Abstract
Haplotypes A and B of ‘Candidatus Liberibacter solanacearum’ (CLso) are associated with diseases of solanaceous plants, especially Zebra chip disease of potato, and haplotypes C, D and E are associated with symptoms on apiaceous plants. To date, one complete genome of haplotype B and two high quality draft genomes of haplotype A have been obtained for these unculturable bacteria using metagenomics from the psyllid vector Bactericera cockerelli. Here, we present the first genomic sequences obtained for the carrot-associated CLso. These two genomic sequences of haplotype C, FIN114 (1.24 Mbp) and FIN111 (1.20 Mbp), were obtained from carrot psyllids (Trioza apicalis) harboring CLso. Genomic comparisons between the haplotypes A, B and C revealed that the genome organization differs between these haplotypes, due to large inversions and other recombinations. Comparison of protein-coding genes indicated that the core genome of CLso consists of 885 ortholog groups, with the pan-genome consisting of 1327 ortholog groups. Twenty-seven ortholog groups are unique to CLso haplotype C, whilst 11 ortholog groups shared by the haplotypes A and B, are not found in the haplotype C. Some of these ortholog groups that are not part of the core genome may encode functions related to interactions with the different host plant and psyllid species.
Introduction
‘Candidatus Liberibacter solanacearum’ (CLso) was first described in connection with diseases of solanaceous crops, including potato, tomato and capsicum in New Zealand and North America [1–7]. Later, the same bacterial species was found in Europe, associated with diseases in the Apiaceae family plants carrot and celery [8–12]. Phylogenetic analysis using the combination of the 16S rRNA, 16S-23S rRNA intergenic spacer region (ISR) and 50S ribosomal protein gene sequences, revealed that the CLso bacteria found in different geographic regions were diverse and could be assigned to five separate clades: haplotype A, B, C, D or E [12,13]. CLso haplotypes A and B are associated with Zebra chip (ZC) disease of potatoes and psyllid yellows of tomato and capsicum [4], and these two haplotypes are transmitted by the tomato/potato psyllid Bactericera cockerelli Šulc (Hemiptera: Triozidae) in a circulative-persistent mode [2,7,14]. CLso haplotype C is associated with carrot yellowing disease in Northern Europe, where it is transmitted by the carrot psyllid, Trioza apicalis Förster [11,15,16]. In addition to findings in Finland, CLso haplotype C has been detected in Sweden, Norway and Germany [17–19]. CLso haplotypes D and E were first described in carrot and celery in Spain, and the psyllid Bactericera trigonica Hodkinson is suspected to act as a vector for these haplotypes in Spain [9,12,20]. Haplotypes D and E have also been found in carrot in France and Morocco [21–23].
In 2011, the complete genome sequence of CLso haplotype B (ZC1) was obtained via metagenomics, using DNA that had been isolated from CLso bacteria collected by immuno-capture from a pooled sample of field-captured tomato/potato psyllids and then amplified by whole genome amplification [24]. CLso ZC1 is the first and only completely assembled CLso genome to date. In 2015, two high quality draft genomes of CLso haplotype A were published. The first assembly, NZ1, was obtained using metagenomics from DNA isolated from one tomato/potato psyllid individual from a greenhouse-reared colony in New Zealand. The DNA for Illumina sequencing was amplified using whole genome amplification. The other CLso haplotype A assembly, HenneA, was obtained from DNA isolated from two tomato/potato psyllid individuals from a greenhouse-reared colony in the United States. The DNA from two psyllid individuals with high CLso titres were combined into one DNA sample for sequencing [25]. Analysis of these complete or high quality draft genomes provided insights into the biology of CLso, and revealed genetic variation between haplotypes A and B. In addition, two draft genome sequences of CLso, R1 and RSTM, were obtained from a tomato plant and a tomato/potato psyllid respectively, in California [26,27]. These two draft genomes are still highly fragmented, R1 consists of 99 contigs and RSTM consists of 26 contigs, which limits their use in genome structure analysis. However, these draft genomes provide information of the genetic variation within CLso. As genome sequences from the other haplotypes (C, D and E) were missing, a more robust analysis could not be undertaken.
Of the five haplotypes of CLso, haplotype C has the most distinct vector, the carrot psyllid Trioza apicalis, which occurs in the temperate and subarctic climate areas in Northern Europe, whereas the identified or suggested vectors of the other haplotypes of CLso belong to genus Bactericera and occur in areas with temperate or tropical climates. To determine if CLso haplotype C is genetically different from haplotypes A and B, we sequenced and assembled the genome of haplotype C and undertook genome comparisons. Two draft genome sequences of CLso haplotype C were obtained from DNA isolated from two carrot psyllid individuals from south-west Finland. One of these haplotype C draft genome sequences, FIN114, was compared with the complete genome sequence of haplotype B (ZC1) and one high quality draft genome sequence of haplotype A (NZ1). Genomic comparisons of these three haplotypes of the same bacterial species revealed the size of both the core and pan genomes, and identified potential haplotype-specific genes that may be involved in the different host plant or psyllid interactions.
Materials and methods
Psyllid and plant samples
All carrot psyllids were captured from the same population in a carrot field in Forssa, south-west Finland in the summer of 2012, with the permission of the land owner. Thereafter, the psyllids were reared on carrot plants (cv. Fontana) in a greenhouse in Jokioinen, Finland. In the transmission experiment, each psyllid individual was released on one carrot seedling enclosed in an insect cage [16]. After three days' exposure, psyllids were removed from the carrot plants, the DNA was extracted from the psyllids using DNeasy Blood and Tissue kit (Qiagen) according to the manufacturer's protocol, and the DNA was then eluted in 30 µl of nuclease-free water. Each DNA sample was further diluted 1/100, and 5 µl per reaction was used for quantitative PCR [16]. DNA samples from two carrot psyllid females 111 and 114 that contained a very high titre of CLso, Ct values 19.25 and 18.93 at sample dilution 10−2 respectively, were used as the material for sequencing (S1 Table). DNA was also extracted from the carrot plants nine weeks after the exposure to psyllids, using the CTAB method as previously described [16]. DNA from the CLso-infected carrots A2F2 and A5F2 exposed to feeding by psyllids 111 and 114, respectively, was used as a PCR template for CLso sequence validation and gap closure. DNA from a healthy control carrot A7C1 grown in an insect proof cage in a greenhouse was used as a CLso-negative PCR template.
Genome sequencing
Whole genome amplification was conducted on the DNA of both the carrot psyllid samples 114 and 111 using a RepliG kit (Qiagen). Library construction and Illumina HiSeq2000 sequencing was conducted by Macrogen Inc. (South Korea). A 432 bp paired-end library was sequenced for sample 114 and a 670 bp paired-end library and a 3 kb mate-pair library were sequenced for sample 111. Part of the DNA sample 114 was also sequenced on two PacBio RS SMRT cells at Expression Analysis Ltd (USA) (S1 Table).
Genome assembly and gap closure
Paired Illumina reads were adaptor-clipped using Mira v4.0.2 [28] and quality clipped using Sickle v1.33 (github.com/najoshi/sickle). Remaining intact read pairs were assembled using idba_ud v1.1.1 [29]. Contigs shorter than 200 nt were discarded. Remaining contigs with a blastn-hit against any published Liberibacter genome among the top 10 best hits, as well as any Liberibacter and related phage sequences available from RefSeq (as at May 2015) were used to extract potential Liberibacter read pairs using Mirabait from the Mira package with default settings. Resulting read pairs were assembled using SPAdes v3.6.1 [30] in MDA mode with k = 27,45,65. Contigs longer than 1kb were edited using Gap5 from the Staden package [31]. The CLso assembly of psyllid 111 DNA was used for predicting the contig joins of the other CLso assembly 114, and the contigs were ordered using Mauve v2.4.0 [32]. The draft genome sequence FIN114 was intensively re-sequenced to obtain high quality sequence that could be used in comparative genomic analyses. Primers binding to the contig ends were designed using Primer3 [33], and these primer pairs (S2 Table) were used to bridge potential gaps between the adjacent contigs of assembly FIN114 by either conventional or long-range PCR using DNA template from the psyllid sample 114. All conventional PCR and long-range PCR amplifications were performed using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) according to the PCR protocol provided by the manufacturer. The PacBio reads were also used to predict joins between the pre-assembled contigs of assembly 114, and those predictions were confirmed by long-range PCR. In addition, to confirm the locations and sequences of the rRNA operons encoding for the 16S, 23S and 5S rRNAs and to determine SNPs or indels in these operons, each of three 5.6 kb gene regions was cloned in pCR-Blunt vector and re-sequenced. Primers (S2 Table) were selected to target the flanking regions beyond each rRNA operon end. All the amplified PCR products were gel-purified and extracted using QIAquick Gel Extraction Kit (Qiagen). The purified PCR products were ligated into pCR-Blunt plasmid vector included in the Zero Blunt PCR Cloning Kit (Thermo Scientific). Ten transformed E. coli Top10 (Thermo Scientific) colonies were picked for each rRNA copy. The re-constructed plasmids of each clone were isolated using QIAprep Spin Miniprep Kit (Qiagen) and analyzed by restriction enzyme digestion using FastDigest EcoRI (Thermo Scientific). The complete sequences of the inserted DNA fragments were obtained by primer walking and Sanger sequencing at Macrogen Europe (The Netherlands). To confirm the presence and locations of putative prophage regions, the joining regions between prophage and bacterial chromosomal sequences were amplified with designed primers (S2 Table) by long-range PCR similarly as described above, and the PCR products were sequenced through primer walking at Macrogen Europe. The quality clipped reads of dataset 111 were mapped against the draft genome sequence of FIN114 using BWA v0.7.12 [34]. The consensus sequence for regions with reads contiguously mapping were extracted. Regions where paired end reads indicated that contigs were joined but differences in the genomes prevented mapping were fixed manually.
Genome annotation
Two draft genome sequences of CLso haplotype C, FIN114 and FIN111, were annotated using the NCBI Prokaryotic Genome Annotation Pipeline (NCBI_PGAP) and deposited in the GenBank under accession numbers LWEB00000000 and LVWB01000000, respectively.
Phylogenetic analysis
To construct a phylogenic tree, four species of ‘Candidatus Liberibacter’ and the species Liberibacter crescens were compared with each other and to twelve closely related Alphaproteobacteria, Agrobacterium tumefaciens, Sinorhizobium meliloti, Brucella melitensis, Brucella abortus, Bartonella quintana, Bartonella bacilliformi, Bartonella henselae, Bartonella vinsonii, Mesorhizobium loti, Mesorhizobium opportunistum, and two Phyllobacterium species. Rhodospirillum rubrum from the order Rhodospirillales of the class Alphaproteobacteria was used as the outgroup (S3 Table). Protein coding genes were clustered into ortholog groups using OrthoMCL v2.0.9 [35]. In total, the annotations of 96 single copy ortholog groups from all the bacterial genomes included in the analysis were manually checked. Any ortholog group related to an unknown function or possible horizontal gene transfer was removed from the set before analysis. Finally, 88 ortholog groups were aligned using Muscle v3.8.31 [36], and then the multiple alignments were trimmed and concatenated into a supermatrix. The best amino acid substitute model for this supermatrix was determined using ProtTest v3.4.1 [37]. Maximum-likelihood tree was constructed using RAxML v8.2.0 [38] and applying ‘PROTGAMMAIWAGF’ setting.
Genome comparisons
As intensive sequence validations and gap closure had been conducted on assembly FIN114, this draft genome sequence was used for genome comparisons. The predicted protein sequences of FIN114 were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway maps to reconstruct the metabolic pathways [39] of CLso haplotype C. The result was compared to the pathways predicted by KEGG for the curated complete ‘Liberibacter’ genomes, including ‘Ca. Liberibacter solanacearum’ (ZC1), ‘Ca. Liberibacter asiaticus’ (psy62, Gxpsy, Ishi-1), ‘Ca. Liberibacter americanus’ (Sao Paulo), ‘Ca. Liberibacter africanus’ (PTSAPSY) and Liberibacter crescens (BT-1). Closer comparisons of the mevalonate pathway between ‘Liberibacter’ and Marinobacterium species was performed through the EcoCyc database [40]. To obtain an overall view of the genome synteny between the three CLso haplotypes, the sequence FIN114 was aligned against the haplotype B ZC1 genome (accession GCA_000183665.1) and the haplotype A NZ1 genome (accession GCA_000968085.1) using progressive Mauve algorithm from Mauve v2.4.0 [32]. Because the genome assemblies of FIN114 and NZ1 are in contigs, the complete genome sequence ZC1 was set as the reference. Long-range PCR was used to confirm the re-arrangement of several genomic regions in FIN114 in relation to ZC1. The local conserved blocks (LCBs) and the guide tree were determined by the progressive Mauve algorithm and visualized using the R package genoPlotR v0.8.4. The annotated protein sequences of assemblies NZ1, ZC1 and FIN114, representing CLso haplotypes A, B and C, respectively, were clustered into ortholog groups using OrthoMCL. Short open reading frames with less than 50 amino acid codons were filtered away. In the subsequent all-blast-all step using Blastp, the required minimum coverage over both query and subject sequence was set at 70%. The ortholog clustering result of OrthoMCL analysis was converted into an orthologs vs. haplotype binary (1/0: present/not present) matrix, and visualized as a Venn diagram using the R package eVenn v2.3.2. Those genes identified as singletons (i.e. genes not assigned to any ortholog group) in each genomic assembly were further filtered by Blastn against the other two CLso genome sequences, and any Blastn-hit with e-value lower than 1e-11 and coverage more than 50% over query was discarded as being a potential homolog. These remaining singletons were then screened by Blastn against NCBI nr database using the same threshold to identify those singletons that show similarity to sequences of other species, and the remaining singletons were considered as FIN114 specific. The same Blastn filtering was applied to the ortholog groups that were present in NZ1 and ZC1, but not in FIN114. Since the integrity of assembly FIN111 was not confirmed by PCR, it was not used in Mauve alignments, but the putative haplotype specific genes or ortholog groups identified by comparisons between FIN114, NZ1 and ZC1 were also compared by reciprocal Blastp (identity above 40% and 70% coverage on both query and subject) against the FIN111 protein dataset. For these haplotype specific genes or ortholog groups, we used Blast2GO [41] to annotate gene functions via a workflow of BLAST, Gene Ontology (GO) mapping, and InterProScan. SignalP v4.1 [42] and SecretomeP v2.0 [43] were used to identify a signal peptide or a non-classical protein secretion signal respectively, as previously described [44]. For the putative protein AYJ09_01490 and its homologs, amino acid sequence alignment was performed using Clustal X [45] and secondary structure prediction was performed using Jpred4 [46]. The complete prophage regions of FIN114, NZ1 [25] and ‘Ca. Liberibacter africanus’ PTSAPSY [47] were predicted using PHASTER [48]. The complete prophage sequences that have been characterized in previous studies, P1 and P2 from ZC1 [24], SC1 and SC2 from ‘Ca. Liberibacter asiaticus’ UF506 [49], FP2 from ‘Ca. Liberibacter asiaticus’ psy62 [50], SP2 from ‘Ca. Liberibacter americanus’ Sao Paulo [51], LC1 and LC2 from Liberibacter crescens BT-1 [52] were extracted from the genome sequences. Because the prophage genomes show mosaic architectures between the different strains, a comparison method that requires long sequence alignments between the genomes could not be used. Instead, pairwise calculation of tetra-nucleotide frequencies (TETRA) between the ‘Liberibacter’ prophage sequences was employed to estimate their relationships, as this analysis is independent of longer sequence alignments. The correlation coefficients of the TETRA of all those selected complete prophage sequences were calculated using Python package pyani (github.com/widdowquinn/pyani) and the result was visualized using R package ggplot2 v2.1.0.
Results
Genome features
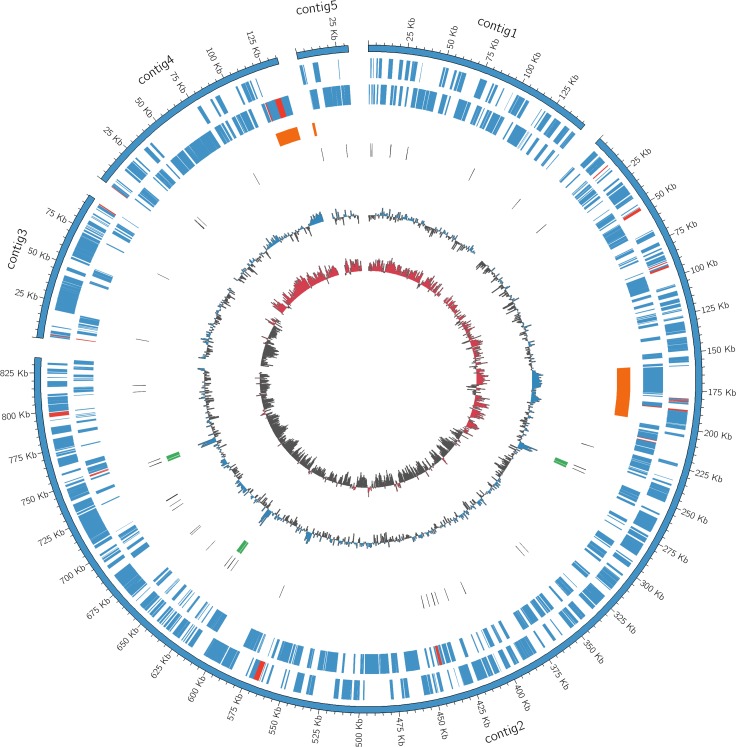
The assemblies FIN114 and FIN111 have 5 and 15 non-redundant contigs respectively, each with 300 times average coverage. The draft genome FIN114 has a GC content of 35.2% and a length of 1.24 Mbp, encoding 1067 predicted proteins. The draft genome FIN111 has a GC content of 34.9% and a length of 1.20 Mbp, encoding 1040 predicted proteins. The average nucleotide identity (ANI) between these two assemblies is 99.87% which shows they are highly similar. The draft genome FIN114 contains one complete prophage region, designated as phage A, in contig 2, and one partial prophage, designated as phage B, between contigs 4 and 5 (Fig 1). The prophage A is 38.3 kb long and has a GC content of 41.0%.
Fig 1. Circular representation of the draft genome sequence of ‘Candidatus Liberibacter solanacearum’ haplotype C FIN114.
The circles represent, from outer to inner, protein coding genes on the forward strand and the reverse strand, prophage regions, tRNA, rRNA, %G+C content and GC-skew. The loci indicated with red color represent the putative haplotype C specific genes.
CLso rRNA operons
Like the previously sequenced ‘Candidatus Liberibacter’ genomes, the CLso haplotype C draft genome FIN114 also contains three rRNA operons. Each of the three rRNA operons, named 16SA, 16SB and 16SC, and their variable flanking sequences were amplified by long-range PCR, and contigs with sizes 6975 bp, 6298 bp and 6825 bp assembled via primer walking and sequencing. After complete sequence alignment and removing the variable flanking sequences, the size of the rRNA operon was determined to be 5670 bp, with almost identical sequence between the three copies. They all include the genes 16S rRNA, tRNA-Ile, tRNA-Ala, 23S rRNA, 5S rRNA and tRNA-Met in the same order. Only one polymorphic site was found within the 23S rRNA sequence where 16SA differs from 16SB and 16SC at position 3848: C/T. The sequences of the three rRNA operons of haplotype C were deposited at GenBank under accession numbers KX431889, KX431890 and KX431891.
Phylogenetic tree
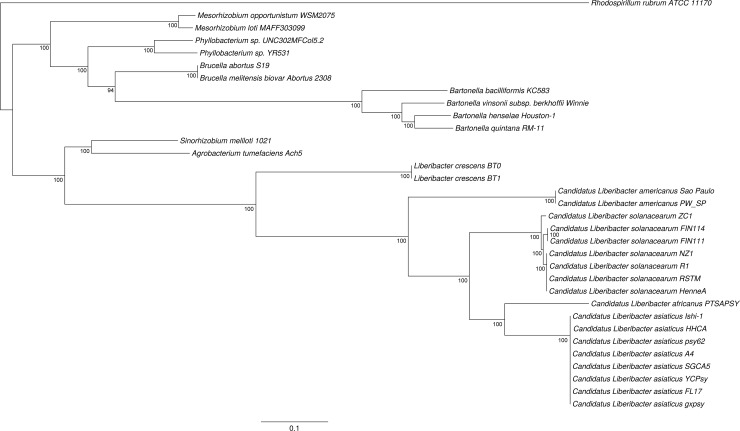
A phylogenic tree for the genus ‘Candidatus Liberibacter’ and related species was constructed using the supermatrix approach and based on 88 single-copy ortholog groups (Fig 2). The tree obtained is robust, with strong bootstrap support. The analysis clearly shows that the ‘Liberibacter’ species are divided into two sub-clades. All the CLso clades and the huanglongbing-associated species ‘Candidatus Liberibacter africanus’ (CLaf), ‘Candidatus Liberibacter asiaticus’ (CLas) and ‘Candidatus Liberibacter americanus’ (CLam) clustered into the plant pathogen sub-clade of ‘Candidatus Liberibacter’. Liberibacter crescens, which is non-pathogenic and culturable, was the only member of the other sub-clade. As expected, CLso haplotype C sequences FIN114 and FIN111 and all the other CLso haplotypes together form the CLso clade, in which the three haplotypes further differentiate into three distinct haplotype clades with strong bootstrap support.
Fig 2. Phylogenetic tree constructed of 88 protein-coding genes from 33 bacterial genome datasets, belonging to genera ‘Candidatus Liberibacter’, Liberibacter, Agrobacterium, Sinorhizobium, Brucella, Bartonella, Mesorhizobium, Phyllobacterium and Rhodospirillum.
The 88 ortholog groups were aligned using Muscle v3.8.31, and these multiple alignments were trimmed and concatenated into a supermatrix. The best amino acid substitute model was determined using ProtTest v3.4.1. Maximum-likelihood tree was constructed using RAxML v8.2.0 with ‘PROTGAMMAIWAGF’ setting. The numbers shown next to the branches indicate the percentage of bootstrap support values (1000 replicates). The branch lengths indicate the evolutionary distance as the number of base substitutions per site.
Prophage sequence
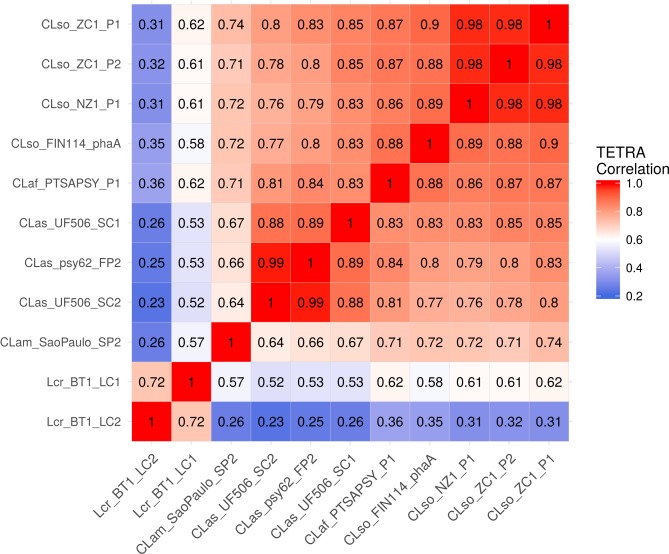
The TETRA correlation coefficient values of 11 complete prophage sequences (Fig 3) show that CLso FIN114 prophage A sequence is highly similar to the prophage sequences from NZ1 (P1) and ZC1 (P1 and P2) with correlation coefficient values between 0.88 to 0.90, and is less correlated to prophage sequences SC1, SC2 and PF2 from CLas with values between 0.77 to 0.83. The prophage sequence of CLaf PTSAPSY is correlated to all CLso prophage sequences with correlation coefficient values from 0.86 to 0.88, and less correlated to CLas prophages. This was unexpected, since the core genome of CLaf is more closely related to CLas than to CLso (Fig 2). All the prophages found in the ‘Ca. Liberibacter’ species and L. crescens belong to order Caudovirales family Podoviridae.
Fig 3. Tetranucleotide frequency correlation coefficients (TETRA) of eleven prophage sequences from ‘Candidatus Liberibacter’ species and Liberibacter crescens.
CLso, ‘Candidatus Liberibacter solanacearum’; CLaf, ‘Candidatus Liberibacter africanus’; CLas, ‘Candidatus Liberibacter asiaticus’; CLam, ‘Candidatus Liberibacter americanus’; and Lcr, Liberibacter crescens.
Differences in genome organization between CLso haplotypes A, B and C
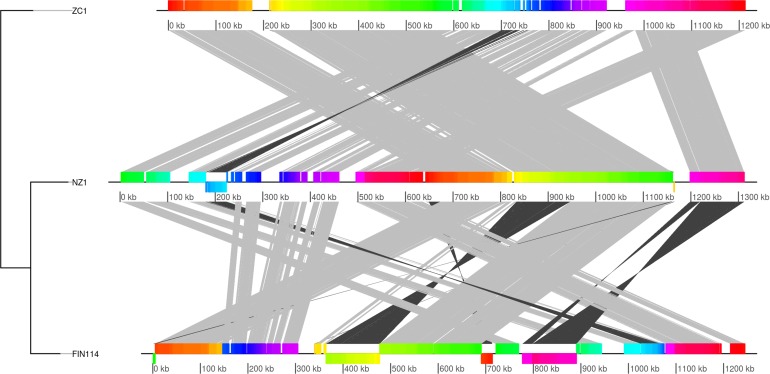
The average nucleotide identity (ANI) is 97.70% between ZC1 and FIN114, and 97.91% between NZ1 and FIN114, which indicates that these lineages belong to the same species. The ANI result agrees with the multi-locus phylogeny tree (Fig 2) showing that haplotype C clade is more closely related to the clade of haplotype A, and agrees with the guide tree of Mauve alignment (Fig 4), which also suggests that NZ1 and FIN114 are more closely related to each other than to ZC1. In general, the Mauve alignment shows synteny between ZC1 and NZ1, except one major rearrangement in the contig 2 of NZ1, which was split into contig 2a and 2b for the Mauve alignment in the previous report [25], and one inverted local conserved block (LCB) in contig 1 of NZ1. In contrast, the Mauve alignment analysis between ZC1 and FIN114 reveals multiple large genome rearrangements (Fig 4). Most of these large rearrangements are located within contig 2, which is the largest contig (834 kbp) of the FIN114 genome sequence. There are multiple large genomic regions within FIN114 contig 2 that are in the reverse orientation, and in different relative positions, to that in ZC1. One large inverted region, starting approximately at nucleotide position 364000, is next to prophage A, and is thus likely to be the result of a phage-mediated rearrangement. For another large inverted region, starting at position 690600, the homologous region in ZC1 has stretches of repetitive sequence on both sides. These differences in the genomic organization in FIN114 in comparison with ZC1 were all confirmed by long-range PCR and Sanger sequencing. The largest inversion, located approximately between positions 770000 and 888000 in FIN114, is between two identical rRNA operons, 16SB and 16SC, and thus the inversion is probably a result of a recombination event between these two rRNA operons. The variable flanking sequences of these two rRNA operons were confirmed by sequencing of the cloned DNA fragments. The genomic region homologous to this inverted region is found in contig 5 of NZ1, and between nucleotides 980000 and 1095000 of the ZC1 genome.
Fig 4. Multiple genome alignment of the ‘Candidatus Liberibacter solanacearum’ haplotypes A, B and C.
Comparison was made between the complete genome sequence of CLso haplotype B ZC1 (top), and the draft genome sequences of haplotype A NZ1 (middle) and haplotype C FIN114 (bottom). Lines connect the homologous local conserved blocks (LCBs) between the genomes, with grey color showing connection between LCBs that are in the same orientation and black color showing connection between LCBs that are in the opposite orientation. The tree shown on the left represents the guide tree of the progressive Mauve alignment.
Haplotype C core genome
There are no significant differences in the core genome gene content between the haplotype B ZC1 genome and the haplotype C assembly FIN114. Thus, CLso haplotype C is likely to have a similar capacity for biosynthesis, metabolism and secretion as the haplotype B. However, new analysis of the biosynthetic and metabolic pathways revealed some differences from the previous analyses of ZC1 [24,53]. Haplotype C harbors genes to synthesize eight amino acids, aspartate, glutamate, glutamine, threonine, lysine, arginine, serine and glycine, either de novo, or from metabolic intermediates, like ZC1. Both ZC1 and FIN114 lack the genes required for the conversion of pyruvate to alanine by transaminase (EC 2.6.1.2) and alanine dehydrogenase (EC 1.4.1.1), suggesting that they are deficient in de novo synthesis of alanine from pyruvate. However, both ZC1 and FIN114 retain the genes encoding for selenocysteine lyase (EC 4.4.1.16) and cysteine desulfurase (EC 2.8.1.7), enabling them to produce alanine through deselenization of selenocysteine, or via desulfuration of cysteine. Both ZC1 and FIN114 still retain the genes for the glycolytic pathway except for the key enzyme glucose-6-phosphate isomerase (EC 5.3.1.9), which converts α-glucose-6-phosphate to β-fructose-6-phosphate. Unlike most of the bacteria that harbor genes encoding the 1-deoxy-D-xylulose-phosphate (DOXP) pathway for isoprenoid biosynthesis, FIN114 and ZC1 have a small gene cluster (AYJ09_00845-AYJ09_00870) which encodes the mevalonate pathway for isoprenoid biosynthesis. This mevalonate pathway is present in the other ‘Candidatus Liberibacter’ species as well, whereas it is not found in the other Rhizobiales [54]. The mevalonate pathway gene cluster of CLso shares the same conserved gene order and also over 50% amino acid identity with that of the members of Gammaproteobacteria genus Marinobacterium, including M. jannaschii DSM 6295 (GenBank accession JHVJ00000000), M. litorale DSM23545 (GenBank accession AUAZ00000000), and M. stanieri S30 (GenBank accession AFPL00000000).
Differences in gene contents between the three haplotypes of CLso
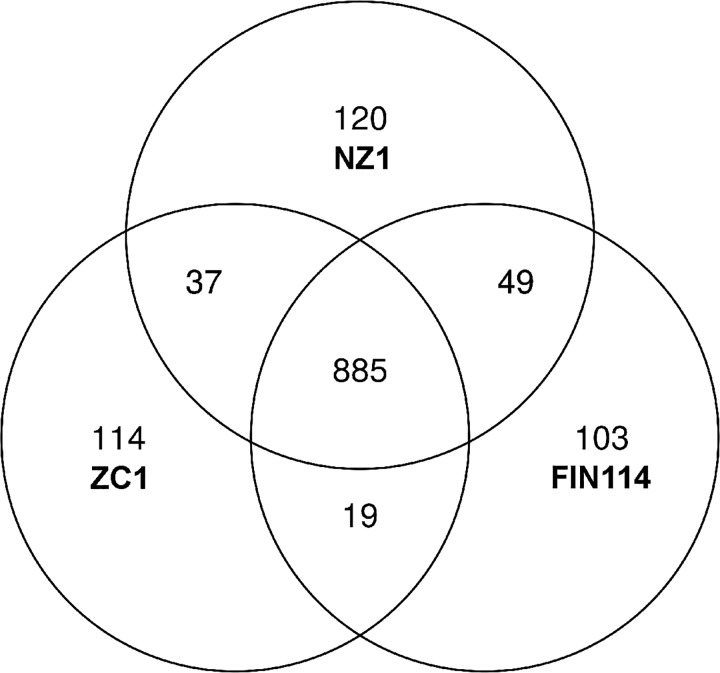
The three CLso genome assemblies NZ1, ZC1 and FIN114, representing the haplotypes A, B and C, respectively, share a core-genome that consists of 885 ortholog groups, and form a CLso species pan-genome that consists of 1327 ortholog groups (Fig 5). Since the OrthoMCL analysis is performed with translated amino acid sequences, the number of ortholog groups can be different from the number of predicted genes, which also includes pseudogenes in the assembly FIN114. After Blastn filtering of the nucleotide sequences, there were 30 protein coding genes that were present in the FIN114 and absent from NZ1 and ZC1 (Table 1). Nineteen of these 30 genes were also found in the assembly FIN111 which lacks the prophage regions. Twenty-four of the 30 genes were located in contig 2, three in contig 3, and another three in contig 4 of FIN114. Two of the 30 genes (AYJ09_03220 and AYJ09_03225) were found in CLas and one (AYJ09_04845) was found in CLaf, and thus 27 genes can be considered as potential CLso haplotype C specific genes. Five of the haplotype C specific genes are putative members of restriction-modification (R-M) systems, and four of them form two complete type II R-M systems, which were not found in the CLso haplotypes A and B. Haplotype A strain NZ1 was also found to harbor two extra strain-specific R-M systems not present in the other ‘Ca. Liberibacter’ genomes. The other 25 genes not present in haplotypes A and B were annotated as hypothetical proteins, of which seven contain a putative secretion signal. Six of the hypothetical protein genes are located within the prophage A region and two are located within the prophage B region of FIN114. One of the genes within the prophage A of FIN114, AYJ09_01490, was found to encode a domain with a strong amino acid sequence homology (30% identity and over 70% query coverage) to the isoform 4G of the eukaryotic translation initiation factor (eIFiso4G) present in plants. The small protein encoded by FIN114 covers a part of the MIF4G domain (S1 Fig) that forms a multi-helix structure and is located on the surface of the eIFiso4G protein according to the predicted 3D structure [55]. Another hypothetical protein (AYJ09_01545) encoded in the prophage A region is a putative transferase with LbetaH domain and has over 50% identity (with an expect value of 1e-33) with a phage-related hypothetical protein of Bartonella species.
Fig 5. Venn diagram showing the results of OrthoMCL analysis with three ‘Candidatus Liberibacter solanacearum’ genomes.
The figures indicate the number of ortholog groups before Blastn filtering.
Table 1. Genes present in ‘Candidatus Liberibacter solanacearum’ haplotype C and absent from haplotypes A and B, with predicted protein products and secretion signals.
| Gene Identifier | Product size (amino acids) | Annotation | Blastn hit to other Liberibacter (1e-11)a | SecretomeP2.0 | SignaIP4.1 (D-cutoff 0.57) |
|---|---|---|---|---|---|
| AYJ09_00740 | 150 | hypothetical protein | - | + | - |
| AYJ09_00930 | 77 | hypothetical protein | - | + | - |
| AYJ09_00935 | 213 | N6 adenine-specific DNA methyltransferase | - | - | - |
| AYJ09_00940 | 236 | type-2 restriction enzyme HaeIII | - | - | - |
| AYJ09_01090 | 149 | hypothetical protein | - | + | - |
| AYJ09_01105 | 361 | HNH endonuclease | - | - | - |
| AYJ09_01490 | 110 | hypothetical protein | - | - | - |
| AYJ09_01505 | 92 | hypothetical protein | - | - | - |
| AYJ09_01510 | 68 | hypothetical protein | - | - | - |
| AYJ09_01535 | 113 | hypothetical protein | - | - | - |
| AYJ09_01540 | 115 | hypothetical protein | - | - | - |
| AYJ09_01545 | 107 | hypothetical protein | - | - | - |
| AYJ09_01670 | 73 | hypothetical protein | - | - | - |
| AYJ09_01675 | 60 | hypothetical protein | - | - | - |
| AYJ09_02650 | 242 | type-2 restriction enzyme eco47II | - | + | - |
| AYJ09_02655 | 333 | C-5 cytosine-specific DNA methyltransferase | - | - | - |
| AYJ09_03215 | 72 | hypothetical protein | - | - | - |
| AYJ09_03220 | 564 | hypothetical protein | + | + | - |
| AYJ09_03225 | 408 | hypothetical protein | + | + | - |
| AYJ09_04045 | 186 | hypothetical protein | - | - | + |
| AYJ09_04050 | 69 | hypothetical protein | - | - | - |
| AYJ09_04230 | 231 | hypothetical protein | - | - | - |
| AYJ09_04235 | 305 | hypothetical protein | - | - | - |
| AYJ09_04245 | 162 | hypothetical protein | - | - | - |
| AYJ09_04430 | 116 | hypothetical protein | - | - | - |
| AYJ09_04435 | 76 | hypothetical protein | - | + | - |
| AYJ09_04835 | 175 | hypothetical protein | - | - | - |
| AYJ09_04845 | 88 | hypothetical protein | + | - | - |
| AYJ09_05395 | 163 | hypothetical protein | - | - | - |
| AYJ09_05425 | 1368 | hypothetical protein | - | - | - |
a Other than ‘Ca. Liberibacter solanacearum’ haplotypes A and B.
There are 271 ortholog groups found in haplotypes A and B, either in both or in at least one, that were not present in haplotype C. After the Blastn filtering, there were twelve ortholog groups that were common to the ZC1 and NZ1 genomes and were not found in FIN114 (Table 2). However, one of these 12 ortholog groups, ortholog group 991 (DJ66_RS00890, CKC_RS03350), was found in FIN111. Eleven of the 12 ortholog groups were annotated as hypothetical proteins: the exception was ortholog group 1003 that encodes for 3-methyladenine DNA glycosylase, which is involved in base excision repair of alkylation damage in DNA. Two small 79 amino acid proteins encoded by ortholog group 986 were found to contain a recognizable N-terminal signal peptide. The ortholog group 22 (DJ66_RS00555, DJ66_RS05330, CKC_RS00980, CKC_RS05675), of which both NZ1 and ZC1 harbor two copies, encodes a hypothetical protein with weak identity (e-value 4e-05) to a conserved domain with unknown function (pfam04859). The proteins encoded by ortholog group 987 share over 40% amino acid sequence identity with a phage-related hypothetical protein of Bartonella species. The proteins encoded by ortholog group 1000 are predicted to belong to the DUF1640 superfamily (pfam07798) with two coiled-coil structures. The gene CKC_RS03570 of the ortholog group 1000 is located in a phage remnant region (779000…784000) in ZC1 and flanked by tRNA-Ser gene, repetitive sequences and two phage-related primase genes. The other gene of this ortholog group 1000 is located within the prophage P2 region in NZ1.
Table 2. Ortholog groups (OG) present in both ‘Candidatus Liberibacter solanacearum’ haplotypes A and B but not in haplotype C.
| OG_ID | Gene 1 | Gene 2 | Gene 3 | Gene 4 | OG annotation | SecretomeP2.0 | SignaIP4.1 (D-cutoff 0.57) |
|---|---|---|---|---|---|---|---|
| OG_22 | LsoZ|CKC_RS00980 | LsoZ|CKC_RS05675 | LsoN|DJ66_RS00555 | LsoN|DJ66_RS05330 | hypothetical protein | - | - |
| OG_37 | LsoZ|CKC_RS00945 | LsoZ|CKC_RS05645 | LsoN|DJ66_RS00585 | hypothetical protein | - | - | |
| OG_39 | LsoZ|CKC_RS01010 | LsoZ|CKC_RS05705 | LsoN|DJ66_RS00525 | hypothetical protein | - | - | |
| OG_985 | LsoN|DJ66_RS00550 | LsoZ|CKC_RS00985 | hypothetical protein | - | - | ||
| OG_986 | LsoN|DJ66_RS00570 | LsoZ|CKC_RS00955 | hypothetical protein | - | + | ||
| OG_987 | LsoN|DJ66_RS00580 | LsoZ|CKC_RS00950 | hypothetical protein | - | - | ||
| OG_990 | LsoN|DJ66_RS00860 | LsoZ|CKC_RS03375 | hypothetical protein | - | - | ||
| OG_991a | LsoN|DJ66_RS00890 | LsoZ|CKC_RS03350 | hypothetical protein | - | - | ||
| OG_998 | LsoN|DJ66_RS01120 | LsoZ|CKC_RS03490 | hypothetical protein | - | - | ||
| OG_999 | LsoN|DJ66_RS01125 | LsoZ|CKC_RS03495 | hypothetical protein | - | - | ||
| OG_1000 | LsoN|DJ66_RS01540 | LsoZ|CKC_RS03570 | DUF1640 domain-containing protein | - | - | ||
| OG_1003 | LsoN|DJ66_RS02910 | LsoZ|CKC_RS05510 | 3-methyladenine DNA glycosylase | - | - |
a OG found in FIN111.
Discussion
In this study, two draft genome sequences of CLso haplotype C, FIN114 and FIN111, were obtained by metagenomics from individual carrot psyllids, and compared with closely related bacterial genomes. Both Illumina and PacBio sequencing were required to obtain the primary assemblies, and then gap closure of assembly FIN114 was accomplished by resequencing of approximately 10% of the genome through PCR and primer walking. Pairwise genomic comparisons by ANI suggest that CLso haplotype C FIN114 genome sequence is more closely related to haplotype A NZ1 than to haplotype B ZC1, and the same clustering was observed in the maximum likelihood phylogenetic tree based on 88 ortholog groups of the bacterial core genome. However, the TETRA correlation coefficients of the prophage regions of ‘Liberibacter’ species revealed that the prophage sequence of haplotype A NZ1 is more closely related to the prophages of haplotype B ZC1 than to the prophage A of FIN114. This suggests that the phage related regions have evolved differently from the bacterial core genome. The maximum likelihood phylogenetic tree shows that the ‘Liberibacter’ clade is clustered within the family Rhizobiaceae, which agrees with the earlier Bayesian phylogeny tree based on 94 conserved single-copy ortholog groups [56]. In contrast to this multi-locus analysis, a phylogenic analysis based on one gene, the 16S rRNA gene, placed the genus ‘Ca. Liberibacter’ outside of the family Rhizobiaceae [57]. Species tree—gene tree conflict may be caused by several reasons, but with closely related species the 16S rRNA gene sequence may not contain enough sequence variation and thus does not give enough information to construct a reliable phylogeny tree. It has been demonstrated that a species tree based on genome-wide datasets gives a better resolution within the class Alphaproteobacteria [58]. Also, when there are several copies of the rRNA genes within a species the copies may not be identical. The three rRNA operon sequences of FIN114 only had one polymorphic site within the 23S rRNA gene. In contrast, 47 polymorphic sites were found within the three rRNA operons in the haplotype B genome [24]. These differences between the three rRNA operons might be explained by the heterogeneous nature of the CLso DNA used for sequencing of the haplotype B genome. The haplotype B genome sequence was derived from a pooled psyllid sample that may have contained a more diverse CLso population than the single psyllid individuals used for sequencing the haplotype C FIN114, the haplotype A NZ1 genome or the two psyllid individuals combined for sequencing HenneA genome. The cloned rRNA operon sequences of haplotype C also differ from the haplotype B at a site that affects the PCR-based detection of CLso. The commonly used 16S reverse primer OI2c (5'-GCCTCGCGACTTCGCAACCCAT-3') was initially designed for detection of ‘Ca. Liberibacter asiaticus’ and ‘Ca. Liberibacter africanus’ [59,60], and it has also been widely used for detection of CLso for years. However, the primer OI2c has one nucleotide mismatch with the CLso haplotype C sequence: 5'-GCCTC(G/A)CGACTTCGCAACCCAT-3'. This mismatch impairs the specificity of PCR amplification when a plant or psyllid DNA sample contains competing bacterial DNA and the titer of CLso haplotype C is low (unpublished data). Therefore, to improve the PCR performance in CLso haplotype C detection, the primer OI2c was replaced with a specific primer Lsc2 [61].
As a result of the intracellular parasitic lifestyle of CLso, its genome has undergone reduction in both size and the gene content, which has reduced its capacity in metabolism and biosynthesis [24,62]. For example, the capacity to undertake carbohydrate metabolism is limited. In general, glucose is the main monosaccharide processed by the glycolytic pathway. However, CLso lacks the key enzyme glucose-6-phosphate isomerase (EC 5.3.1.9), suggesting it cannot convert α-glucose-6-phosphate to β-fructose-6-phosphate. However, it may bypass this step via the pentose phosphate pathway [24]. The genome of Liberibacter crescens contains the gene encoding for glucose-6-phosphate isomerase, whereas all the unculturable plant disease-associated ‘Candidatus Liberibacter’ species lack this gene [53]. Loss of this gene may be associated with a change in the bacterial lifestyle, when the bacterium became an obligate parasite. However, these bacteria have retained the capability of converting β-D-glucose to β-D-glucose-6-phosphate and generating β-D-fructose-6-phosphate via the pentose phosphate pathway, to bypass the upstream step of the glycolytic pathway. Thus, after the gene loss event these ‘Ca. Liberibacter’ species relied on β-D-glucose for glycolysis instead of α-D-glucose. Besides glycolysis, β-D-glucose is also a substrate for callose and cellulose biosynthesis in plants. Callose deposition and cell wall enhancement at pathogen infection sites are common responses of pattern-triggered immunity (PTI) of plants. Callose deposition has also been observed in response to CLso flg22 peptide when infiltrated into leaves of tobacco, tomato and potato plants [63]. It is possible that the ‘Ca. Liberibacter’ species specifically use β-D-glucose as their main source of reduced carbon for glycolysis to reduce the amount of free β-D-glucose that could serve as the substrate for PTI response-related plant cell wall fortification. CLso also has reduced capacity to synthesize amino acids, vitamins and cofactors due to gene losses in several biosynthetic pathways. In general, free-living bacteria may utilize variable carbon sources derived from central metabolism as precursors for amino acid biosynthesis. Commonly used precursors include pyruvate, oxaloacetate, 2-ketoglutarate, 3-phosphoglycerate, phosphoenolpyruvate, erythrose-4-phosphate and ribose-5-phosphate [64]. However, both the haplotype B ZC1 and haplotype C FIN114 genomes only retain the genes for utilization of oxaloacetate, 2-ketoglutarate and 3-phosphoglycerate as precursors for de novo synthesis of amino acids. Although ZC1 and FIN114 lack the genes for de novo synthesis of alanine, they harbor genes for converting selenocysteine and cysteine into alanine. The enzyme selenocysteine lyase is also found in another insect-transmitted plant phloem-inhabiting bacterium, Candidatus Phytoplasma asteris, which has an even smaller genome (860kbp) than CLso [65], suggesting this enzymatic activity is important for bacterial survival.
CLso species harbor a gene cluster encoding for the mevalonate pathway that may have been acquired through an ancient horizontal gene transfer. The closest homologs for these genes were found in Marinobacterium species that were isolated from marine environments and seawater [66–70]. Predatory bacteria belonging to Alphaproteobacteria and Deltaproteobacteria also harbor genes of this specific mevalonate pathway, which gives an advantage over the DOXP pathway by conserving energy when the substrate (aceto)acetyl-coA can be derived from the prey bacteria [71]. As obligate parasites, CLso and the other ‘Ca. Liberibacter’ species also share many other genetic signatures with the obligate predatory bacteria, including reduced capacity in metabolic and synthetic pathways. All of these bacteria have multiple von Willebrand factor (vWF) type A domain containing proteins (AYJ09_00680, AYJ09_02790, AYJ09_04130 in FIN114) that may facilitate cell to cell adhesion, and several proteases and peptidases that are related to the prey cell modification and degradation [71,72]. For example, CLso FIN114 possesses genes for M23 zinc metalloprotease (AYJ09_02825) and RTX toxin (AYJ09_02810). Within plants, ‘Ca. Liberibacter’ species are restricted to the phloem sieve cells, suggesting that they may derive nutrients directly from the sieve cell cytoplasm, while in psyllids they are likely to derive nutrients either from the psyllid cells or from the psyllid mutualist bacteria, such as Candidatus Carsonella ruddii [73].
The observed differences at both the genome and gene level between the haplotype C and haplotypes A and B that infect solanaceous plants may be related to the adaptation of the bacteria to different host plants and different psyllid species. Genes only present in the haplotype C were found, and among them five R-M system related genes were identified. Previously, a lineage-specific R-M system has been identified in Staphylococcus aureus [74]. R-M systems are common in many bacteria, and their role may not be limited to protecting the bacterial genome against phage infection. In some cases, they can also behave as mobile genetic elements and contribute to the reshaping of the bacterial genome [75]. Although the biological functions of the strain-specific R-M systems are still unclear, their presence suggests that the genomes of the strains harboring them could have methylation patterns different from the other strains. This could reduce the possibility of horizontal gene transfer between the strains in case two strains of the same species co-infect the same host organism. Of the other haplotype C-specific genes, eight genes annotated as hypothetical proteins were located within the prophage-related regions. These genes could be potential candidates for genes encoding functions required in the plant-bacteria or insect-bacteria interactions, since prophage regions have been previously found to contribute to the bacterial virulence. In ‘Ca. Liberibacter asiaticus’ UF506 the prophage SC2 carries a small gene SC2_gp095 that was identified to encode a peroxidase enzyme. Transgenic expression studies revealed that this enzyme is secreted and that it may have an effector function in the host plant, suppressing H2O2-mediated defense signaling [44]. In CLas samples from Florida, homologs of the two prophage regions FP1 and FP2 have also been found to form a recombinant, incomplete prophage variant iFP3. The presence of iFP3 was correlated with blotchy mottle symptoms observed in the plants, suggesting that it could be involved in disease development [76]. Thus, the prophages found in CLso could also contribute to the genome plasticity and carry genes involved in the bacteria-host interactions.
One of the haplotype C specific genes (AYJ09_01490) was found to be homologous to the MIF4G domain of eIFiso4G that is involved in virus-plant interactions in different plant species [55,77]. Thus, the phage-encoded small protein found in CLso haplotype C might play a role in the interaction with the host plant. Another hypothetical protein (AYJ09_01545) shared over 50% amino acid identity with a phage-related hypothetical protein from a Bartonella species. Bartonella, which belongs to Alphaproteobacteria and is closely related to ‘Ca. Liberibacter’ species, is an intracellular parasite and pathogen of humans and animals, and transmitted by insects and ticks. Thus, it is possible that this hypothetical protein found in CLso haplotype C could be involved in the psyllid transmission.
Of the ortholog groups that are present in the haplotypes A and B, and absent from the haplotype C, some may encode proteins that would be disadvantageous for the interactions of the haplotype C with the carrot plant or carrot psyllids, e.g. by acting as avirulence factors. Alternatively, some of these ortholog groups could encode proteins that are involved in specific interactions with the solanaceous host plants or the psyllid B. cockerelli. In this respect, the 11 ortholog groups that are present in both ZC1 and NZ1 but not in FIN114 or FIN111, are worth closer examination. These ortholog groups were annotated as hypothetical proteins, except the ortholog group 1003 that encodes for a DNA repair enzyme 3-methyladenine DNA glycosylase. This enzyme has been well characterized in E. coli, and in general, it confers protection to DNA by limiting mutagenic and clastogenic events [78–80]. For now, the substrate range of the 3-methyladenine DNA glycosylase encoded by ZC1 and NZ1 has not been tested, but the absence of this gene in FIN114 suggests that haplotype C might have a reduced DNA repair capacity and higher mutation frequency than the haplotypes A and B. The two copies (CKC_RS00980 and CKC_RS05675) of the ortholog group 22 in ZC1 were recently recognized as potential effectors, and CKC_RS05675 was shown to have a higher expression level in the haplotype B than in the haplotype A, possibly modifying the interaction with the psyllid B. cockerelli [81]. The putative small secreted proteins encoded by ortholog group 986 could also be considered as candidate effectors. The proteins encoded by ortholog group 987 are homologous to a phage-related hypothetical protein of Bartonella. As Bartonella species are insect-transmitted animal pathogens, these proteins might contribute to the bacteria-insect interaction. The proteins encoded by ortholog group 1000 and containing a DUF1640 domain are likely to have a phage origin. A DUF1640 superfamily protein of bacteriophage AKFV33, a putative biocontrol agent for Shiga toxin-producing Escherichia coli O157:H7, encodes a tail fiber protein which may play a role in bacterial surface recognition and adhesion [82].
In this study, two genomic sequences of CLso haplotype C were assembled and analyzed, and discussed in relation to the obligate parasitic lifestyle of CLso. The comparative genome analysis including three different haplotypes of CLso may help to identify potential haplotype-specific effectors. Since haplotype C is transmitted by a different psyllid vector and has a different plant host range than the haplotypes A and B, the differences found in both the gene content and genome organization may explain some of the differences in the interactions with plants and psyllids. Besides the mining of novel gene candidates, the new haplotype C genome sequences are also useful for applied research and diagnostics.
Supporting information
(DOCX)
(DOCX)
(DOCX)
Alignment of sequences corresponding to AYJ09_01490 were performed using Clustal X and the secondary structure was predicted using Jpred 4. S.lyc, S.tub, S.pen, O.gla represent sequences from Solanum lycopersicum, Solanum tuberosum, Solanum pennellii, Oryza glaberrima, respectively, and 'X1/X2' represent different isoforms of eIF4G. The amino acid residues highlighted with colors have high conservation (above 30%) of similarity regarding the hydrophobicity character. The secondary structure predictions, helix (red) or coil (black), are displayed on the bottom row.
(TIF)
Acknowledgments
We thank Senja Tuominen at Luke for technical assistance and Anneli Virta at Luke for sequencing, and the CSC-IT Center for Science Ltd Finland for providing computing services.
Data Availability
All relevant data are within the paper and its Supporting Information files. All the genomic and sub-genomic DNA sequence files are available in the NCBI GenBank database (accession numbers LWEB00000000, LVWB00000000, KX431889, KX431890 and KX431891).
Funding Statement
Funding for this project was provided by the Ministry of Agriculture and Forestry of Finland (project numbers 1651/311/2011, 2052/312/2011 and 1842/312/2013), Marjatta & Eino Kolli Foundation, China Scholarship Council, the New Zealand Institute for Plant & Food Research Limited, Better Border Biosecurity and the Australian Government via the Plant Biosecurity Cooperative Research Centre. The authors declare that one co-author (TS) provides bioinformatics services as a commercial freelancer under the name Herne Genomik, Neustadt, Germany. The funders provided support in the form of salaries and grants for authors [AN, MH, JW, ST, GS, TS] but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1. Abad JA, Bandla M, French-Monar RD, Liefting LW, and Clover GRG. First report of the detection of 'Candidatus Liberibacter' species in zebra chip disease-infected potato plants in the United States. Plant Dis. 2009;93: 108–109. [DOI] [PubMed] [Google Scholar]
- 2.Hansen AK, Trumble JT, Stouthamer R, and Paine TD. A new huanglongbing species, "Candidatus liberibacter psyllaurous," found to infect tomato and potato, is vectored by the psyllid Bactericera cockerelli (Sulc). Appl Environ Microbiol. 2008;74: 5862–5865. 10.1128/AEM.01268-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liefting LW, Perez-Egusquiza ZC, Clover GRG, and Anderson JAD. A new 'Candidatus Liberibacter' species in Solanum tuberosum in New Zealand. Plant Dis. 2008;92: 1474. [DOI] [PubMed] [Google Scholar]
- 4.Liefting LW, Sutherland PW, Ward LI, Paice KL, Weir BS, and Clover GRG. A new 'Candidatus Liberibacter' species associated with diseases of solanaceous crops. Plant Dis. 2009;93: 208–214. [DOI] [PubMed] [Google Scholar]
- 5.Liefting LW, Weir BS, Pennycook SR, and Clover GRG. 'Candidatus Liberibacter solanacearum', associated with plants in the family Solanaceae. Int J Syst Evol Microbiol. 2009;59: 2274–2276. 10.1099/ijs.0.007377-0 [DOI] [PubMed] [Google Scholar]
- 6.Lin H, Doddapaneni H, Munyaneza JE, Civerolo EL, Sengoda VG, Buchman JL, and et al. Molecular characterization and phylogenetic analysis of 16S rRNA from a new 'Candidatus Liberibacter' strain associated with zebra chip disease of potato (Solanum tuberosum L.) and the potato psyllid (Bactericera cockerelli Sulc). J Plant Pathol. 2009;91: 215–219. [Google Scholar]
- 7.Secor GA, Rivera VV, Abad JA, Lee IM, Clover GRG, Liefting LW, and et al. Association of 'Candidatus Liberibacter solanacearum' with zebra chip disease of potato established by graft and psyllid transmission, electron microscopy, and PCR. Plant Dis. 2009;93: 574–583. [DOI] [PubMed] [Google Scholar]
- 8.Alfaro-Fernández A, Cebrián MC, Villaescusa FJ, de Mendoza AH, Ferrandiz JC, Sanjuan S, and et al. First report of 'Candidatus Liberibacter solanacearum' in carrot in mainland Spain. Plant Dis. 2012;96: 582. [DOI] [PubMed] [Google Scholar]
- 9.Alfaro-Fernández A, Siverio F, Cebrián MC, Villaescusa FJ, and Font MI. 'Candidatus Liberibacter solanacearum' associated with Bactericera trigonica-affected carrots in the Canary Islands. Plant Dis. 2012;96: 581. [DOI] [PubMed] [Google Scholar]
- 10.Munyaneza JE, Fisher TW, Sengoda VG, Garczynski SF, Nissinen A, and Lemmetty A. First report of "Candidatus Liberibacter solanacearum" associated with psyllid-affected carrots in Europe. Plant Dis. 2010;94: 639. [DOI] [PubMed] [Google Scholar]
- 11.Munyaneza JE, Fisher TW, Sengoda VG, Garczynski SF, Nissinen A, and Lemmetty A. Association of "Candidatus Liberibacter solanacearum" with the psyllid, Trioza apicalis (Hemiptera: Triozidae) in Europe. J Econ Entomol. 2010;103: 1060–1070. [DOI] [PubMed] [Google Scholar]
- 12.Teresani GR, Bertolini E, Alfaro-Fernández A, Martínez C, Tanaka FAO, Kitajima EW, and et al. Association of 'Candidatus Liberibacter solanacearum' with a vegetative disorder of celery in Spain and development of a real-time PCR method for its detection. Phytopathology. 2014;104: 804–811. 10.1094/PHYTO-07-13-0182-R [DOI] [PubMed] [Google Scholar]
- 13.Nelson WR, Fisher TW, and Munyaneza JE. Haplotypes of "Candidatus Liberibacter solanacearum" suggest long-standing separation. Eur J Plant Pathol. 2011;130: 5–12. [Google Scholar]
- 14.Sengoda VG, Cooper WR, Swisher KD, Henne DC, and Munyaneza JE. Latent period and transmission of "Candidatus Liberibacter solanacearum" by the potato psyllid Bactericera cockerelli (Hemiptera: Triozidae). PLoS One. 2014;9: e93475 10.1371/journal.pone.0093475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munyaneza JE, Lemmetty A, Nissinen AI, Sengoda VG, and Fisher TW. Molecular detection of aster yellows phytoplasma and "Candidatus Liberibacter Solanacearum" in carrots affected by the psyllid Trioza apicalis (Hemiptera: Triozidae) in Finland. J Plant Pathol. 2011;93: 697–700. [Google Scholar]
- 16.Nissinen AI, Haapalainen M, Jauhiainen L, Lindman M, and Pirhonen M. Different symptoms in carrots caused by male and female carrot psyllid feeding and infection by 'Candidatus Liberibacter solanacearum'. Plant Pathol. 2014;63: 812–820. [Google Scholar]
- 17.Munyaneza JE, Sengoda VG, Stegmark R, Arvidsson AK, Anderbrant O, Yuvaraj JK, and et al. First report of "Candidatus Liberibacter solanacearum" associated with psyllid-affected carrots in Sweden. Plant Dis. 2012;96: 453. [DOI] [PubMed] [Google Scholar]
- 18.Munyaneza JE, Sengoda VG, Sundheim L, and Meadow R. First report of "Candidatus Liberibacter solanacearum" associated with psyllid-affected carrots in Norway. Plant Dis. 2012;96: 454. [DOI] [PubMed] [Google Scholar]
- 19.Munyaneza JE, Swisher KD, Hommes M, Willhauck A, Buck H, and Meadow R. First report of 'Candidatus Liberibacter solanacearum' associated with psyllid-infested carrots in Germany. Plant Dis. 2015;99: 1269. [Google Scholar]
- 20.Nelson WR, Sengoda VG, Alfaro-Fernández AO, Font MI, Crosslin JM, and Munyaneza JE. A new haplotype of "Candidatus Liberibacter solanacearum" identified in the Mediterranean region. Eur J Plant Pathol. 2013;135: 633–639. [Google Scholar]
- 21.Bertolini E, Teresani GR, Loiseau M, Tanaka FAO, Barbé S, Martínez C, and et al. Transmission of 'Candidatus Liberibacter solanacearum' in carrot seeds. Plant Pathol. 2015;64: 276–285. [Google Scholar]
- 22.Loiseau M, Gamier S, Boirin V, Merieau M, Leguay A, Renaudin I, and et al. First report of 'Candidatus Liberibacter solanacearum' in carrot in France. Plant Dis. 2014;98: 839. [DOI] [PubMed] [Google Scholar]
- 23.Tahzima R, Maes M, Achbani EH, Swisher KD, Munyaneza JE, and De Jonghe K. First report of 'Candidatus Liberibacter solanacearum' on carrot in Africa. Plant Dis. 2014;98: 1426. [DOI] [PubMed] [Google Scholar]
- 24.Lin H, Lou BH, Glynn JM, Doddapaneni H, Civerolo EL, Chen CW, and et al. The complete genome sequence of 'Candidatus Liberibacter solanacearum', the bacterium associated with potato zebra chip disease. PLoS One. 2011;6: e19135 10.1371/journal.pone.0019135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson SM, Johnson CP, Lu AY, Frampton RA, Sullivan KL, Fiers MWEJ, and et al. Genomes of 'Candidatus Liberibacter solanacearum' haplotype A from New Zealand and the United States suggest significant genome plasticity in the species. Phytopathology. 2015;105: 863–871. 10.1094/PHYTO-12-14-0363-FI [DOI] [PubMed] [Google Scholar]
- 26.Zheng Z, Clark N, Keremane M, Lee R, Wallis C, Deng X, and et al. Whole-genome sequence of "Candidatus Liberibacter solanacearum" strain R1 from California. Genome Announc. 2014;2: e01353–14. 10.1128/genomeA.01353-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu F, Deng X, Liang G, Wallis C, Trumble JT, Prager S, and et al. De novo genome sequence of "Candidatus Liberibacter solanacearum" from a single potato psyllid in California. Genome Announc. 2015;3: e01500–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chevreux B, Wetter T, and Suhai S. Genome sequence assembly using trace signals and additional sequence information. German Conference on Bioinformatics. 1999;99: 45–56. [Google Scholar]
- 29.Peng Y, Leung HCM, Yiu SM, and Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28: 1420–1428. 10.1093/bioinformatics/bts174 [DOI] [PubMed] [Google Scholar]
- 30.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, and et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonfield JK, and Whitwham A. Gap5-editing the billion fragment sequence assembly. Bioinformatics. 2010;26: 1699–1703. 10.1093/bioinformatics/btq268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darling ACE, Mau B, Blattner FR, and Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14: 1394–1403. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koressaar T, and Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23: 1289–1291. 10.1093/bioinformatics/btm091 [DOI] [PubMed] [Google Scholar]
- 34.Li H, and Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, and et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011: 6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darriba D, Taboada GL, Doallo R, and Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27: 1164–1165. 10.1093/bioinformatics/btr088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatakis A, Ludwig T, and Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;2: 456–463. [DOI] [PubMed] [Google Scholar]
- 39.Kanehisa M, and Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, and et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 2013;41: D605–D612. 10.1093/nar/gks1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, and Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21: 3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 42.Petersen TN, Brunak S, von Heijne G, and Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8: 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 43.Bendtsen JD, Jensen LJ, Blom N, von Heijne G, and Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Engineering Design and Selection. 2004;17: 349–356. [DOI] [PubMed] [Google Scholar]
- 44.Jain M, Fleites LA, and Gabriel DW. Prophage-encoded peroxidase in 'Candidatus Liberibacter asiaticus' is a secreted effector that suppresses plant defenses. Mol Plant-Microbe Interact. 2015;28: 1330–1337. 10.1094/MPMI-07-15-0145-R [DOI] [PubMed] [Google Scholar]
- 45.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, and et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23: 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 46.Drozdetskiy A, Cole C, Procter J, and Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43: W389–W394. 10.1093/nar/gkv332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin H, Pietersen G, Han C, Read DA, Lou B, Gupta G, and et al. Complete genome sequence of "Candidatus Liberibacter africanus," a bacterium associated with citrus huanglongbing. Genome Announc. 2015;3: e00733–15. 10.1128/genomeA.00733-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang YJ, and et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44: W16–W21. 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang SJ, Flores-Cruz Z, Zhou LJ, Kang BH, Fleites LA, Gooch MD, and et al. 'Ca. Liberibacter asiaticus' carries an excision plasmid prophage and a chromosomally integrated prophage that becomes lytic in plant infections. Mol Plant-Microbe Interact. 2011;24: 458–468. 10.1094/MPMI-11-10-0256 [DOI] [PubMed] [Google Scholar]
- 50.Zhou LJ, Powell CA, Hoffman MT, Li WB, Fan GC, Liu B, and et al. Diversity and plasticity of the intracellular plant pathogen and insect symbiont "Candidatus Liberibacter asiaticus" as revealed by hypervariable prophage genes with intragenic tandem repeats. Appl Environ Microbiol. 2011;77: 6663–6673. 10.1128/AEM.05111-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wulff NA, Zhang S, Setubal JC, Almeida NF, Martins EC, Harakava R, and et al. The complete genome sequence of 'Candidatus Liberibacter americanus', associated with citrus huanglongbing. Mol Plant-Microbe Interact. 2014;27: 163–176. 10.1094/MPMI-09-13-0292-R [DOI] [PubMed] [Google Scholar]
- 52.Leonard MT, Fagen JR, Davis-Richardson AG, Davis MJ, and Triplett EW. Complete genome sequence of Liberibacter crescens BT-1. Standards in Genomic Sciences. 2012;7: 271–283. 10.4056/sigs.3326772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fagen JR, Leonard MT, McCullough CM, Edirisinghe JN, Henry CS, Davis MJ, and et al. Comparative genomics of cultured and uncultured strains suggests genes essential for free-living growth of Liberibacter. PLoS One. 2014;9: e84469 10.1371/journal.pone.0084469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuykendall LD, Shao JY, and Hartung JS. Conservation of gene order and content in the circular chromosomes of 'Candidatus Liberibacter asiaticus' and other Rhizobiales. PLoS One. 2012;7: e34673 10.1371/journal.pone.0034673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albar L, Bangratz-Reyser M, Hebrard E, Ndjiondjop MN, Jones M, and Ghesquière A. Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J. 2006;47: 417–426. 10.1111/j.1365-313X.2006.02792.x [DOI] [PubMed] [Google Scholar]
- 56.Duan YP, Zhou LJ, Hall DG, Li WB, Doddapaneni H, Lin H, and et al. Complete genome sequence of citrus huanglongbing bacterium, 'Candidatus Liberibacter asiaticus' obtained through metagenomics. Mol Plant-Microbe Interact. 2009;22: 1011–1020. 10.1094/MPMI-22-8-1011 [DOI] [PubMed] [Google Scholar]
- 57.Willems A. The Family Phyllobacteriaceae In: The Prokaryotes: Alphaproteobacteria and Betaproteobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 355–418. [Google Scholar]
- 58.Williams KP, Sobral BW, and Dickerman AW. A robust species tree for the Alphaproteobacteria. J Bacteriol. 2007;189: 4578–4586. 10.1128/JB.00269-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jagoueix S, Bové JM, and Garnier M. The phloem-limited bacterium of greening disease of citrus is a member of the alpha-subdivision of the Proteobacteria. Int J Syst Bacteriol. 1994;44: 379–386. 10.1099/00207713-44-3-379 [DOI] [PubMed] [Google Scholar]
- 60.Jagoueix S, Bové JM, and Garnier M. PCR detection of the two 'Candidatus' liberobacter species associated with greening disease of citrus. Molecular and Cellular Probes. 1996;10: 43–50. 10.1006/mcpr.1996.0006 [DOI] [PubMed] [Google Scholar]
- 61.Haapalainen M, Kivimäki P, Latvala S, Rastas M, Hannukkala A, Jauhiainen L, and et al. Frequency and occurrence of the carrot pathogen ‘Candidatus Liberibacter solanacearum’ haplotype C in Finland. Plant Pathol. 2016; [Google Scholar]
- 62.Lin H, and Gudmestad NC. Aspects of pathogen genomics, diversity, epidemiology, vector dynamics, and disease management for a newly emerged disease of potato: zebra chip. Phytopathology. 2013;103: 524–537. 10.1094/PHYTO-09-12-0238-RVW [DOI] [PubMed] [Google Scholar]
- 63.Hao GX, Pitino M, Ding F, Lin H, Stover E, and Duan YP. Induction of innate immune responses by flagellin from the intracellular bacterium, 'Candidatus Liberibacter solanacearum'. BMC Plant Biol. 2014; 14: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim B. H., and Gadd G. M. Biosynthesis and microbial growth In: Bacterial Physiology and Metabolism. Cambridge: Cambridge University Press; 2008. pp. 126–201 [Google Scholar]
- 65.Oshima K, Kakizawa S, Nishigawa H, Jung HY, Wei W, Suzuki S, and et al. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat Genet. 2004;36: 27–29. 10.1038/ng1277 [DOI] [PubMed] [Google Scholar]
- 66.Baumann P, Bowditch RD, Baumann L, and Beaman B. Taxonomy of marine Pseudomonas species: P. stanieri sp. nov.; P. perfectomarina sp. nov., nom. rev.; P. nautica: and P. doudoroffii. Int J Syst Bacteriol. 1983;33: 857–865. [Google Scholar]
- 67.Bowditch RD, Baumann L, and Baumann P. Description of Oceanospirillum kriegii sp. nov. and O. jannaschii sp. nov. and assignment of two species of Alteromonas to this genus as O. commune comb. nov. and O. vagum comb. nov. Curr Microbiol. 1984;10: 221–229. [Google Scholar]
- 68.Choi YU, Kwon YK, Ye BR, Hyun JH, Heo SJ, Affan A, and et al. Draft genome sequence of Marinobacterium stanieri S30, a strain isolated from a coastal lagoon in Chuuk state in Micronesia. J Bacteriol. 2012;194: 1260 10.1128/JB.06703-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim H, Choo YJ, Song J, Lee JS, Lee KC, and Cho JC. Marinobacterium litorale sp. nov. in the order Oceanospirillales. Int J Syst Evol Microbiol. 2007;57: 1659–1662. 10.1099/ijs.0.64892-0 [DOI] [PubMed] [Google Scholar]
- 70.Satomi M, Kimura B, Hamada T, Harayama S, and Fujii T. Phylogenetic study of the genus Oceanospirillum based on 16S rRNA and gyrB genes: emended description of the genus Oceanospirillum, description of Pseudospirillum gen. nov., Oceanobacter gen. nov and Terasakielia gen. nov and transfer of Oceanospirillum jannaschii and Pseudomonas stanieri to Marinobacterium as Marinobacterium jannaschii comb. nov and Marinobacterium stanieri comb. nov. Int J Syst Evol Microbiol. 2002;52: 739–747. 10.1099/00207713-52-3-739 [DOI] [PubMed] [Google Scholar]
- 71.Pasternak Z, Pietrokovski S, Rotem O, Gophna U, Lurie-Weinberger MN, and Jurkevitch E. By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J. 2013;7: 756–769. 10.1038/ismej.2012.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Z, Kadouri DE, and Wu M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics. 2011;12: 453 10.1186/1471-2164-12-453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thao ML, Moran NA, Abbot P, Brennan EB, Burckhardt DH, and Baumann P. Cospeciation of psyllids and their primary prokaryotic endosymbionts. Appl Environ Microbiol. 2000;66: 2898–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waldron DE, and Lindsay JA. Sau1: a novel line age-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J Bacteriol. 2006;188: 5578–5585. 10.1128/JB.00418-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, and Uchiyama I. Shaping the genome—restriction-modification systems as mobile genetic elements. Curr Opin Genet Dev. 1999;9: 649–656. [DOI] [PubMed] [Google Scholar]
- 76.Zhou L, Powell CA, Li W, Irey M, and Duan Y. Prophage-mediated dynamics of 'Candidatus Liberibacter asiaticus' populations, the destructive bacterial pathogens of citrus huanglongbing. PLoS One. 2013;8: e82248 10.1371/journal.pone.0082248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yoshii M, Nishikiori M, Tomita K, Yoshioka N, Kozuka R, Naito S, and et al. The Arabidopsis cucumovirus multiplication 1 and 2 loci encode tip translation initiation factors 4E and 4G. J Virol. 2004;78: 6102–6111. 10.1128/JVI.78.12.6102-6111.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O'Brien PJ, and Ellenberger T. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. Journal of Biological Chemistry. 2004;279: 26876–26884. 10.1074/jbc.M403860200 [DOI] [PubMed] [Google Scholar]
- 79.Wyatt MD, Allan JM, Lau AY, Ellenberger TE, and Samson LD. 3-Methyladenine DNA glycosylases: structure, function, and biological importance. Bioessays. 1999;21: 668–676. [DOI] [PubMed] [Google Scholar]
- 80.Thomas L, Yang CH, and Goldthwait DA. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982;21: 1162–1169. [DOI] [PubMed] [Google Scholar]
- 81.Yao JX, Saenkham P, Levy J, Ibanez F, Noroy C, Mendoza A, and et al. Interactions "Candidatus Liberibacter solanacearum"—Bactericera cockerelli: haplotype effect on vector fitness and gene expression analyses. Front Cell Infect Microbiol. 2016;6: 62 10.3389/fcimb.2016.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Niu YD, Stanford K, Kropinski AM, Ackermann HW, Johnson RP, She YM, and et al. Genomic, proteomic and physiological characterization of a T5-like bacteriophage for control of Shiga toxin-producing Escherichia coli O157: H7. PLoS One. 2012;7: e34585 10.1371/journal.pone.0034585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
Alignment of sequences corresponding to AYJ09_01490 were performed using Clustal X and the secondary structure was predicted using Jpred 4. S.lyc, S.tub, S.pen, O.gla represent sequences from Solanum lycopersicum, Solanum tuberosum, Solanum pennellii, Oryza glaberrima, respectively, and 'X1/X2' represent different isoforms of eIF4G. The amino acid residues highlighted with colors have high conservation (above 30%) of similarity regarding the hydrophobicity character. The secondary structure predictions, helix (red) or coil (black), are displayed on the bottom row.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. All the genomic and sub-genomic DNA sequence files are available in the NCBI GenBank database (accession numbers LWEB00000000, LVWB00000000, KX431889, KX431890 and KX431891).