---|---|
| A Pardanani [14] | 2013 | Leukemia | CNL | USA | 35 | WHO classification | 17% | PCR and Sanger sequencing | D868N D870D G827R G870S |
| Daichi Inoue [15] | 2015 | Leukemia | MDS | Taiwan | 386 | WHO classification | 9% | PCR and Sanger sequencing | D868N E858K G870S S867R |
| F Damm [6] | 2013 | Leukemia | CMML | France | 195 | WHO classification | 6% | PCR and Sanger sequencing | D868N G870S G870D E858K D868H S869G S869R I871S D908N |
| F Thol [12] | 2013 | Leukemia | AML and MDS | German | 326 | WHO classification | 2% | PCR and Sanger sequencing | D868N G870S S854A S869N |
| Hideki Makishima [5] | 2013 | Nat Genet | MM | Japan | 727 | WHO classification | - | PCR and Sanger sequencing | D868N G870S I871T D868T D880N S869N D880E |
| Hou [10] | 2014 | American Journal of Hematology | MDS | Taiwan | 430 | WHO classification | 3% | PCR and Sanger sequencing | D868N G870S E858K S867R I871T |
| M Meggendorfer [16] | 2013 | Leukemia | CMML and aCML | Italy | 227 | WHO classification | 12% | PCR and Sanger sequencing | G870S I871T G870D |
| Michelle A. Elliott [7] | 2015 | American Journal of Hematology | CNL | USA | 13 | WHO classification | 31% | - | G870D D868N G872R |
| Mrinal M. Patnaik [17] | 2015 | American Journal of Hematology | CMML | USA | 36 | WHO classification | 3% | - | - |
| RR Laborde [11] | 2015 | Leukemia | CMML | USA | 179 | - | 4% | PCR and Sanger sequencing | D868N D868Y G870S I871T |
| Vera Adema [18] | 2015 | British Journal of Haematology | MDS | Barcelona | 25 | WHO classification | 50% | PCR and Sanger sequencing | - |
| Cui [19] | 2014 | Chinese Journal of Hematology | CNL | China | 8 | WHO classification | 50% | PCR and Sanger sequencing | D847N D868N G870S I871T |
WHO: World Health Organization
MM: Myeloid Malignancies
CNL: Chronic neutrophilic leukemia
aCML: Atypical chronic myeloid leukemia
MDS: Myelodysplastic syndromes
CMML: Chronic myelomonocytic leukemia
Quality of studies
Individual studies scored well on the NOS; 10 studies scored 8 out of 9 stars, 1 study scored 7 out of 9 stars, and 1 with 6 out of 9 stars (Table 2). Overall, all studies included in this meta-analysis were of high quality.
Table 2. Quality assessment of individual study.
| Study | Selection | Comparability | Outcome | Score | |||||
|---|---|---|---|---|---|---|---|---|---|
| Representativeness of exposed cohort | Selection of non-exposed cohort | Ascertainment of exposure | Outcome not present at start | Assessment of outcome | Follow-up length | Follow-up adequacy | |||
| A Pardanani (2013) [14] | * | * | * | * | * | * | * | * | 8 |
| Daichi Inoue (2015) [15] | * | * | * | * | * | * | * | * | 8 |
| F Damm (2013) [6] | * | * | * | * | * | * | * | * | 8 |
| F Thol (2013) [12] | * | * | * | * | * | * | * | * | 8 |
| Hideki Makishima (2013) [5] | * | * | * | * | * | * | * | * | 8 |
| Hou (2014) [10] | * | * | * | * | * | * | * | * | 8 |
| M Meggendorfer (2013) [16] | * | * | * | * | * | * | * | * | 8 |
| Michelle A. Elliott (2015) [7] | * | * | * | * | * | * | 6 | ||
| Mrinal M. Patnaik (2015) [17] | * | * | * | * | * | * | * | 7 | |
| RR Laborde (2015) [11] | * | * | * | * | * | * | * | * | 8 |
| Vera Adema (2015) [18] | * | * | * | * | * | * | * | * | 8 |
| Cui (2014) [19] | * | * | * | * | * | * | * | * | 8 |
Relationship between SETBP1 mutation and MDS prognosis
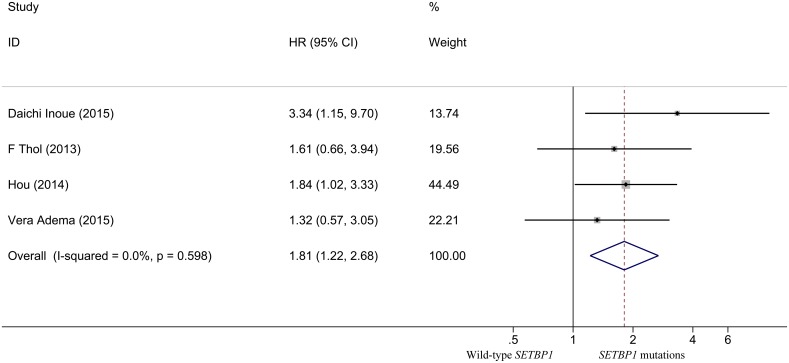
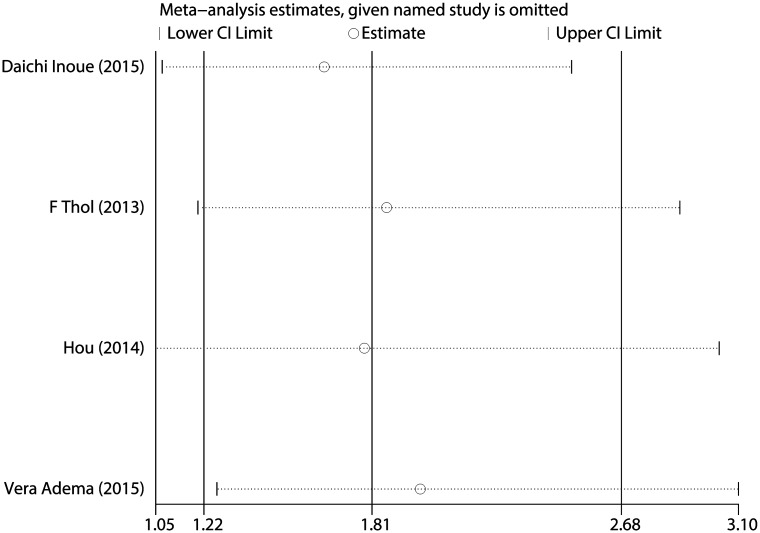
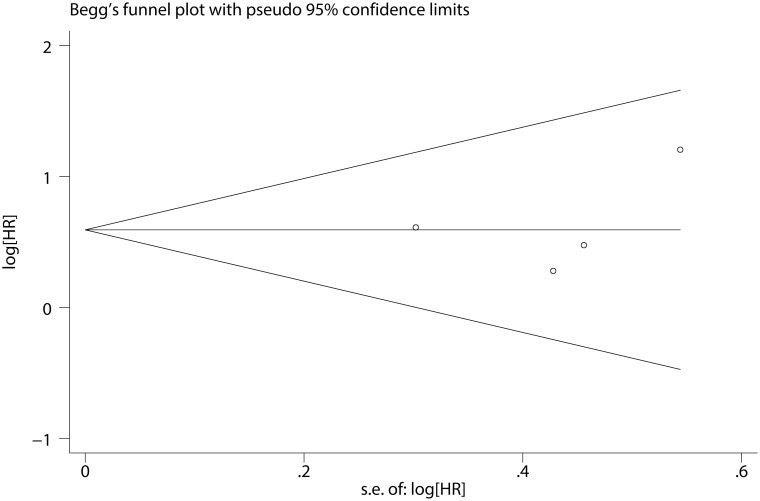
The quantitative analysis for patients with MDS showed no significant heterogeneity between studies (I2 = 0.0%, P = 0.598). Fixed-effects model showed the pooled HR for the OS comparing SETBP1 mutation versus wild-type SETBP1 was 1.808 (95% CI: 1.218–2.685, P = 0.001) (Fig 2), suggesting that SETBP1 mutation may be a factor for the poor prognosis in MDS patients. Sensitivity analysis showed no individual studies significantly change the pooled HR (Fig 3), indicating that the results were reliable and stable. The Begg's (P = 0.734) and Egger's (P = 0.668) tests showed no significant publication bias (Fig 4 and Table 3).
Fig 2. Forest plot of the HR and 95% CI for OS in MDS patients.
Fig 3. Sensitivity analysis for all included MDS studies.
Fig 4. Begg’s funnel plot of the prognostic significance of SETBP1 mutation in MDS patients.
Table 3. Meta-analysis of the prognostic effect of SETBP1 mutations compared with wild type SETBP1 in MDS, CNL and CMML patients.
| Study | HR | Lower Limit | Upper Limit | P (HR) | I2 | P (Heterogeneity) | P (Begg's Test) | P (Egger's test) |
|---|---|---|---|---|---|---|---|---|
| CMML | 2.223 | 1.493 | 3.308 | <0.001 | 30.4% | 0.219 | 0.462 | 0.568 |
| CNL | 1.773 | 0.877 | 3.582 | 0.111 | 0.0% | 0.498 | 0.296 | 0.375 |
| MDS | 1.808 | 1.218 | 2.685 | 0.001 | 0.0% | 0.598 | 0.734 | 0.668 |
Relationship between SETBP1 mutation and CMML prognosis
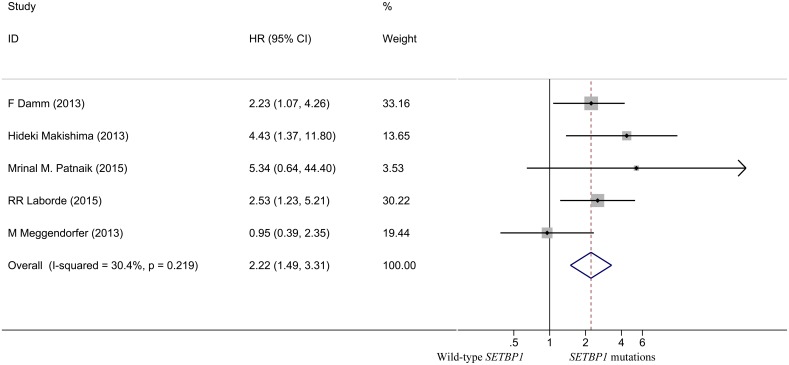
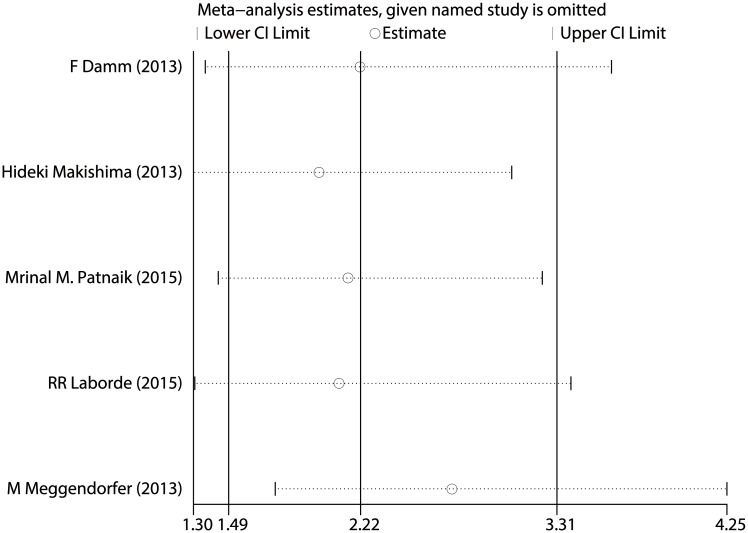
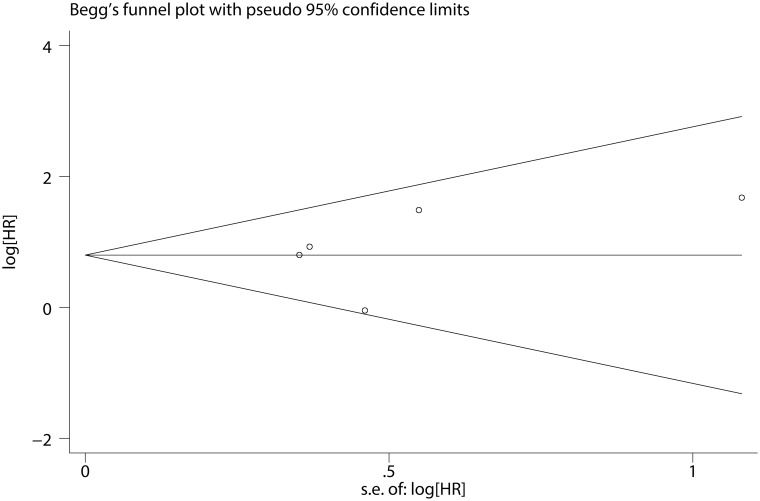
As shown in Fig 5, the pooled HR for OS in CMML patients was 2.223 (95% CI: 1.493–3.308, P <0.001), suggesting that SETBP1 mutation may also be a factor of the poor prognosis in CMML patients. No heterogeneity between studies was observed (I2 = 30.4%, P = 0.219) and fixed-effects model was applied. Sensitivity analysis showed that the results were consistent (Fig 6). The Begg's (P = 0.462) and Egger's (P = 0.568) tests also revealed no evidence of publication bias (Fig 7 and Table 3).
Fig 5. Forest plot of the HR and 95% CI for OS in CMML patients.
Fig 6. Sensitivity analysis for all included CMML studies.
Fig 7. Begg’s funnel plot of the prognostic significance of SETBP1 mutation in CMML patients.
Relationship between SETBP1 mutation and CNL prognosis
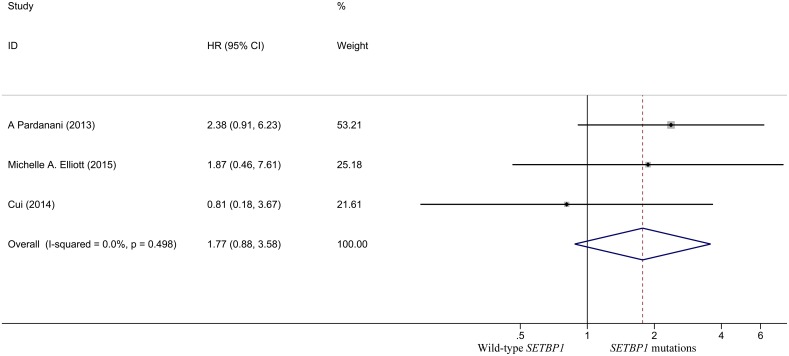
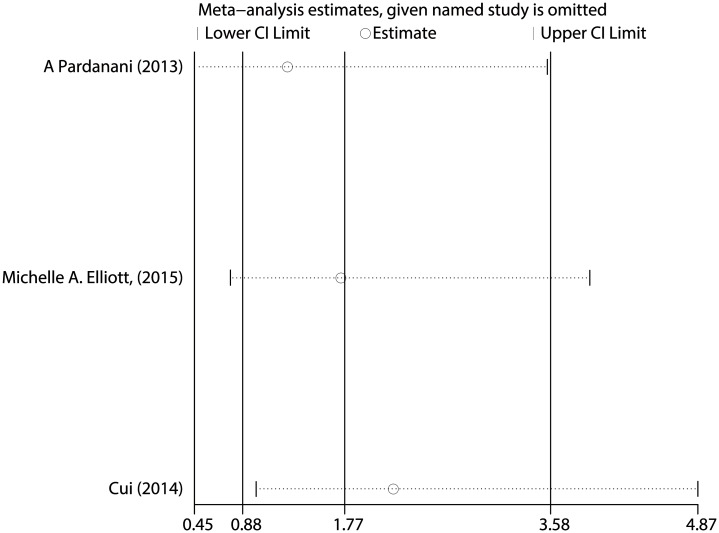
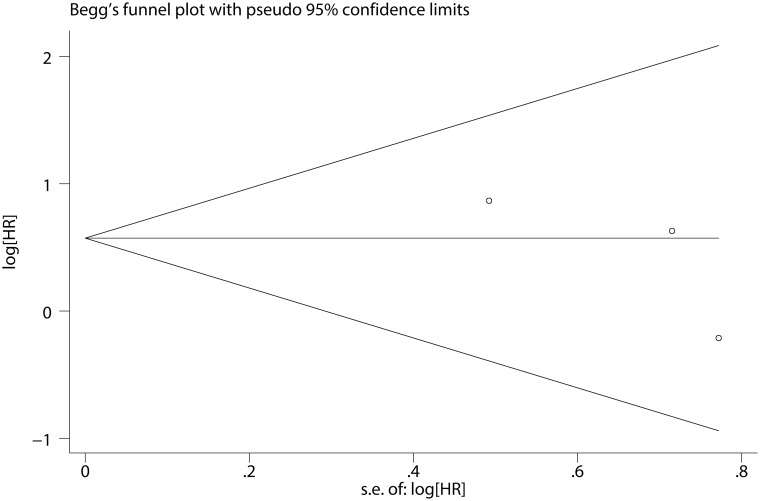
The pooled HR for the OS in CNL patients was 1.773 (95% CI: 0.877–3.582, P = 0.111) (Fig 8), and showed no statistically significant difference in the prognosis between SETBP1 mutation and wildtype CNL patients. There was no heterogeneity between studies (I2 = 0.0%, P = 0.498) and fixed-effects model was applied. Sensitivity analysis showed that the results were consistent (Fig 9). There was no evidence of publication bias (Begg’s test, P = 0.296; Egger’s test, P = 0.375) (Fig 10 and Table 3).
Fig 8. Forest plot of the HR and 95% CI for OS in CNL patients.
Fig 9. Sensitivity analysis for all included CNL studies.
Fig 10. Begg’s funnel plot of the prognostic significance of SETBP1 mutation in CNL patients.
Discussion
MDS, CMML, and CNL are chronic myeloid malignancies involved in the hematopoietic system and can be separated into three categories according to the 2008 World Health Organization (WHO) classification: myelodysplastic syndrome (MDS), myeloproliferative neoplasm (CNL), and myelodysplastic/myeloproliferative neoplasms overlap (CMML). These diseases show abnormalities in myeloid progenitor proliferation and differentiation. Genetic mutations are major risk factors for these diseases. Recently, mutations of the SETBP1 gene have been reported in patients with MDS [10, 12, 15, 18], CMML [5, 6, 11, 16, 17], and CNL [7, 14, 19]. However, the role of SETBP1 mutations in these diseases remains to be elucidated.
The major aim of this meta-analysis was to investigate the relationship between SETBP1 mutations and the prognosis in patients with MDS, CMML, or CNL. This meta-analysis suggests that SETBP1 mutations are strongly associated with a poorer survival in patients with MDS (HR = 1.808, 95% CI (1.218–2.685), P = 0.001) and CMML (HR = 2.223, 95% CI (1.493–3.308), P<0.001), but not in patients with CNL (HR = 1.773, 95% CI (0.877–3.582), P = 0.111). No heterogeneities between studies were observed, indicating that the SETBP1 mutations are not influenced by variables such as age and sex. Furthermore, Laborde et al. has also previously demonstrated that there were no significant correlations between SETBP1 mutations and age, and gender in CMML patients [11]. Our results show that the prognostic impact of SETBP1 mutations is similar to that of ASXL1 mutations, which is one of the strongest independent negative prognostic factor [13, 20] in MDS and CMML patients (HR = 1.45, 95% CI (1.24–1.70)) [21].
Although the role of SETBP1 remains unclear, Inoue et al. recently demonstrated that SETBP1 mutations augmented leukemic transformation and subsequently adverse survival in MDS patients with ASXL1 mutations [15]. SETBP1 mutations targeted the substrate recognition domain of the E3 ubiquitin ligase and inhibited ubiquitin binding [2, 15]. Loss of ubiquitination affected the degradation of SETBP1, leading to a gain-of-function effect. Increased SETBP1 expressions suppressed the PP2A activity, activated the Akt pathway, and enhanced the expression of posterior Hoxa genes in ASXL1-mutant cells [15]. This data suggest that SETBP1 mutations are potential drivers of other mutations that lead to poor prognosis. However, the statistically significant association between SETBP1 with other mutations is controversial. Damm et al. reported that the SETBP1 mutations observed in about 6% of patients with CMML are likely associated with ASXL1 mutations [6], while Thol et al. demonstrated that no association was found between ASXL1 and SETBP1 mutations in his MDS patients [12]. Therefore, this context remains to be determined.
This meta-analysis has a few limitations. First, the relatively small sample sizes of some of the included studies may result in an overestimation of the effect size when compared with larger sample size studies. Second, not all of the HRs with 95% CI values were obtained directly from the included studies. Some studies provided Kaplan-Meier curves rather than the HRs with 95% CI for OS. Therefore, some of the HRs and 95% CI values were obtained from the Kaplan-Meier curves, which might lead to less accurate data. Third, as per discussed, there could be association between SETBP1 and other mutations. In the current meta-analysis however, we did not demonstrate the association of SETBP1 mutations with other mutations as there were limited studies available. Moreover, our analyses indicated that were was no heterogeneity between studies and the sensitivity analyses showed the robustness of our results.
Conclusion
In conclusion, our meta-analysis showed that SETBP1 mutations have unfavourable prognosis in MDS and CMML patients. These findings may help physicians to evaluate the risk levels and prognosis of the disease for appropriate treatments.
Supporting information
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Cristobal I, Blanco FJ, Garcia-Orti L, Marcotegui N, Vicente C, Rifon J, et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115(3):615–25. Epub 2009/12/08. 10.1182/blood-2009-06-227363 [DOI] [PubMed] [Google Scholar]
- 2.Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2013;45(1):18–24. Epub 2012/12/12. 10.1038/ng.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, Steehouwer M, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42(6):483–5. 10.1038/ng.581 [DOI] [PubMed] [Google Scholar]
- 4.Schinzel A, Giedion A. A syndrome of severe midface retraction, multiple skull anomalies, clubfeet, and cardiac and renal malformations in sibs. Am J Med Genet. 1978;1(4):361–75. 10.1002/ajmg.1320010402 [DOI] [PubMed] [Google Scholar]
- 5.Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y, et al. Somatic SETBP1 mutations in myeloid malignancies. Nat Genet. 2013;45(8):942–6. Epub 2013/07/09. 10.1038/ng.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damm F, Itzykson R, Kosmider O, Droin N, Renneville A, Chesnais V, et al. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia. 2013;27(6):1401–3. Epub 2013/02/28. 10.1038/leu.2013.35 [DOI] [PubMed] [Google Scholar]
- 7.Elliott MA, Pardanani A, Hanson CA, Lasho TL, Finke CM, Belachew AA, et al. ASXL1 mutations are frequent and prognostically detrimental in CSF3R-mutated chronic neutrophilic leukemia. Am J Hematol. 2015;90(7):653–6. 10.1002/ajh.24031 [DOI] [PubMed] [Google Scholar]
- 8.Mason CC, Khorashad JS, Tantravahi SK, Kelley TW, Zabriskie MS, Yan D, et al. Age-related mutations and chronic myelomonocytic leukemia. Leukemia. 2016;30(4):906–13. 10.1038/leu.2015.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merlevede J, Droin N, Qin T, Meldi K, Yoshida K, Morabito M, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7:10767 10.1038/ncomms10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou HA, Kuo YY, Tang JL, Chou WC, Yao M, Lai YJ, et al. Clinical implications of the SETBP1 mutation in patients with primary myelodysplastic syndrome and its stability during disease progression. American journal of hematology. 2014;89(2):181–6. Epub 2013/10/16. 10.1002/ajh.23611 [DOI] [PubMed] [Google Scholar]
- 11.Laborde RR, Patnaik MM, Lasho TL, Finke CM, Hanson CA, Knudson RA, et al. SETBP1 mutations in 415 patients with primary myelofibrosis or chronic myelomonocytic leukemia: independent prognostic impact in CMML. Leukemia. 2013;27(10):2100–2. Epub 2013/04/06. 10.1038/leu.2013.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thol F, Suchanek KJ, Koenecke C, Stadler M, Platzbecker U, Thiede C, et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia. 2013;27(10):2072–5. Epub 2013/05/08. 10.1038/leu.2013.145 [DOI] [PubMed] [Google Scholar]
- 13.Patnaik MM, Itzykson R, Lasho TL, Kosmider O, Finke CM, Hanson CA, et al. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: a two-center study of 466 patients. Leukemia. 2014;28(11):2206–12. 10.1038/leu.2014.125 [DOI] [PubMed] [Google Scholar]
- 14.Pardanani A, Lasho TL, Laborde RR, Elliott M, Hanson CA, Knudson RA, et al. CSF3R T618I is a highly prevalent and specific mutation in chronic neutrophilic leukemia. Leukemia. 2013;27(9):1870–3. 10.1038/leu.2013.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue D, Kitaura J, Matsui H, Hou HA, Chou WC, Nagamachi A, et al. SETBP1 mutations drive leukemic transformation in ASXL1-mutated MDS. Leukemia. 2015;29(4):847–57. Epub 2014/10/14. 10.1038/leu.2014.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meggendorfer M, Bacher U, Alpermann T, Haferlach C, Kern W, Gambacorti-Passerini C, et al. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia. 2013;27(9):1852–60. Epub 2013/05/01. 10.1038/leu.2013.133 [DOI] [PubMed] [Google Scholar]
- 17.Patnaik MM, Wassie EA, Lasho TL, Hanson CA, Ketterling R, Tefferi A. Blast transformation in chronic myelomonocytic leukemia: Risk factors, genetic features, survival, and treatment outcome. Am J Hematol. 2015;90(5):411–6. 10.1002/ajh.23962 [DOI] [PubMed] [Google Scholar]
- 18.Adema V, Larrayoz MJ, Calasanz MJ, Palomo L, Patino-Garcia A, Agirre X, et al. Correlation of myelodysplastic syndromes with i(17)(q10) and TP53 and SETBP1 mutations. British journal of haematology. 2015;171(1):137–41. Epub 2015/02/27. 10.1111/bjh.13355 [DOI] [PubMed] [Google Scholar]
- 19.Cui Y, Li B, Jiang Q, Xu Z, Qin T, Zhang P, et al. [CSF3R, ASXL1,SETBP1, JAK2 V617F and CALR mutations in chronic neutrophilic leukemia]. Zhonghua xue ye xue za zhi = Zhonghua xueyexue zazhi. 2014;35(12):1069–73. Epub 2014/12/30. 10.3760/cma.j.issn.0253-2727.2014.12.005 [DOI] [PubMed] [Google Scholar]
- 20.Itzykson R, Kosmider O, Renneville A, Gelsi-Boyer V, Meggendorfer M, Morabito M, et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31(19):2428–36. 10.1200/JCO.2012.47.3314 [DOI] [PubMed] [Google Scholar]
- 21.Lin Y, Zheng Y, Wang ZC, Wang SY. Prognostic significance of ASXL1 mutations in myelodysplastic syndromes and chronic myelomonocytic leukemia: A meta-analysis. Hematology. 2016;21(8):454–61. 10.1080/10245332.2015.1106815 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.