Abstract
Clostridium perfringens enterotoxin (CPE) has a unique mechanism of action that results in the formation of large, sodium dodecyl sulfate-resistant complexes involving tight junction proteins; those complexes then induce plasma membrane permeability alterations in host intestinal epithelial cells, leading to cell death and epithelial desquamation. Previous deletion and point mutational studies mapped CPE receptor binding activity to the toxin's extreme C terminus. Those earlier analyses also determined that an N-terminal CPE region between residues D45 and G53 is required for large complex formation and cytotoxicity. To more finely map this N-terminal cytotoxicity region, site-directed mutagenesis was performed with recombinant CPE (rCPE). Alanine-scanning mutagenesis produced one rCPE variant, D48A, that failed to form large complexes or induce cytotoxicity, despite having normal ability to bind and form the small complex. Two saturation variants, D48E and D48N, also had a phenotype resembling that of the D48A variant, indicating that both size and charge are important at CPE residue 48. Another alanine substitution rCPE variant, I51A, was highly attenuated for large complex formation and cytotoxicity, but rCPE saturation variants I51L and I51V displayed a normal large complex formation and cytotoxicity phenotype. Collectively, these mutagenesis results identify a core CPE sequence extending from residues G47 to I51 that directly participates in large complex formation and cytotoxicity.
Less than 5% of all Clostridium perfringens isolates, mostly belonging to type A, produce a 35-kDa enterotoxin (CPE) with a unique 319-amino-acid sequence (5, 20). Those enterotoxigenic type A isolates cause C. perfringens type A food poisoning, which currently ranks as the third most commonly identified form of food poisoning in the United States (29). In addition, CPE-positive C. perfringens type A isolates have been increasingly linked to other human gastrointestinal illnesses, such as antibiotic-associated and sporadic diarrhea (1, 3).
CPE's unique mechanism of action starts with its binding to one or more protein receptors, which possibly include certain members of the claudin family of tight junction structural proteins, present on the apical surface of intestinal epithelial cells (6, 17). This binding promotes formation of a sodium dodecyl sulfate (SDS)-sensitive, ∼90-kDa CPE complex that apparently includes either an unidentified ∼50-kDa protein (41) or a claudin aggregate. That small complex then associates with additional proteins to form an SDS-resistant, CPE-containing complex of ∼155 kDa (43). Formation of the ∼155-kDa complex is concurrent with the onset of plasma membrane permeability alterations in a CPE-treated cell, suggesting a cause-and-effect relationship (41, 43). Those permeability alterations activate Ca2+-dependent cell death pathways, with low CPE doses triggering a classical caspase 3/7-mediated apoptosis and high CPE doses inducing oncosis (4). Consequent morphological damage exposes the basolateral surfaces of both dying and adjacent healthy cells, providing CPE access to basolateral receptors and the tight junction protein occludin to form an additional SDS-resistant CPE complex of ∼200 kDa. Formation of that complex triggers the internalization of occludin (35), which probably disrupts tight junctions and alters paracellular permeability in polarized enterocytes (25).
Some information is available concerning how CPE interacts with cellular proteins. Receptor binding activity localizes to the enterotoxin's C terminus, as (i) deletion fragments lacking only the five C-terminal CPE amino acids cannot bind to either rabbit brush border membranes (BBMs) or sensitive cells (11, 14, 19) and (ii) synthetic peptides or recombinant fragments corresponding to the 30 C-terminal CPE amino acids completely inhibit native enterotoxin binding to BBMs (12). Since C-terminal enterotoxin fragments are not cytotoxic (11, 12, 14), some N-terminal CPE sequences are important for cytotoxicity. However, extreme N-terminal CPE sequences are not involved in cytotoxicity, since (i) tryptic or chymotryptic treatment removes the 25 or 36 N-terminal CPE amino acids, respectively, increasing cytotoxic activity by two- to threefold (8, 9, 13) and (ii) a recombinant CPE (rCPE) fragment lacking the first 44 amino acids of the native enterotoxin also exhibits a two- to threefold increase in large complex formation and cytotoxicity (19). Further deletion of N-terminal rCPE sequences produces an rCPE53-319 fragment that maintains wild-type binding activity, but neither forms a large complex nor elicits a cytotoxic response (19). That observation, coupled with the cytotoxic activity of the rCPE45-319 fragment, implicates a region between D45 and G53 of native CPE as being essential for cytotoxicity. This conclusion is supported by random mutagenesis studies (18), where a G49D random mutant formed the small complex and had normal receptor binding activity but could not form large complexes or elicit 86Rb release.
The present study has fine mapped the D45-to-E54 N-terminal cytotoxicity region of CPE by using a systematic site-directed mutagenesis approach coupled with careful phenotypic characterization of the resultant rCPE variants. Via this approach, two residues crucial for the enterotoxin's action have now been identified.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The XL-1 Blue strain of Escherichia coli was used for all mutagenesis work. Plasmids used or produced in this study are listed in Table 1. The plasmid construct pJKFLt-1 (19), which served as the template for introducing all mutations, consists of the complete cpe open reading frame inserted into the multiple cloning site of the pTrcHis A vector (Invitrogen, Carlsbad, Calif.). This vector encodes a 32-amino-acid tag that includes a His6 sequence fused to the N terminus of rCPE, enabling affinity enrichment of rCPE or its derivatives (see below). Luria-Bertani medium (liquid and solid) was used to culture bacteria for mutant isolation or plasmid preparation, while SOB medium was used to culture rCPE-expressing E. coli cultures for affinity enrichment of rCPE. Ampicillin (100 μg/ml) was added to all cultures for plasmid maintenance and, where appropriate, 0.5 mM isopropyl-β-d-thiogalactopyranoside was used to induce expression of rCPE. All bacterial cultures were grown overnight at 37°C, unless otherwise indicated.
TABLE 1.
Mutagenesis primers
| rCPE species expressed | Primer sequencea | Plasmid |
|---|---|---|
| D45A | 5′-GGATTATATGTAATAGCTAAAGGAGATGGTTGGATATTAGGGGAACCC-3′ | pJSD45A |
| 3′-GGGTTCCCCTAATATCCAACCATCTCCTTTAGCTATTACATATAATCC-5′ | ||
| K46A | 5′-GGATTATATGTAATAGATGCAGGAGATGGTTGGATATTAGGGGAACCC-3′ | pJSK46A |
| 3′-GGGTTCCCCTAATATCCAACCATCTCCTGCATCTATTACATATAATCC-5′ | ||
| G47A | 5′-GGATTATATGTAATAGATAAAGCAGATGGTTGGATATTAGGGGAACCC-3′ | pJSG47A |
| 3′-GGGTTCCCCTAATATCCAACCATCTGCTTTATCTATTACATATAATCC-5′ | ||
| D48A | 5′-GGATTATATGTAATAGATAAAGGAGCTGGTTGGATATTAGGGGAACCC-3′ | pJSD48A |
| 3′-GGGTTCCCCTAATATCCAACCAGCTCCTTTATCTATTACATATAATCC-5′ | ||
| G49A | 5′-GATTATATGTAATAGATAAAGGAGATGCTTGGATATTAGGGGAAC-3′ | pJSG49A |
| 3′-GTTCCCCTAATATCCAAGCATCTCCTTTATCTATTACATATAATC-5′ | ||
| W50A | 5′-GTAATAGATAAAGGAGATGGTGCGATATTAGGGGAACCCTCAG-3′ | pJSW50A |
| 3′-CTGAGGGTTCCCCTAATATCGCACCATCTCCTTTATCTATTAC-5′ | ||
| I51A | 5′-GTAATAGATAAAGGAGATGGTTGGGCATTAGGGGAACCCTCAG-3′ | pJSI51A |
| 3′-CTGAGGGTTCCCCTAATGCCCAACCATCTCCTTTATCTATTAC-5′ | ||
| L52A | 5′-GATAAAGGAGATGGTTGGATAGCAGGGGAACCCTCAGTAGTTTC-3′ | pJSL52A |
| 3′-GAAACTACTGAGGGTTCCCCTGCTATCCAACCATCTCCTTTATC-5′ | ||
| G53A | 5′-GGAGATGGTTGGATATTAGCGGAACCCTCAGTAGTTTC-3′ | pJSG53A |
| 3′-GAAACTACTGAGGGTTCCGCTAATATCCAACCATCTCC-5′ | ||
| E54A | 5′-GGAGATGGTTGGATATTAGGGGCACCCTCAGTAGTTTC-3′ | pJSE54A |
| 3′-GAAACTACTGAGGGTGCCCCTAATATCCAACCATCTCC-5′ | ||
| D48E | 5′-GGATTATATGTAATAGATAAAGGAGAGGGTTGGATATTAGGGGAACCC-3′ | pJSD48E |
| 3′-GGGTTCCCCTAATATCCAACCCTCTCCTTTATCTATTACATATAATCC-5′ | ||
| D48N | 5′-TTATATGTAATAGATAAAGGAAATGGTTGGATATTAGGGGAACCC-3′ | pJSD48N |
| 3′-GGGTTCCCCTAATATCCAACCATTTCCTTTATCTATTACATATAA-5′ | ||
| W50Y | 5′-GTAATAGATAAAGGAGATGGTTACATATTAGGGGAACCCTCAG-3′ | pJSW50Y |
| 3′-CTGAGGGTTCCCCTAATATGTAACCATCTCCTTTATCTATTAC-5′ | ||
| W50F | 5′-GTAATAGATAAAGGAGATGGTTTTATATTAGGGGAACCCTCAG-3′ | pJSW50F |
| 3′-CTGAGGGTTCCCCTAATATAAAACCATCTCCTTTATCTATTAC-5′ | ||
| 151L | 5′-GTAATAGATAAAGGAGATGGTTGGCTATTAGGGGAACCCTCAG-3′ | pJSI51L |
| 3′-CTGAGGGTTCCCCTAATAGCCAACCATCTCCTTTATCTATTAC-5′ | ||
| 151V | 5′-GTAATAGATAAAGGAGATGGTTGGGTATTAGGGGAACCCTCAG-3′ | pJSI51V |
| 3′-CTGAGGGTTCCCCTAATACCCAACCATCTCCTTTATCTATTAC-5′ |
Mutations are underlined.
Site-directed mutagenesis.
Single point mutations were introduced into the cpe gene of pJKFLt-1 by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) as instructed by the manufacturer. Briefly, primers were designed with the desired mutation (Table 1), and a thermocycling reaction permitted in vitro synthesis of the plasmid DNA with the high-fidelity polymerase PfuTurbo. Parental template DNA was digested away by the methylase-specific DNase, DpnI, and the resultant mutated plasmid mixture was used to transform XL-1 Blue supercompetent E. coli cells.
Nucleotide sequencing.
Plasmids subjected to site-directed mutagenesis reactions were isolated from E. coli transformants with the QIAGEN plasmid mini kit (QIAGEN, Valencia, Calif.). The cpe open reading frame in each mutated plasmid was then sequenced to confirm the presence of the desired mutation and ensure that no unintended mutations had occurred during mutagenesis. Using the primers listed in Table 2, both strands of the complete cpe open reading frame were sequenced by the University of Pittsburgh Genomics and Proteomics Core Laboratories. Resulting sequences were analyzed using BioEdit software (10).
TABLE 2.
Sequencing primers
| Primer name | Primer sequence |
|---|---|
| TreSeq-1F | 5′-GGACAGCAAATCGGTCGG-3′ |
| 2-F | 5′-GGTACCTTAAGCCAATCA-3′ |
| PH-310-F | 5′-GATTTAGCTGCTACAGAAAGA-3′ |
| PH-309-R | 5′-TGATCAATATTCCTAAGCTATCTGCAG-3′ |
| 6-R | 5′-CCGGAATTCTATATGGAAGGAGAAATTAATGC-3′ |
Affinity enrichment of rCPE variants.
Using TALON metal affinity resin (BD Biosciences Clontech, Palo Alto, Calif.), rCPE variants were affinity enriched from SOB cultures containing ampicillin (100 μg/ml) as previously described (18). After enrichment, rCPE variants were stored at −80°C, which did not affect their biologic activities (data not shown). The amount of each rCPE species recovered after affinity enrichment was then determined by quantitative Western immunoblotting, as previously described (18), except that quantitative imaging of blots was performed using the Gel Doc imaging system (Bio-Rad, Hercules, Calif.) and Quantity One software (Bio-Rad).
CaCo-2 cell culture.
CaCo-2 cells were maintained in Eagle minimal essential medium (Sigma, St. Louis, Mo.) with 10% fetal bovine serum (Mediatech, Herndon, Va.), 1% nonessential amino acids (Sigma), 100 U of ampicillin/ml, and 100 μg of streptomycin/ml. All cultures were grown at 37°C with 5% atmospheric CO2.
Cytotoxicity of rCPE species.
Biological activity of each affinity-enriched rCPE species was initially screened using CaCo-2 monolayers. Six-well dishes were inoculated with 2.5 × 105 cells in 5 ml and then grown to confluency (typically 4 to 5 days). Cultures were washed once with prewarmed Hank's balanced salt solution (HBSS) and then treated with 2.5 μg of an rCPE species/ml. After 60 min of toxin treatment, cultures were visualized with a Zeiss Axiovert 25 inverted microscope with a 10× magnification objective lens. Images were captured with a Canon Powershot G5 digital camera and Canon Utilities RemoteCapture software (Canon, Inc., Lake Success, N.Y.) and then processed using Adobe Photoshop software (Adobe Systems, Inc., San Jose, Calif.).
86Rb release assay.
Membrane permeability alterations induced by each rCPE species were assayed by measuring 86Rb release from labeled CaCo-2 cells, as previously described (34). Briefly, CaCo-2 cells grown to confluency in 24-well plates were radiolabeled for 4 h at 37°C with 4 μCi of 86Rb (specific activity, 6.4 mCi/mg; Perkin-Elmer, Boston, Mass.) per well. The labeled cultures were washed twice with HBSS and then treated with an rCPE species (at the specified concentrations) added to 2 ml of HBSS. After 15 min of treatment at 37°C, culture supernatants were carefully collected and the radiation present in the supernatants was determined with a Cobra Quantum gamma counter (Perkin-Elmer). CPE-induced effects were then expressed as the percentage of maximal 86Rb release, using the following equation: [(experimental release − spontaneous release)/(maximal release − spontaneous release)] × 100. Spontaneous release (86Rb release in the absence of CPE) and maximal release (86Rb levels present in wells at the start of an experiment) were determined as described previously (34). Table 3 values, indicating the concentration of each rCPE species required to elicit a 50% maximal release, were calculated graphically from three independent trials with each rCPE species.
TABLE 3.
Summary of rCPE variant phenotypes
| rCPE species | Concn needed for:
|
Small complex formation | Large complex formation | |
|---|---|---|---|---|
| 50% maximal 86Rb release (μg/ml) | 50% [125I]CPE binding inhibition (μg/ml) | |||
| rCPE | 0.57 ± 0.19 | 3.69 ± 0.52 | + | + |
| D45A | 0.24 ± 0.04 | 1.83 ± 0.54 | + | + |
| K46A | 0.82 ± 0.07 | 2.89 ± 0.86 | + | + |
| G47A | 0.92 ± 0.63 | 5.52 ± 0.52 | + | + |
| D48A | >50 | 3.31 ± 0.78 | + | − |
| G49A | 0.83 ± 0.23 | 2.45 ± 0.59 | + | + |
| W50A | 1.35 ± 0.64 | 4.13 ± 0.90 | + | +a |
| I51A | >50 | 2.59 ± 0.16 | + | − |
| L52A | 0.68 ± 0.21 | 1.45 ± 0.04 | + | + |
| G53A | 0.52 ± 0.15 | 3.20 ± 0.74 | + | + |
| E54A | 0.38 ± 0.19 | 2.47 ± 0.56 | + | + |
| D48E | >50 | 1.92 ± 0.05 | + | − |
| D48N | >50 | 2.21 ± 0.16 | + | − |
| W50Y | 0.65 ± 0.4 | 2.68 ± 0.45 | + | + |
| W50F | 0.9 ± 0.2 | 2.60 ± 1.41 | + | + |
| I51L | 0.9 ± 0.3 | 3.23 ± 0.18 | + | + |
| I51V | 1.6 ± 1.2 | 2.82 ± 0.57 | + | + |
The W50A variant produces an atypical large complex (Fig. 6).
Binding analysis of rCPE species.
Small-intestine BBMs were isolated from female New Zealand White rabbits as described previously (33). [125I]CPE was prepared using IODO-GEN-precoated iodination tubes (Pierce, Rockford, Ill.), with 2.5 mCi of Na125I (specific activity, 17.0 mCi/mg; ICN) for 1 μg of native CPE. As described previously (18), BBMs (100 μg) were preincubated for 20 min in 200 μl of phosphate-buffered saline (PBS) containing various concentrations (0.01 to 50 μg/ml) of each competing rCPE species. [125I]CPE (0.5 μg) was then added to each sample for an additional 20 min at room temperature. After washing, [125I]CPE bound to the BBMs was quantified using a Cobra Quantum gamma counter. Data are presented as the percentage of total binding, where total (100%) binding was determined by incubating BBMs with [125I]CPE in the absence of any competing rCPE species. Table 3 binding values represent the concentration of each rCPE species required to inhibit 50% of the total [125I]CPE binding from two independent trials, each done in triplicate. The previously described rCPE random point variant W226Stop (18) was affinity enriched (as described above) and used as a negative control in these binding experiments.
Small complex formation.
The ability of rCPE variants to form the small complex was measured as previously described (18). Briefly, BBMs (200 μg) were treated for 15 min at 4°C (to inhibit formation of CPE large complexes [27]) with 5 μg of rCPE dissolved in 500 μl of PBS. Those BBMs were then pelleted, washed, and extracted for 30 min at 4°C with 40 μl of PBS containing 1% Triton X-100 (Sigma). After microcentrifugation, 10 μl of 5× native loading buffer (5 mM Tris-HCl, 38 mM glycine, 33% glycerol, 0.25 mg of bromphenol blue) was added to the extraction supernatants. These samples were electrophoresed at 5 mA on a 6% native gel, and small complexes were electrotransferred onto Immobilon-P polyvinylidene difluoride membrane (Millipore Corporation, Bedford, Mass.) by using Gershoni's transfer buffer (15.6 mM Tris-HCl, 120 mM glycine; pH 8.3). Western blotting to detect the CPE-containing small complex was then performed as described for affinity enrichment quantification.
Large complex formation.
Large complex formation by each rCPE species was assayed as described elsewhere (35) using confluent CaCo-2 cell cultures grown in a 100-mm dish. Briefly, CaCo-2 cultures were washed once with HBSS, gently dislodged from the plate with a cell scraper, and harvested by centrifugation. After one additional wash with HBSS, 2.5 × 106 harvested cells were resuspended in 200 μl of HBSS. An rCPE species was then added at a final concentration of 2.5 μg/ml, and samples were incubated for 45 min at 37°C with shaking. After microcentrifugation at 4°C, pelleted cells were washed with HBSS containing protease inhibitors (10 μg of aprotinin/ml, 10 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride; Sigma) and then resuspended in 45 μl of ice-cold HBSS containing the protease inhibitors specified above. Benzonase nuclease (12.5 U; Novagen, Inc., Darmstadt, Germany) was added to each sample for 10 min at room temperature. Fifty microliters of 2× Laemmli buffer was added to each sample, which was then loaded onto a 4% acrylamide SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel that was run overnight for ∼19 h at 5 mA before electrotransfer and CPE Western immunoblotting as described above.
Limited trypsin proteolysis assay.
As previously described (18), a 400-ng aliquot of each affinity-enriched rCPE species was brought to a final volume of 100 μl with PBS. Reactions mixtures were prepared in quadruplicate and digested at 25°C for 0, 5, 15, or 30 min with 4 ng of trypsin (Sigma). Immediately after completion of the digestion period, 100 μl of 2× Laemmli buffer with 5% β-mercaptoethanol was added to each reaction mixture and the samples were boiled for 5 min to terminate the proteolytic digestion. A 50-μl aliquot of each sample was then loaded onto a 10% acrylamide SDS-PAGE gel, followed by Western blotting with anti-CPE antibody as described above for the rCPE quantification.
RESULTS
Mutagenesis of the N-terminal cytotoxicity region.
Since previous rCPE deletion mutagenesis studies (19) had implicated the region between residues D45 and G53 of the native enterotoxin as being important for cytotoxicity, the present study first targeted this region for alanine scanning site-directed mutagenesis (Fig. 1). By sequentially introducing single alanine mutations with the QuikChange kit (see Materials and Methods), the contribution of each individual amino acid from D45 to E54 was assessed in a fully toxic rCPE background. All mutations were confirmed by nucleotide sequencing of both strands of the mutated cpe gene.
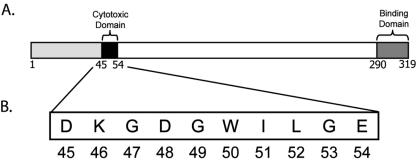
FIG. 1.
Functional map of CPE. (A) A previous deletion mutagenesis study of CPE (19) identified two distinct functional domains, including (i) an ∼10-amino-acid region near the N terminus that is essential for cytotoxicity and large complex formation and (ii) a receptor binding region located at the extreme C terminus of the toxin. (B) The amino acid sequence of the N-terminal CPE 45-54 cytotoxicity region targeted for mutagenesis in this study.
Assessment of morphological damage induced by rCPE species.
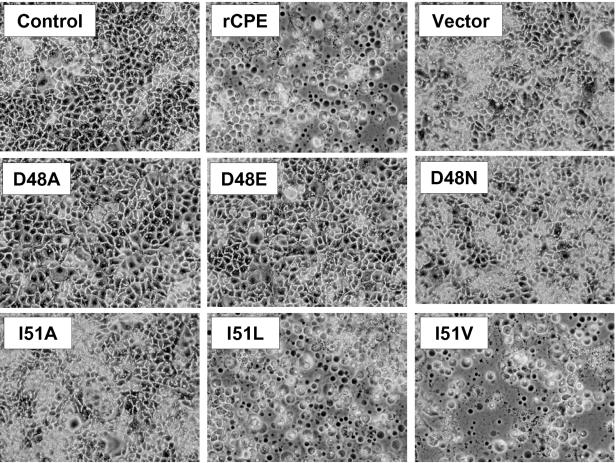
As an initial screen for toxicity, each affinity-enriched rCPE species (2.5 μg/ml) in HBSS was applied to confluent CaCo-2 cell cultures for 1 h. In this assay, CaCo-2 monolayers treated with rCPE developed typical CPE-induced changes in cell morphology (Fig. 2), including cell rounding, nucleus condensation, membrane blebbing, and cellular detachment (4). In contrast, no detectable damage was observed in monolayers treated for 1 h with HBSS alone or HBSS containing mock affinity-enriched preparations from E. coli lysates transformed with the empty vector, pTrcHis A (Fig. 2). Damage to CaCo-2 cell monolayers was readily apparent using all affinity-enriched, alanine-substituted rCPE variants except for D48A and I51A (Fig. 2), suggesting that those two N-terminal CPE residues are critical for cytotoxicity.
FIG. 2.
Morphological damage of CaCo-2 cells by rCPE variants. To screen variant rCPE species for cytotoxicity, confluent CaCo-2 cultures were treated with 2.5 μg of the indicated rCPE species/ml for 60 min at 37°C. Treatment buffer alone was added to cells in the control panel, while buffer containing a mock affinity enrichment preparation of lysates from E. coli cells transformed with the pTrcHis A empty vector was used to treat cells in the vector panel. After treatment, cells were photomicrographed at 130× total magnification.
Plasma membrane permeability alterations induced by the alanine-substituted rCPE variants.
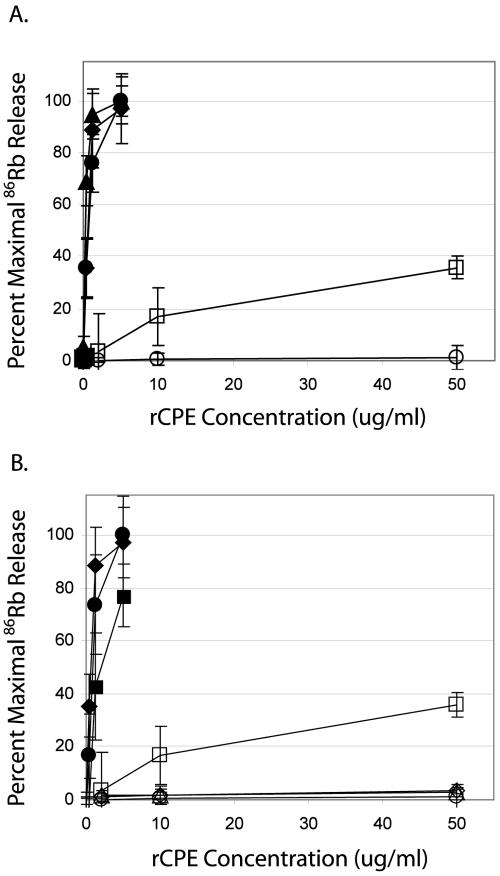
The N-terminal 45-53 region of CPE under examination in this study is required (19) for the plasma membrane permeability alterations responsible for CPE-induced killing of mammalian cells (15, 23, 24, 26, 37). Therefore, the ability of each alanine-substituted rCPE variant to induce plasma membrane permeability changes in CaCo-2 cells was assessed in a 86Rb release assay. Consistent with morphological damage screening results (Fig. 2), D48A and I51A were the only rCPE alanine-substituted variants that exhibited a substantially reduced ability to cause 86Rb release (Table 3; Fig. 3A). Even at 50 μg/ml, D48A elicited no detectable 86Rb release from CaCo-2 cells, while I51A induced only a slight release (∼35% of maximal release) at that very high toxin concentration, i.e., the D48A and I51A variants are >160-fold less active than rCPE in this assay. All remaining alanine variant rCPEs had a <3-fold variation in their toxicity relative to rCPE.
FIG. 3.
86Rb release from CaCo-2 cells induced by rCPE variants. Confluent CaCo-2 cells in 24-well plates were labeled with 4 μCi of 86Rb per well for 4 h and then treated with increasing concentrations of rCPE species. 86Rb released into the culture medium after 15 min of rCPE treatment was collected. To correct for background release, the data were converted to the percentage of maximal 86Rb release (see Materials and Methods). (A) Depicted are the following rCPE species: rCPE (♦), D45A (▴), D48A (○), I51A (□), and L52A (•). (B) 86Rb release experiments with rCPE saturation variants with the following rCPE species: rCPE (♦), D48A (○), D48E (▵), D48N (⋄), I51A (□), I51L (•), and I51V (▪). All data points represent means of three independent experiments, and error bars represent the standard deviations.
Competitive binding abilities of the alanine rCPE variants.
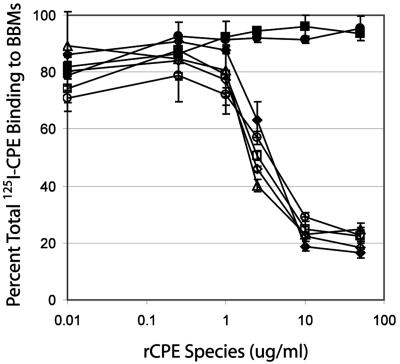
To determine why the D48A and I51A rCPE variants exhibit attenuated cytotoxicity, the ability of each alanine-substituted rCPE variant to perform early steps in CPE action was assessed. Since CPE action initiates with binding of the enterotoxin to a cellular surface receptor(s) and previous structure-function analyses had mapped CPE receptor binding activity to its C-terminal domain (19), the introduction of alanine substitutions in the N-terminal 45-54 region was not expected to affect binding activity. In a well-established BBM competitive binding assay (18, 19) that measures the ability of an rCPE species to inhibit binding of 125I-labeled native CPE to BBMs, this expectation was confirmed, i.e., all alanine-substituted rCPE variants generated in this study had similar abilities as rCPE to inhibit [125I]CPE binding to BBMs (see Table 3 for summary data with all variants and Fig. 4 for representative data with selected rCPE species of particular interest). This assay was verified by the binding-deficient random point rCPE variant W226Stop which, as previously noted (18), failed to inhibit [125I]CPE binding to BBMs (Fig. 4). In addition to their competitive binding properties in the BBM assay, the D48A and I51A rCPE variants also both inhibited [125I]CPE binding to CaCo-2 cells, similar to rCPE (data not shown). Collectively, these binding activity results support previous rCPE or CPE studies indicating that the N terminus of the enterotoxin is not involved in binding (12-14, 19, 25, 28) and imply that the D48 and I51 residues of the enterotoxin are important for some postbinding step in CPE's action.
FIG. 4.
Competitive binding of rCPE variants. BBMs were preincubated with increasing concentrations (0.01 to 50 μg/ml) of each rCPE species before the addition of [125I]CPE (2.5 μg/ml). The amount of [125I]CPE bound after treatment was quantified and converted to the percentage of total binding ([125I]CPE bound to BBMs in absence of a competitor). rCPE species shown are as follows: rCPE (♦), D48A (○), D48E (▴), D48N (⋄), I51A (□), the previously described (18) binding-deficient rCPE random point variant W226Stop (▪), and a mock affinity enrichment preparation with lysates from E. coli cells transformed with the pTrcHis A empty vector (•). Data points represent the average of duplicate independent experiments run in triplicate, and error bars represent the standard deviations.
Small complex formation by alanine-substituted rCPE variants.
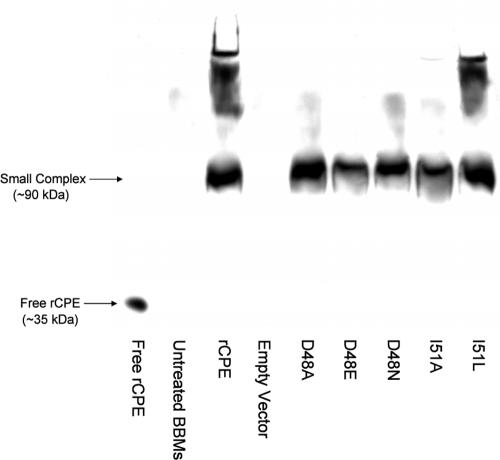
Upon binding to sensitive cells, CPE localizes in an SDS-sensitive complex of ∼90 kDa (41, 42) that likely represents CPE bound to its receptor(s). It was expected, therefore, that the abilities of the alanine rCPE variants to form the small complex should parallel their binding activities (Fig. 4; Table 3). When small complex formation by each rCPE variant species was assessed by treating BBMs with each rCPE species at 4°C, all rCPE alanine variants were, as predicted, able to form the small complex (see Table 3 for summary results with all variants and Fig. 5 for a representative experiment). Although these experimental manipulations were performed at 4°C, which is inhibitory for large complex formation (27), limited amounts of large complex formation were still observed by using active rCPE species (Fig. 5; note the immunoreactive material migrating above the small complex in the rCPE lane of the gel). No large complex formation was apparent in lanes containing extracts from BBMs treated with the D48A or I51A rCPE variants (Fig. 5).
FIG. 5.
Small complex formation by rCPE variants. BBMs were treated at 4°C with 5 μg of rCPE species per sample; Triton X-100 extracts of these samples were then separated on a 6% acrylamide native gel. After electrotransfer, small complex formation by each rCPE variant was evaluated by Western immunoblotting with rabbit polyclonal anti-CPE antiserum preabsorbed with BBMs. The migration of 20 ng of rCPE (∼39 kDa) is denoted by the arrow labeled free rCPE, while the migration of the classic ∼90-kDa small complex is depicted with the small complex arrow. The untreated lane represents BBMs treated with PBS only, while the empty vector lane corresponds to BBMs treated with a mock affinity enrichment preparation with lysates from E. coli cells transformed with the pTrcHis A empty vector.
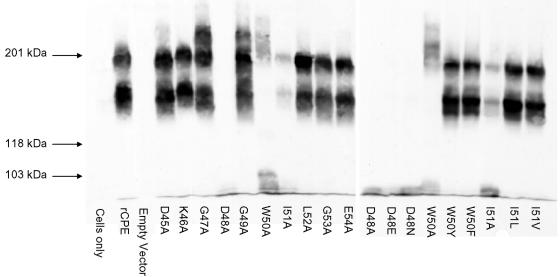
Large complex formation by rCPE alanine variants.
At 37°C, the small CPE complex becomes localized in two large SDS-resistant complexes (35, 43), including an ∼155-kDa complex (or perhaps higher-order aggregates thereof) that likely represents a pore or channel responsible for the membrane permeability alterations that kill CPE-treated cells. When experiments were performed to analyze the large-complex-forming ability of each alanine rCPE variant prepared in this study, several intriguing observations were made (Fig. 6). The most important result was that the completely inactive D48A variant formed no detectable amount of SDS-resistant, high-Mr material, while the highly attenuated I51A variant formed only small amounts of the classic ∼155- and ∼200-kDa CPE complexes, i.e., impaired large complex formation by these two variants correlated with their attenuated cytotoxic activities, as measured by morphological damage (Fig. 2) or 86Rb release (Fig. 3). In addition, Fig. 6 results indicate that all rCPE alanine variants eliciting 86Rb release levels comparable to that of rCPE (Table 3; Fig. 3A) could also form SDS-resistant, high-Mr material. However, the high-Mr material formed by several toxic rCPE alanine variants differed qualitatively and quantitatively from the classical ∼155- and ∼200-kDa CPE complexes formed by rCPE. For example, rCPE variants G47A and G49A produced appreciable levels of the classical ∼155- and ∼200-kDa CPE complexes, along with some even-higher-Mr immunoreactive material. In contrast, the W50A rCPE variant, which was only slightly attenuated in 86Rb release experiments (Table 3), made little ∼155-kDa CPE complex but did form higher-Mr immunoreactive material roughly similar in size to the ∼200-kDa large complex produced by rCPE. In addition, substantially greater amounts of immunoreactive material were present at, or near, the dye front of lanes loaded with SDS extracts of CaCo-2 cells treated with the W50A rCPE variant compared to that with rCPE (or other toxic rCPE alanine-substituted variants).
FIG. 6.
Large complex formation by rCPE variants. Confluent CaCo-2 cells harvested from 100-mm culture dishes were treated with 2.5 μg of each rCPE species/ml. After 45 min, the treated cells were extracted with 1% SDS and loaded onto a 4% acrylamide SDS-PAGE gel. Complexes were then electrotransferred and Western immunoblotted with rabbit polyclonal anti-CPE serum. “Cells only” and “empty vector” lanes represent, respectively, CaCo-2 cells treated with either HBSS or mock affinity enrichment preparations of lysates from E. coli cells transformed with pTrcHis A.
Analysis of gross conformational changes in rCPE alanine variants.
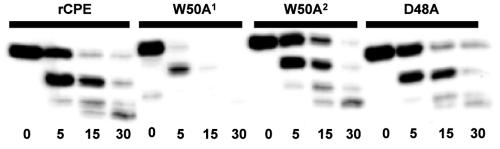
The above results indicated that the D48A and I51A rCPE variants are both highly attenuated due to an impaired ability to form large complexes. To address whether those two rCPE alanine variants lost the ability to form large complexes because of gross conformational changes resulting from their alanine substitution, limited trypsin proteolysis was performed. As described previously (18), the rCPE protein normally exposes only a few of the 24 trypsin digestion sites present in the primary enterotoxin amino acid sequence, while misfolded rCPE variants have more sites exposed and thus are more sensitive to limited digestion with trypsin.
With the exception of an initial affinity enrichment batch of W50A, all rCPE variants prepared in this study displayed trypsin digestion patterns similar to that of rCPE (a representative blot is shown in Fig. 7). These results suggest that the attenuation of the D48A and I51A rCPE variants is specifically due to the loss of the native amino acid at D48 and I51, rather than from gross conformational changes occurring in those two rCPE variants. As mentioned above, the first affinity enrichment preparation of the W50A rCPE variant was nontoxic and displayed a trypsin digestion pattern suggestive of a substantially misfolded protein (Fig. 7, W50A1). Therefore, that inactive batch of W50A served as a control, verifying the ability of the trypsin proteolysis assay (Fig. 7) to detect misfolded rCPE. Note that all subsequent affinity enrichment preparations of W50A displayed a trypsin digestion pattern similar to that of rCPE (Fig. 7, W50A2); those later W50A preparations were also active for receptor binding, small and large complex formation, morphological damage, and 86Rb release (Table 3).
FIG. 7.
Assessment of gross conformational changes in rCPE variants. rCPE species (400 ng) were incubated with trypsin (1 ng) at 25°C for the time periods indicated below each lane. Digestions were stopped by the addition of Laemmli buffer and boiling for 5 min. Samples from each reaction mixture were loaded on 10% acrylamide gels and separated by SDS-PAGE, followed by Western blotting with rabbit anti-CPE polyclonal antibody. The blot depicted in this figure represents a trypsin digestion of rCPE, a nontoxic affinity enrichment preparation of W50A (W50A1), a toxic affinity enrichment preparation of W50A (W50A2), and D48A (see Results for explanation of W50A activity variation).
Saturation mutagenesis of residues important for CPE action.
Alanine-scanning mutagenesis identified CPE residues D48 and I51 as being important for CPE toxicity. In addition, the W50 residue of CPE appears to play some structural role for the protein, given the batch-to-batch variation in protein folding and activity observed with the W50A rCPE variant. Therefore, these three CPE residues were subjected to saturation mutagenesis to determine which of their biochemical properties are important for CPE function. These additional studies involved introducing more conservative substitutions at each of the three amino acids by using the QuikChange kit to produce the rCPE variants D48E, D48N, W50F, W50Y, I51L, and I51V (Table 1). All saturation mutations were confirmed by nucleotide sequencing, and each saturation rCPE variant was expressed in E. coli for subsequent affinity enrichment.
The D48A rCPE variant was the only alanine-substituted rCPE lacking any detectable cytotoxic activity, even using extremely high toxin concentrations (Table 3; Fig. 2, 3A, and 6). In addition, this variant exhibited no gross conformational changes (Fig. 7) and had wild-type binding and small complex activities (Fig. 4 and 5), indicating some property of the D48 residue is specifically important for formation of SDS-resistant, large CPE complexes. To elucidate the property (size or charge) important for toxicity at CPE amino acid 48, the rCPE variants D48E and D48N were engineered and tested for cytotoxicity. These rCPE saturation variants both failed to elicit any morphological damage (Fig. 2) or 86Rb release from CaCo-2 cells, even at 50 μg/ml (Fig. 3B), matching earlier results observed with the D48A rCPE variant. Also resembling the D48A rCPE variant, the D48E and D48N rCPE variants could bind and form the SDS-sensitive small complexes in CaCo-2 cells (Fig. 4 and 5), but both of these variants were unable to form either of the two SDS-resistant large complexes made by rCPE (Fig. 6), explaining their lack of cytotoxicity in the 86Rb release experiments (Fig. 3B).
Activity characterization studies showed that the I51A rCPE variant is highly attenuated but maintains a small amount of cytotoxic activity (Table 3; Fig. 3A and 6). Therefore, conservative mutations were engineered into rCPE at this amino acid residue to determine whether restoring the aliphatic hydrocarbon side chain properties of isoleucine at CPE residue 51 is sufficient to obtain normal toxicity. The resulting variant rCPE species, I51L and I51V, displayed the same cytotoxicity as rCPE in the CaCo-2 cell morphology and 86Rb release assays (Fig. 2 and 3B). Consistent with that observation, both rCPE saturation variants also had unimpaired receptor binding activity (Table 3), could form both the small complex (Table 3) and the classic ∼155- and ∼200-kDa large complexes (Fig. 6), and displayed trypsin digestion patterns similar to that of rCPE, i.e., these variants apparently lack gross conformational changes.
As the W50A rCPE variant displayed some interesting phenotypes that could provide insights into CPE structure, conservative substitutions more closely resembling the tryptophan residue were engineered at CPE amino acid 50. CPE activity characterization revealed that the resulting W50Y and W50F variants are fully active, i.e., they bind, form small and large complexes, and elicit 86Rb release at levels comparable to those of rCPE (Table 3; Fig. 3B). While the W50A rCPE variant formed an atypical large complex species similar in size to the ∼200-kDa complex made by rCPE (Fig. 6), the rCPE saturation variants W50Y and W50F both formed the classic ∼155- and ∼200-kDa large complexes (Fig. 6). Finally, the W50Y and W50F variants consistently exhibited similar trypsin sensitivity as rCPE, indicating they had not undergone gross conformational changes.
DISCUSSION
Previous deletion and random point mutagenesis studies implicated the N-terminal CPE region between D45 and G53 in formation of the large CPE-containing complexes that are responsible for CPE-induced toxicity. Our present alanine-scanning mutagenesis study has now identified several specific amino acid residues in the CPE 45-54 region that are important for large CPE complex formation and toxicity. In particular, two alanine substitution mutants, i.e., D48A and I51A, were found to be completely or strongly (respectively) attenuated for large CPE complex formation and cytotoxicity. The nontoxic phenotype of these two rCPE variants appears specifically attributable to their inability to form the large CPE complexes, since (i) both variants exhibited normal binding and small complex-forming abilities and (ii) neither variant appeared to have undergone gross conformational distortions, based on their trypsin sensitivities. More conservative saturation mutagenesis of the D48 residue of rCPE created two variants, D48E and D48N, that were also completely deficient in large complex formation and toxicity. Neither of those D48 saturation variants appeared to be grossly misfolded, since they exhibited a normal ability to bind to receptors and form the small complex and also exhibited trypsin sensitivity similar to that of rCPE. Those observations suggest that both the size and charge characteristics of CPE residue D48 are important for formation of the large complex and toxicity, strongly suggesting that ionic interactions and spatial constraints of the D48 residue play a role in large complex formation.
Similar saturation mutagenesis of the I51 residue in CPE showed that, in contrast to the sharply attenuated large-complex-forming ability and toxicity of the I51A rCPE variant, I51L and I51V rCPE variants form large complexes and kill CaCo-2 cells similarly to rCPE. These data indicate that the length of the aliphatic side chain is an important feature of I51 and that hydrophobic interactions or steric positioning of the I51 residue are important for toxicity. Further supporting this conclusion is the reduced, but still significant, activity of the I51A rCPE variant, i.e., the size of the nonpolar functional group at CPE amino acid 51 appears roughly proportional to activity.
The G47A and G49A rCPE variants are both active yet display an atypical large complex phenotype (Fig. 6) that includes, besides the classical ∼155- and ∼200-kDa large complexes, formation of additional SDS-resistant material with a higher molecular mass than the ∼200-kDa large complex (Fig. 6). Despite their formation of atypical large complexes, no major differences in other CPE activities (binding, small complex formation, 86Rb release) were detected for either the G47A or G49A variant, indicating that their slightly abnormal large-complex-forming properties had a limited effect on toxicity. The novel immunoreactive material formed by the G47A and G49A rCPE variants could simply represent anomalous aggregation, as both variants displayed (in the absence of any CaCo-2 proteins) an unusual propensity to aggregate on SDS-PAGE gels even after heating and reduction (data not shown). This aggregation tendency may be attributable to the substituted Ala residue in these two variants, restraining (due to the increased freedom of rotation about phi and psi angles) the normal protein flexibility of Gly residues at CPE residues 47 and 49. However, the atypical large complex formation phenotype of G47A and G49A is clearly related to the location of these two glycine residues in CPE, as the G53A rCPE variant created in this study formed only the classical two CPE large complexes. It is also intriguing that the G47 and G49 residues sandwich the D48 residue that was determined in this study to be so functionally important.
As noted in Results, our first W50A rCPE variant preparation was inactive and appeared to be improperly folded, although subsequent preparations of this variant produced active toxin lacking gross conformational changes as measured by limited trypsin proteolysis (Fig. 7). However, even active W50A rCPE variant preparations formed little or no detectable ∼155-kDa complex, although they did produce the atypical high-Mr material also seen with the G47A and G49A rCPE variants (Fig. 6). In addition, substantially greater amounts of immunoreactive material, which could represent dissociated high-Mr CPE complexes, were present at or near the dye front of large-complex gel lanes loaded with extracts of W50A-treated CaCo-2 cells (Fig. 6). If the W50A rCPE variant were structurally sensitive (as noted above), then the large complex formed by this variant might be functional and form a ∼155-kDa complex that is more sensitive to the SDS used for extraction in large-complex experiments. Since the W50Y and W50F saturation variants formed large complexes resembling those of wild-type rCPE (Fig. 6), these observations collectively suggest that an aromatic amino acid is preferred at CPE residue 50 to maintain normal structural integrity.
Prior to beginning this study, we noted the presence of a KGD motif at CPE amino acids 46 to 48 (Fig. 1) and hypothesized that this three-amino-acid sequence might explain large complex formation, since RGD and KGD sequences are known protein binding motifs (2, 31). However, the unimpaired cytotoxic activities observed for rCPE variants K46A and G47A now preclude the CPE amino acids 46 to 48 (as a complete KGD motif) being important for CPE action. A GXXXG sequence also exists in the CPE N-terminal cytotoxicity region, extending from G49 to G53 (Fig. 1). This was interesting, given that GXXXG motifs have been implicated in dimerization of the Helicobacter pylori vacuolating toxin (21, 22). However, the GXXXG motif of the CPE N-terminal region does not appear important for toxicity, since G-to-A mutations did not alter the pore-forming ability of rCPE (Table 3), in contrast to the attenuation of oligomerization and membrane channel formation observed following some G-to-A mutations in VacA (22).
Considering our G47, D48, G49, W50, and I51 rCPE variant data together, it is notable that at least one substitution introduced at each of these five amino acid residues during this study produced an atypical phenotype. While G47A, G49A, and W50A were active rCPE variants, each displayed a large complex phenotype different from rCPE (Fig. 6). As also mentioned, identical amino acid substitutions introduced outside rCPE residues 47 to 51 did not produce these atypical phenotypes. Taken together, these observations suggest that N-terminal CPE residues G47 to I51 represent a critical core sequence important for cytotoxic activity. This conclusion is also fully consistent with previous random point mutagenesis work (18), which produced a G49D rCPE variant that has an inactive phenotype similar to the D48A rCPE variant generated in the present study. The precise role of this core cytotoxicity sequence appears to involve mediation of formation of the ∼155-kDa large complex required for CPE to induce membrane permeability alterations (27). Although CPE shares no protein sequence homology with other bacterial toxins, it does possess several characteristics reminiscent of the β-barrel pore-forming toxin (β-PFT) family (16). These β-PFTs typically have a high β-sheet composition and penetrate lipid bilayers by oligomerizing and collectively inserting their transmembrane stem domains into the cellular membrane. Oligomerization of some β-PFTs involves N-terminal sequences that serve as a “latch” domain, linking two neighboring monomers (e.g., Staphylococcus aureus alpha-hemolysin and LukF hemolysin [30, 36, 38-40]). CPE also has a high β-sheet composition (7, 32) and a β-PFT-like ability to induce membrane permeability changes in sensitive cells (24) and forms pores or channels in artificial membranes (15, 37). Our present data appear consistent with the possibility that the CPE N-terminal core sequence identified in this study acts as an N-terminal latch that mediates the protein-protein interactions needed for membrane permeability alterations.
The insights gained from this study can be applied to a model in which CPE amino acids 47 to 51 act as a latch domain. The size and charge requirement of CPE residue 48, along with the aliphatic hydrocarbon preference at position 51, could permit ionic and/or hydrophobic interactions between the CPE 45-51 putative latch sequence and a latching site on either another CPE molecule or a host membrane protein. The glycines at CPE residues 47 and 49, which flank the critical D48 residue, may provide flexibility that properly frames this latch so it can interact with its binding partner. The latch domain of the S. aureus alpha-hemolysin has been predicted to involve ionic and hydrophobic interactions between the monomers of its oligomeric pore (36), and so it is plausible that the core sequence identified in this study in CPE mediates similar interactions. Disrupting these sequences, or altering their biochemical properties, may negatively affect CPE's interactions with its binding partner, thereby leading to less efficient or less stable large complex formation.
Finally, previous observations from our laboratory indicate that, in intact CaCo-2 monolayers, the ∼155-kDa large complex forms soon after CPE treatment. However, the ∼200-kDa large complex forms much more slowly in that system (34) because CPE needs basolateral exposure to gain access to the tight junction protein occludin, which is a component of the ∼200-kDa CPE complex but not the ∼155-kDa CPE complex. The large-complex experiments performed in this study used isolated CaCo-2 cells that were harvested from a cell culture plate by scraping, thereby disrupting and exposing their tight junctions. This method allowed us to rapidly assess both ∼155-kDa and ∼200-kDa complex formation by our rCPE variants. It is therefore notable that none of the rCPE variants generated in the present study could form the ∼155-kDa CPE complex without forming the ∼200-kDa complex, or vice versa. That observation suggests that the CPE 47-51 latch region is important for formation of both CPE large complexes, which could indicate that either the ∼155-kDa complex formation is a precursor for ∼200-kDa formation or that formation of both CPE large complexes is independent but involves similar protein-protein interactions. This and many other questions remain regarding the CPE structure-function relationship. In particular, it will be informative to learn the positioning of the CPE 47-51 sequence within the three-dimensional structure of the CPE protein. Attempts to solve the CPE protein structure are currently under way.
Acknowledgments
This work was supported by Public Health Service grant AI19844 from the National Institute of Allergy and Infectious Diseases.
We thank Ganes Chakrabarti, Derek J. Fisher, and Sameera Sayeed for their technical and editorial assistance during the preparation of the manuscript.
Editor: J. T. Barbieri
REFERENCES
- 1.Abrahao, C., R. J. Carman, H. Hahn, and O. Liesenfeld. 2001. Similar frequency of detection of Clostridium perfringens enterotoxin and Clostridium difficile toxins in patients with antibiotic-associated diarrhea. Eur. J. Clin. Microbiol. Infect. Dis. 20:676-677. [DOI] [PubMed] [Google Scholar]
- 2.Arnaout, M. A., S. L. Goodman, and J. P. Xiong. 2002. Coming to grips with integrin binding to ligands. Curr. Opin. Cell Biol. 14:641-651. [DOI] [PubMed] [Google Scholar]
- 3.Brett, M. M., J. C. Rodhouse, T. J. Donovan, G. M. Tebbutt, and D. N. Hutchinson. 1992. Detection of Clostridium perfringens and its enterotoxin in cases of sporadic diarrhoea. J. Clin. Pathol. 45:609-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarti, G., X. Zhou, and B. A. McClane. 2003. Death pathways activated in CaCo-2 cells by Clostridium perfringens enterotoxin. Infect. Immun. 71:4260-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czeczulin, J. R., P. C. Hanna, and B. A. McClane. 1993. Cloning, nucleotide sequencing, and expression of the Clostridium perfringens enterotoxin gene in Escherichia coli. Infect. Immun. 61:3429-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujita, K., J. Katahira, Y. Horiguchi, N. Sonoda, M. Furuse, and S. Tsukita. 2000. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 476:258-261. [DOI] [PubMed] [Google Scholar]
- 7.Granum, P. E., and O. Harbitz. 1985. A circular-dichrosim study of the enterotoxin from Clostridium perfringens type A. J. Food Biochem. 9:137-146. [Google Scholar]
- 8.Granum, P. E., and M. Richardson. 1991. Chymotrypsin treatment increases the activity of Clostridium perfringens enterotoxin. Toxicon 29:898-900. [DOI] [PubMed] [Google Scholar]
- 9.Granum, P. E., J. R. Whitaker, and R. Skjelkvale. 1981. Trypsin activation of enterotoxin from Clostridium perfringens type A: fragmentation and some physicochemical properties. Biochim. Biophys. Acta 668:325-332. [DOI] [PubMed] [Google Scholar]
- 10.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 11.Hanna, P. C., and B. A. McClane. 1991. A recombinant C-terminal toxin fragment provides evidence that membrane insertion is important for Clostridium perfringens enterotoxin cytotoxicity. Mol. Microbiol. 5:225-230. [DOI] [PubMed] [Google Scholar]
- 12.Hanna, P. C., T. A. Mietzner, G. K. Schoolnik, and B. A. McClane. 1991. Localization of the receptor-binding region of Clostridium perfringens enterotoxin utilizing cloned toxin fragments and synthetic peptides. The 30 C-terminal amino acids define a functional binding region. J. Biol. Chem. 266:11037-11043. [PubMed] [Google Scholar]
- 13.Hanna, P. C., E. U. Wieckowski, T. A. Mietzner, and B. A. McClane. 1992. Mapping of functional regions of Clostridium perfringens type A enterotoxin. Infect. Immun. 60:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanna, P. C., A. P. Wnek, and B. A. McClane. 1989. Molecular cloning of the 3′ half of the Clostridium perfringens enterotoxin gene and demonstration that this region encodes receptor-binding activity. J. Bacteriol. 171:6815-6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardy, S. P., C. Ritchie, M. C. Allen, R. H. Ashley, and P. E. Granum. 2001. Clostridium perfringens type A enterotoxin forms mepacrine-sensitive pores in pure phospholipid bilayers in the absence of putative receptor proteins. Biochim. Biophys. Acta 1515:38-43. [DOI] [PubMed] [Google Scholar]
- 16.Heuck, A. P., R. K. Tweten, and A. E. Johnson. 2001. Beta-barrel pore-forming toxins: intriguing dimorphic proteins. Biochemistry 40:9065-9073. [DOI] [PubMed] [Google Scholar]
- 17.Katahira, J., N. Inoue, Y. Horiguchi, M. Matsuda, and N. Sugimoto. 1997. Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J. Cell Biol. 136:1239-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokai-Kun, J. F., K. Benton, E. U. Wieckowski, and B. A. McClane. 1999. Identification of a Clostridium perfringens enterotoxin region required for large complex formation and cytotoxicity by random mutagenesis. Infect. Immun. 67:5634-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kokai-Kun, J. F., and B. A. McClane. 1997. Deletion analysis of the Clostridium perfringens enterotoxin. Infect. Immun. 65:1014-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McClain, M. S., P. Cao, and T. L. Cover. 2001. Amino-terminal hydrophobic region of Helicobacter pylori vacuolating cytotoxin (VacA) mediates transmembrane protein dimerization. Infect. Immun. 69:1181-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McClain, M. S., H. Iwamoto, P. Cao, A. D. Vinion-Dubiel, Y. Li, G. Szabo, Z. Shao, and T. L. Cover. 2003. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. J. Biol. Chem. 278:12101-12108. [DOI] [PubMed] [Google Scholar]
- 23.McClane, B. A. 1989. Characterization of calcium involvement in the Clostridium perfringens type A enterotoxin-induced release of 3H-nucleotides from Vero cells. Microb. Pathog. 6:17-28. [DOI] [PubMed] [Google Scholar]
- 24.McClane, B. A. 1994. Clostridium perfringens enterotoxin acts by producing small molecule permeability alterations in plasma membranes. Toxicology 87:43-67. [DOI] [PubMed] [Google Scholar]
- 25.McClane, B. A. 2000. Clostridium perfringens enterotoxin and intestinal tight junctions. Trends Microbiol. 8:145-146. [DOI] [PubMed] [Google Scholar]
- 26.McClane, B. A., and J. L. McDonel. 1980. Characterization of membrane permeability alterations induced in Vero cells by Clostridium perfringens enterotoxin. Biochim. Biophys. Acta 600:974-985. [DOI] [PubMed] [Google Scholar]
- 27.McClane, B. A., and A. P. Wnek. 1990. Studies of Clostridium perfringens enterotoxin action at different temperatures demonstrate a correlation between complex formation and cytotoxicity. Infect. Immun. 58:3109-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mietzner, T. A., J. F. Kokai-Kun, P. C. Hanna, and B. A. McClane. 1992. A conjugated synthetic peptide corresponding to the C-terminal region of Clostridium perfringens type A enterotoxin elicits an enterotoxin-neutralizing antibody response in mice. Infect. Immun. 60:3947-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsen, S. J., L. C. MacKinnon, J. S. Goulding, N. H. Bean, and L. Slutsker. 2000. Surveillance for foodborne disease outbreaks—United States, 1993-1997. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 49:1-62. [PubMed] [Google Scholar]
- 30.Olson, R., H. Nariya, K. Yokota, Y. Kamio, and E. Gouaux. 1999. Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel. Nat. Struct. Biol. 6:134-140. [DOI] [PubMed] [Google Scholar]
- 31.Ruoslahti, E. 1996. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12:697-715. [DOI] [PubMed] [Google Scholar]
- 32.Salinovich, O., W. L. Mattice, and E. W. Blakeney, Jr. 1982. Effects of temperature, pH and detergents on the molecular conformation of the enterotoxin of Clostridium perfringens. Biochim. Biophys. Acta 707:147-153. [DOI] [PubMed] [Google Scholar]
- 33.Sigrist, H., P. Ronner, and G. Semenza. 1975. A hydrophobic form of the small-intestinal sucrase-isomaltase complex. Biochim. Biophys. Acta 406:433-446. [DOI] [PubMed] [Google Scholar]
- 34.Singh, U., L. L. Mitic, E. U. Wieckowski, J. M. Anderson, and B. A. McClane. 2001. Comparative biochemical and immunocytochemical studies reveal differences in the effects of Clostridium perfringens enterotoxin on polarized CaCo-2 cells versus Vero cells. J. Biol. Chem. 276:33402-33412. [DOI] [PubMed] [Google Scholar]
- 35.Singh, U., C. M. Van Itallie, L. L. Mitic, J. M. Anderson, and B. A. McClane. 2000. CaCo-2 cells treated with Clostridium perfringens enterotoxin form multiple large complex species, one of which contains the tight junction protein occludin. J. Biol. Chem. 275:18407-18417. [DOI] [PubMed] [Google Scholar]
- 36.Song, L., M. R. Hobaugh, C. Shustak, S. Cheley, H. Bayley, and J. E. Gouaux. 1996. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science 274:1859-1866. [DOI] [PubMed] [Google Scholar]
- 37.Sugimoto, N., M. Takagi, K. Ozutsumi, S. Harada, and M. Matsuda. 1988. Enterotoxin of Clostridium perfringens type A forms ion-permeable channels in a lipid bilayer membrane. Biochem. Biophys. Res. Commun. 156:551-556. [DOI] [PubMed] [Google Scholar]
- 38.Valeva, A., J. Pongs, S. Bhakdi, and M. Palmer. 1997. Staphylococcal alpha-toxin: the role of the N-terminus in formation of the heptameric pore—a fluorescence study. Biochim. Biophys. Acta 1325:281-286. [DOI] [PubMed] [Google Scholar]
- 39.Walker, B., and H. Bayley. 1995. Key residues for membrane binding, oligomerization, and pore forming activity of staphylococcal alpha-hemolysin identified by cysteine scanning mutagenesis and targeted chemical modification. J. Biol. Chem. 270:23065-23071. [DOI] [PubMed] [Google Scholar]
- 40.Walker, B., M. Krishnasastry, L. Zorn, and H. Bayley. 1992. Assembly of the oligomeric membrane pore formed by staphylococcal alpha-hemolysin examined by truncation mutagenesis. J. Biol. Chem. 267:21782-21786. [PubMed] [Google Scholar]
- 41.Wieckowski, E. U., A. P. Wnek, and B. A. McClane. 1994. Evidence that an approximately 50-kDa mammalian plasma membrane protein with receptor-like properties mediates the amphiphilicity of specifically bound Clostridium perfringens enterotoxin. J. Biol. Chem. 269:10838-10848. [PubMed] [Google Scholar]
- 42.Wnek, A. P., and B. A. McClane. 1983. Identification of a 50,000 Mr protein from rabbit brush border membranes that binds Clostridium perfringens enterotoxin. Biochem. Biophys. Res. Commun. 112:1099-1105. [DOI] [PubMed] [Google Scholar]
- 43.Wnek, A. P., and B. A. McClane. 1989. Preliminary evidence that Clostridium perfringens type A enterotoxin is present in a 160,000-Mr complex in mammalian membranes. Infect. Immun. 57:574-581. [DOI] [PMC free article] [PubMed] [Google Scholar]