Abstract
Although interleukin (IL)-23 receptor (IL-23R) plays an important role in the pathogenesis of multiple cancers, its association with cancer risk is inconsistent across different studies. We therefore conducted a meta-analysis with the aim of resolving the relationship among the 3 common polymorphisms of IL-23R (rs6682925, rs10889677, rs1884444) and cancer risk.
Case-control studies evaluating the association between IL-23R polymorphisms (rs6682925, rs10889677, rs1884444) and cancer risk were searched in the PubMed, Web of Science, and CNKI databases.
Data were included in the meta-analysis if they were from original studies adopting a case-control design investigating the association between IL-23R polymorphisms and risk of any cancer; all cancer cases must have been confirmed by histology or pathology, and controls selected from noncancer individuals. Case-only studies and review papers were excluded.
Odds ratios (ORs) and 95% confidence intervals (CIs) were used to evaluate the relationship of IL-23R polymorphisms (rs6682925, rs10889677, rs1884444) with cancer risk. A random-effects model or fixed-effects model was used depending on the heterogeneity of the data.
Ultimately, 15 studies, involving 8784 cancer patients and 10,321 cancer-free controls, were included in our meta-analysis. In the overall analysis, the rs10889677 polymorphism was associated with breast cancer (BC) under the allelic, homozygous, dominant, and heterozygous models. Rs1884444 polymorphism was relevant to hepatocellular carcinoma (HCC) under the homozygous, recessive, and allelic models. However, no evidence of a relationship between IL-23R polymorphisms (rs6682925, rs10889677, rs1884444) and cancer risk was found in the overall population.
Our meta-analysis provides no evidence supporting a global association of IL-23R polymorphisms (rs6682925, rs10889677, rs1884444) with the risk of cancer. However, rs10889677 may be associated with BC susceptibility and rs1884444 had association with HCC risk. Further large and well-designed studies are warranted to confirm this finding.
INTRODUCTION
Cytokines are critical coordinators of the immune response necessary for resolving bacterial and viral assaults on the immune system. In particular, members of the IL-12 family of cytokines are key players in the regulation of T-cell responses, comprising the only heterodimeric cytokines, including IL-12, IL-23, IL-27, and IL-35. In this family, there is a balanced dichotomy of T-cell regulation, in which IL-12 and IL-23 are positive regulators and IL-27 and IL-35 are negative regulators.1,2IL-12 and IL-23 bind to the β1 receptor of T-cells and natural killer cells via their shared p40 subunit. Together, IL-23R and IL-12R β1 comprise the IL-23R complex in IL-23-responsive cells.3 The IL-23R gene is located on chromosome 1p31 and encodes a subunit of the IL-23 receptor. Interleukin (IL)-23 is a pro-inflammatory cytokine comprised of the IL-12 p40 and IL-23 p19 subunits. It is mainly secreted by macrophages and dendritic cells, and can promote autoimmunity through T-cell-mediated inflammation by affecting the T helper 17 (Th17) cell response.4 Th17 cells are a recently discovered proinflammatory CD4+-effector T-cell population that contribute to pathogen clearance and tissue inflammation by expressing high levels of the proinflammatory cytokine IL-17.5IL-17, which is an inflammatory cytokine, plays an important role in the regulation of leukocyte migration in the inflammatory reaction.6
The novel inflammation pathway IL-23/IL-17 axis has proven to serve as a useful biomarker for renal disease activity and for predicting the response to immunosuppressive treatment.7IL-23R affects the IL23/IL17 axis by increasing the expression and production of IL-17A and IL-17F in Th17 cells.8 In a murine melanoma model, Tang et al9 demonstrated that high-mobility group box 1 stimulated the production of IL-23 in a RAGE-dependent manner. Some studies found that genetic variants of IL-23R may contribute to the pathological development from hepatitis to HCC,10,11 and that hepatitis B virus could induce hepatitis by increasing IL-23 expression in a mannose receptor/endocytosis-dependent or -independent manner, and result in liver damage through the IL-23/IL-17 axis.12 Besides Th17 cells, some innate immunity-like T-cells such as TCR γδ 17 and iNKT17 cells have been found to play a key role in the IL-23/IL-17 pathway and to potentially have a vital function in the development of spondyloarthritis-related pathology.13IL-23 signals work through the IL-23 receptor (IL-23R), which is also comprised of 2 subunits, IL12R1 and IL-23R, and is unique to IL-23.14 Moreover, previous studies have indicated that IL-23R exerts an immunosurveillance function via CD8+ T-cells and accelerates tumor growth.15 These findings suggest that IL-23R may play an important role in cancer development and progression.
Recently, some case-control studies have investigated the association of IL-23R polymorphisms with the risk of cancer, including esophageal squamous cell carcinoma, bladder cancer, acute myeloid leukemia, gastric cancer, ovarian cancer, breast cancer, lung cancer, colorectal cancer, and nasopharyngeal cancer.16–25 However, the results from different studies remain controversial. Wrobel et al26 found a correlation between the IL-17F rs763780 polymorphism and AML, but no relationship between IL-23R and AML was observed. However, Qian et al27 reported that genetic variants of IL-23R may contribute to AML risk. Based on these findings, the effects of IL-23R polymorphisms (rs6682925, rs10889677, rs1884444) have been widely discussed, but no conclusive relationships have been determined. Therefore, we conducted a meta-analysis to achieve a more comprehensive evaluation of the association among 3 IL-23R polymorphisms (rs6682925, rs10889677, rs1884444) with cancer risk.
METHODS
Publication Search
A comprehensive literature search was conducted using the following search terms: “Interleukin-23 receptor” or “IL-23R,” “polymorphism” or “SNP,” “cancer,” and “tumor.” The PubMed, Web of Science, and Chinese National Knowledge Infrastructure (CNKI) databases were searched up to April 1, 2015. Only articles published in English were eligible for inclusion. Furthermore, the reference lists of all eligible articles, including review articles, were also checked to find additional relevant publications. This study was approved by the ethics committee of Xi’an Jiaotong University.
Selection Criteria
The following criteria were used to select eligible studies for further meta-analysis: (1) original studies; (2) case-control design investigating the association between IL-23R polymorphisms and risk of any cancer; and (3) all cancer cases were confirmed by histology or pathology, and the controls were selected from noncancer individuals. Case-only studies and review papers were excluded. If 2 or more studies contained overlapping cases or controls, the study with the largest sample size was included in the meta-analysis.
Data Extraction
Articles were reviewed independently by 2 authors, and any discrepant data were discussed by all authors to reach a consensus. For each included study, the raw data and demographic information, including first author, publication year, country of origin, ethnicity, source of controls, total number of cases and controls, cancer type, and genotypes, were independently extracted. Different ethnic groups were categorized as Caucasian, Asian, and “mixed.”
Data Synthesis
Using the genotype and allele frequencies in cases and controls, we applied crude odds ratios (ORs) with corresponding 95% confidence intervals (CIs) to evaluate the associations between IL-23R (rs6682925, rs10889677, rs1884444) polymorphisms and cancer risk. The Z test was used to test the significance of all pooled ORs and a P value <5% was considered significant.
Five different genetic models were used in the meta-analysis to assess the association: allelic comparison of B versus A, homozygous comparison of BB versus AA, dominant comparison of AB+BB versus AA, recessive comparison of BB versus AA+AB, and heterozygous comparison of AB versus AA (A: the major allele, B: the minor allele). Statistical heterogeneity among studies was assessed with the Q and I2 statistic.28 The meta-regression and stratified analyses were also used to analyze the heterogeneity. Publication bias was evaluated with a funnel plot and further assessed by Egger's linear regression test, and P < 0.05 was considered statistically significant. All statistical analyses were carried out with the software STATA (Version 11.0; Stata Corp, College Station, TX).
RESULTS
Characteristics of the Included Studies
The literature search for this meta-analysis started in January 2010 and ended in April 2015 through primary literature retrieval in the PubMed, Web of Science, and CNKI databases. As shown in Figure 1, a total of 134 studies were identified that evaluated IL-23R polymorphisms (rs6682925, rs10889677, rs1884444), and 41 of these studies were excluded as duplicate publications. According to the guidelines of Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA), after manually screening the titles and abstracts, 51 studies were ultimately excluded. After reading the full texts of the remaining 42 articles, 27 were excluded due to lack of complete necessary data (20 articles) or because of reporting unrelated IL-23 polymorphisms (7 articles). Finally, a total of 15 studies with 8784 cases and 10,321 cancer-free controls were found to meet the inclusion criteria for assessing the influence of the rs6682925, rs10889677, and rs1884444 polymorphisms on cancer risk. All of the included eligible studies were published in English.
FIGURE 1.
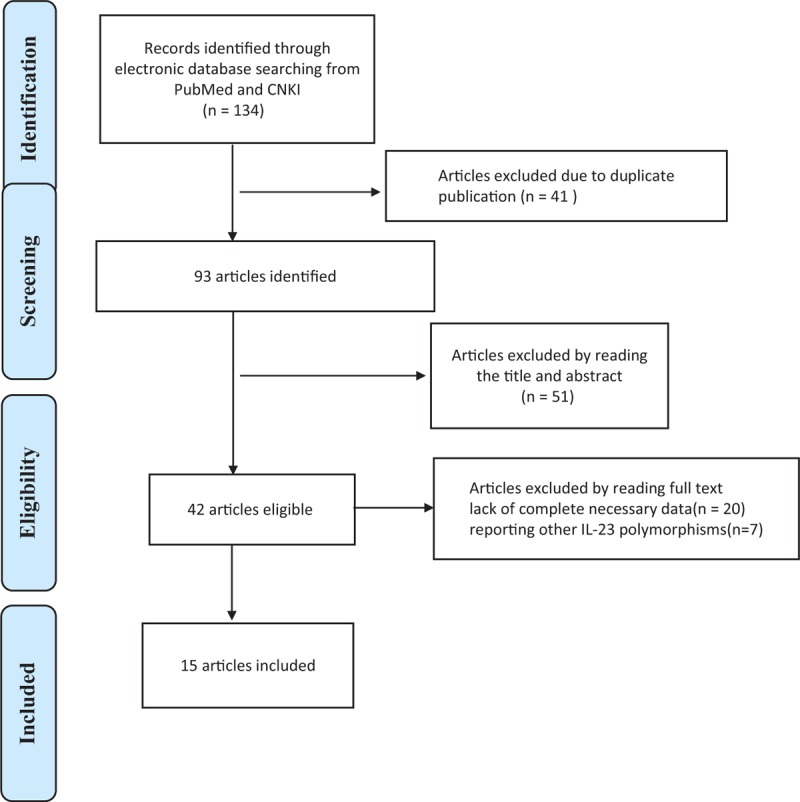
Flow diagram of included studies for the meta-analysis. CNKI = China National Knowledge Infrastructure.
Among the eligible 15 studies, 2 were carried out in Caucasians from Iran and Tunis. Thirteen studies were based on subjects with an Asian background and all were performed in China. All studies were case–control studies, including 2 hepatocellular carcinoma (HCC) studies, 2 colorectal cancer studies, 2 breast cancer (BC) studies, 2 gastric cancer studies, 1 bladder cancer study, 1 esophageal squamous cell carcinoma study, 1 acute myeloid leukemia study, 1 oral cancer study, 1 ovarian cancer, 1 esophageal cancer study, and 1 nonsmall-cell lung cancer. There were 5 hospital-based studies and 10 population-based studies. The main characteristics of the included studies are listed in Tables 1 and 2.
TABLE 1.
Characteristics of the Studies Included in the Meta-Analysis
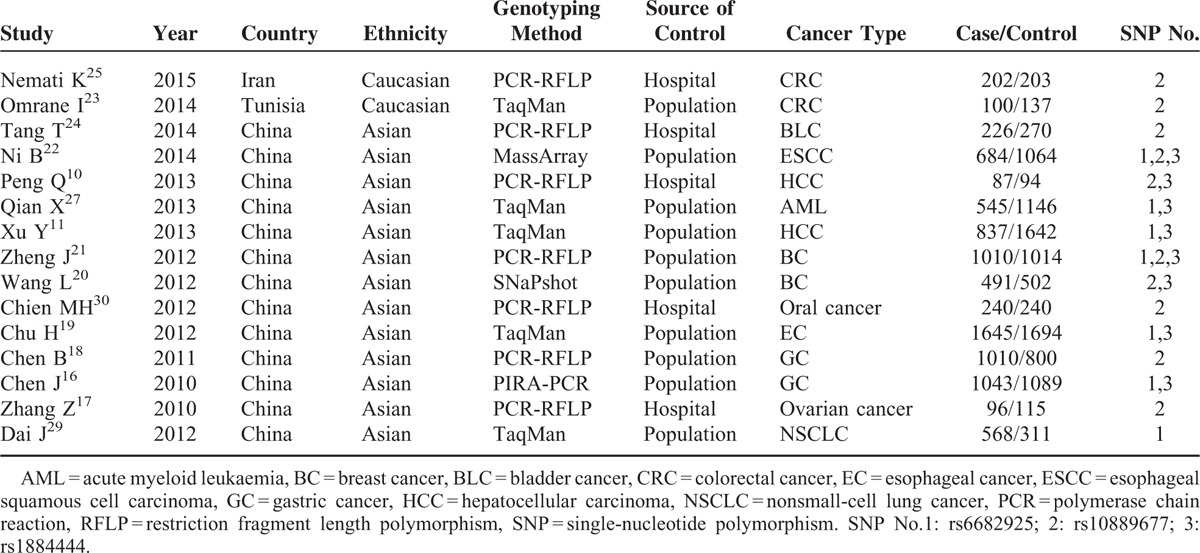
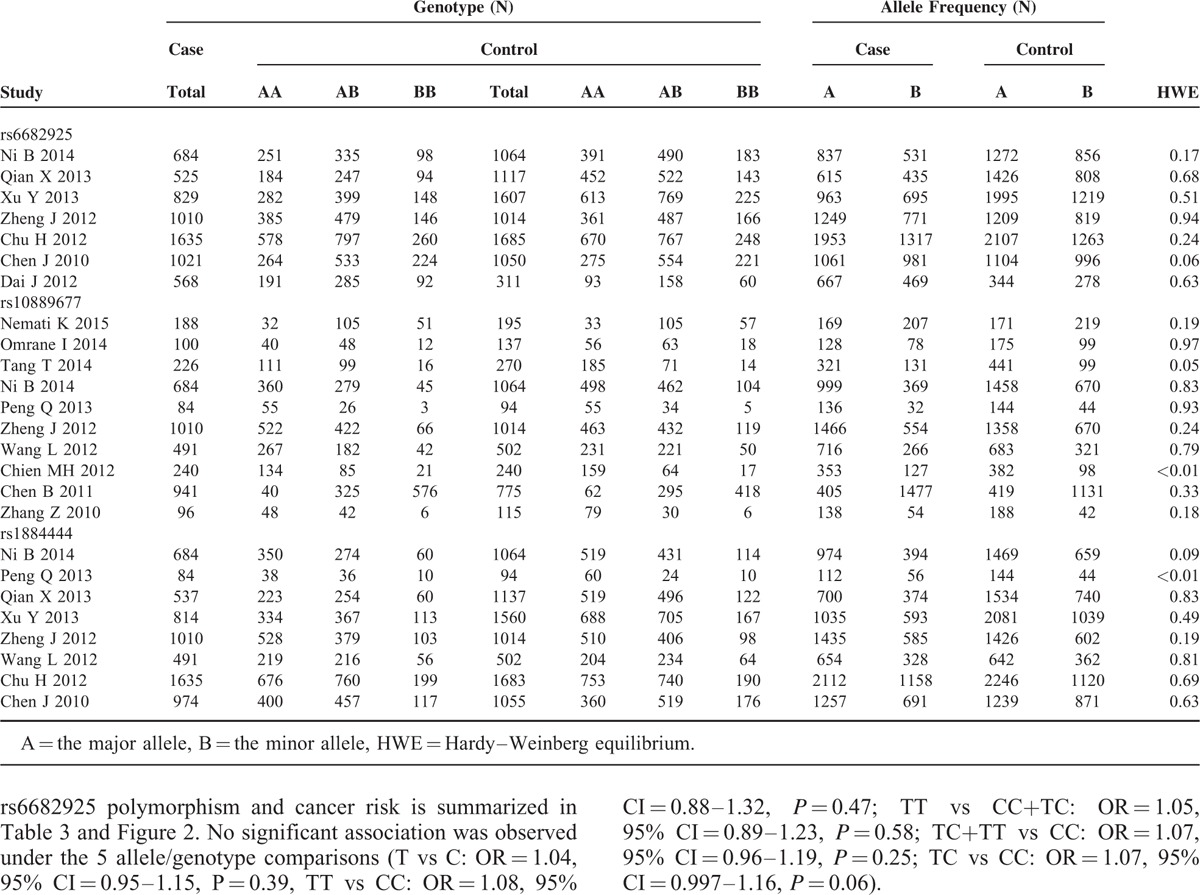
TABLE 2.
IL-23R Polymorphisms Genotype Distribution and Allele Frequency in Cases and Controls
Meta-Analysis of the rs6682925 Polymorphism and Cancer Risk
There were 7 studies with 6272 cases and 7848 controls for rs6682925. Evaluation of the association between the rs6682925 polymorphism and cancer risk is summarized in Table 3 and Figure 2. No significant association was observed under the 5 allele/genotype comparisons (T vs C: OR = 1.04, 95% CI = 0.95–1.15, P = 0.39, TT vs CC: OR = 1.08, 95% CI = 0.88–1.32, P = 0.47; TT vs CC+TC: OR = 1.05, 95% CI = 0.89–1.23, P = 0.58; TC+TT vs CC: OR = 1.07, 95% CI = 0.96–1.19, P = 0.25; TC vs CC: OR = 1.07, 95%CI = 0.997–1.16, P = 0.06).
TABLE 3.
Meta-Analysis Results
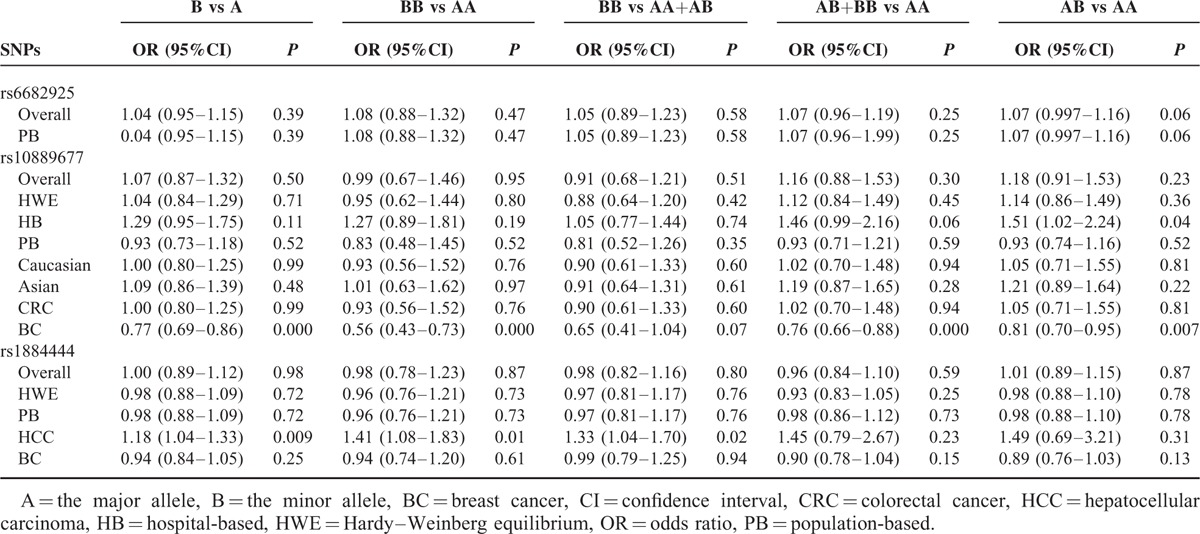
FIGURE 2.
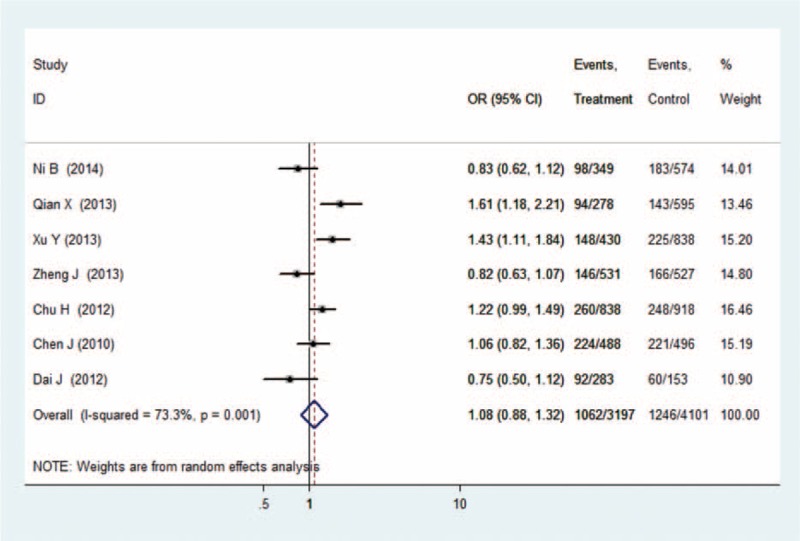
Forest plot of cancer risk related to rs6682925 polymorphism under TT versus CC genetic model. C = the major allele in rs6682925 polymorphism, CI = confidence interval, OR = odds ratio, T = the minor allele in rs6682925 polymorphism.
Meta-Analysis of the rs10889677 Polymorphism and Cancer Risk
There were 10 studies with 4060 cases and 4406 controls evaluating the effect of rs10889677 on cancer risk. As shown in Table 3 and Figure 3, there was no association observed in the overall population under the 5 allele/genotype comparisons. Moreover, after omitting the study which was not according with the Hardy–Weinberg equilibrium (HWE), the results were in accordance with the overall population. And in the subgroup analyses based on ethnicity, we failed to find any significant association in Caucasians or Asians (Table 3). However, in the subgroup analyses by cancer type, a significant association was found between rs10889677 polymorphism and BC risk under the allelic, homozygous, dominant, and heterozygous models (C vs A: OR = 0.77, 95% CI = 0.69–0.86, P = 0.000; CC vs AA: OR = 0.56, 95% CI = 0.43–0.73, P = 0.000; AC+CC vs AA: OR = 0.76, 95% CI = 0.66–0.88, P = 0.000; AC vs AA: OR = 0.81, 95% CI = 0.70–0.95, P = 0.007).
FIGURE 3.
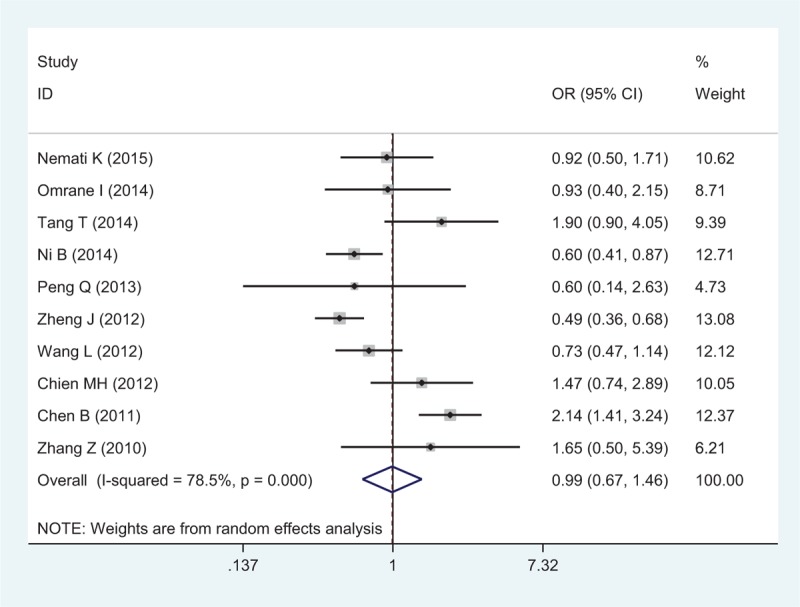
Forest plot of cancer risk related to rs10889677 polymorphism under CC versus AA genetic model. A = the major allele in rs10889677 polymorphism, C = the minor allele in rs10889677 polymorphism, CI = confidence interval, OR = odds ratio.
Meta-Analysis of the rs1884444 Polymorphism and Cancer Risk
Eight studies with 6229 cases and 8109 controls were used to evaluate the relationship between the rs1884444 polymorphism with cancer risk, which is summarized in Table 3 and Figure 4. In the overall analysis, no association was detected under all the genetic models. Further analysis of the studies which were in agreement with HWE also showed no association between rs1884444 polymorphism and cancer risk generally. However, we found that rs1884444 was significantly associated with HCC risk based on the allelic model, homozygous genetic model, and recessive genetic model (T vs G: OR = 1.18, 95% CI = 1.04–1.33, P = 0.009; TT vs GG: OR = 1.41, 95% CI = 1.08–1.83, P = 0.01; TT vs GG+TG: OR = 1.33, 95% CI = 1.04–1.70, P = 0.02). Furthermore, in the stratified analysis according to the source of controls, no association was observed in the hospital-based or population-based group (Table 3).
FIGURE 4.
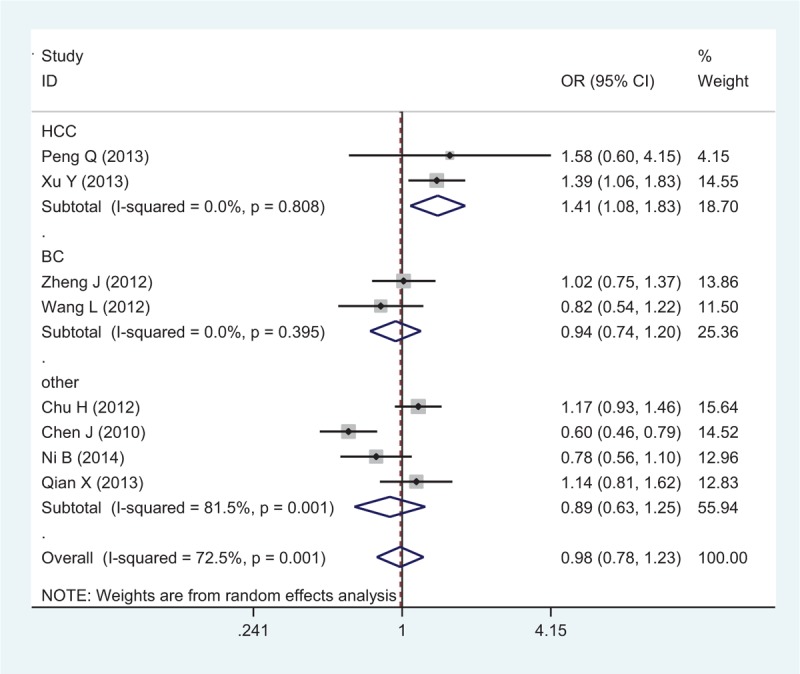
Forest plot of cancer risk related to rs1884444 polymorphism under TT versus GG genetic model. CI = confidence interval, G = the major allele in rrs1884444 polymorphism, OR = odds ratio, T = the minor allele in rs1884444 polymorphism.
Sensitivity Analysis
Sensitivity analyses were performed by sequential removal of each eligible study to assess the influence of each individual study on the pooled OR for the respective comparisons of the rs6682925, rs10889677, and rs1884444 polymorphisms. The omission of any study did not have a significant effect on the results, indicating that the results of this meta-analysis are statistically reliable (Fig. 5).
FIGURE 5.
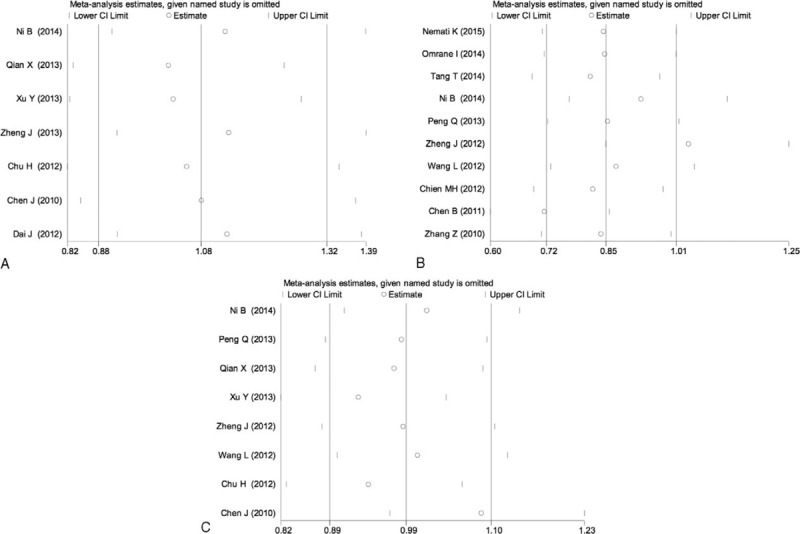
Sensitivity analysis of the heterozygous model in IL-23R polymorphisms: rs6682925 (A), rs10889677 (B), rs1884444 (C) under the homozygous model.
Heterogeneity Analysis and Publication Bias
The Q test and I2 value were used to test the variation in the data caused by heterogeneity. The results of the heterogeneity test are shown in Table 4. A random-effects model was applied when the P value of heterogeneity tests was ≤0.1, and the fixed-effects model was used for P ≥ 0.1. There was heterogeneity among studies in both the overall comparisons and the subgroup analyses for all 3 polymorphisms evaluated (rs6682925, rs10889677, and rs1884444). To explore the potential sources of heterogeneity across studies, we analyze the latent factors by the meta-regression analysis. The results shown no evidence of heterogeneity coming from the source of control (rs6682925: P = 0.30; rs10889677: P = 0.10; rs1884444: P = 0.06), ethnicity (P = 0.30, 0.68, and 0.68, respectively), and year of publication (P = 0.89, 0.36, and 0.14, respectively). Then, we assessed the pooled ORs under all comparisons via subgroup and sensitivity analyses. In the subgroup by race, the heterogeneity of rs10889677 polymorphism was significant in the Asian studies. When stratified by source of control, the heterogeneity of rs1884444 polymorphism in population-based studies was significant in all genetic models.
TABLE 4.
Heterogeneity-Analysis Results
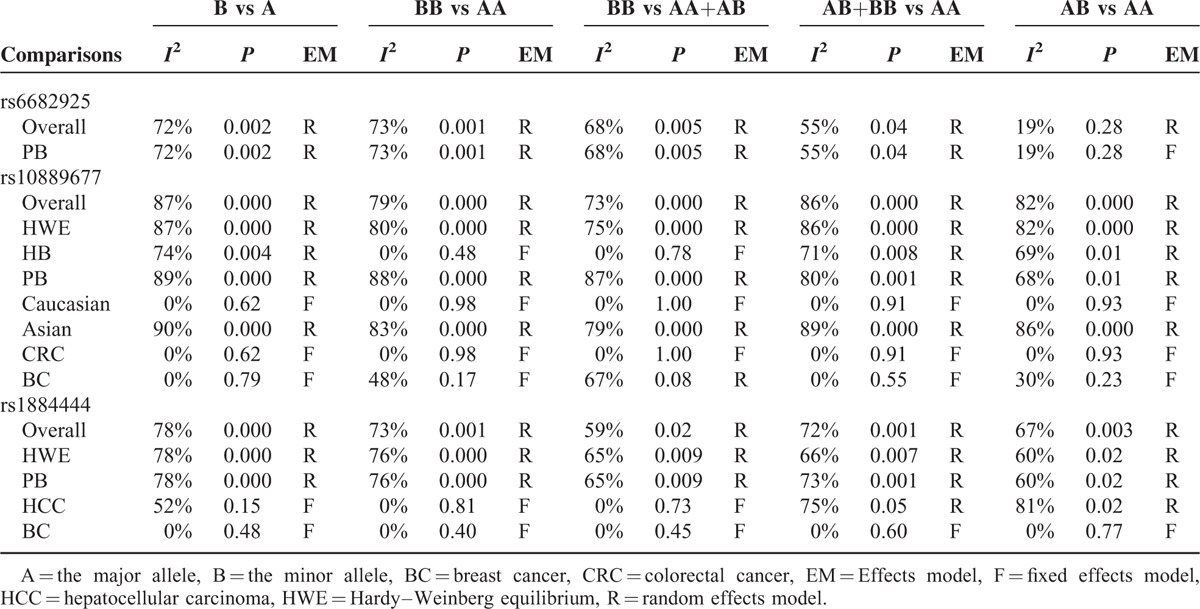
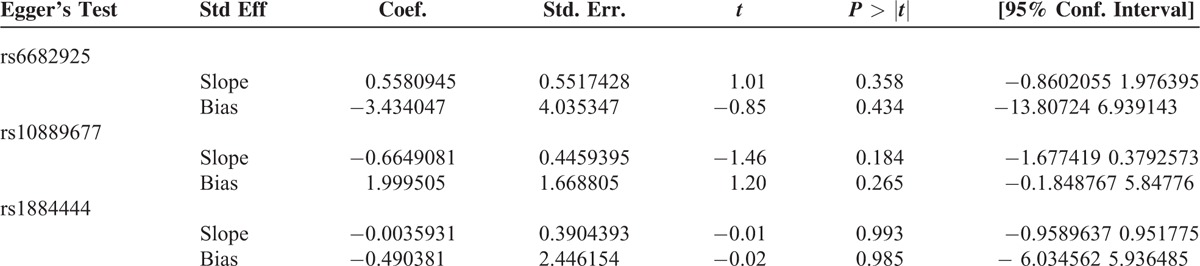
We constructed a funnel plot and performed Egger's test to assess the extent of publication bias in our dataset. As shown in Figure 6, the funnel plots failed to reveal any obvious asymmetry for the 3 polymorphisms in the overall population, and the results of Egger's test revealed no publication bias (Table 5). Therefore, publication bias was not a significant factor affecting the results of this meta-analysis.
FIGURE 6.
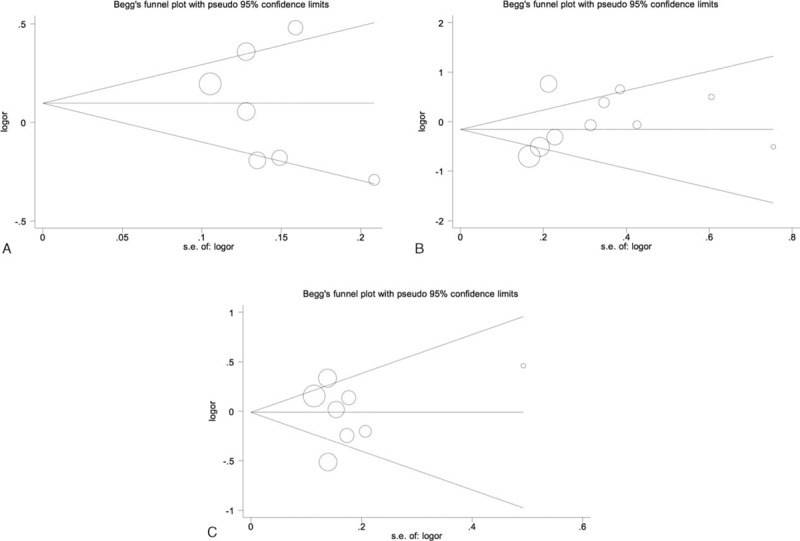
Begg funnel plot for publication bias test of IL-23R polymorphisms: rs6682925 (A), rs10889677 (B), rs1884444 (C) under the homozygous model.
TABLE 5.
Egger's Test for Publication Bias Test of IL-23R Polymorphisms Under the Homozygous Model
DISCUSSION
In the present stratified meta-analysis based on cancer type, there was no significant association between rs6682925, rs10889677, or rs1884444 and cancer risk in the overall population. Our meta-analysis involved 15 independent case-control studies involving 8784 cancer patients and 10,321 cancer-free controls, and the results showed that the rs6682925 polymorphism was not associated with the susceptibility to any cancer and rs10889677 polymorphism may only increase BC susceptibility. Contradictory results have been obtained among individual studies on these associations. Qian et al27 found that rs6682925 TC/CC variant genotypes were associated with an increased risk of acute myeloid leukemia, and this polymorphism was also proposed to have predictive value for nonsmall-cell lung cancer clinical outcomes.29 Individuals with at least 1 variant C allele of the rs10889677 polymorphism showed a higher risk of developing oral cancer and tumor lymph node metastasis compared with patients carrying the wild-type A/A and C/C homozygous genotypes, and IL23R was suggested to play an important role in the susceptibility and prognosis of ovarian cancer in the Chinese population.30 A recent meta-analysis indicated individuals with AC and CC genotype of rs10889677 polymorphism may decrease risk of multiple solid tumors (P < 0.001).31 Zheng et al21 demonstrated rs10889677A > C genotype could affect T-cell proliferation rate, the proportion of Tregs and IL-23R expression, which further influenced cancer susceptibility. Furthermore, IL-23 promoted the expression of IL-17, which is mainly generated by γδ T-cells, thereby accelerating tumor growth through IL-6 induction to activate STAT3 in cancers. The present meta-analysis revealed an increased risk of HCC in carriers of the rs1884444 polymorphism; indeed, significant associations were observed between rs1884444 and HCC risk in 3 genotype models, but not the dominant and heterozygous models, in the overall population.
Though there was heterogeneity among the studies for the 3 polymorphisms, the meta-regression and subgroup analyses indicated that the “source of control,” “ethnicity,” and “cancer type” could explain the heterogeneity. All of the studies included in this meta-analysis met our inclusion criteria and no evidence of publication bias was found. Nevertheless, several limitations of this meta-analysis should be acknowledged. First, the meta-analysis was performed at the study level only, and owing to lack of detailed information from the included studies, we were unable to analyze potential correlative factors such as sex, age, life-style habits, and environmental factors, which are generally considered to contribute to increasing cancer risks. Second, some studies evaluated a specific subtype of cancer, such as the study of Dai et al,29 which was based only on nonsmall-cell lung cancer. Third, we aimed to explore the distinction between Caucasians and Asians, but the subgroup of the 3 polymorphisms involved relatively fewer data in the Caucasian group. Our results did not find any difference between different ethnics, and this conclusion may have some bias. Fourth, meta-analysis is the statistical analysis of large collection of analysis from individual studies for the purpose of integrating the findings, but there were few studies evaluated the association between rs10889677 and bladder cancer, HCC, oral cancer and other cancer, rs1884444 and esophageal squamous cell carcinoma, acute myeloid leukemia, esophageal cancer, and other cancer. So we fail to get the relationship about them. Therefore, further large-scale multicenter studies combined with biochemical and statistical approach are warranted to validate the association between IL-23R and cancer risk.
In conclusion, the current evidence does not support a significant association between the rs6682925, rs10889677, and rs1884444 polymorphisms of IL-23R with cancer susceptibility in the overall population, although rs10889677 appears to influence the susceptibility to BC and rs1884444 may increase the risk of HCC. However, available prospective data are still sparse. In addition, further studies investigating the effect of gene–gene and gene–environment interactions are clearly needed to better understand the association between these 3 polymorphisms and cancer risk.
ACKNOWLEDGMENT
We also thank the Editage for the professional language editing.
Footnotes
Abbreviations: BC = breast cancer, CI = confidence interval, CNKI = China National Knowledge Infrastructure, CRC = colorectal cancer, HCC = hepatocellular carcinoma, HWE = Hardy-Weinberg equilibrium, IL-23R = Interleukin-23 receptor, OR = odds ratio, Th17 = T helper 17.
X-HL and Z-MD contributed equally to this study and share joint first authorship.
Funding: this study was supported by National Natural Science Foundation, China (No. 81471670); China Postdoctoral Science Foundation (No. 2014M560791; 2015T81037); the Fundamental Research Funds for the Central Universities, China (No. 2014qngz-04); and specialized Research Fund of the Second Affiliated Hospital of Xi’an Jiaotong University, China (RC [GG] 201203).
The authors have no conflict of interest to disclose.
REFERENCES
- 1.Gee K, Guzzo C, Che Mat NF, et al. The IL-12 family of cytokines in infection, inflammation and autoimmune disorders. Inflamm Allergy Drug Targets 2009; 8:40–52. [DOI] [PubMed] [Google Scholar]
- 2.Vignali DA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nat Immunol 2012; 13:722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parham C, Chirica M, Timans J, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol 2002; 168:5699–5708. [DOI] [PubMed] [Google Scholar]
- 4.Ahern PP, Izcue A, Maloy KJ, et al. The interleukin-23 axis in intestinal inflammation. Immunol Rev 2008; 226:147–159. [DOI] [PubMed] [Google Scholar]
- 5.Romagnani S. Human Th17 cells. Arthritis Res Ther 2008; 10:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He D, Wu L, Kim HK, et al. CD8+ IL-17-producing T cells are important in effector functions for the elicitation of contact hypersensitivity responses. J Immunol 2006; 177:6852–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zickert A, Amoudruz P, Sundstrom Y, et al. IL-17 and IL-23 in lupus nephritis—association to histopathology and response to treatment. BMC Immunol 2015; 16:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith JA, Colbert RA. Review: the interleukin-23/interleukin-17 axis in spondyloarthritis pathogenesis: Th17 and beyond. Arthritis Rheumatol 2014; 66:231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Q, Li J, Zhu H, et al. Hmgb1-IL-23-IL-17-IL-6-Stat3 axis promotes tumor growth in murine models of melanoma. Mediators Inflamm 2013; 2013:713859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Q, Qin Y, Chen Z, et al. Correlation between interleukin23 receptor gene polymorphisms and risk of hepatitis B virus infection in patients. Mol Med Rep 2013; 8:613–620. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Liu Y, Pan S, et al. IL-23R polymorphisms, HBV infection, and risk of hepatocellular carcinoma in a high-risk Chinese population. J Gastroenterol 2013; 48:125–131. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Zhou J, Zhang B, et al. Hepatitis B virus induces IL-23 production in antigen presenting cells and causes liver damage via the IL-23/IL-17 axis. PLoS Pathog 2013; 9:e1003410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Venken K, Elewaut D. IL-23 responsive innate-like T cells in spondyloarthritis: the less frequent they are, the more vital they appear. Curr Rheumatol Rep 2015; 17:30. [DOI] [PubMed] [Google Scholar]
- 14.Poole EM, Curtin K, Hsu L, et al. Genetic variability in IL23R and risk of colorectal adenoma and colorectal cancer. Cancer Epidemiol 2012; 36:e104–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langowski JL, Zhang X, Wu L, et al. IL-23 promotes tumour incidence and growth. Nature 2006; 442:461–465. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Lu Y, Zhang H, et al. A nonsynonymous polymorphism in IL23R gene is associated with risk of gastric cancer in a Chinese population. Mol Carcinog 2010; 49:862–868. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Zhou B, Zhang J, et al. Association of interleukin-23 receptor gene polymorphisms with risk of ovarian cancer. Cancer Genet Cytogenet 2010; 196:146–152. [DOI] [PubMed] [Google Scholar]
- 18.Chen B, Zeng Z, Xu L, et al. IL23R +2199A/C polymorphism is associated with decreased risk of certain subtypes of gastric cancer in Chinese: a case-control study. Cancer Epidemiol 2011; 35:165–169. [DOI] [PubMed] [Google Scholar]
- 19.Chu H, Cao W, Chen W, et al. Potentially functional polymorphisms in IL-23 receptor and risk of esophageal cancer in a Chinese population. Int J Cancer 2012; 130:1093–1097. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, Liu W, Jiang W, et al. A miRNA binding site single-nucleotide polymorphism in the 3’-UTR region of the IL23R gene is associated with breast cancer. PLoS One 2012; 7:e49823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng J, Jiang L, Zhang L, et al. Functional genetic variations in the IL-23 receptor gene are associated with risk of breast, lung and nasopharyngeal cancer in Chinese populations. Carcinogenesis 2012; 33:2409–2416. [DOI] [PubMed] [Google Scholar]
- 22.Ni B, Chen S, Xie H, et al. Functional polymorphisms in interleukin-23 receptor and susceptibility to esophageal squamous cell carcinoma in Chinese population. PLoS One 2014; 9:e89111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omrane I, Baroudi O, Bougatef K, et al. Significant association between IL23R and IL17F polymorphisms and clinical features of colorectal cancer. Immunol Lett 2014; 158:189–194. [DOI] [PubMed] [Google Scholar]
- 24.Tang T, Xue H, Cui S, et al. Association of interleukin-23 receptor gene polymorphisms with risk of bladder cancer in Chinese. Fam Cancer 2014; 13:619–623. [DOI] [PubMed] [Google Scholar]
- 25.Nemati K, Golmoghaddam H, Hosseini SV, et al. Interleukin-17FT7488 allele is associated with a decreased risk of colorectal cancer and tumor progression. Gene 2015; 561:88–94. [DOI] [PubMed] [Google Scholar]
- 26.Wrobel T, Gebura K, Wysoczanska B, et al. IL-17F gene polymorphism is associated with susceptibility to acute myeloid leukemia. J Cancer Res Clin Oncol 2014; 140:1551–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian X, Cao S, Yang G, et al. Potentially functional polymorphism in IL-23 receptor and risk of acute myeloid leukemia in a Chinese population. PLoS One 2013; 8:e55473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neupane B, Beyene J. Multivariate meta-analysis of genetic association studies: a simulation study. PLoS One 2015; 10:e0133243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai J, Hu Z, Dong J, et al. Host immune gene polymorphisms were associated with the prognosis of non-small-cell lung cancer in Chinese. Int J Cancer 2012; 130:671–676. [DOI] [PubMed] [Google Scholar]
- 30.Chien MH, Hsin CH, Chou LS, et al. Interleukin-23 receptor polymorphism as a risk factor for oral cancer susceptibility. Head Neck 2012; 34:551–556. [DOI] [PubMed] [Google Scholar]
- 31.Zhou S, Ruan Y, Yu H, et al. Functional IL-23R rs10889677 genetic polymorphism and risk of multiple solid tumors: a meta-analysis. PLoS One 2013; 8:e80627. [DOI] [PMC free article] [PubMed] [Google Scholar]


