Abstract
Influenza B viral infection is of great importance, but the epidemiological and phylogenetic characteristics of influenza B infection in severe acute respiratory infection (SARI) cases are still unclear.
The clinical information of 2816 SARI cases and 467,737 influenza-like illness (ILI) cases in Beijing area from September 2014 to April 2015 were collected and analyzed. Among them, 91 influenza B viruses isolated from SARI cases were sequenced.
The overall yield rate of influenza A/B infection was 14.21% and 27.77% in sampled SARI and ILI cases, respectively. Compared with influenza A infection, the frequency of influenza B infection in SARI cases was higher in younger patients. Phylogenetic analysis suggested that most tested hemagglutination genes belonged to Yamagata lineage Clade 3, which were similar with current circulating viruses but different with 2014 to 2015 influenza season vaccine strain (Clade 2). Importantly, HA-Y3/NA-V4 intralineage reassorting was identified in Beijing area for the first time, which can act as a possible risk factor of SARIs.
The influenza activity and virus types/subtypes/lineages among SARI patients were well correlated with that of ILI cases. Furthermore, the potential risk of reassorted influenza B virus infection should not be overlooked.
INTRODUCTION
Influenza viruses are negative-sense, single-stranded RNA viruses, which can be divided into 3 types (influenza A, B, and C) based on the antigenic specificity of the nucleoprotein and matrix protein.1,2 For influenza B, 2 antigenically and genetically distinct lineages, defined by the reference strains B/Victoria/2/87 (Victoria) and B/Yamagata/16/88 (Yamagata), have been identified since 1980s.3 Both lineages are slowly evolving and cocirculate globally, causing outbreaks yearly.4
Although influenza B infection is less frequent than influenza A infection on a worldwide scale, the disease burden of influenza B infection cannot be overlooked, particularly in Asia. More than half of all influenza-associated mortality was associated with influenza B in temperate and subtropical cities in China from 2003 to 2008.5 During 2010 to 2012, 0.058% deaths in Southern China were caused by influenza B, which was higher than influenza A/H3N2 or A/H1N1 09 pdm.6 Influenza B virus infections were also responsible for approximately one-quarter of influenza-associated hospitalization in Hong Kong within the same period.7 Therefore, the disease bundle of influenza B virus highlights the need for continuous surveillance in particular to severe cases, to predict the potential outbreak of pandemics and design effective vaccination strategy.
A regional surveillance for severe acute respiratory infection (SARI) has been established by Beijing Center for Disease Prevention and Control since July 2014. According to the World Health Organization's recommendation, this system is designed to monitor the respiratory pathogens infections among inpatients. In total, 11 sentinel hospitals in 11 districts around Beijing area were involved in this program.
So far, the epidemiological and phylogenetic characteristics of influenza B infection in SARI cases were still unclear: the regions which run well-designed SARI surveillance system were limited worldwide. Moreover, prior studies mainly focused on the demography and clinical features, instead of virological characteristics of laboratory-confirmed influenza (LCI) infections.8–11 Therefore, the objective of this retrospective study is to identify epidemiological and phylogenetic characteristics of influenza B infection in SARI cases. Influenza-like illness (ILI) was also enrolled and assessed, as a mild control group in this study.
METHODS
Specimens and Information Collection
Ethical approval was obtained from the institutional review board and human research ethics committee of the Beijing Center for Disease Prevention and Control. Written informed consents were obtained from each participant. This retrospective study included patients with SARI and ILI in Beijing area from September 2014 to April 2015.
The surveillance for SARI included 11 inpatient departments in local hospitals located in urban and suburban districts of Beijing area (Figure 1). The enrolment criteria for SARI cases included: inpatients with a temperature greater than 38 °C and cough; onset of clinical symptoms within 10 days.12 We also enrolled ILI cases as mild controls in this study. The sentinel sites included 11 outpatient departments in local hospitals located in urban and suburban districts of Beijing area. The enrolment criteria for ILI cases included: outpatients seeking medical care at the designated sentinel hospitals for ILI, defined as an acute respiratory infection with a temperature greater than 38 °C and cough or sore throat; onset of ILI clinical symptoms within 10 days; and no antiviral treatment applied.12
FIGURE 1.
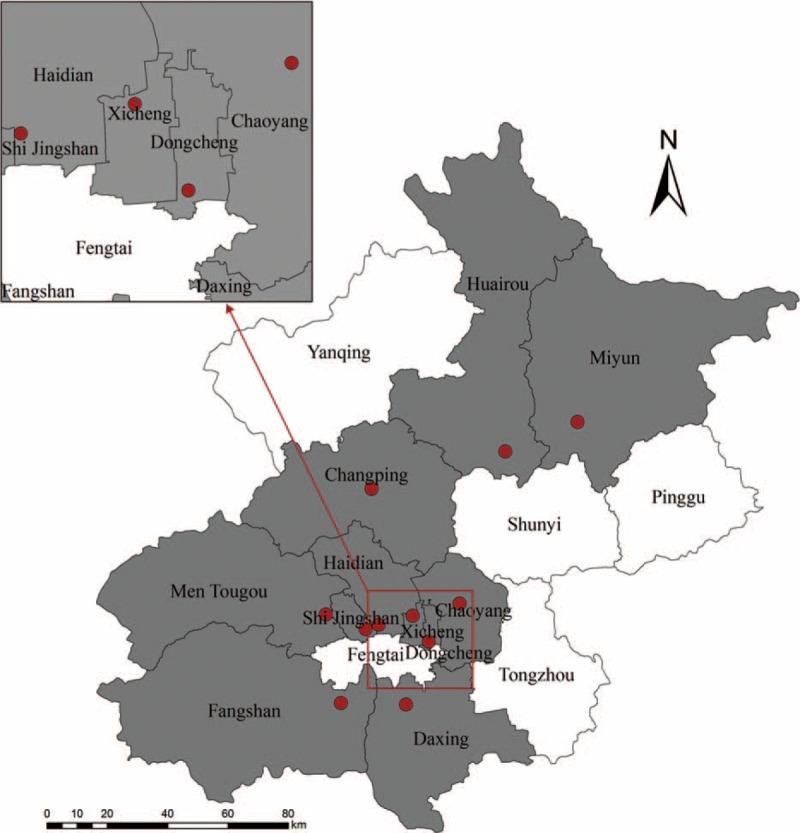
Map of Severe Acute Respiratory Infection sentinel sites in Beijing area. The sentinel hospitals are marked as red dots, and the districts which had at least one sentinel hospitals are in grey.
The numbers of SARI or ILI cases were reported by sentinel site every week. Nasopharyngeal swabs, throat swabs, or sputum were collected from the enrolled patients. Specimens were kept in 3 mL of virus transport medium at 4 °C and tested within 24 hours. Meanwhile, information questionnaires, including the demographic information, vaccine inoculation, etc. were completed by participating physicians at the same time.
Virus Identification and Isolation
All specimens were typed and subtyped by real-time RT-PCR according to the protocol of the Chinese National Influenza Centre.13 Positive specimens were cultured in Madin–Darby canine kidney cells for 4 to 5 days to isolate the influenza virus. Hemagglutination test (HA) and Hemagglutination Inhibition test were then performed to verify the subtype or lineages of the strains.13
Viral RNA Extraction, One-step Real-time Polymerase Chain Reaction and Gene Sequencing
A total of 91 influenza B positive specimens were randomly selected and then enrolled for sequencing. Influenza viral RNA was extracted using QIAmp Viral Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instruction. Reverse transcription and amplification of HA and NA genes were carried out using the one-step reverse transcription-polymerase chain reaction Kit (Qiagen) with primers described previously.14 Polymerase chain reaction products were selected and purified using EZNA Gel Extraction Kit (Omega, Norcross, GA), then sequenced by ABI Prism 3130xl automated sequencer (Applied Biosystems, Foster City, CA).
Phylogenetic Analyses of Various Genotypes of Influenza B Virus
Nucleotide and deduced amino acid sequences of the HA and NA genes were assembled using MEGA software (ver. 6.0.4).15 The nucleotide sequences of the influenza viruses included in this study (91 sequences for HA gene and 81 sequences for NA gene) have been submitted to GenBank (accession numbers: KT383491-KT383662). A total of 77 HA and 66 NA sequences derived from 2014 to 2015 ILI isolates in Northern China and vaccine strains were downloaded from Global Initiative on Sharing All Influenza Data website (www.gisaid.org) and used as global background in this study. The sequences were aligned using the MUSCLE program in MEGA (http://www.megasoftware.net/) with manual adjustment. Maximum likelihood phylogeny trees were inferred by using BEAST (http://beast.bio.ed.ac.uk/) program and employing the HKY substitution model/strict clock model with a gamma-distributed rate parameter.16
Statistical Analysis
Data was analyzed using SPSS 15.0 (SataCorp, College Station, TX) and Prism 5 software (GraphPad, La Jolla, San Diego, CA). Difference between groups were evaluated using Pearson χ2 or Fisher exact test, and odds ratio (ORs) together with 95% confidence intervals were estimated. Meanwhile, demographic and clinical characteristics between different groups were adjusted for age using logistic regression, to calculate the adjusted ORs (aORs) with corresponding 95% confidence interval.
RESULTS
Influenza Virus Activity in Severe Acute Respiratory Infection and Influenza-like Illness Cases From September 2014 to April 2015
A total of 2816 SARI cases and 467,737 ILI cases were reported by sentinel hospitals from September 2014 to April 2015. Among all the reported cases, 2195 (76.72%) SARI cases and 14,006 (2.99%) ILI cases were randomly surveyed, sampled, and tested. The overall yield rate of LCI (influenza A/B) was 14.21% and 27.77% in sampled SARI and ILI cases, respectively. Subtyping test showed that 125 of 2195 (5.69%) tested SARI cases and 1386 of 14,006 (9.90%) tested ILI cases were influenza B positive. The yield rate of LCI in SARI cases were fitted well with that in ILI cases during 2014 to 2015 influenza season (r = 0.899, P < 0.001, by Spearman correlation; Figure 2A). Particularly, the yield rate of influenza B in SARI cases varied from 0% to 52.32% in 2014 to 2015 influenza season. Most Influenza B infection occurred in the latter period of influenza season (February 2015 to April 2015), which showed no difference in SARI and ILI cases (Figure 2B and 2C). Only 6 influenza B Victoria lineage infections were identified in this study, including 5 ILI individuals and 1 SARI case, which suggested the predominated influenza B Yamagata lineage infection in Beijing area.
FIGURE 2.
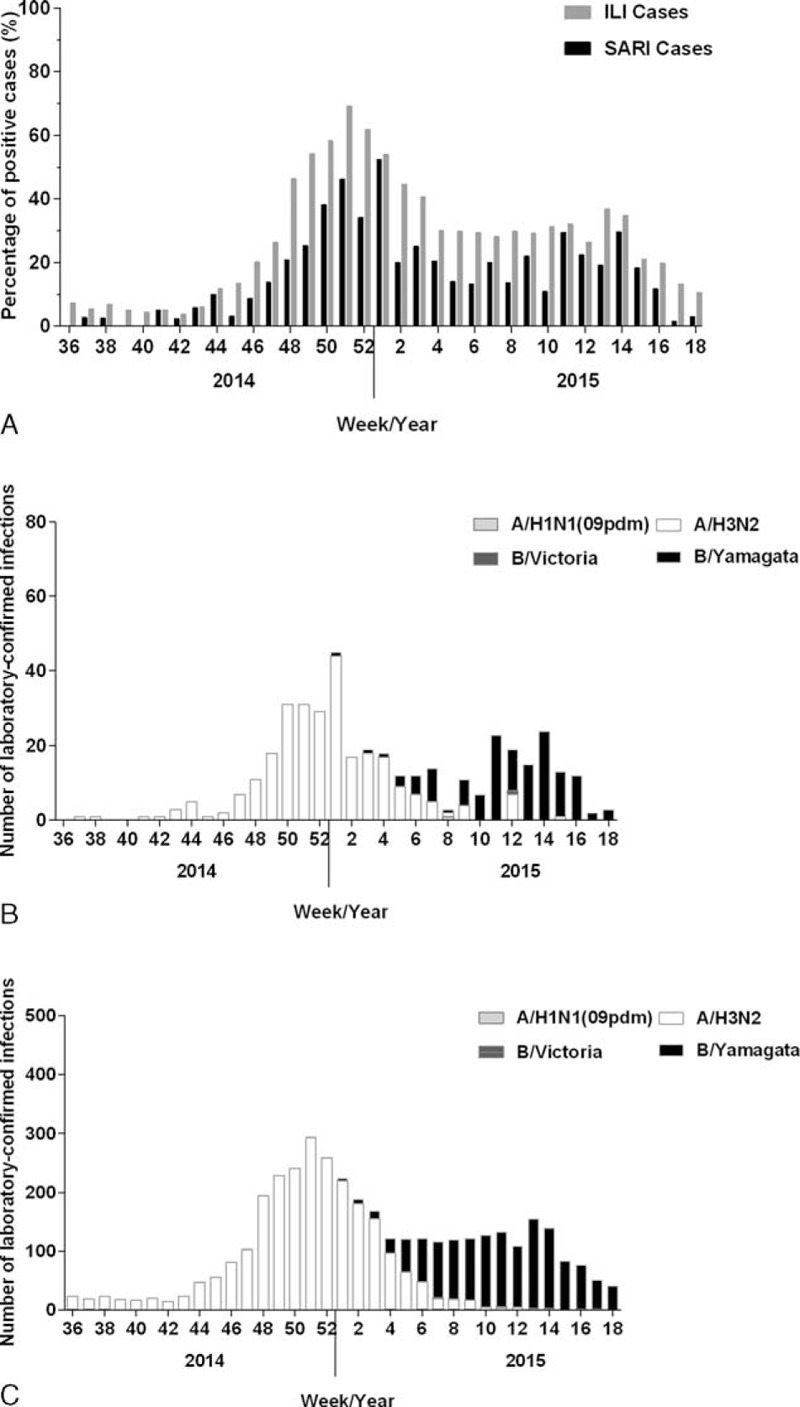
The influenza virus activity in SARI and ILI cases in Beijing areas from September 2014 to April 2015. The data were collected from sentinel hospitals and summarized every week. A, The LCI yield rate in SARI and ILI cases from September 2014 to April 2015. B, Week number of LCI by A/H1N1 (09 dpm) subtype, A/H3N2 subtype, B/Victoria, and B/Yamagata lineages in SARI cases. C, Week number of LCI by A/H1N1 (09 dpm) subtype, A/H3N2 subtype, B/Victoria, and B/Yamagata lineages in ILI cases. SARI = severe acute respiratory infection, ILI = Influenza-like illness, LCI = laboratory-confirmed influenza.
Demographic Characteristics and Vaccine Inoculation of Influenza B Positive Severe Acute Respiratory Infection Cases
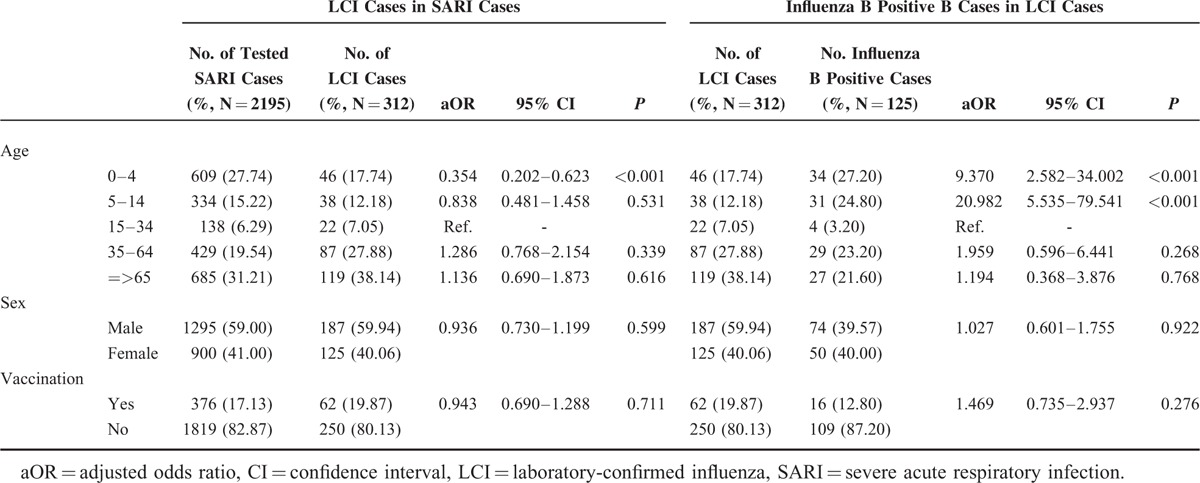
The demographic data and influenza vaccination information of 2195 surveyed SARI cases were collected and analyzed in Table 1. It showed that the yield rate of LCI in SARI patients between 0 and 4 years old was significantly lower than other age groups (aOR 0.354, P < 0.001). We also, however, found that influenza B infection more frequently occurred in younger LCI cases rather than older patients (aOR 9.370, P < 0.001 and aOR 20.982, P < 0.001, for 0–4 age group and 5–14 age group, respectively). No statistically significant difference between SARI and LCI cases or between LCI and influenza B positive cases was observed in sex distribution. Influenza vaccination showed no significant relationship with influenza infection in SARI patients.
TABLE 1.
Demographic and Clinical Characteristics for Severe Acute Respiratory Infection Patients
Phylogentic Analysis of Influenza B Yamagata Lineage From Severe Acute Respiratory Infection Cases
To determine the genetic characteristics of influenza B Yamagata lineage isolated from SARI and ILI cases, we first compared the nucleotide homology and deduced amino acid sequence of 91 isolates from SARI cases with 77 ILI cases and current vaccine strains. It showed that the HA genes in SARI isolates had lower amino acid similarity that ranged from 95.93% to 98.12%, compared with that of the vaccine strain (B/Massachusetts/2/2012) recommended for 2014 to 2015 northern hemisphere influenza season. Although the similarity between HA genes in vaccine strain and HA genes in ILI isolates was higher, with a range from 97.95% to 98.12%. We further analyzed the complete sequences of HA and NA genes of tested isolates, circulating strains and 2012 to 2015 vaccine strains and then evaluated the phylogenetic relationship among them. All the isolates belonged to Clade 3, which was the major circulating Yamagata lineage genotype but different with B/Massachusetts/2/2012 vaccine strain belonged to Clade 2. We, however, noticed that the isolates can be further be distinguished into 2 genotypes namely B/Beijinghuairou/14068/2014-like strains and B/Beijingxicheng/15060/2015-like strains. All of the 14B/Beijinghuairou/14068/2014-like strains isolated from SARI cases and showed significant genetic distances compared with both B/Massachusetts/2/2012 vaccine strain and B/Phuket/3073/2013 vaccine strain (Figure 3).
FIGURE 3.
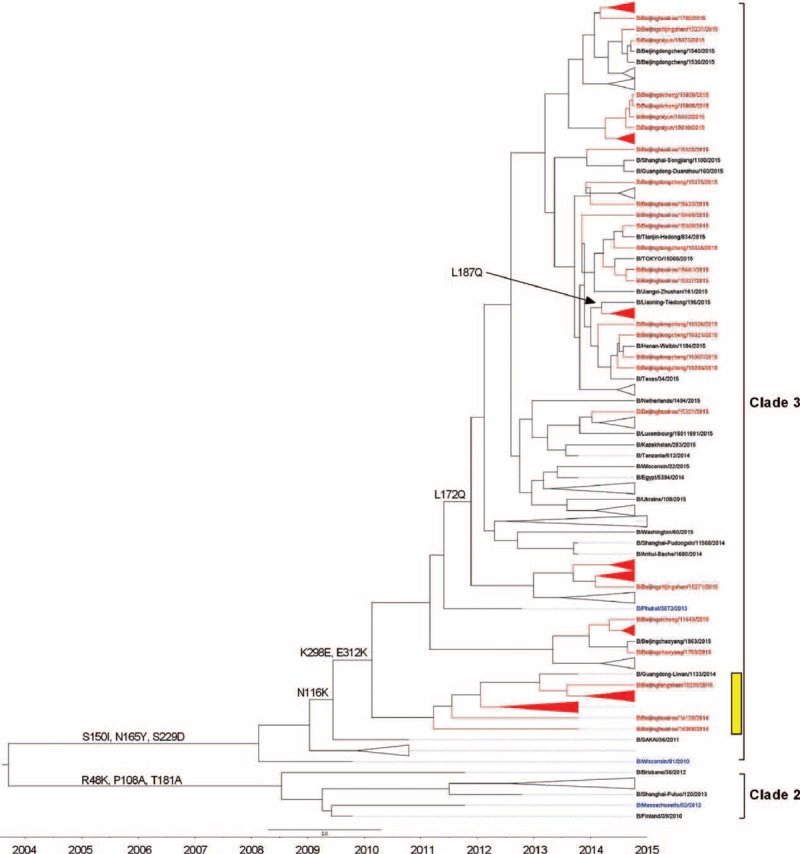
Phylogentic analysis of HA gene of influenza B-Yamagata lineage virus from severe acute respiratory infection and influenza-like illness cases. The phylogenetic trees were constructed using BEAST program and employing the general-time reversal HKY substitution model/strict clock model with a gamma-distributed rate parameter. The viruses isolated from severe acute respiratory infection cases were shown in red, and the vaccine strains were shown in blue. HA-Y3/NA-V4 reassortants were marked in yellow.
The genetic analysis for NA genes shows similar results to HA. The NA genes in these strains isolated from SARI and ILI cases shared nucleotide similarity that ranged from 87.88% to 99.19% and 98.79% to 99.29%, respectively, compared with B/Massachusetts/2/2012 vaccine strain. Furthermore, the NA genes of 18 Yamagata lineage B/Beijinghuairou/14068/2014-like strains, which were isolated from SARI cases, clustered with the Victoria lineage (B/Fujian-luzhou/1272/2008-like, Clade 4), indicating the intra-lineage reassortment (HA-Y3/NA-V4). Except for these 18 isolates, other tested strains showed little phylogenetic distinction: they were closely related to each other and to B/Phuket/3073/2013 vaccine strain (Figure 4).
FIGURE 4.
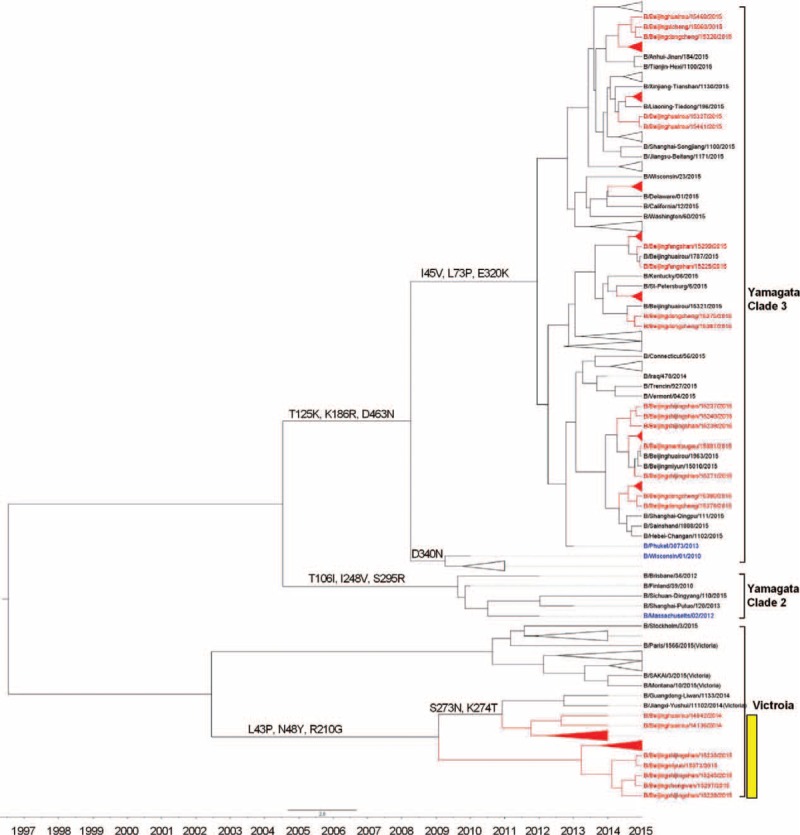
Phylogentic analysis of NA gene of influenza B-Yamagata lineage virus from severe acute respiratory infection and influenza-like illness cases. The phylogenetic trees were constructed using BEAST program and employing the general-time reversal HKY substitution model/strict clock model with a gamma-distributed rate parameter. The viruses isolated from severe acute respiratory infection cases were shown in red, and the vaccine strains were shown in blue. HA-Y3/NA-V4 reassortants were marked in yellow.
Phylogentic Analysis of Influenza B Victoria Lineage From Severe Acute Respiratory Infection Case
In this study, only 1 influenza B Victoria lineage-positive SARI cases and 5 influenza B Victoria lineage-positive ILI case were found. The SARI case was presented by a 3–year-old boy with no influenza vaccination in the last 2 years. All 6 isolates were closely related to B/Brisbane/60/2008-like virus (Clade 1A), with nucleotide similarity that ranging from 98.55% to 99.70%, and 98.14% to 99.65%, for HA gene and NA gene, respectively.
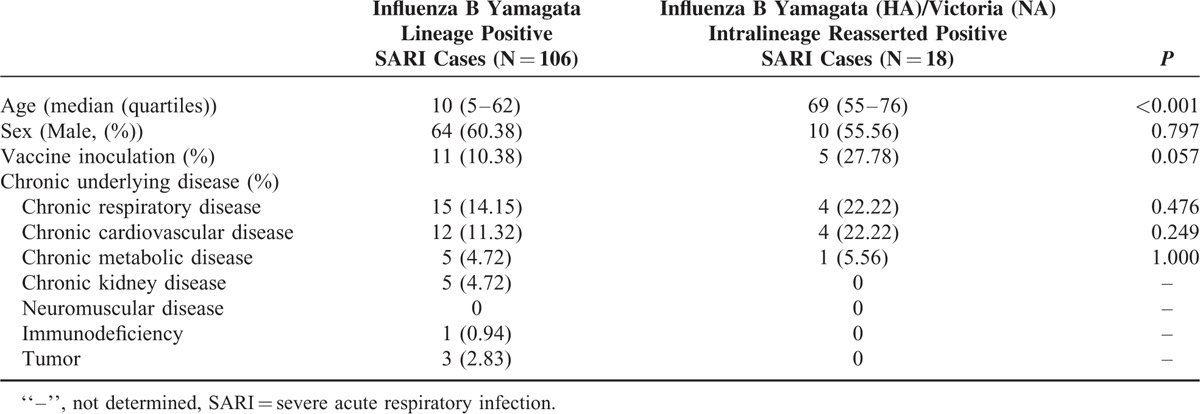
Clinical Feature of Intralineage Reassorted Influenza B Virus Infected Severe Acute Respiratory Infection Cases
Overall 18 of 91 (19.78%) influenza B positive SARI patients were identified with reassortants infections. All of them were patients older than 55 years of age. Although infections caused by none-reassorted influenza B virus were more common in young SARI cases. Moreover, the vaccination rate was higher in intralineage reassorted Influenza B virus infected cases than others (27.78% versus 10.38%). No significant difference was found between these 2 groups either by sex or underlying disease (Table 2).
TABLE 2.
Clinical Feature of Intralineage Reasserted Influenza B Virus Infected Severe Acute Respiratory Infection Cases
DISCUSSION
The study described here analyzed the epidemiology, virology, and clinical data of influenza B infection from the SARI surveillance system in Beijing area from September 2014 to April 2015. Our data showed that the influenza activity among SARI patients was well correlated with that of ILI cases, which peaked in the winter-spring period. Of note, the epidemic peak presented in SARI cases was approximately 1 or 2 weeks later than that in ILIs. For example, the first peak in ILIs in 2014 to 2015 influenza season showed in week 51, 2014, which was mainly caused by influenza A/H3N2 infection. Two weeks later, a peak in SARIs was observed. This pattern was repeated in the second epidemic peak in March 2015, which was caused by influenza B infection. The delay can be explained as the progression of disease from mild ILI to severe respiratory infection. Though the delay existed, the activity data from SARI surveillance is still of value in predicting the rise and drop of influenza infection as did from ILIs.17 Moreover, the predominant type/subtype/lineage identified by the SARI surveillance system was well fitted among that identified by the ILI system. Different from the cocirculating of Victoria and Yamagata lineages in tropical or subtropical areas of Asia,18 Yamagata lineages was dominant, in Beijing area.
Similar to prior reports, our study found that approximately 55.88% SARI patients with LCI were children of age less than 5 or elderly with age greater than 65, which indicated the heaviest burden of severe influenza disease within these 2 age groups.9,19 Further genotyping showed that influenza B virus predominantly infects children and young teenagers under the age of 15 (52.00%), in contrast to influenza A, especially A/H3N2 in this study. These findings are also consisted with previous study.20,21 When it comes to influenza B Yamagata infection specifically, previous studies have shown that patients.21–23 We, however, noticed that approximately 51.61% influenza B Yamagata infected SARI cases were under 15 years of age, which showed significant difference. It is worth noticing that the studies mentioned above only investigated adults,23 or performed in tropical or subtropical areas, where B Yamagata lineage and Victoria lineage were cocirculating.21,22 Our result pointed that the age structure of the patients infected by Yamagata lineage in Yamagata lineage-dominant areas, such as East Asia and Northeast Asia, may be different, and the disease burden of influenza B infection in children and teenagers cannot be overlooked.
An important finding of this study is that we identified the intralineage reassorting in Beijing area for the first time, and have shown that quite a proportion of SARI cases were caused by the reassorted strain. Phylogenetic analysis suggested that influenza B Yamagata lineage in Beijing areas might be introduced from multiple origins: from local circulating, southeast of China, or overseas. Meanwhile, the B Yamagata lineage strains isolated from SARI cases showed more variability compared with isolates derived from mild ILI cases. In this study, 18 intralineage reassorted B/Beijinghuairou/14068/2014-like strains were identified from samples of SARI cases. For these strains, the HA gene was probably generated from B/Nanjing/1338/2011-like strains, reassorting with the NA gene derived from B/Sydney/3/2009-like strains. Molecular clock analysis made it clear that this HA-Y3/NA-V4 reassorting should not be completed earlier than 2011. Such Yamagata (HA)/Victoria (NA) intralineage reassorting was observed in Beijing area for the first time, and also rare in previous reports.21,23
It is notable that reassorted strains contributed nearly one-fifth of SARI cases in this study. All of the cases showed a unique cluster in elderly people, and no significant difference in underlying diseases from controls. In contrast, no such reassorting infection was reported in ILI cases in Northern China yet. Thus, the reassorted strains infection may be a risk factor of SARI. Owing to the limited number of sentinel hospitals and sequenced strains, following studies on distribution and evaluation, however, is needed.
The influenza vaccine has been widely recognized for its effectiveness to prevent pandemics and seasonal epidemics.24 Although 2 factors should be assessed before evaluating the vaccine effectiveness (VE) in large population: the rate of influenza vaccine inoculation and the antigenic difference between vaccine strains and circulating strains.19,25 The rate of influenza vaccine inoculation in Beijing area should be the highest in China, because the city provides free vaccination for citizens over 60 years of age and subsidizing half of the cost of the vaccination for primary and high school students, since 2007.26 In this study, the vaccination with seasonal influenza vaccine in the prior year was approximately 17.13% in all SARI cases and approximately 19.87% in SARI cases with LCI. This rate was comparable with previous study, which showed the coverage rates of influenza vaccination among Beijing residents aged above 18 year old was approximately 18.60% in 2010,26 but was considerably higher than that in the national wide (1.1%).9 We proposed that the inoculation rate of SARI patients with influenza B infection should be lower than those without infection, if the VE run well. Indeed, the inoculation rate was slightly lower in influenza B positive SARI cases, though no significant difference was observed (12.80% versus 16.67%, P = 0.097, by χ2 test).
The antigenic and genetic difference between vaccine strains and current local strains is another main concern for evaluating VE. Influenza trivalent vaccine recommended by WHO that were used in Northern Hemisphere from 2014 to 2015 influenza season contained a component of the Yamagata lineage B/Massachusetts/2/2012-like virus.27 Unfortunately, we found some mismatch between circulating Yamagata lineage strains and vaccine strains. All of the isolates in this study were identified as Clade 3 strains, whereas B/Massachusetts/2/2012-like vaccine strain belongs to Clade 2, which means a limited protective effect against circulating strains in the vaccinated population.28 Another concern is that trivalent vaccine did not provide protection against influenza B Victoria lineage. In fact, the limit coverage of vaccine strains had been discussed in areas where Victoria and Yamagata lineages cocirculated,29 it did not become an issue for Beijing areas in the past years. The protective effect of the trivalent vaccines, however, needs to be reassessed, because of its large genetic distance with Yamagata (HA)/Victoria (NA) intralineage reassorted strains identified in this study. We showed that the SARI patients with intralineage reassorted Influenza B have a higher vaccination rate compared with other influenza B virus infected SARI cases (27.78% versus 10.38%), indicating the VE against reassorted B infection is limited. To resolve this dilemma, WHO updated the influenza vaccines on February 2015: B/Massachusetts/2/2012-like strain (Clade 2) was replaced by B/Phuket/3073/2013-like strain (Clade 3), and B/Brisbane/60/2008-like Victoria lineage strain was included.19 This update coincided with our data, because most isolates showed high similarity and homology with B/Phuket/3073/2013-like strain and Victoria lineage vaccine should share certain antigenicity with intralineage reassorted strains.
Acknowledgements
We are grateful for the valuable contribution of sentinel hospitals and local Centers for Disease Prevention and Control. We are also grateful to Oh DY (Victorian Infectious Diseases Reference Laboratory, Melbourne, Australia) for his helpful advice.
Footnotes
Abbreviations: ILI = Influenza-like illness, LCI = laboratory-confirmed influenza, SARI = Severe Acute Respiratory Infection.
This work was supported by the Capital Health Research and Development of Special (2014–1–1011), Beijing Municipal Science and Technology Commission (D141100003114002), Beijing Natural Science Foundation (7152075), and Beijing Young Top-notch Talent Project (2014000021223ZK36).
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.World Health Organization. A revision of the system of nomenclature for influenza viruses: a WHO memorandum. Bull World Health Organ 1980; 58:585–591. [PMC free article] [PubMed] [Google Scholar]
- 2.Dowdle WR, Galphin JC, Coleman MT, et al. A simple double immunodiffusion test for typing influenza viruses. Bull World Health Organ 1974; 51:213–215. [PMC free article] [PubMed] [Google Scholar]
- 3.Rota PA, Wallis TR, Harmon MW, et al. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 1990; 175:59–68. [DOI] [PubMed] [Google Scholar]
- 4.Matsuzaki Y, Sugawara K, Takashita E, et al. Genetic diversity of influenza B virus: the frequent reassortment and cocirculation of the genetically distinct reassortant viruses in a community. J Med Virol 2004; 74:132–140. [DOI] [PubMed] [Google Scholar]
- 5.Feng L, Shay DK, Jiang Y, et al. Influenza-associated mortality in temperate and subtropical Chinese cities, 2003-2008. Bull World Health Organ 2012; 90:279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Fu C, Li K, et al. Influenza associated mortality in Southern China, 2010–2012. Vaccine 2014; 32:973–978. [DOI] [PubMed] [Google Scholar]
- 7.Chan PK, Chan MC, Cheung JL, et al. Influenza B lineage circulation and hospitalization rates in a subtropical city, Hong Kong, 2000–2010. Clin Infect Dis 2013; 56:677–684. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, Huang J, Huai Y, et al. The substantial hospitalization burden of influenza in central China: surveillance for severe, acute respiratory infection, and influenza viruses, 2010–2012. Influenza Other Respir Viruses 2014; 8:53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Z, Feng L, Carolyn GM, et al. Characterizing the epidemiology, virology, and clinical features of influenza in China's first severe acute respiratory infection sentinel surveillance system, February 2011–October 2013. BMC Infect Dis 2015; 15:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brottet E, Vandroux D, Gauzere BA, et al. Influenza season in Reunion dominated by influenza B virus circulation associated with numerous cases of severe disease, France, 2014. Euro Surveill 2014; 19:20916. [DOI] [PubMed] [Google Scholar]
- 11.Breiman RF, Cosmas L, Njenga M, et al. Severe acute respiratory infection in children in a densely populated urban slum in Kenya, 2007–2011. BMC Infect Dis 2015; 15:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization. WHO Global Epidemiological Surveillance Standards for Influenza. Geneva: World Health Organization; 2014. [2015-8-1]. [Google Scholar]
- 13.Chinese National Influenza Center. Influenza Surveillance Program for Mainland China, 2010. Beijing: Chinese National Influenza Center; 2010. [2015-8-1]. [Google Scholar]
- 14.Zhou B, Lin X, Wang W, et al. Universal influenza B virus genomic amplification facilitates sequencing, diagnostics, and reverse genetics. J Clin Microbiol 2014; 52:1330–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drummond AJ, Suchard MA, Xie D, et al. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 2012; 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Wu S, Macintyre CR, et al. Using an adjusted Serfling regression model to improve the early warning at the arrival of peak timing of influenza in Beijing. PLoS One 2015; 10:e119923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng X, Tan Y, He M, et al. Epidemiological dynamics and phylogeography of influenza virus in southern China. J Infect Dis 2013; 207:106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization. Influenza (Seasonal). Geneva: World Health Organization; 2014. [2015-8-1]. [Google Scholar]
- 20.Lunelli A, Rizzo C, Puzelli S, et al. Understanding the dynamics of seasonal influenza in Italy: incidence, transmissibility and population susceptibility in a 9-year period. Influenza Other Respir Viruses 2013; 7:286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan Y, Guan W, Lam TT, et al. Differing epidemiological dynamics of influenza B virus lineages in Guangzhou, southern China, 2009–2010. J Virol 2013; 87:12447–12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Socan M, Prosenc K, Ucakar V, et al. A comparison of the demographic and clinical characteristics of laboratory-confirmed influenza B Yamagata and Victoria lineage infection. J Clin Virol 2014; 61:156–160. [DOI] [PubMed] [Google Scholar]
- 23.International Network for Strategic Initiatives in Global HIV Trials (INSIGHT). FLU 003: Baseline Characteristics by Influenza Subtype, 2015-10-30, Richard T, Dominic D, Marcelo HL, et al. International Network for Strategic Initiatives in Global HIV Trials (INSIGHT). FLU 003: Baseline Characteristics by Influenza Subtype, 2015-10-30, Twin Cities: International Network for Strategic Initiatives in Global HIV Trials; 2015. http://insight.ccbr.umn.edu/index.php?study=insight&page=&menu=FLU_studies. [Google Scholar]
- 24.Peasah SK, Azziz-Baumgartner E, Breese J, et al. Influenza cost and cost-effectiveness studies globally: a review. Vaccine 2013; 31:5339–5348. [DOI] [PubMed] [Google Scholar]
- 25.Carrat F, Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine 2007; 25:6852–6862. [DOI] [PubMed] [Google Scholar]
- 26.Wu SS, Yang P, Li HY, et al. The coverage rate and obstructive factors of influenza vaccine inoculation among residents aged above 18 years in Beijing from 2007 to 2010[in Chinese]. Zhonghua Yu Fang Yi Xue Za Zhi 2011; 45:1077–1081. [PubMed] [Google Scholar]
- 27.World Health Organization. Recommended composition of influenza virus vaccines for use in the northern hemisphere 2015-16 influenza season and development of candidate vaccine viruses for pandemic preparedness. Geneva: World Health Organization; 2015. [2015-8-10]. [Google Scholar]
- 28.Caini S, Huang QS, Ciblak MA, et al. Epidemiological and virological characteristics of influenza B: results of the Global Influenza B Study. Influenza Other Respir Viruses 2015; 9:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meier G, Gregg M, Poulsen NB. Cost-effectiveness analysis of quadrivalent influenza vaccination in at-risk adults and the elderly: an updated analysis in the UK. J Med Econ 2015; 1:1–16. [DOI] [PubMed] [Google Scholar]


