Abstract
Pseudomonas aeruginosa uses a type III secretion system to promote development of severe disease, particularly in patients with impaired immune defenses. While the biochemical and enzymatic functions of ExoU, ExoS, and ExoT, three effector proteins secreted by this system, are well defined, the relative roles of each protein in the pathogenesis of acute infections is not clearly understood. Since ExoU and ExoS are usually not secreted by the same strain, it has been difficult to directly compare the effects of these proteins during infection. In the work described here, several isogenic mutants of a bacterial strain that naturally secretes ExoU, ExoS, and ExoT were generated to carefully evaluate the relative contribution of each effector protein to pathogenesis in a mouse model of acute pneumonia. Measurements of mortality, bacterial persistence in the lung, and dissemination indicated that secretion of ExoU had the greatest impact on virulence while secretion of ExoS had an intermediate effect and ExoT had a minor effect. It is of note that these results conclusively show for the first time that ExoS is a virulence factor. Infection with isogenic mutants secreting wild-type ExoS, ExoS defective in GTPase-activating protein (GAP) activity, or ExoS defective in ADP-ribosyltransferase activity demonstrated that the virulence of ExoS was largely dependent on its ADP-ribosyltransferase activity. The GAP activity of this protein had only a minor effect in vivo. The relative virulence associated with each of these type III effector proteins may have important prognostic implications for patients infected with P. aeruginosa.
Pseudomonas aeruginosa is a gram-negative pathogen that causes a variety of serious infections predominantly in immunocompromised patients. To promote severe illness, P. aeruginosa uses a type III secretion system to inject toxic effector proteins into the cytoplasm of eukaryotic cells. To date, four effector proteins have been described in P. aeruginosa: ExoU, ExoS, ExoT, and ExoY (9, 18, 59-62). ExoU is a potent cytotoxin with phospholipase A2 activity (43, 49). ExoS and ExoT are bifunctional enzymes that have 75% amino acid identity (60) and encode both GTPase-activating protein (GAP) and ADP-ribosyltransferase (ADPRT) activities (15, 24-26, 33). ExoY is an adenylate cyclase (62).
The overall importance of type III secretion as a virulence mechanism of P. aeruginosa has been well established. Early studies showed that ExsA, a transcriptional activator of the type III secretion system (11, 61), is essential for full virulence in animal models of acute pneumonia (28, 34, 58). More recently, these conclusions have been validated by the use of mutants with specific disruptions in genes encoding portions of the type III secretion apparatus (54). In addition, type III secretion has been associated with more severe clinical disease in human patients (16, 47).
Several studies have analyzed mutants with deletions in individual effector-encoding genes to begin to address the roles of each effector protein in virulence. Efforts have been focused on the three effector proteins most closely linked to virulence: ExoU, ExoS, and ExoT. Deletion of exoU significantly reduced overall virulence and prevented development of severe pathology in the lung (9, 18). Similarly, early work suggested that ExoS contributed to dissemination in a burn model and to pathology in a lung model of disease (36-38). However, the mutant strain used in these studies was later shown to have pleiotropic effects on type III secretion. The transposon insertion in this strain was in an operon encoding a portion of the secretion apparatus, thus preventing secretion of ExoS, as well as all other type III secreted proteins (61). A more recent study with a mutant with a specific deletion in the exoS structural gene failed to detect a contribution of ExoS to virulence in a burn model (22). Thus, the role of this effector protein in disease is unclear. Like that of exoS, targeted disruption of the exoT gene showed no effect on virulence in a burn model (22). However, a minor role for ExoT in pathogenesis was revealed when this protein was examined in the absence of ExoU. Comparison of an exoU mutant to an exoT-exoU double mutant showed that ExoT affects dissemination of bacteria from the lung to the liver (14). Importantly, interpretation of the relative contributions to virulence of each effector protein has been difficult because the mutants compared in these studies were from a variety of parent strain backgrounds with differing type III secretion characteristics.
The significant genotypic and phenotypic heterogeneity in type III secretion that exists among clinical isolates of P. aeruginosa has complicated the interpretation of virulence studies in this organism. Nearly all strains contain the genes for at least a portion of the secretion apparatus, as well as for one or more effector proteins (8). Unlike type III secreted effector proteins in some other bacteria, three of the four effector proteins in P. aeruginosa are variable traits that are not encoded by all strains. In one study of a large collection of clinical isolates, 100% encoded exoT, 89% exoY, 72% exoS, and 28% exoU (8). Interestingly, very few strains encoded both exoS and exoU. In addition, the presence of genes encoding effector proteins does not necessarily predict the secretion phenotype of clinical isolates in vitro (6, 16, 47). In a collection of respiratory isolates from patients with ventilator-associated pneumonia, only 77% of the strains had detectable type III secretion in vitro although 100% encoded genes associated with type III secretion (16). Of the strains that were capable of secretion in vitro, approximately 60% secreted ExoS while about 40% secreted ExoU. As a result of the exclusive relationship between ExoU and ExoS, virulence studies on these proteins have been performed with different strains, making it difficult to compare the relative contributions of each effector protein to disease pathogenesis.
In the experiments described here, we directly compared the contributions of ExoU, ExoS, and ExoT to virulence by analyzing each effector protein in the same isogenic background. We chose a clinical isolate that naturally secreted these three effector proteins for our studies and constructed mutants that secreted ExoS alone, ExoT alone, or ExoU alone. This strategy allowed us to avoid the complications of differences in strain background and to prevent masking of the effects of one effector protein by another. Thus, this is the first direct comparison of the effects of ExoU and ExoS in disease pathogenesis. Our results indicate that ExoU has the greatest effect on virulence of the type III secreted proteins whereas ExoT has only a small effect. We also show unambiguously that ExoS is an important virulence factor in the lung, resolving the confusion over the role of this protein.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and mutants used in this study are listed in Table 1. PA99 is a P. aeruginosa clinical isolate that naturally carries the exoU, exoS, and exoT genes but lacks the gene for exoY (8). P. aeruginosa strains PA103 (31) and 388 (3) were used for PCR amplification of exoT and exoS, respectively. Bacterial strains were streaked from frozen cultures onto Luria-Bertani (LB) agar or Vogel-Bonner minimal (VBM) agar (57). For infections, overnight cultures of P. aeruginosa grown in 5 ml of MINS medium (39) at 37°C were diluted into fresh medium and regrown into exponential phase.
TABLE 1.
Bacterial strains and mutants used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| P. aeruginosa strains | ||
| PA99 | Secretes ExoS, ExoT, and ExoU; clinical isolate | 8 |
| PA99S | Secretes only ExoS; PA99ΔexoTexoU | This work |
| PA99T | Secretes only ExoT; PA99null complemented with mini-CTX1-ExoT | This work |
| PA99U | Secretes only ExoU; PA99ΔexoSexoT | This work |
| PA99null | Secretes no known effectors, makes functional secretion apparatus; PA99ΔexoSexoTexoU | This work |
| PA99secr− | Secretes no effectors, does not make secretion apparatus; PA99ΔpscJ | This work |
| PA99null+S | Secretes only ExoS; PA99ΔexoSexoT exoU + mini-CTX1-ExoS | This work |
| PA99null+S(R146A) | Secretes only ExoS(R146A); no GAP activity; PA99ΔexoSexoTexoU +mini-CTX1-ExoS-R146A | This work |
| PA99null+S(E379A/E381A) | Secretes only ExoS (E379A/381A); no ADPRT activity: PA99ΔexoS exoTexoU+mini-CTX1-ExoS-E379A/E381A | This work |
| PA103 | Secretes ExoT and ExoU; laboratory isolate | 31 |
| 388 | Secretes ExoS and ExoT; laboratory isolate | 3 |
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lac1qZ ΔM15 Tn10 (Tetr)] | Stratagene |
| GM2163 | F−ara-14 leuB6 fhuA31 lacY1 tsx-78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 rpsL136 (Strr) dam-13::Tn9 (Camr) xylA5 mtl-1 thi-1 mcrB1 hsdR2 | New England Biolabs |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZ ΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsI (Strr) endA1 nupG | Invitrogen |
| S17.1 | thi pro hsdR recA RP4-2 (Tet::Mu)(Km::Tn7) | 53 |
| Plasmids | ||
| Mini-CTX1 | Plasmid for chromosomal gene integration into attB locus; Tetr | H. Schweizer; 21 |
| Mini-CTX1-ExoS | exoS from strain 388 in mini-CTX1; Tetr | This work |
| Mini-CTX1-ExoS-R146A | GAP point mutant in mini-CTX1-ExoS; Tetr | This work |
| Mini-CTX1-ExoS-E379A/E381A | ADPRT point mutant in mini-CTX1-ExoS; Tetr | This work |
| Mini-CTX1-ExoT | exoT from PA103 in mini-CTX1; Tetr | This work |
| pCM104 | ΔexoT in pEX100T (in frame); Apr (Cbr)a | This work |
| pCM111 | ΔpscJ in pEX100T (in frame); Apr (Cbr) | This work |
| pEX100T | Plasmid for homologous recombination; Apr | 52 |
| pFlp2 | Plasmid carrying recombinase to excise mini-CTX1 plasmid sequences; Apr (Cbr) | H. Schweizer; 21 |
| pGS003 | ΔexoS in pEX100T (in frame); Apr (Cbr) | This work |
| pGS013 | ΔexoU in pEX100T (in frame); Apr (Cbr) | This work |
| pGS015 | ΔexoU in pEX100T with xylE aacC1 cassette inserted into the SalI site of pGS013; Apr (Cbr) Gmr | This work |
Apr in E. coli, Cbr in P. aeruginosa.
Escherichia coli strains XL1-Blue, GM2163, and TOP10 were used for cloning and propagation of plasmids. E. coli strain S17.1 was used for mating of constructs into P. aeruginosa.
Antibiotics were used when necessary at the following concentrations: ampicillin at 50 μg/ml, gentamicin at 15 μg/ml, and kanamycin at 25 μg/ml for plasmids in E. coli; carbenicillin at 500 μg/ml, gentamicin at 100 μg/ml, and tetracycline at 100 μg/ml for P. aeruginosa.
Bacterial mutants.
Isogenic in-frame deletion mutants of strain PA99 were constructed via allelic replacement by the method of Schweizer and Hoang (52). Briefly, in-frame deletions in the genes of interest were constructed in plasmid pEX100T, which is competent for replication in E. coli but not in P. aeruginosa. Final constructs were transformed into E. coli strain S17.1 prior to mating with P. aeruginosa. Exconjugants with the deletion allele integrated into the corresponding chromosomal gene locus were selected by growth on VBM agar plates supplemented with carbenicillin. Growth on VBM agar supplemented with 5% sucrose was then used to counterselect for bacteria in which the vector sequences had been excised by a second recombination event. When the gene of interest was disrupted with a gentamicin resistance cassette, exconjugants were selected with carbenicillin and gentamicin while counterselection was performed with 5% sucrose and gentamicin. Correct construction of all deletion mutants was confirmed by PCR amplification of the gene of interest and Western blotting of culture supernatants with the appropriate antisera (see below and Fig. 1) (51).
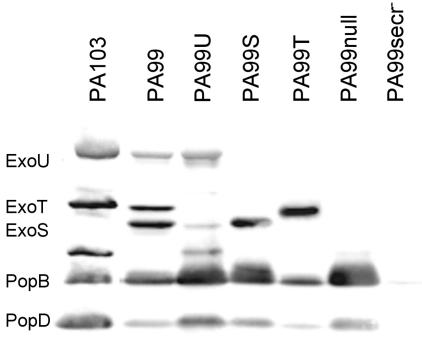
FIG. 1.
Type III effector proteins secreted by wild-type and isogenic mutant forms of strain PA99. Each strain was grown in low-calcium medium to induce type III secretion. Immunoblot analysis with a mixture of antibodies to ExoS, ExoT, ExoU, PopB, and PopD was then performed on the culture supernatants from each strain. The locations of bands representing ExoU, ExoT, ExoS, PopB, and PopD are shown to the left. The band that migrates between ExoS and PopB is a product of ExoU breakdown (data not shown).
To make a strain secreting only ExoS (referred to as PA99S), exoU and exoT were sequentially deleted from wild-type strain PA99 with plasmids pGS013 and pCM104, respectively, for allelic replacement. To generate pGS013, exoU-containing plasmid pAH806 (18) was digested with BsmI to remove sequences flanking exoU and self-ligated to yield pAH808. This plasmid was digested with XmaI and SalI, treated with T4 polymerase, and self-ligated to create a ΔexoU allele containing an internal in-frame deletion of nucleotides 361 to 1962 (of 2063 total nucleotides). The resulting construct, pGS012, was then digested with NruI and MscI and treated with T4 polymerase to generate a 1.7-kb ΔexoU-containing fragment. This fragment was subsequently purified and ligated into pEX100T that had been digested with SmaI, resulting in pGS013. After mating pGS013 into PA99 to generate PA99ST, exoT was deleted with plasmid pCM104. To generate pCM104, exoT was PCR amplified from strain PA103. (All of the PCR primers used in these experiments are listed in Table 2.) The 1.8-kb amplification product was ligated into pCR-Blunt (Invitrogen, Carlsbad, Calif.) in accordance with the manufacturer's instructions, creating pCM101. The exoT-containing 1.8-kb EcoRI-EcoRI fragment was purified, treated with T4 polymerase, ligated into SmaI-digested pEX100T, and transformed into E. coli strain GM2163. To create the in-frame exoT deletion, this construct was digested with XmaI and PpuMI to remove nucleotides 108 to 1334 (of a total of 1,373 nucleotides) of exoT. The 6.4-kb ΔexoT-containing fragment was purified by gel electrophoresis, treated with T4 polymerase, and religated to create pCM104. pCM104 was then mated with PA99ST to generate PA99S.
TABLE 2.
Primers used for PCR amplification
| Primer | Purpose | Primer sequence (5′→3′)a |
|---|---|---|
| exoT5 | Amplification of exoT | ATATCCCATCGGGTTCTCCGCCCCGG |
| exoT-CM-2 | Amplification of exoT | CGTTTCGTTATGCAGGAAGC |
| exoSl-F | Amplification of exoS | TCTGAATTCTTCCAGGCGGGTGAACATCA |
| exoSl-R | Amplification of exoS | TTTAGATCTCACCCTGGTATCCAAGGCGA |
| 70.6 | Amplification of exoU | GCAGCCTATCGTGCAAG |
| 70.9 | Amplification of exoU | AGCGTTAGTGACGTGCG |
| pscJ-5′ | Amplification of pscJ | CAGTCTCGAAGAACTCTCCG |
| pscJ-3′ | Amplification of pscJ | TGCGCCGTACCCGCGCACCG |
| R146A-forward | Mutagenesis of exoS | CGGAGATGGGGCGCTGGCTTCGCTGAGCACCGC |
| R146A-reverse | Mutagenesis of exoS | GCGGTGCTCAGCGAAGCCAGCGCCCCATCTCCG |
| E379A/E381A-forward | Mutagenesis of exoS | CGAACTACAAGAATGCAAAAGCGATTCTCTATAACAAAG |
| E379A/E381A-reverse | Mutagenesis of exoS | TTGTTATAGAGAATCGCTTTTGCATTCTTGTAGTTCGAT |
Nucleotides altered for insertion of point mutations are in bold and underlined.
To make a strain secreting only ExoU (referred to as PA99U), exoS and exoT were sequentially deleted from wild-type PA99 with plasmids pGS003 and pCM104, respectively, for allelic replacement. First, a 2.0-kb fragment containing exoS was PCR amplified from strain 388, ligated into pCR-Blunt, and transformed into TOP10 cells to create pGS001. The exoS-containing 2.0-kb EcoRI-EcoRI fragment was purified by gel electrophoresis, treated with T4 polymerase, and ligated into pEX100T previously digested with SmaI. This construct was digested with SmaI and Bpu1102I, treated with T4 polymerase, and religated to create an in-frame deletion by removal of nucleotides 397 to 1180 (of a total of 1,361 nucleotides) of exoS. The resulting construct was designated pGS003. After mating pGS003 into PA99 to create PA99TU, exoT was deleted with pCM104 as described above to generate PA99U.
To make a strain that assembled a functional secretion apparatus but secreted none of the known effector proteins (referred to as PA99null), exoU was deleted from strain PA99U (described above) with plasmid pGS015. To generate pGS015, a 1.5-kb SalI-SalI fragment containing an aacC1 gentamicin resistance cassette was purified from pX1918G (52) and ligated into pGS013 that had been linearized with SalI. pGS015 was then mated into PA99U with gentamicin for selection of exconjugants to generate PA99null.
To make a strain secreting only ExoT (referred to as PA99T), we first attempted to sequentially delete exoU and exoS from PA99. However, we were unsuccessful in constructing this double mutant despite multiple attempts. Therefore, we adopted the alternate strategy of integrating a wild-type copy of the exoT gene, with its endogenous promoter, into the chromosome of the PA99null mutant by using the approach of Hoang et al. (21). The 1.8-kb exoT-containing EcoRI-EcoRI fragment of pCM101 (described above) was ligated into mini-CTX1 (21) that had been digested with EcoRI. The resulting construct, mini-CTX1-ExoT, was mated into PA99null and integrated into the chromosomal attB site, and vector sequences were excised by a recombinase expressed by pFlp2 (21).
To make a strain that did not assemble the secretion apparatus or secrete any type III secreted proteins (referred to as PA99secr−), we deleted pscJ from PA99 with plasmid pCM111 for allelic exchange. The pscJ gene encodes PscJ, an essential component of the type III secretion apparatus. First, a 1.3-kb DNA fragment encoding pscJ was PCR amplified from PA99, ligated into pCR-Blunt, and transformed into E. coli strain GM2163. This construct was digested with BbsI, treated with T4 polymerase, and religated to create a pscJ allele (designated ΔpscJ) with an in-frame deletion of nucleotides 1 to 620 (of 746 total nucleotides). The 0.7-kb ΔpscJ-containing EcoRI-EcoRI fragment was then treated with T4 polymerase and ligated into SmaI-digested pEX100T to create pCM111, which was mated into PA99 to generate PA99secr−.
PA99-derived strains expressing ExoS with mutated enzymatic domains were constructed as follows. A 2.0-kb exoS-containing EcoRI-EcoRI fragment from pGS001 (described above) was cloned into mini-CTX1 (21), which was previously digested with EcoRI, to create mini-CTX1-ExoS. Specific mutations known to eliminate the GAP activity (R146A) or ADPRT activity (E379A/E381A) were introduced into mini-CTX1-ExoS by the QuikChange II XL site-directed mutagenesis approach (Stratagene, La Jolla, Calif.) (12). The resulting plasmids were designated mini-CTX1-ExoS-R146A and mini-CTX1-ExoS-E379A/E381A, respectively. Each of these constructs was individually conjugated into P. aeruginosa strain PA99null and integrated into the chromosomal attB site in accordance with the method of Hoang et al. (21). Thus, enzymatically deficient forms of ExoS were expressed under control of the endogenous exoS promoter from a single-copy chromosomal insertion. These strains were designated PA99null+S, PA99null+S(R146A), and PA99null+S(E379/381A). Immunoblot analyses of culture supernatants were performed to ensure that each protein was secreted in approximately equal amounts in vitro (Fig. 1). Note that these mutant forms of ExoS have been previously shown to be translocated into mammalian cells by the P. aeruginosa type III secretion system (12).
Immunoblot analysis.
The type III effector proteins secreted by P. aeruginosa strains were determined by immunoblot analysis as previously described (51). Briefly, strains were grown in MINS medium for approximately 17 h at 37°C with vigorous shaking. Bacterial supernatants were obtained from 4-ml cultures by centrifugation at 6,000 × g at 4°C for 20 min. Proteins in supernatants were precipitated by the addition of ammonium sulfate to a final concentration of 55% (wt/vol). Following incubation on ice for 2 h, the precipitated material was collected by centrifugation at 13,000 × g and 4°C for 20 min. The pellet was boiled in 500 μl of 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer for 5 min, and 75 μl of each sample was subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (30). Proteins were then electrotransferred to nitrocellulose (Schleicher & Schuell, Inc., Keene, N.H.) and exposed to mixtures of polyclonal antisera against ExoS, ExoT, ExoU, PopB, and PopD (16, 18, 48). (PopB and PopD are type III secreted proteins that make up the translocation complex [19, 59].) Goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) diluted 1:3,000 was used as a secondary antibody. Proteins were visualized by incubating the membranes in 225 μM coumaric acid (Sigma Chemical Co., St. Louis, Mo.)-1.25 mM 3-aminophthalhydrazide (Sigma)-0.009% hydrogen peroxide (Fisher Scientific Co., Pittsburgh, Pa.) in 100 mM Tris (pH 8.5) for 1 min and then recording luminescence with a ChemiImager 5500 (Alpha Innotech Corp., San Leandro, Calif.).
Mouse model of acute pneumonia.
Studies of acute pneumonia were conducted with the mouse model described by Comolli et al. (4). Briefly, bacterial cultures were grown overnight in MINS medium at 37°C with shaking (250 rpm) and then diluted and regrown to exponential phase. Bacteria were collected by centrifugation and resuspended in phosphate-buffered saline (PBS; Invitrogen) to the appropriate concentration. Six- to seven-week-old female BALB/c mice were anesthetized by intraperitoneal injection of a mixture of ketamine (100 mg/ml) and xylazine (20 mg/ml). Mice were inoculated with 1.2 × 106 CFU of bacteria in 50 μl of PBS, as determined by optical density. Inocula were confirmed by plating of serial dilutions onto VBM agar. Preliminary experiments with this dose indicated that severe illness in response to the wild-type PA99 strain (secreting ExoS, ExoT, and ExoU) began to occur between 22 and 28 h postinfection. For each experiment, at least five mice per strain were infected for each time point.
At 18 h postinfection, mice were scored for severity of illness on an objective scale developed in our laboratory. Individual mice were scored as follows in three categories, with lower scores reflecting increasing disease: respiration (3, normal rapid breaths; 2, slow shallow breaths; 1, slow labored breaths; 0, moribund), appearance (3, smooth fur; 2, ruffled fur; 1, matted fur; 0, moribund), and activity (3, running and very active; 2, huddled and lethargic; 1, hunched or unsteady gait; 0, moribund). Each animal therefore received a total score between 9 (completely healthy) and 0 (moribund). In all experiments, mice were sacrificed when severe illness developed and received a score of 0 in each category.
For determination of bacterial numbers in individual organs, mice were reanesthetized and sacrificed at 18 h postinfection. Lungs, spleens, and livers were aseptically removed and individually weighed prior to homogenization in 5 ml of PBS. The bacterial load in each organ was determined following plating of serial dilutions on VBM agar and incubation at 37°C for 24 h.
For determination of the 50% lethal dose (LD50), mice were anesthetized with inhaled methoxyflurane and intranasally infected with a range of bacterial inocula. In all experiments, mice were sacrificed when severe illness developed and were scored as dead. Mice were monitored for a total of 7 days. A minimum of 10 mice were used for each strain. The LD50 was calculated by the method of Reed and Muench (45).
For survival experiments, groups of five mice per day were infected with each strain as described above. In all experiments, mice were sacrificed when severe illness developed and were scored as dead. Survival was monitored for 48 h after infection. The experiment was performed twice, and results were pooled.
All experiments were approved by and performed in accordance with the guidelines of the Northwestern University Animal Care and Use Committee.
Statistical methods.
Analyses of bacterial load differences were performed by analysis of variance, followed by multiple unplanned comparisons with the Tukey-Kramer HSD test with an α of 0.05. For comparison of mutant forms of ExoS, bacterial load differences were tested with the Student t test. Prior to analysis, all colonization data were natural log transformed so as to fit a normal distribution. The use of parametric tests on transformed colonization data was justified by analysis of a large set of control data that confirmed that colonization data were log normally distributed. Chi-squared tests were performed on health score data, frequencies of dissemination, and Kaplan-Meier survival plots. P < 0.05 was considered significant.
RESULTS
Relative contributions of ExoS, ExoT, and ExoU to virulence in a mouse model of acute pneumonia. To determine the relative roles of the type III effector proteins of P. aeruginosa in disease, we first screened a collection of clinical isolates for a strain that naturally secreted ExoU, ExoS, and ExoT. One such strain, designated PA99, was found to secrete all three of these effector proteins when grown under conditions that induced type III secretion (Fig. 1). PA99 does not secrete the fourth effector protein, ExoY, or contain the gene that encodes this protein (data not shown). Evaluation of the cytotoxic, invasive, and virulence properties of PA99 compared to a collection of clinical isolates indicated that PA99 was representative of characterized P. aeruginosa strains (data not shown).
To facilitate our studies of the relative effects of type III secreted proteins in virulence, isogenic bacterial mutants that secreted only ExoU, ExoS, or ExoT were constructed and designated PA99U, PA99S, and PA99T, respectively (Table 1). In addition, a mutant with all three effector genes disrupted was generated and designated PA99null. Although this mutant does not secrete any known effector proteins, it assembles a functional type III secretion apparatus. Finally, a mutant with a defective secretion apparatus owing to disruption of the pscJ gene was generated and designated PA99secr−. All mutants grew at rates identical to that of wild-type strain PA99 (data not shown), and each secreted the expected type III effector proteins in appropriate amounts when grown under secretion-inducing conditions (Fig. 1). These mutant strains were used to compare the relative contributions of the effector proteins to overall virulence in a mouse model of acute pneumonia.
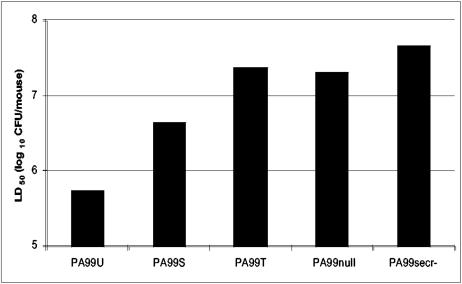
The effect of each protein on virulence was quantified with a modified LD50 assay. PA99-derived strains that did not secrete the known type III secreted effector proteins were associated with modest virulence. PA99secr−, a strain lacking a functional secretion system, had an LD50 of 4.7 × 107 CFU (Fig. 2). Likewise, PA99null, which does not secrete any of the known type III secreted effector proteins but assembles a functional secretion apparatus, had an LD50 of 2.1 × 107 CFU. Secretion of ExoU caused the greatest increase in virulence compared to PA99null; in infections with PA99U, 40-fold fewer bacteria (5.5 × 105 CFU) were required to cause 50% mortality. Interestingly, secretion of ExoS alone was also sufficient to increase virulence, showing clearly that ExoS is a virulence factor of P. aeruginosa. PA99S was associated with a four- to fivefold decrease in the LD50 (4.4 × 106 CFU) compared to that of PA99null. In contrast, secretion of only ExoT had little effect on virulence as measured by LD50 (2.4 × 107 CFU).
FIG. 2.
Virulence of strains expressing individual type III secreted effector proteins in a mouse model of acute pneumonia. Bars represent the bacterial dose needed to cause mortality in 50% of infected mice. Shorter bars indicate increased virulence. Values were calculated on the basis of at least 10 mice per LD50. Data are shown as the average log10 number of CFU per mouse.
Secretion of ExoU or ExoS but not ExoT is sufficient for bacterial persistence in the lung.
To investigate the process by which type III secretion may affect disease severity, we monitored infected BALB/c mice for signs of disease and measured the number of bacteria present in the lungs at various times after intranasal infection with 1.2 × 106 bacteria. After 3 h, none of the animals displayed overt signs of illness, such as decreased activity or ruffled fur. At this time point, the CFU counts of bacteria in the lung were identical for all of the strains tested (data not shown). However, after 18 h, mice infected with either the ExoU-secreting strain (PA99U) or the ExoS-secreting strain (PA99S) displayed ruffled fur, decreased activity, and decreased respiratory rates compared to mice infected with PA99null or PA99secr− (health scores [mean ± standard error of the mean]: 8.0 ± 0.63 for PA99U and 6.8 ± 0.20 for PA99S versus 9.0 ± 0.0 for PA99null and PA99secr− [P < 0.05]). Consistent with this observation, PA99null and PA99secr− were present in small numbers in the lungs of infected mice at this time point (3.5 × 103 and 3.3 × 103 CFU/lung, respectively) whereas approximately 110 times as many bacteria were present in the lungs of animals infected with PA99U (3.9 × 105 CFU/lung [P < 0.05]) (Fig. 3). Similarly, infection with PA99S increased the bacterial load in the lungs approximately 95-fold relative to that of PA99null (3.2 × 105 CFU/lung [P < 0.05]). In contrast, secretion of ExoT was not sufficient to cause obvious signs of disease (health score [mean ± standard error of the mean]: 9.0 ± 0.0) or to allow bacteria to persist in the lung; the pulmonary bacterial load following infection with PA99T (2.5 × 103 CFU/lung) was similar to that following infection with PA99null or PA99secr−. On the basis of this evidence, we conclude that ExoU and ExoS, but not ExoT, contribute to the ability of P. aeruginosa to persist in the lung through 18 h postinfection.
FIG. 3.
Persistence of strains expressing individual type III secreted effector proteins in the lung through 18 h postinfection. Each symbol represents the bacterial load in the lung of an individual animal after 18 h. Bars represent the median value. §, Significant difference from PA99null (P < 0.05).
ExoU, ExoS, or ExoT is sufficient for dissemination of bacteria to extrapulmonary sites.
Previous reports have suggested a role for type III secreted effector proteins in the spread of bacteria from the lung into the bloodstream and to peripheral sites (14, 28, 29, 58). Thus, we investigated whether ExoU, ExoS, or ExoT was sufficient for dissemination to the liver and spleen. For each bacterial mutant, we determined the percentage of animals in which dissemination occurred, as well as the number of bacteria present in the liver and spleen after 18 h of infection. Viable PA99U bacteria were cultured from the liver and spleen more frequently than PA99null or PA99secr− bacteria (P < 0.05) (Table 3). Secretion of ExoS or ExoT was also associated with more frequent spread of bacteria to the liver (P < 0.05), although the incidence of bacterial spread to the spleen did not reach statistical significance (P = 0.23). In addition, PA99U was able to achieve statistically significantly higher bacterial numbers in the liver compared to PA99null and PA99secr− (P < 0.05) and showed a trend toward greater bacterial numbers in the spleen. Secretion of ExoS or ExoT showed a trend toward an increased bacterial burden in the liver but not in the spleen.
TABLE 3.
Dissemination of bacteria to the spleen and liver at 18 h postinfection
| Strain | Spleen
|
Liver
|
||
|---|---|---|---|---|
| No. of mice with culture-positive organs/total (%) | Median log10 no. of CFU/organ (range) | No. of mice with culture-positive organs/total (%) | Median log10 no. of CFU/organ (range) | |
| PA99U | 3/5 (60)a | 2.9 (0.0-3.7) | 4/5 (80)a | 3.2 (0.0-4.7)a |
| PA99S | 2/5 (40) | 0.0 (0.0-3.6) | 3/5 (60)a | 2.2 (0.0-2.7) |
| PA99T | 2/7 (29) | 0.0 (0.0-1.7) | 4/7 (57)a | 2.0 (0.0-2.6) |
| PA99null | 0/5 (0) | 0.0 (0.0-0.0) | 1/5 (20) | 0.0 (0.0-1.7) |
| PA99secr− | 1/10 (10) | 0.0 (0.0-2.0) | 0/10 (10) | 0.0 (0.0-0.0) |
P < 0.05 compared to PA99null and PA99secr−.
The ADPRT domain of ExoS is required for full virulence in mice.
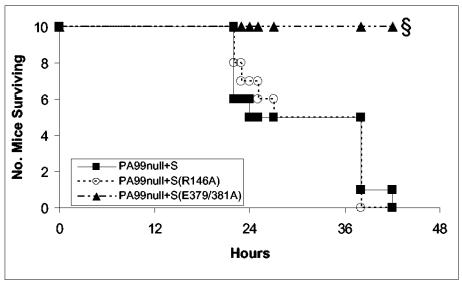
Whereas the roles of the enzymatic activities of ExoU and ExoT in virulence have been partially characterized in previous studies (14, 43, 49), it is unclear whether the enzymatic activities of ExoS contribute to its ability to cause disease. ExoS encodes both GAP and ADPRT activities; each of these activities affects a variety of cellular processes via modification of several eukaryotic host proteins. To determine which enzymatic activities of ExoS contribute to its effects on virulence, we constructed the following three bacterial strains in the PA99null background: (i) PA99null+S, which has a single-copy wild-type allele of exoS under control of its endogenous promoter inserted into the chromosomal attB site; (ii) PA99null+S(R146A), which secretes ExoS with a mutated GAP domain (15); and (iii) PA99null+S(E379A/E381A), which secretes ExoS with a mutated ADPRT domain (32, 44). The effects of these mutations on virulence were analyzed by infecting groups of 10 mice with each strain and monitoring survival over 48 h (Fig. 4). Following infection with 9.2 × 106 CFU, which is approximately three times the LD50 of a strain secreting only wild-type ExoS, PA99null+S and PA99null+S(R146A) each resulted in 100% mortality. In contrast, infection with PA99null+S(E379/381A) resulted in 100% survival over the same time period (P < 0.0001), with little apparent clinical disease as assessed by health score (data not shown). This indicates that the ADPRT activity of ExoS is responsible for most of the virulence associated with ExoS during acute pneumonia.
FIG. 4.
Virulence of strains expressing wild-type or mutated ExoS in a mouse model of acute pneumonia. Lines represent the number of animals surviving in each experimental group over time (n = 10 per strain, pooled from two separate experiments). §, Significant difference from PA99null+S (P < 0.0001).
The ADPRT activity of ExoS affects bacterial persistence in the lung.
We next compared the pulmonary bacterial load of strains encoding targeted mutations in each enzymatic domain of ExoS (Fig. 5). Inactivation of the ADPRT domain [strain PA99null+S(E379/381A)] resulted in a 50-fold reduction in the number of viable bacteria in the lung after 18 h, compared to that of a strain secreting wild-type ExoS (PA99null+S) (P < 0.0001). In contrast, inactivation of GAP activity [strain PA99null+S(R146A)] yielded a much smaller, statistically insignificant, decrease in the pulmonary bacterial load. Together, these data show that the ADPRT activity of ExoS is responsible for most of the ExoS-dependent bacterial persistence in the lung and that the GAP activity of ExoS is less important during acute infection.
FIG. 5.
Persistence of strains expressing wild-type or mutated ExoS in the lung through 18 h postinfection. Each symbol represents the bacterial load in the lung of an individual animal after 18 h. Bars represent the median value. §, Significant difference from PA99null+S (P < 0.0001).
DISCUSSION
Results obtained with our carefully controlled model of acute pneumonia indicate that the P. aeruginosa effector protein ExoU has the greatest impact on disease severity, that ExoS has an intermediate effect, and that ExoT has only a small effect on virulence. The relative virulence of these effector proteins is important since clinical isolates of P. aeruginosa commonly fall into one of the following three phenotypic categories: (i) those that secrete ExoU and ExoT, (ii) those that secrete ExoS and ExoT, and (iii) those that do not secrete type III proteins (16, 47). Integration of our present data with these groupings of clinical isolates suggests a hierarchy of virulence among P. aeruginosa strains causing acute pulmonary infections. Because ExoU has the greatest contributions to pathogenesis in our model system, in general those strains that secrete ExoU and ExoT may be the most virulent strains. Since ExoS has a significant but slightly smaller effect on virulence in our model, those strains that secrete ExoS and ExoT may be intermediate in virulence. Those strains that are incapable of type III secretion and therefore do not secrete ExoU, ExoS, and ExoT may be the least virulent. To more definitively address the contributions of the secreted effector proteins to the virulence of P. aeruginosa, studies assessing the consequences of secretion of effector proteins in various combinations and possible interactions between different effector proteins in the host cell are necessary. Nonetheless, this hierarchy of virulence is supported by previous studies in which clinical isolates secreting ExoU and ExoT tended to be the most virulent in mouse models of pneumonia (47, 51) and in human patients (16, 47). Although these distinctions may not be absolute because strains also differ in other bacterially encoded virulence factors, such categorization of the type III secretion phenotype of P. aeruginosa isolates has clinical utility. For example, patients infected with or even colonized by P. aeruginosa strains that secrete ExoU may require more aggressive therapy than those infected with strains that do not secrete this toxin. Additional studies of patients infected with P. aeruginosa are necessary to more fully elucidate the clinical implications of these findings.
In the mouse model of acute pneumonia, secretion of ExoU was associated with a 40-fold decrease in the LD50. That secretion of ExoU in the lung augmented virulence is consistent with prior reports that associated ExoU with increased mortality and lung pathology (1, 9, 18, 29). In vitro, ExoU rapidly kills a number of different cell types, including epithelial cells, macrophages, and fibroblasts (1, 2, 7, 9, 10, 17, 18, 23, 29, 50). More recent work has shown that ExoU-dependent cell death is a result of its phospholipase A2 activity (43, 49). Thus, the most likely explanation for the large increase in virulence associated with ExoU is that secretion of this protein leads to the death of a variety of cell types in the lung. Killing of immune cells may directly prevent clearance of P. aeruginosa, whereas overall tissue destruction may create an environment that hinders optimal phagocytic cell trafficking and function. Alternatively, ExoU-mediated host cell lysis may lead to an exaggerated inflammatory response that itself causes additional tissue injury. Finally, ExoU-dependent cytotoxicity may compromise the epithelial barrier of the lungs, allowing locally produced cytokines to leak into the systemic circulation and cause sepsis (29). In support of this, the number of bacteria in the lungs of PA99U-infected mice did not differ from that of bacteria in the lungs of PA99S-infected mice at 18 h postinfection.
To our knowledge, this is the first study that conclusively shows that ExoS is a virulence factor in P. aeruginosa infection. Prior studies suggesting that ExoS contributes to pathology were limited in their conclusions by the use of a pleiotropic mutant with an overall defect in type III secretion (36-38, 61). A subsequent study by Holder and colleagues found that disruption of the exoS gene in a strain encoding exoS, exoT, and exoY had little impact on virulence in a burn model of infection (22). Our study, which compared the virulence of a strain secreting only ExoS to an isogenic strain secreting no known type III effector proteins, demonstrated a significant contribution of ExoS to the pathogenesis of acute P. aeruginosa pneumonia. Secretion of ExoS was associated with increased mortality, decreased bacterial clearance from the lung, and more frequent bacterial dissemination to peripheral organs. The reason our findings differ from those of Holder and colleagues is unclear, but the difference may be due to functional redundancy of ExoS, ExoT, or ExoY or to differences in the effects of ExoS in burns compared to acute pneumonia.
To further understand how secretion of ExoS contributes to disease, the enzymatic activities of ExoS responsible for its effects on virulence were examined. The N terminus of ExoS encodes GAP activity that inactivates Rho GTPases, leading to cytoskeletal alterations (15, 27). Functional GAP activity has been shown to play a major role in blocking internalization of bacteria by both phagocytic and nonphagocytic cells in vitro (13, 46). The C terminus of ExoS encodes ADPRT activity, which has numerous deleterious effects on host cells in vitro, including induction of long-term cell rounding, as well as decreased DNA synthesis, cellular adherence, and viability (12, 13, 41, 42, 46). Previous work with cell culture model systems suggested that the ADPRT activity of ExoS has greater cellular effects than GAP activity (12). Interestingly, the consequences of ExoS intoxication in our in vivo experiments correlated with those previously observed with in vitro model systems. The effects of GAP activity were limited, suggesting that GAP-mediated antiphagocytic functions of ExoS may be less important during acute infection than anticipated. Rather, the ADPRT activity of ExoS accounted for the majority of the ExoS-dependent contribution to virulence.
Surprisingly, ExoS and ExoT, which have 75% amino acid identity (60), had markedly different effects on virulence during acute pneumonia. The biochemical similarities between these two proteins had suggested that they exhibit functional redundancy in vivo, as they do in some in vitro assays (5, 14). However, in the experiments described here, ExoS and ExoT were not interchangeable during infection in vivo but instead had profoundly different effects on disease pathogenesis. Secretion of ExoS was associated with relatively low LD50s, bacterial persistence in the lungs, and dissemination. In contrast, secretion of ExoT was not associated with a significant change in mortality or bacterial load in the lungs but did lead to a modest increase in bacterial dissemination. In this regard, our results agree with those of Garrity-Ryan and colleagues, who observed that in competition assays secretion of ExoT was associated with small increases in bacterial dissemination to the liver but not with persistence in the lungs of mice with acute pneumonia (14). In these same assays, the ADPRT activity and not the GAP activity of ExoT was responsible for the virulence associated with this toxin (14). Thus, both ExoS and ExoT appear to affect virulence through the ADP-ribosylation of various host proteins. Differences in the particular targets modified by the ADPRT activities of these proteins may explain why ExoS has a greater effect on virulence than does ExoT. ExoS covalently modifies a wide variety of cellular proteins, including Ras, RalA, and Rab5 (12, 20, 35, 40, 46, 56). In contrast, ExoT modifies the Crk-I and Crk-II kinases (55). Further work is necessary to determine the mechanisms by which ADP-ribosylation of different substrates by ExoS and ExoT results in their differential effects on virulence. In particular, understanding why secretion of ExoS is sufficient for bacterial persistence in the lungs while secretion of ExoT is not will be of importance.
Our results also suggest that the secretion apparatus itself or unidentified effector proteins play at most a minor role in the pathogenesis of strain PA99. PA99null, a mutant with a functional type III secretion system but with deletions in the genes encoding ExoU, ExoS, and ExoT, was not significantly more virulent than PA99secr−, a mutant that fails to assemble a functional type III secretion apparatus. Comparison of infections with PA99null and PA99secr− revealed only a small difference between their LD50s and no differences in the bacterial loads in any of the organs tested. Thus, ExoU, ExoS, and ExoT account for nearly all of the virulence associated with the type III secretion system of strain PA99.
In summary, a panel of isogenic bacterial mutants in a clinical isolate of P. aeruginosa (PA99) that naturally secretes ExoU, ExoS, and ExoT was used to carefully evaluate the relative role of each effector protein in disease pathogenesis. This approach demonstrated that ExoU has a major effect, ExoS has an intermediate effect, and ExoT has a minor effect on virulence in acute pneumonia. This is the first study to conclusively show that ExoS is a virulence factor and that its pathogenic effects are dependent on its ADRPT activity. Surprisingly, ExoS and ExoT were associated with different effects in the lung, despite their similar enzymatic activities whereas ExoU and ExoS were both individually sufficient to allow bacterial persistence in the lung and dissemination even though they have distinct enzymatic activities. Understanding the relative virulence associated with ExoU, ExoS, and ExoT may have important clinical implications for patients infected with P. aeruginosa.
Acknowledgments
We thank Aaron Shaver for statistical assistance, Daniel Ramirez and Karla Satchell for technical assistance, and other members of the Hauser laboratory for advice and review of the manuscript. We thank Herbert Schweizer for providing the pEX100T and mini-CTX1 vectors.
This work was supported by the NIH (AI053674 to A.R.H., AI07476-07 to C.M.S., and ES013082 to C.M.S. and A.R.H.), Philip Morris USA Inc. and Philip Morris International (A.R.H.), and the American Lung Association (A.R.H.).
Editor: D. L. Burns
REFERENCES
- 1.Allewelt, M., F. T. Coleman, M. Grout, G. P. Priebe, and G. B. Pier. 2000. Acquisition of expression of the Pseudomonas aeruginosa ExoU cytotoxin leads to increased bacterial virulence in a murine model of acute pneumonia and systemic spread. Infect. Immun. 68:3998-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apodaca, G., M. Bomsel, R. Lindstedt, J. Engel, D. Frank, K. Mostov, and J. Wiener-Kronish. 1995. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation defective host cells are resistant to bacterial killing. Infect. Immun. 63:1541-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorn, M. J., O. R. Pavlovskis, M. R. Thompson, and B. H. Iglewski. 1979. Production of exoenzyme S during Pseudomonas aeruginosa infection in burned mice. Infect. Immun. 24:837-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowell, B. A., D. Y. Chen, D. W. Frank, A. J. Vallis, and S. M. J. Fleiszig. 2000. ExoT of cytotoxic Pseudomonas aeruginosa prevents uptake by corneal epithelial cells. Infect. Immun. 68:403-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dacheux, D., B. Toussaint, M. Richard, G. Brochier, J. Croize, and I. Attree. 2000. Pseudomonas aeruginosa cystic fibrosis isolates induce rapid, type III secretion-dependent, but ExoU-independent, oncosis of macrophages and polymorphonuclear neutrophils. Infect. Immun. 68:2916-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, D. J., T. C. Kuo, M. Kwong, R. Van, and S. M. Fleiszig. 2002. Mutation of csk, encoding the C-terminal Src kinase, reduces Pseudomonas aeruginosa internalization by mammalian cells and enhances bacterial cytotoxicity. Microb. Pathog. 33:135-143. [DOI] [PubMed] [Google Scholar]
- 8.Feltman, H., G. Schulert, S. Khan, M. Jain, L. Peterson, and A. R. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 9.Finck-Barbançon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. J. Fleiszig, C. Wu, L. Mende-Mueller, and D. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 10.Fleiszig, S. M. J., T. S. Zaidi, M. J. Preston, M. Grout, D. J. Evans, and G. B. Pier. 1996. Relationship between cytotoxicity and corneal epithelial cell invasion by clinical isolates of Pseudomonas aeruginosa. Infect. Immun. 64:2288-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank, D., and B. H. Iglewski. 1991. Cloning and sequence analysis of a trans-regulatory locus required for exoenzyme S synthesis in Pseudomonas aeruginosa. J. Bacteriol. 173:6460-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraylick, J. E., J. R. La Rocque, T. S. Vincent, and J. C. Olson. 2001. Independent and coordinate effects of ADP-ribosyltransferase and GTPase-activating activities of exoenzyme S on HT-29 epithelial cell function. Infect. Immun. 69:5318-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frithz-Lindsten, E., Y. Du, R. Rosqvist, and A. Forsberg. 1997. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol. Microbiol. 25:1125-1139. [DOI] [PubMed] [Google Scholar]
- 14.Garrity-Ryan, L., B. Kazmierczak, R. Kowal, J. Comolli, A. Hauser, and J. N. Engel. 2000. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goehring, U.-M., G. Schmidt, K. J. Pederson, K. Aktories, and J. T. Barbieri. 1999. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem. 274:36369-36372. [DOI] [PubMed] [Google Scholar]
- 16.Hauser, A. R., E. Cobb, M. Bodí, D. Mariscal, J. Vallés, J. N. Engel, and J. Rello. 2002. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 17.Hauser, A. R., and J. N. Engel. 1999. Pseudomonas aeruginosa induces type III secretion-mediated apoptosis of macrophages and epithelial cells. Infect. Immun. 67:5530-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauser, A. R., P. J. Kang, and J. Engel. 1998. PepA, a novel secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 19.Hauser, A. R., P. J. Kang, S. J. M. Fleiszig, K. Mostov, and J. Engel. 1998. Defects in type III secretion correlate with internalization of Pseudomonas aeruginosa by epithelial cells. Infect. Immun. 66:1413-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henriksson, M. L., C. Sundin, A. L. Jansson, A. Forsberg, R. H. Palmer, and B. Hallberg. 2002. Exoenzyme S shows selective ADP-ribosylation and GTPase-activating protein (GAP) activities towards small GTPases in vivo. Biochem. J. 367:617-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 22.Holder, I. A., A. N. Neely, and D. W. Frank. 2001. Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns 27:129-130. [DOI] [PubMed] [Google Scholar]
- 23.Kang, P. J., A. R. Hauser, G. Apodaca, S. Fleiszig, J. Wiener-Kronish, K. Mostov, and J. N. Engel. 1997. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol. Microbiol. 24:1249-1262. [DOI] [PubMed] [Google Scholar]
- 24.Kazmierczak, B. I., and J. N. Engel. 2002. Pseudomonas aeruginosa ExoT acts in vivo as a GTPase-activating protein for RhoA, Rac1, and Cdc42. Infect. Immun. 70:2198-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight, D. A., V. Finck-Barbancon, S. M. Kulich, and J. T. Barbieri. 1995. Functional domains of Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 63:3182-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krall, R., G. Schmidt, K. Aktories, and J. T. Barbieri. 2000. Pseudomonas aeruginosa ExoT is a Rho GTPase-activating protein. Infect. Immun. 68:6066-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krall, R., J. Sun, K. J. Pederson, and J. T. Barbieri. 2002. In vivo rho GTPase-activating protein activity of Pseudomonas aeruginosa cytotoxin ExoS. Infect. Immun. 70:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudoh, I., J. P. Wiener-Kronish, S. Hashimoto, J.-F. Pittet, and D. Frank. 1994. Exoproduct secretions of P. aeruginosa strains influence severity of alveolar epithelial injury. Am. J. Physiol. 267(5 Pt. 1):L551-L556. [DOI] [PubMed] [Google Scholar]
- 29.Kurahashi, K., O. Kajikawa, T. Sawa, M. Ohara, M. A. Gropper, D. W. Frank, T. R. Martin, and J. P. Wiener-Kronish. 1999. Pathogenesis of septic shock in Pseudomonas aeruginosa pneumonia. J. Clin. Investig. 104:743-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Liu, P. V. 1966. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. II. Effects of lecithinase and protease. J. Infect. Dis. 116:112-116. [DOI] [PubMed] [Google Scholar]
- 32.Liu, S., S. M. Kulich, and J. T. Barbieri. 1996. Identification of glutamic acid 381 as a candidate active site residue of Pseudomonas aeruginosa exoenzyme S. Biochemistry 35:2754-2758. [DOI] [PubMed] [Google Scholar]
- 33.Liu, S., T. L. Yahr, D. W. Frank, and J. T. Barbieri. 1997. Biochemical relationships between the 53-kilodalton (Exo53) and 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa. J. Bacteriol. 179:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McElroy, M., J. Pittet, J. Wiener-Kronish, and L. Dobbs. 1994. A specific alveolar epithelial type I cell marker detects injury to the alveolar epithelial barrier in a model of acute lung injury induced by Pseudomonas aeruginosa. Prog. Respir. Res. 27:165-168. [Google Scholar]
- 35.McGuffie, E. M., J. E. Fraylick, D. J. Hazen-Martin, T. S. Vincent, and J. C. Olson. 1999. Differential sensitivity of human epithelial cells to Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 67:3494-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicas, T., and B. H. Iglewski. 1985. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can. J. Microbiol. 31:387-392. [DOI] [PubMed] [Google Scholar]
- 37.Nicas, T. I., D. W. Frank, P. Stenzel, and B. Iglewski. 1985. Role of exoenzyme S in chronic Pseudomonas aeruginosa lung infections. Eur. J. Clin. Microbiol. 4:175-179. [DOI] [PubMed] [Google Scholar]
- 38.Nicas, T. I., and B. H. Iglewski. 1985. Contribution of exoenzyme S to the virulence of Pseudomonas aeruginosa. Antibiot. Chemother. 36:40-48. [DOI] [PubMed] [Google Scholar]
- 39.Nicas, T. I., and B. H. Iglewski. 1984. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect. Immun. 45:470-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olson, J. C., J. E. Fraylick, E. M. McGuffie, K. M. Dolan, T. L. Yahr, D. W. Frank, and T. S. Vincent. 1999. Interruption of multiple cellular processes in HT-29 epithelial cells by Pseudomonas aeruginosa exoenzyme S. Infect. Immun. 67:2847-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pederson, K. J., and J. T. Barbieri. 1998. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas aeruginosa exoenzyme S is cytotoxic to eukaryotic cells. Mol. Microbiol. 30:751-759. [DOI] [PubMed] [Google Scholar]
- 42.Pederson, K. J., R. Krall, M. J. Riese, and J. T. Barbieri. 2002. Intracellular localization modulates targeting of ExoS, a type III cytotoxin, to eukaryotic signalling proteins. Mol. Microbiol. 46:1381-1390. [DOI] [PubMed] [Google Scholar]
- 43.Phillips, R. M., D. A. Six, E. A. Dennis, and P. Ghosh. 2003. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J. Biol. Chem. 278:41326-41332. [DOI] [PubMed] [Google Scholar]
- 44.Radke, J., K. J. Pederson, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S is a biglutamic acid ADP-ribosyltransferase. Infect. Immun. 67:1508-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reed, L., and J. Muench. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 46.Rocha, C. L., J. Coburn, E. A. Rucks, and J. C. Olson. 2003. Characterization of Pseudomonas aeruginosa exoenzyme S as a bifunctional enzyme in J774A.1 macrophages. Infect. Immun. 71:5296-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawa, T., T. Yahr, M. Ohara, K. Kurahashi, M. A. Gropper, J. P. Wiener-Kronish, and D. W. Frank. 1999. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat. Med. 5:392-398. [DOI] [PubMed] [Google Scholar]
- 51.Schulert, G. S., H. Feltman, S. D. P. Rabin, C. G. Martin, S. E. Battle, J. Rello, and A. R. Hauser. 2003. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 188:1695-1706. [DOI] [PubMed] [Google Scholar]
- 52.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 53.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 54.Smith, R. S., M. C. Wolfgang, and S. Lory. 2004. An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun. 72:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun, J. C., and M. J. Bevan. 2003. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300:339-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vincent, T. S., J. E. Fraylick, E. M. McGuffie, and J. C. Olson. 1999. ADP-ribosylation of oncogenic Ras proteins by Pseudomonas aeruginosa exoenzyme S in vivo. Mol. Microbiol. 32:1054-1064. [DOI] [PubMed] [Google Scholar]
- 57.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithinase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 58.Wiener-Kronish, J. P., T. Sakuma, I. Kudoh, J. F. Pittet, D. Frank, L. Dobbs, M. L. Vasil, and M. A. Matthay. 1993. Alveolar epithelial injury and pleural empyema in acute Pseudomonas aeruginosa pneumonia in anesthetized rabbits. J. Appl. Physiol. 75:1661-1669. [DOI] [PubMed] [Google Scholar]
- 59.Yahr, T., L. M. Mende-Mueller, M. B. Friese, and D. W. Frank. 1997. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 179:7165-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yahr, T. L., J. T. Barbieri, and D. W. Frank. 1996. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J. Bacteriol. 178:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yahr, T. L., J. Goranson, and D. W. Frank. 1996. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III secretion pathway. Mol. Microbiol. 22:991-1003. [DOI] [PubMed] [Google Scholar]
- 62.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]