Abstract
A bacteriophage encoding the Shiga toxin 2c variant (Stx2c) was isolated from the human Escherichia coli O157 strain CB2851 and shown to form lysogens on the E. coli K-12 laboratory strains C600 and MG1655. Production of Stx2c was found in the wild-type E. coli O157 strain and the K-12 lysogens and was inducible by growing bacteria in the presence of ciprofloxacin. Phage 2851 is the first reported viable bacteriophage which carries an stx2c gene. Electron micrographs of phage 2851 showed particles with elongated hexagonal heads and long flexible tails resembling phage lambda. Sequence analysis of an 8.4-kb region flanking the stx2c gene and other genetic elements revealed a mosaic gene structure, as found in other Stx phages. Phage 2851 showed lysis of E. coli K-12 strains lysogenic for Stx phages encoding Stx1 (H19), Stx2 (933W), Stx (7888), and Stx1c (6220) but showed superinfection immunity with phage lambda, presumably originating from the similarity of the cI repressor proteins of both phages. Apparently, phage 2851 integrates at a different chromosomal locus than Stx2 phage 933W and Stx1 phage H19 in E. coli, explaining why Stx2c is often found in combination with Stx1 or Stx2 in E. coli O157 strains. Diagnostic PCR was performed to determine gene sequences specific for phage 2851 in wild-type E. coli O157 strains producing Stx2c. The phage 2851 q and o genes were frequently detected in Stx2c-producing E. coli O157 strains, indicating that phages related to 2851 are associated with Stx2c production in strains of E. coli O157 that were isolated in different locations and time periods.
Shiga toxin (Stx) (verocytotoxin)-producing strains of Escherichia coli O157 (STEC O157) are important human pathogens that can cause severe disease, such as hemorrhagic colitis and hemolytic uremic syndrome (HUS) (22). The ability of STEC O157 to cause disease is directly related to production of Stxs by the bacteria, and the genes for Stxs are located on bacteriophages, which are harbored on the bacterial chromosome (15, 33, 35, 45, 48, 53). Two major types of Shiga toxins called Stx1 and Stx2, whose genes show 58% identity on the DNA level, were found in STEC O157 strains (18). Further analysis revealed the presence of Stx2 variants called Stx2c or Stx2vha in some STEC O157 strains, and the nucleotide sequences of the corresponding stx genes were analyzed (18, 30, 32). Stx2 and its variants Stx2c and Stx2vha show >99% identity in the genes coding for the A subunit and ∼96% identity in those coding for the B subunit of the toxin (18, 30). The nucleotide sequences of the B subunits of Stx2c (43) and Stx2vha (originally designated VTx2ha) (17) genes are identical, and the proteins were described as variants of the Stx2 toxin family.
Many O157 strains were found to produce more than one type of Stx, and combinations of different toxin types are frequent in STEC O157, strains which were from geographically different places and isolated at different time periods (4, 34, 38, 43, 52). Subtyping of stx genes by PCR and endonuclease digestion of PCR products revealed that stx2c or stx2vha variant genes are frequently found in STEC O157 strains from different geographical origins (9, 19, 30, 34, 38, 52). The stx2c allele was most common among stx genes in STEC O157 strains that were isolated in Australia, Belgium, and Germany and is often present in combination with stx1 or stx2 genes (4, 11, 12, 36).
Epidemiological studies have indicated that the virulence of STEC O157 strains for humans is related to the type of Stx which is produced by the strains. Production of Stx2 was found to be closely associated with HUS and bloody diarrhea in STEC O157 and non-O157 strains (4, 5, 8, 13). The role of Stx2c in disease is less clear because it is often produced together with Stx1 or Stx2 in STEC O157 strains. Nishikawa et al. (34) reported that O157 strains carrying only the stx2vha/stx2c gene are less associated with bloody diarrhea than stx2-positive strains. However, it was not clear whether this is due to differences in toxin binding to the eukaryotic target cells or the amount of toxin which is produced by either strain (34).
Bacteriophages encoding Stx1 or Stx2 were successfully isolated from STEC O157 strains, and their complete nucleotide sequences were determined (37, 40, 41). In contrast, viable phages carrying stx2c genes were not reported to be isolated from STEC O157 or non-O157 STEC strains, and the genetic analysis of stx2c genes and their adjacent sequences were obtained only from studies of prophage DNA (10, 43, 50). The complete nucleotide sequence of an Stx2c- or Stx2vha-encoding bacteriophage is not known, and both variant genes were not present in the two STEC O157 strains whose complete genomic sequences were analyzed (15, 35). Since not much is known about Stx2c bacteriophages, we were interested in comparing them with other well-characterized Stx phages. For this purpose, we isolated a viable Stx2c-encoding bacteriophage from a human STEC O157 strain and compared its morphology, biological properties, and toxin production with those of other Stx-encoding phages. A genetic analysis of an 8.4-kb DNA region encompassing the stx2c gene and the genes involved in regulation of Stx2c production was performed by nucleotide sequencing of Stx2c phage DNA.
MATERIALS AND METHODS
Bacteria and bacteriophages.
E. coli O157 and non-O157 strains were from the laboratory collection of the Robert Koch Institute. The non-sorbitol-fermenting STEC strain CB2851 (O157:NM) was chosen as the source of the Stx2c-encoding bacteriophage because it carries only the stx2c determinant and is negative for other stx genes (7). Strain CB2851 was isolated in 1993 from the feces of a 4-year-old girl with diarrhea in Germany. The 82 E. coli O157 strains that were investigated for stx genotype and for the presence of the bacteriophage 2851-associated q, o, and antB genes came originally from Canada, Denmark, Germany, The Netherlands, the United Kingdom, and the United States and were described previously (4, 6).
The E. coli K-12 strain C600 and its derivative strains C600(H19), which carries the Stx1 bacteriophage H19, and C600(W34), which is lysogenic for the Stx2 bacteriophage 933W, were described previously (47). Strain CSH67 (R5) was used as a source for bacteriophage lambda for the preparation of lysates and transduction of lambda to strain C600 (28). The origin and source of Stx phages 6220 (stx1c) and 7888 (stx) are described elsewhere (24, 47).
Detection of Stx and subtyping of genes encoding Stx.
Production of Stx was determined by the verocell test and by the VTEC-RPLA test (a reverse-passive latex agglutination test for the detection of verocytotoxin 1 [VT1] [Stx1] and VT2 [Stx2]) (7). Detection of stx-specific sequences was performed by PCR, and subtyping of stx genes was done by endonuclease digestion of PCR products, as previously described (5). An stx-specific PCR product of 897 (stx1 family) or 905 (stx2 family) bp was obtained using common primers for all stx genes (26), and restriction fragment typing of stx genes was performed with HincII and AccI as described previously (3).
Induction and isolation of bacteriophages.
Induction of temperate bacteriophages from E. coli was performed with ciprofloxacin (54). A single colony of strain CB2851 was inoculated into 30 ml of Luria broth and grown under aeration for 3 h at 37°C. Ciprofloxacin (final concentration, 0.15 μg/ml) was added to the growing culture, which was further incubated overnight. The bacteria were sedimented by centrifugation, and the culture fluid was filtered through 0.45-μm-pore-size membranes (Schleicher and Schüll, Dassel, Germany). Dilutions of the supernatant were titrated on the E. coli K-12 strain C600. Isolation of single plaques and preparation of high-titer phage stocks were performed as described previously (47).
Phage sensitivity tests and lysogenization of E. coli K-12 strains with Stx phages.
The E. coli K-12 strains C600 and MG1655 (2) were used as bacteriophage-free recipient strains for transduction and selection of lysogens. Lysogenization of strains was performed as described previously (47), and phage lysogens were identified by the presence of stx2c-related sequences and production of verotoxin, as described previously (5) (Fig. 1). Sensitivity to various phages was investigated by spotting phage lysates on bacterial test strains as previously described (47). The plating efficiencies of phages were determined by plating serial dilutions of phage lysates on E. coli test strains, followed by calculating the number of PFU per milliliter after overnight incubation at 37°C. The relative plating efficiency of a phage strain was calculated as the number of PFU per milliliter obtained with the test strain divided by the number of PFU per milliliter obtained on strain C600, which was used for the propagation of bacteriophages.
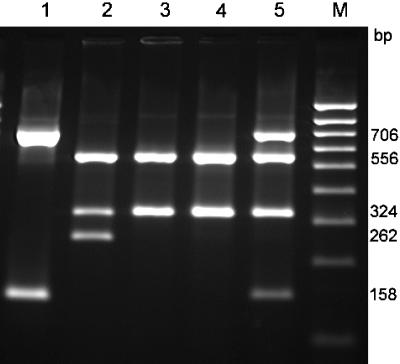
FIG. 1.
Subtyping of stx genes in STEC O157 strains and in C600 transductants. HincII digest of stx-specific PCR products. Lanes: 1, C600 (H19) (stx1); 2, E32511 (stx2 and stx2c); 3, CB2851 (stx2c); 4, C600 (phage 2851) (stx2c); 5, C600 phage H19 and phage 2851 (stx1 and stx2c); M, DNA Hyperladder II (range, 100 to 1,000 bp; Bioline GmbH, Luckenwalde, Germany).
Induction of Stx2c production with ciprofloxacin.
Overnight cultures of bacteria grown in tryptic soy broth (TSB) were diluted to 107 bacteria/ml in flasks containing TSB or TSB with ciprofloxacin (5 ng/ml). The cultures were incubated for 6 h at 37°C under aeration for induction of Stx production, as described previously (54). After incubation, the number of living bacteria was determined by plating serial dilutions on Luria broth-agar plates, which were incubated overnight at 37°C. Stx2c production was measured by testing serial twofold dilutions of the bacterial-culture supernatant with the VTEC-RPLA assay as described previously (7).
Electron microscopy.
Phages were isolated in CsCl step gradients according to standard protocols (39). Phage suspensions were prepared by negative staining with 1% uranyl acetate on carbon films according to the method of Steven et al. (46) and examined in a Philips CM100 transmission electron microscope.
Isolation of phage DNA.
High-titer phage lysates (>109 PFU/ml) were incubated with 1 μg each of DNase and RNase C (QIAGEN, Hilden, Germany)/ml for 30 min at 37°C. Phage DNA was prepared with the QIAGEN Lambda kit according to the instructions of the manufacturer.
Cross-hybridization of phage genomes.
Restriction endonuclease-digested phage DNA fragments were separated in 0.8% agarose gels and transferred to Hybond N+ membrane (Amersham Pharmacia Biotech, Freiburg, Germany) by capillary blotting. Hybridization was performed at 65°C as described earlier (47). For preparing gene probes, phage DNAs were digested with AccI and labeled using a fluorescein labeling kit (NEN Life Science, Boston, Mass.) according to the manufacturer's recommendations.
PCR.
Amplification of DNA by PCR was performed in 30 cycles as indicated in Table 1. Annealing temperatures were calculated with Mac Vector software (see below).
TABLE 1.
Oligonucleotide primer pairs used in this study
| PCR producta | Primer | Sequence (5′-3′) | Product size (bp) | Genetic element | Reference or source |
|---|---|---|---|---|---|
| 1 | CIVTF | CACAACGGAACAACTCTCAT | 454 | cI | 20 |
| CIV2R | ATACCTTGGTATTGATAACA | ||||
| 2 | 50 | CTAACGAGATTACCGACAGTC | 710 | o | AJ413274 |
| 51 | AAACTTCCCTTCCCGAAC | ||||
| 3 | 52 | TGACCAGCAAAAACTGCC | 836 | p | AJ413274 |
| 53 | TGACTCAGGGAGAATACCG | ||||
| 4 | 54 | TCGGTATCGTTCATCGTCC | 799 | orf31/ninB | AJ413274 |
| 55 | CCTTCCCTTTTCGTTGTG | ||||
| 5 | 46-B | TCACCCATAACCAGATTCC | 643 | antA | AJ413274 |
| 47-B | TCCGTCAGATGCTTGAAGGC | ||||
| 6 | 48 | GGCACAGAACCAGATTACATC | 579 | antB | AJ413274 |
| 49 | TGAAGGCGGCAACTCTTTG | ||||
| 7 | 58 | GTGAAAATCGGAGTGAACAAGGTG | 387 | o | AJ413274 |
| 59 | AAACGCCCTGACATACGCTCTG | ||||
| 8 | 56 | GCTCAAAACACCTTAGCAAGTAGC | 311 | antB | AJ413274 |
| 57 | CGGATGTAATCTGGTTCTGTGCC | ||||
| 9 | 5 | GGGCATAAGAGAACTAAACCTCACC | 441 | q | AJ605767 |
| 6 | AACGCACTATCCAAAATCGGG | ||||
| 62 | CAGGTGGATGTGATGGTTTGTTC | 578 | Stx2 phage integration site (right) | NC_004914 | |
| 61 | TGCGTGCTGGGCTTTATCTTCC | AP002554 | |||
| 60 | TGAATACGGGGATG(A/C)GTTGACTG | 652 | Stx2 phage integration site (left) | NC_004914 | |
| 63 | CGCTACGGAATAGAGATAACACGAG | AP002554 |
Figure 4.
Nucleotide sequencing.
PCR products purified with the QIAquick PCR Purification kit (QIAGEN) and genomic DNA of phage 2851 were used for sequencing. Sequencing reactions were carried out using dye terminator chemistry (PE Applied Biosystems, Darmstadt, Germany) and separated on an automated DNA sequencer (ABI PRISM 3100 Genetic Analyzer). The sequences were analyzed using Lasergene (DNASTAR, Madison, Wis.) and Mac Vector (Oxford Molecular Group, Campbell, Calif.) software to assemble and align the sequences and to determine putative open reading frames (ORFs). Sequence similarity searching of the current version of GenBank at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/) was accomplished with the BLASTN, BLASTP, or BLASTX algorithm (1).
Nucleotide sequence accession number.
The nucleotide sequence of the Stx2c-encoding region of phage 2851 with a size of 8,417 bp was submitted to the EMBL data library under accession number AJ605767.
RESULTS AND DISCUSSION
Host range of phage 2851 and immunity to other Stx phages and to bacteriophage lambda.
Phage lysates from the Stx2c-positive E. coli O157 strain CB2851 were prepared as described in Materials and Methods. Infection with phage 2851 resulted in lysis of the E. coli K-12 laboratory strains C600 and MG1655, showing small clear plaques; however, no lysis was observed with E. coli wild-type O157 or non-O157 STEC and Stx-negative strains. C600 and MG1655 derivative strains lysogenized with phage 2851 were isolated and shown to be cytotoxic to Vero cells and to react positively in the VTEC-RPLA assay for Stx2-type toxins. PCR-restriction fragment length polymorphism subtyping of stx genes present in lysogenized E. coli K-12 strains revealed the presence of an stx2c gene, as found in the parental strain, CB2851 (Fig. 1).
We were further interested to see if the presence of other Stx phages in the E. coli K-12 strain C600 would have an influence on sensitivity to subsequent infection with bacteriophage 2851. For this purpose, we constructed a series of C600 derivative strains which were lysogenic for phages encoding Stx1 (phage H19), Stx2 (phage 933W), Stx (phage 7888), and Stx1c (phage 6220) and for bacteriophage lambda. Lysates of the above-mentioned phages were propagated on E. coli C600 and were used in spot tests for sensitivity assays. Phage 2851 showed lysis and plaque formation on C600 strains lysogenic for phages H19, 933W, 7888, and 6220 but did not plate on C600 lysogenic for phage 2851 or on C600 lambda lysogen. In order to investigate if phages 2851 and lambda show superinfection immunity to each other, we compared the plating efficiencies of these phages on the respective C600 lysogenic strains (Table 2). The plating efficiencies of phage lambda and phage 2851 were both reduced 105- to 106-fold on C600 strains lysogenic for either phage. These findings might indicate that phages lambda and 2851 possess related cI repressor proteins that confer superinfection immunity on phages of the same type in lysogenic E. coli strains. This possibility was further explored by nucleotide sequence analysis of the cI gene of phage 2851, which was found to be highly similar to the lambda cI gene in regions encoding functional regions of the repressor (see below).
TABLE 2.
Plating efficiencies of phages lambda and 2851 on C600 lysogenic with either phage
| Phage lysate | Plating efficiency of phage lysate on strain:
|
||
|---|---|---|---|
| C600 | C600 (phage 2851) | C600 (lambda) | |
| Phage 2851 | 1.0 | <1.0 × 10−8 | 2.5 × 10−5 |
| Lambda | 1.0 | 1.6 × 10−6 | <1.0 × 10−8 |
Induction of Stx production in E. coli wild-type and K-12 strains.
Induction of Stx2c production by ciprofloxacin was investigated with the parental O157 strain CB2851 and with an E. coli C600 derivative strain lysogenic for the Stx2c bacteriophage 2855. The minimal concentration of ciprofloxacin that inhibited growth of the bacteria was determined to be 25 ng/ml. Addition of 5 ng of ciprofloxacin per ml of culture did not significantly inhibit the growth of wild-type E. coli O157 or E. coli K-12 strains but had a stimulating effect on the production of Stx2c. Stx2c production was enhanced 8- to 16-fold in CB2851 and 4-fold in the E. coli C600 phage 2851 lysogens compared to that in noninduced cultures (Table 3). These results show that the production of Stx2c is inducible by ciprofloxacin. Similar findings were made for Stx2 in previously published studies (23, 54).
TABLE 3.
Enhancement of Stx2c production by ciprofloxacin
| Strain | Characteristics | Ciprofloxacin (5 ng/ml)a | Titer (±SD)/mlb (10a) | VTEC-RPLA test resultc |
|---|---|---|---|---|
| CB2851 | O157:[H7]; stx2c | − | 3.35 ± 0.2 | 1:8-1:16 |
| CB2851 | O157:[H7]; stx2c | + | 2.73 ± 0.7 | 1:128 |
| C600 (phage 2851) | E. coli K-12; stx2c | − | 1.51 ± 0.2 | 1:8-1:16 |
| C600 (phage 2851) | E. coli K-12; stx2c | + | 1.28 ± 0.5 | 1:32-1:64 |
−, TSB without ciprofloxacin; +, TSB supplemented with ciprofloxacin.
Viable bacteria after 6 h of incubation in TSB at 37°C.
Highest dilution of culture supernatant that is positive in the VTEC-RPLA test with the VT2 (Stx2) reagent. Results from two separate experiments.
Morphology and genome size of phage 2851.
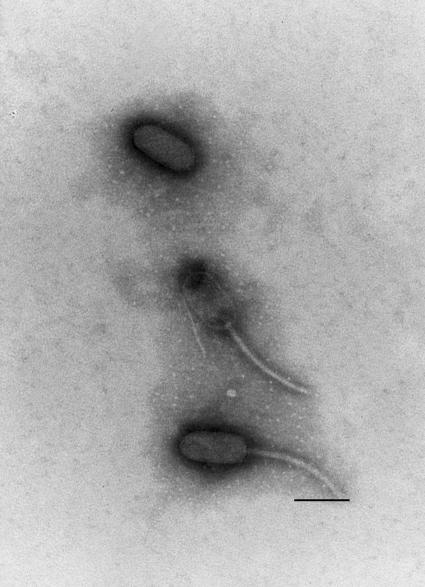
Phage lysates were purified in CsCl step gradients as described previously (47) and examined by transmission electron microscopy. Electron micrographs depicted phage particles with elongated hexagonal heads and long flexible tails, resembling phage lambda in their morphology (16) (Fig. 2). The mean dimensions of the head were 106 nm long and 53 nm wide, and the tail was ∼165 nm long and 8 nm wide. Isolated DNA of the phage was subjected to enzyme restriction digestion and was estimated to be ∼53 ± 1.5 kb. On the basis of its morphology and the DNA type of nucleic acid, phage 2851 was grouped in the phage family Siphoviridae.
FIG. 2.
Electron micrograph of CsCl-purified phage 2851 particles. Bar = 100 nm.
Cross-hybridization studies with other Stx phages and phage lambda.
Cross-hybridization studies of AccI-digested phage 2851 DNA with total genomic DNA of phage lambda, the Stx1-encoding phage H19, and the Stx2-encoding phage 933W were carried out to determine the potential relationship of phage 2851 to these phages. The results are shown in Fig. 3. Clear hybridization signals were obtained with DNA fragments ranging from 1 to 20 kb in size. All phages investigated showed significant cross-hybridization to each other, differing only in size and numbers of hybridizing bands. However, a possible closer genetic relationship among the different types of Stx phages was not detectable by DNA hybridization of total phage genomes. In order to obtain more information on the genetic relationships among phage 2851, lambda, and lambdoid Stx phages, we decided to sequence a larger part of the genomic DNA flanking the stx2c gene of phage 2851.
FIG. 3.
Restriction patterns and cross-hybridizations of DNAs of phages lambda (Stx negative), H19 (Stx1), 933W (Stx2), and 2851 (Stx2c). Shown are AccI digests of phage DNAs. Lanes 1, lambda; lanes 2, H19; lanes 3, 933W; lanes 4, CB2851. (A) Restriction pattern. (B) Southern hybridization using genomic DNA of lambda cut with AccI as a probe. (C) Southern hybridization using H19 DNA cut with AccI as a probe. (D) Southern hybridization using 933W DNA cut with AccI as a probe. (E) Southern hybridization using 2851 DNA cut with AccI as a probe.
Sequence analysis of the stx2c region of phage 2851.
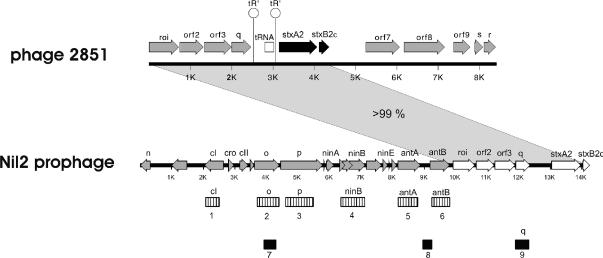
An 8,417-bp DNA region spanning the stx2c gene of phage 2851 was sequenced by primer walking and PCR amplification. The physical map of the stx2c region of phage 2851 is presented in Fig. 4.
FIG. 4.
Structures of the stx2c genes and flanking regions of phage 2851 and prophage Nil2. The grey shaded area indicates the overlapping sequenced DNA (identity > 99%). The hatched boxes (no. 1 to 6) represent genetic elements of phage 2851 DNA (Table 1). The black boxes (no. 7 to 9) represent PCR products derived from phage 2851 DNA used for diagnostic PCR (Table 1). tR, putative terminators involved in antitermination by Q.
The coding sequence of the variant Stx2c toxin of the E. coli O157 reference strain E32511 differs from that of the prototypical Stx2 toxin as found in bacteriophage 933W by three amino acid replacements in the B subunit, whereas the A subunits in both toxins are identical (43). Comparison of the Stx2c-encoding nucleotide sequence of phage 2851 to the nucleotide sequence of the stx2c gene present in the prototype strain E32511 revealed 100% identity in the coding region of the toxin B subunit, thus confirming the stx2c genotype of the phage. In the coding region of the toxin A subunit, we found five nucleotide changes to the sequence of strain E32511, leading to one amino acid change (an aspartic acid residue in position 277 instead of a glutamic acid residue). The region upstream and downstream of the stx2c gene shows a genetic map similar to that of other Stx-encoding phages. Upstream of the stx2c gene, three tRNA genes, two putative transcription terminators, and the coding region of the Q antiterminator are located. Downstream of the stx2c gene are two putative open reading frames (orf7 and orf8), which in other Stx phages (like 933W) form only one large continuous reading frame coding for a hypothetical protein. Further downstream are genes encoding lysis proteins S and R, suggesting that the stx2c gene is expressed late in the lytic cycle of the bacteriophage. The sequenced region encompasses three more open reading frames upstream of the Q gene which encode a Roi homologue, a NinG homologue (orf2), and a putative serine/threonine protein kinase (orf3). The last protein is frequently encoded in Stx phages and prophages and was recently described as similar to a eukaryotic tyrosine (Tyr) kinase (49). Table 4 summarizes the data from nucleotide sequence analysis and the results of the BLAST analysis of the deduced ORFs. The presence of the late antiterminator Q, with a close relationship to the E. coli lambdoid bacteriophage 21 Q (similarity, 96.9%), which was found only rarely in Stx phages, is remarkable. So far, only the Stx2 phage LC59 has been reported to harbor a closely related q gene (29).
TABLE 4.
Summary of sequence data
| ORF | Gene | Putative function | 5′ end | 3′ end | No. of aaa | Closest relationshipb | e value | % Identityc | % Similarityd |
|---|---|---|---|---|---|---|---|---|---|
| 1 | roi | DNA binding | 11 | 739 | 242 | (CAC95101) Roi protein [prophage bacteriophage Nil2] | 1e-131 | (241/242) 99.6 | (241/242) 99.6 |
| 2 | orf2 | Recombination endonuclease | 739 | 1344 | 201 | (CAC95102) recombination endonuclease [bacteriophage Nil2] | 1e-117 | (201/201) 100 | (201/201) 100 |
| 3 | orf3 | Serine/threonine protein kinase | 1341 | 2012 | 223 | (AAN59918) hypothetical protein [bacteriophage LC159] | 1e-131 | (223/223) 100 | (223/223) 100 |
| 4 | q | Antiterminator Q | 2003 | 2491 | 162 | (CAC95104) late antiterminator [prophage bacteriophage Nil2] | 4e-90 | (161/162) 99.4 | (162/162) 100 |
| (CAB39993) Q protein [bacteriophage 21] | 5e-84 | (154/162) 95.1 | (157/162) 96.9 | ||||||
| ileZ | tRNA-Ile | 2805 | 2880 | (AF125520) ileZ [bacteriophage 933W] | 2e-34 | 100 | |||
| argN | tRNA-Arg | 2882 | 2958 | (AF654845) argN [bacteriophage LC159] | 5e-34 | 100 | |||
| argO | tRNA-Arg | 2972 | 3048 | (M59432) arg [E. coli 32511] | 5e-35 | 100 | |||
| 5 | stxAa | Shiga toxin A subunit | 3141 | 4100 | 319 | (BAA33759) Shiga toxin 2 A subunit [E. coli] | 1e-179 | (319/319) 100.0 | (319/319) 100 |
| 6 | stxB2c | Shiga toxin B subunit | 4112 | 4381 | 89 | (A60279) Shiga toxin 2c B subunit [E. coli 32511] | 2e-45 | (89/89) 100 | (89/89) 100 |
| 7 | orf7 | Hypothetical | 5245 | 6087 | 280 | (CAC05543) hypothetical protein [E. coli] | 1e-160 | (274/280) 97.9 | (278/280) 99.3 |
| 8 | orf8 | Hypothetical | 6175 | 7182 | 335 | (CAC05552) hypothetical protein [E. coli] | 0 | (333/335) 99.4 | (334/335) 99.7 |
| 9 | orf9 | Hypothetical | 7365 | 7811 | 148 | (NP_049503) hypothetical protein [bacteriophage 933W] | 2e-77 | (145/148) 98.0 | (147/148) 99.3 |
| 10 | s | Lysis protein | 7888 | 8103 | 71 | (NP_050543) S protein [bacteriophage VT2-Sa] | 2e-34 | (71/71) 100.0 | (71/71) 100.0 |
| 11 | r | Endolysin (partial) | 8108 | 8416 | 103 | (NP_613035) endolysin [Stx2-converting bacteriophage 1] | 9e-54 | (103/103) 100.0 | (103/103) 100.0 |
aa, amino acids.
(accession number) putative function [microorganism or plasmid].
Numbers in parentheses represent the number of amino acids from which the identity is calculated (whole-length identity).
Number of amino acids and whole-length similarity.
Immunity and replication genes present in phage 2851.
Based on similarity to DNA sequences of Stx-encoding phages or prophages available in the database, we used published oligonucleotide primers or designed new primers for PCR to determine additional genetic elements of phage 2851 outside the sequenced stx2c region. The list of primer pairs used for PCR amplification is in Table 1. Special focus was given to the sequence of an Stx2c-encoding prophage sequence (prophage Nil2; accession number AJ413274), which partially overlapped with our sequence (the stretch from roi to the stxB2c gene). We were able to amplify more genetic elements upstream of the roi gene based on primers that were deduced from the Nil2 sequence. The identity of the sequences of phage 2851 and the Nil2 prophage in the overlapping sequence from roi to the stxB2c gene, as well as in the PCR products, is >99%. Figure 4 shows the regions amplified using the primer pairs given in Table 1 below the Nil2 prophage sequence. These findings may indicate that the order of the genetic elements in phage 2851 upstream of the roi gene is the same as in the Nil2 prophage sequence.
The genetic elements identified in phage 2851 DNA by PCR and sequence analysis of the PCR products belong to the immunity and replication region. The PCR product derived from the repressor gene cI using the primers CIVTF and CIV2R is 100% identical to the corresponding regions of the cI genes of some other Stx phages (41). The deduced cI repressor is also very similar to the cI repressor of bacteriophage HK97 (79% similarity) (21) and bacteriophage lambda (71% similarity) (accession number J02459). Remarkably, the C-terminal domains of the repressor proteins, which are responsible for dimerization of the active repressors, are identical in lambda and phage 2851. In the N-terminal region of cI2851, an amino acid sequence with similarity to the sequence of helices 2 and 3 of cIlambda involved in DNA binding can be identified, thus suggesting a likely explanation for the superinfection immunity between the two phages (see above). The replication genes o and p are closely related to the corresponding genes of the lambdoid, Stx-negative E. coli phage HK97 (21) but have not yet been described in other Stx phages, with the exception of the Nil2 prophage sequence.
Downstream of the replication gene p and upstream of the roi gene are a number of genetic elements in the Nil2 prophage sequence which belong to the nin (N independence) region (25). This region, which is named after deletions in the archetypical phage lambda, is frequently present in lambdoid phages, though most of the ORFs are not yet functionally assigned. We identified some genetic elements (antA, antB, and orf31/ninB) in the phage 2851 genome by PCR analysis using different primer pairs (Table 1).
Integration site of phage 2851 in CB2851 and C600.
The reported integration site for a number of Stx2 phages in E. coli is in the wrbA locus (44). Based on published sequences, we deduced primers 60 and 61 from the sequences of the Stx2-converting bacteriophage II (GenBank accession number NC_004914) (41) and phage 933W (37) and primers 62 and 63 from the chromosomal integration site in the wrbA locus (accession number AP002554) (Table 1). In order to explore whether the Stx2 bacteriophage 933W and the Stx2c phage 2851 use the same integration site in the chromosomes of wild-type O157 strains and lysogenized E. coli K-12 strains, we performed PCRs for amplification of DNA fragments encompassing bacterial and phage DNA segments on the left (primers 60 and 63) and the right (primers 61 and 62) sides of the integrated prophages. In the case of phage 933W, we found amplicons of the expected sizes in the E. coli O157 reference strain EDL933W and in C600 lysogenized with phage 933W. No amplicons were obtained from strain C600 lysogenized with phage 2851 or from the E. coli O157 strain CB2851, indicating that the Stx2 and the Stx2c phage do not share the same integration site in E. coli O157 and K-12 host strains. This result might explain why stx2c and stx2 genes are often found together in E. coli O157 strains (4, 38, 43, 52). Similar findings were made for stx1, which frequently occurs in combination with the stx2 or stx2c gene in STEC O157 strains, and we succeeded in isolating E. coli C600 transductants that harbored both Stx1 (H19)- and Stx2c (2851)-encoding phages in the chromosome of the E. coli K-12 host (Fig. 1, lane 5).
Detection of phage 2851-specific gene sequences in wild-type E. coli O157 and association with strains carrying the stx2c gene.
Database searching using the BLASTN algorithm indicated few genetic elements of phage 2851 and the Nil2 prophage that might be specifically associated with the stx2c allele and thus putative candidates for diagnostic PCR. PCR primers binding in the following DNA regions were determined: (i) replication gene o; (ii) the 5′ region of the antB gene, including the upstream region; and (iii) the q gene (Table 1). The PCR products for diagnostic PCR are indicated in Fig. 4 (PCR products 7, 8, and 9). C600 strains lysogenic for Stx phages H19 (Stx1), 933W (Stx2), 7888 (Stx), and 6220 (Stx1c) and the corresponding wild-type strains H19, EDL933, CB7888, and CB6220, which were the sources of these phages, were all negative in PCRs for the q, antB, and o gene regions. In contrast, C600 lysogenic for bacteriophage 2851 (Stx2c) and the parental strain CB2851 were positive in all three PCR assays.
We were further interested in the presence of phage 2851-specific q2851, o2851, and antB2851 sequences in E. coli O157 strains and in their association with the stx2c gene. For this purpose, 82 E. coli O157:H7 strains that were isolated in six different countries between 1983 and 1999 were investigated by PCR for q2851, o2851, and antB2851 genes (Table 5). Seven of the 82 strains were negative for production of Stx and for stx genes, and these all tested negative for q2851, o2851, and antB2851 sequences. The stx2c gene was present in 35 E. coli O157 strains, and 34 (97.1%) of these were positive for the q2851 gene and 32 (91.4%) were positive for the o2851 gene. The antB2851 sequence was detected in only nine (25.7%) of the stx2c-positive strains. However, while the antB2851 sequence was present in all five stx2c strains that were isolated in the United States, Denmark, and the United Kingdom, it was absent in 26 (86.6%) of the 30 stx2c-positive E. coli O157 strains that came from Germany. This finding could indicate that a variant Stx2c phage that differs from 2851 in the antB sequence has spread through Stx2c-producing E. coli O157 strains in Germany. However, this needs to be confirmed by future studies.
TABLE 5.
Association of Φ2851 q, o, and ant genes with stx genes in wild-type E. coli O157:H7 strains
|
|
No. of strains in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| stx gene | O157:H7 strains
|
q2851 gene PCR
|
02851 gene PCR
|
antB2851 gene PCR
|
||||
| Total no. | Origin (no. of strains)a | Positive | Negative | Positive | Negative | Positive | Negative | |
| Negative | 8b | D (6), NL (2) | 0 | 8 | 0 | 8 | 0 | 8 |
| stx1 | 1 | D (1) | 1 | 0 | 1 | 0 | 1 | 0 |
| stx2 | 24c | D (23), USA (1) | 3 | 21 | 2 | 22 | 2 | 22 |
| stx1 + stx2 | 14d | USA (9), CAN (3), UK (1), D (1) | 0 | 14 | 1 | 13 | 1 | 13 |
| stx2c | 4e | D (4) | 3 | 1 | 3 | 1 | 2 | 2 |
| stx1 + stx2c | 26f | D (25), USA (1) | 26 | 0 | 24 | 2 | 3 | 23 |
| stx2 + stx2c | 5g | USA (1), DK (2), UK (1), D (1) | 5 | 0 | 5 | 0 | 4 | 1 |
| All | 82 | 38 | 44 | 36 | 46 | 13 | 69 | |
Country of origin: D, Germany; NL, The Netherlands; UK, United Kingdom; DK, Denmark; USA, United States. CAN, Canada.
All strains were negative for verotoxicity (verocell test) and for stx genes (PCR).
Two strains from Germany were positive for q2851, O2851, and the antB2851 gene; one strain was positive only for the q2851 gene but negative for o2851 and antB2851.
One strain from the United States was negative for q2851 but positive for o2851 and antB2851.
Two strains were positive for q2851, o2851, and antB2851 genes; one was positive only for q2851 and o2851 genes; and another strain was negative for q2851, o2851, and antB2851 genes.
All strains were positive for the q2851 gene. Among the German strains, the o gene was missing in 2 isolates and the antB2851 gene was missing in 23 isolates. The strain from the United States was positive for all q2851, o2851, and antB2851 genes.
All strains were positive for q2851 and o2851 genes; the antB2851 gene was missing in the German isolate.
Phage q2851, o2851, and antB2851 sequences were detected in only 4 (10.3%) of the 39 O157 strains that carried stx genes other than stx2c (Table 5). Eleven strains from four countries—Denmark (two strains), Germany (six strains), United States (two strains), and the United Kingdom (the stx2c prototype strain E32511) (43, 53)—were positive for all sequences, q2851, o2851, and antB2851; three of these strains were negative for stx2c. Taken together, these data indicate that the stx2c gene is associated with bacteriophages similar to phage 2851 in most STEC O157 strains that were isolated in different countries and at different time periods.
Primers 5 and 6 (Table 1), which were used for amplification of part of the coding sequence of the q gene of phage 2851, are also specific for the q gene (qV20) found in the Stx2vhd (Stx2c) prophage of E. coli O157 strain V20 (14) (GenBank accession number AB071845) and for the Stc2c Nil2 prophage (GenBank accession number CAC95104). Whereas the Nil2 prophage q sequence is 100% identical to the q2851 gene sequence, the nucleotide sequences of the published qV20 gene (AB071845) and the q2851 gene are only 98% identical. The differences can be shown by digesting the 441-bp q gene-specific PCR products with BsrD1. This enzyme cuts at two positions in the qV20 gene (AB071845) and at only one position in the q2851 gene. Primers 5 and 6 gave size-specific PCR products in 38 of 82 E. coli O157 strains investigated. Nucleotide sequencing of some of the PCR products and digestion of all PCR products with BsrD1 revealed the presence of the bacteriophage 2851-specific q2851 gene in all 37 O157 strains, thus differing from the Japanese E. coli O157 strain V20 (data not shown).
Conclusion.
This is the first report of the isolation of a viable Stx2c-encoding bacteriophage from an STEC O157 strain that is transduceable and stably inherited in E. coli K-12 laboratory strains.
The stx2c allele was originally described as a second toxin-encoding locus in the E. coli O157 strain E32511 and was shown to differ from the prototypical stx2 gene of phage 933W by three amino acids in the B subunit of the toxin. After cleavage of the signal peptide, the mature B subunit differs by two amino acids from the B subunit of Stx2 of bacteriophage 933W. At least one amino acid exchange is present in the toxin binding domain directed to the eukaryotic cell surface receptor Gb3 (27). Altered receptor binding properties were shown to influence the toxicity and virulence of Stx2-related toxins (27). On the other hand, the regulation of the different stx genes, including stx2c, seems to be very similar. The finding that patients infected with Stx2-producing O157 strains are at higher risk to develop severe sequelae, like HUS and bloody diarrhea, than those infected with only Stx2c-producing strains therefore cannot be related to the induction of toxin production.
Comparison of the genomic sequences of Stx2c phage 2851 to the genomes of other lambdoid phages confirmed that it belongs to the family of mosaic Stx phages (42) possessing some genetic elements closely related to the corresponding genes of the lambdoid coliphage HK97, e.g., the repressor gene cI, the replication genes o and p, and the roi gene (21). The sequence analysis suggests that expression of stx2c genes is regulated by Q antitermination and is enhanced by induction of phage lytic growth, as was shown for other Stx phages (31, 51). Phage 2851 resembles other Stx phages in the genetic locations and the regulation and inducibility of genes encoding Stx production (42, 54).
In contrast to other Stx phages, phage 2851 is more similar to the toxin-negative bacteriophage lambda in its morphology, genome size, and superinfection immunity properties. On the other hand, E. coli K-12 strains lysogenized with bacteriophage 2851 were not immune to superinfection with other Stx phages, and phage 2851 does not share the same chromosomal integration site with protoype phages carrying stx1 and stx2 genes. These results might explain why the stx2c determinant is frequently found together with stx1 or stx2 genes that are carried by other phages in E. coli O157 strains, as in the prototype strain E32511, which carries an stx2 and an stx2c gene (43, 53). E32511 was isolated from a HUS patient in the United Kingdom in 1983 and shows the same o, p, and antB genes as strain CB2851, which was isolated 10 years later from a patient in Germany. Our findings that the q2851 and o2851 genes are associated with Stx2c-positive STEC O157 strains from different countries and time periods indicate that phages similar to 2851 are spread globally among strains of STEC O157. Further research is needed to explore the relationship of lambdoid Stx phages with nontoxigenic lambda-like bacteriophages, which are harbored in the chromosome of wild-type E. coli and other Enterobacteriaceae, and with the ecological and genetic mechanisms which contribute to the conversion of harmless prophages into vectors encoding potent pathogenicity factors causing disease in humans.
Acknowledgments
We are grateful to Walther Messer, Max Planck Institute for Molecular Genetics, Berlin, Germany, for kindly donating the lambda lysogenic strain CSH67.
Editor: J. T. Barbieri
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann, B. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 3.Bastian, S. N., I. Carle, and F. Grimont. 1998. Comparison of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res. Microbiol. 149:457-472. [DOI] [PubMed] [Google Scholar]
- 4.Beutin, L., S. Kaulfuss, T. Cheasty, B. Brandenburg, S. Zimmermann, K. Gleier, G. A. Willshaw, and H. R. Smith. 2002. Characteristics and association with disease of two major subclones of Shiga toxin (Verocytotoxin)-producing strains of Escherichia coli (STEC) O157 that are present among isolates from patients in Germany. Diagn. Microbiol. Infect. 44:337-346. [DOI] [PubMed] [Google Scholar]
- 5.Beutin, L., G. Krause, S. Zimmermann, S. Kaulfuss, and K. Gleier. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin, L., M. A. Montenegro, I. Orskov, F. Orskov, J. Prada, S. Zimmermann, and R. Stephan. 1989. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J. Clin. Microbiol. 27:2559-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutin, L., S. Zimmermann, and K. Gleier. 1996. Rapid detection and isolation of Shiga-like toxin (verocytotoxin)-producing Escherichia coli by direct testing of individual enterohemolytic colonies from washed sheep blood agar plates in the VTEC-RPLA assay. J. Clin. Microbiol. 34:2812-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boerlin, P., S. A. McEwen, F. Boerlin-Petzold, J. B. Wilson, R. P. Johnson, and C. L. Gyles. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caprioli, A., I. Luzzi, A. Gianviti, H. Russmann, and H. Karch. 1995. Pheno-genotyping of verotoxin 2 (VT2)-producing Escherichia coli causing haemorrhagic colitis and haemolytic uraemic syndrome by direct analysis of patients' stools. J. Med. Microbiol. 43:348-353. [DOI] [PubMed] [Google Scholar]
- 10.Datz, M., C. Janetzki-Mittmann, S. Franke, F. Gunzer, H. Schmidt, and H. Karch. 1996. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl. Environ. Microbiol. 62:791-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eklund, M., K. Leino, and A. Siitonen. 2002. Clinical Escherichia coli strains carrying stx genes: stx variants and stx-positive virulence profiles. J. Clin. Microbiol. 40:4585-4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fegan, N., and P. Desmarchelier. 2002. Comparison between human and animal isolates of Shiga toxin-producing Escherichia coli O157 from Australia. Epidemiol. Infect. 128:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 14.Hamabata, T., T. Tanaka, A. Ozawa, T. Shima, T. Sato, and Y. Takeda. 2002. Genetic variation in the flanking regions of Shiga toxin 2 gene in Shiga toxin-producing Escherichia coli O157:H7 isolated in Japan. FEMS Microbiol. Lett. 215:229-236. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix, R. W., J. W. Roberts, F. W. Stahl, and R. A. Weisberg. 1983. Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Ito, H., A. Terai, H. Kurazono, Y. Takeda, and M. Nishibuchi. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47-60. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, M. P., J. W. Newland, R. K. Holmes, and A. D. O'Brien. 1987. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb. Pathog. 2:147-153. [DOI] [PubMed] [Google Scholar]
- 19.Jelacic, J. K., T. Damrow, G. S. Chen, S. Jelacic, M. Bielaszewska, M. Ciol, H. M. Carvalho, A. R. Melton-Celsa, A. D. O'Brien, and P. I. Tarr. 2003. Shiga toxin-producing Escherichia coli in Montana: bacterial genotypes and clinical profiles. J. Infect. Dis. 188:719-729. [DOI] [PubMed] [Google Scholar]
- 20.Johansen, B. K., Y. Wasteson, P. E. Granum, and S. Brynestad. 2001. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 147:1929-1936. [DOI] [PubMed] [Google Scholar]
- 21.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 22.Karmali, M. A. 1989. Infection by verocytotoxin-producing Escherichia coli. Clin. Microbiol. Rev. 2:15-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 2000. Toxin gene expression by Shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 6:458-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch, C., S. Hertwig, R. Lurz, B. Appel, and L. Beutin. 2001. Isolation of a lysogenic bacteriophage carrying the stx1OX3 gene, which is closely associated with Shiga toxin-producing Escherichia coli strains from sheep and humans. J. Clin. Microbiol. 39:3992-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leason, K. R., and D. I. Friedman. 1988. Analysis of transcription termination signals in the nin region of bacteriophage lambda: the roc deletion. J. Bacteriol. 170:5051-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, Z., H. Kurazono, S. Yamasaki, and Y. Takeda. 1993. Detection of various variant verotoxin genes in Escherichia coli by polymerase chain reaction. Microbiol. Immunol. 37:543-548. [DOI] [PubMed] [Google Scholar]
- 27.Ling, H., N. S. Pannu, A. Boodhoo, G. D. Armstrong, C. G. Clark, J. L. Brunton, and R. J. Read. 2000. A mutant Shiga-like toxin IIe bound to its receptor Gb(3): structure of a group II Shiga-like toxin with altered binding specificity. Struct. Fold. Des. 8:253-264. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Muniesa, M., M. de Simon, G. Prats, D. Ferrer, H. Panella, and J. Jofre. 2003. Shiga toxin 2-converting bacteriophages associated with clonal variability in Escherichia coli O157:H7 strains of human origin isolated from a single outbreak. Infect. Immun. 71:4554-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakao, H., K. Kimura, H. Murakami, T. Maruyama, and T. Takeda. 2002. Subtyping of Shiga toxin 2 variants in human-derived Shiga toxin-producing Escherichia coli strains isolated in Japan. FEMS Immunol. Med. Microbiol. 34:289-297. [DOI] [PubMed] [Google Scholar]
- 31.Neely, M. N., and D. I. Friedman. 1998. Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene 223:105-113. [DOI] [PubMed] [Google Scholar]
- 32.Newland, J. W., R. J. Neill, A. D. O'Brien, and R. K. Holmes. 1987. Nucleotide sequence analysis and comparision of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol. Lett. 44:109-114. [Google Scholar]
- 33.Newland, J. W., N. A. Strockbine, S. F. Miller, A. D. O'Brien, and R. K. Holmes. 1985. Cloning of Shiga-like toxin structural genes from a toxin converting phage of Escherichia coli. Science 230:179-181. [DOI] [PubMed] [Google Scholar]
- 34.Nishikawa, Y., Z. Zhou, A. Hase, J. Ogasawara, T. Cheasty, and K. Haruki. 2000. Relationship of genetic type of Shiga toxin to manifestation of bloody diarrhea due to enterohemorrhagic Escherichia coli serogroup O157 isolates in Osaka City, Japan. J. Clin. Microbiol. 38:2440-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 36.Pierard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plunkett, G., D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russmann, H., H. Schmidt, J. Heesemann, A. Caprioli, and H. Karch. 1994. Variants of Shiga-like toxin II constitute a major toxin component in Escherichia coli O157 strains from patients with haemolytic uraemic syndrome. J. Med. Microbiol. 40:338-343. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Sato, T., T. Shimizu, M. Watarai, M. Kobayashi, S. Kano, T. Hamabata, Y. Takeda, and S. Yamasaki. 2003. Distinctiveness of the genomic sequence of Shiga toxin 2-converting phage isolated from Escherichia coli O157:H7 Okayama strain as compared to other Shiga toxin 2-converting phages. Gene 309:35-48. [DOI] [PubMed] [Google Scholar]
- 41.Sato, T., T. Shimizu, M. Watarai, M. Kobayashi, S. Kano, T. Hamabata, Y. Takeda, and S. Yamasaki. 2003. Genome analysis of a novel Shiga toxin 1 (Stx1)-converting phage which is closely related to Stx2-converting phages but not to other Stx1-converting phages. J. Bacteriol. 185:3966-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt, H. 2001. Shiga-toxin-converting bacteriophages. Res. Microbiol. 152:687-695. [DOI] [PubMed] [Google Scholar]
- 43.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H- strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaikh, N., and P. I. Tarr. 2003. Escherichia coli O157:H7 Shiga toxin-encoding bacteriophages: integrations, excisions, truncations, and evolutionary implications. J. Bacteriol. 185:3596-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith, H. R., N. P. Day, S. M. Scotland, R. J. Gross, and B. Rowe. 1984. Phage-determined production of vero cytotoxin in strains of Escherichia coli serogroup O157. Lancet i:1242-1243. [DOI] [PubMed] [Google Scholar]
- 46.Steven, A. C., B. L. Trus, J. V. Maizel, M. Unser, D. A. Parry, J. S. Wall, J. F. Hainfeld, and F. W. Studier. 1988. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J. Mol. Biol. 200:351-365. [DOI] [PubMed] [Google Scholar]
- 47.Strauch, E., R. Lurz, and L. Beutin. 2001. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect. Immun. 69:7588-7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strockbine, N. A., L. R. Marques, J. W. Newland, H. W. Smith, R. K. Holmes, and A. D. O'Brien. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect. Immun. 53:135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyler, J. S., and D. I. Friedman. 2004. Characterization of a eukaryotic-like tyrosine protein kinase expressed by the Shiga toxin-encoding bacteriophage 933W. J. Bacteriol. 186:3472-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner, P. L., J. Livny, M. N. Neely, D. W. Acheson, D. I. Friedman, and M. K. Waldor. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957-970. [DOI] [PubMed] [Google Scholar]
- 52.Willshaw, G. A., H. R. Smith, T. Cheasty, P. G. Wall, and B. Rowe. 1997. Vero cytotoxin-producing Escherichia coli O157 outbreaks in England and Wales, 1995: phenotypic methods and genotypic subtyping. Emerg. Infect. Dis. 3:561-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willshaw, G. A., H. R. Smith, S. M. Scotland, A. M. Field, and B. Rowe. 1987. Heterogeneity of Escherichia coli phages encoding Vero cytotoxins: comparison of cloned sequences determining VT1 and VT2 and development of specific gene probes. J. Gen. Microbiol. 133:1309-1317. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. Acheson. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]