Abstract
The heme oxygenase-1 (Hmox1; HO-1) pathway was tested for defense of mitochondrial quality control in cardiomyocyte-specific Hmox1 KO mice (HO-1[CM]–/–) exposed to oxidative stress (100% O2). After 48 hours of exposure, these mice showed persistent cardiac inflammation and oxidative tissue damage that caused sarcomeric disruption, cardiomyocyte death, left ventricular dysfunction, and cardiomyopathy, while control hearts showed minimal damage. After hyperoxia, HO-1(CM)–/– hearts showed suppression of the Pgc-1α/nuclear respiratory factor-1 (NRF-1) axis, swelling, low electron density mitochondria by electron microscopy (EM), increased cell death, and extensive collagen deposition. The damage mechanism involves structurally deficient autophagy/mitophagy, impaired LC3II processing, and failure to upregulate Pink1- and Park2-mediated mitophagy. The mitophagy pathway was suppressed through loss of NRF-1 binding to proximal promoter sites on both genes. These results indicate that cardiac Hmox1 induction not only prevents heme toxicity, but also regulates the timing and registration of genetic programs for mitochondrial quality control that limit cell death, pathological remodeling, and cardiac fibrosis.
Hyperoxia induces cardiac inflammation, mitochondrial damage and cardiomyocyte death in mice with inducible, cardiac-specific deletion of heme oxygenase-1.
Introduction
The heme oxygenase-1 (HO-1; Hmox1) enzyme system is among the most important inducible mechanisms for cell protection against oxidative damage in the cardiovascular system. During oxidative stress, HO-1 eliminates cellular free heme (1), a powerful oxidant (2, 3), and produces the physiological gas carbon monoxide (CO) (4), which has potent antiinflammatory and antiapoptotic effects and stimulates mitochondrial biogenesis (5, 6). Moreover, administration of low-level CO gas or CO-releasing molecules mitigates many pathological injuries that affect energy metabolism in cells and tissues of experimental animals (7, 8). Hmox1-deficient heart cells accumulate lethal molecular oxidant damage, and embryonic Hmox1 deletion in mice has a high prenatal mortality, with survivors showing growth retardation, iron overload, organ fibrosis, and inflammatory tissue damage (9). Hmox1-deficient mice are highly susceptible to ischemia/reperfusion (I/R) injury and, after hypoxia, show evidence of right ventricular infarction (10, 11). This enzyme system has thus been a focus of attention in several cardiovascular diseases, including heart failure (HF) (12, 13), which is accompanied by oxidant-mediated hypertrophy, dilation, fibrosis, and metabolic disturbances that reduce contractile function (14, 15). The rare human HO-1 deficiency state and certain polymorphisms of the human HO-1 gene promoter are also associated with the development of cardiovascular disease (16, 17).
Moreover, HO-1 is mechanistically important for cardiomyocyte function through its ability to regulate mitochondrial health because most (95%) of the heart’s ATP production is derived from oxidative phosphorylation (OXPHOS) (18, 19). Indeed, even sublethal mitochondrial damage can impair cardiac ATP provision and ventricular contraction. Persistent mitochondrial damage is a major source of oxidants (20) and a major factor in the pathogenesis of many cardiovascular diseases (21, 22). The induction of HO-1 leads to increased cellular CO production, which generates a redox signal for the induction of mitochondrial biogenesis through stimulation of mitochondrial ROS production (5, 23). HO-1 has also been implicated in the induction of macroautophagy, (24) and if this capacity includes mitophagy, the enzyme would be involved in the regulation of the entire mitochondrial quality control cycle, which enables accurate and precise mitochondrial turnover (24–26). Disruption of mitophagy is proinflammatory and prooxidant (27, 28), and impaired autophagy compromises bioenergetics and leads to cell death, chronic inflammatory heart disease, and HF progression (29, 30). Accordingly, mitophagy has a putative cardioprotective role (31, 32).
The induction of mitophagy by cell stress involves the PINK1/Parkin pathway (33). PINK1, a serine/threonine kinase, normally localizes to mitochondria, where it is actively degraded by mitochondrial processing peptidase and presenilin-associated rhomboid-like protease (34). Loss of membrane potential blocks PINK1 importation, and its accumulation on the outer mitochondrial membrane allows the recruitment of Parkin (PARK2), an E3 ubiquitin ligase (34). PINK1 first phosphorylates mitofusin 2 (MFN2), which acts as a mitochondrial receptor for PARK2 (35). It then phosphorylates ubiquitin, which activates PARK2’s E3 ubiquitin ligase activity (36, 37). Activated PARK2 causes nonclassical ubiquitination of several outer mitochondrial membrane proteins, targeting the organelles for lysosomal degradation (38). Adaptor proteins such as p62/SQSTM1 bind ubiquitinated proteins and bind LC3 on the autophagosome (39). PARK2 also acts as a mediator of classical proteasomal degradation of mitochondrial proteins (40).
Physiologically, PARK2-deficient mice show impaired recovery of cardiac contractility after sepsis (41) and impaired mitophagy after myocardial infarction, resulting in poorer survival and larger infarct sizes relative to WT (31). Similarly, PINK1-deficient hearts are more susceptible to I/R injury compared with WT (32), and PINK1 overexpression in cardiac cells inhibits simulated I/R-mediated cell death (32). PINK1 deficiency in mice leads to ventricular dysfunction and hypertrophy (42), and by 2 months of age, these hearts show oxidative stress, reduced respiration, and fibrosis (32, 42). In contrast, PARK2−/− mice have normal mitochondrial and cardiac function (31). This means that PINK1 is essential for clearing damaged mitochondria, and there may also be other functions for PINK1 in myocytes.
Oxidative stress also contributes to adaptation to myocardial ischemia and can be induced by pretreatment with hyperoxia (>95% O2), a potentially valuable means of clinical preconditioning (43). O2 preconditioning attenuates I/R injury in rat hearts by decreasing infarct size and improving postischemic heart function (44, 45). This suggests that oxidative stress may not only induce antioxidant protection, but may also generate a protective mitochondrial status through HO-1 induction and downstream support of mitochondrial quality control (25, 46). However, O2 is also toxic at high concentrations over prolonged periods of time, which sets limits on its therapeutic use.
We generated KO mice containing myocyte-specific inducible HO-1 and exposed them to 100% O2 in order to induce the HO-1 response and to stimulate HO-1–mediated mitochondrial biogenesis and assess its linkage to the molecular regulation of mitophagy. We hypothesized that the absence of HO-1 would block the redox-sensitive transcriptional response for mitochondrial biogenesis during the oxidative stress of 100% O2 exposure, thereby revealing its linkage with mitophagy through PINK1 and PARK2 gene regulation. Such information would inform HO-1/CO–based therapeutics and ways that the induction of mitophagy could be used to treat HF and perhaps other degenerative heart diseases.
Results
Development of HO-1 KO mice and exposure to 100% O2.
Cardiac-specific Hmox1 (HO-1) KO mice were generated by crossing Hmox1fl/fl (HO-1fl/fl) mice with transgenic mice expressing MerCreMer under the control of the myosin heavy chain (Myh6) promoter to permit tamoxifen-inducible deletion of the Hmox1 allele in the adult. HO-1fl/fl/MerCreMer (HO-1fl/fl/Myh6-CreER) mice were born at the expected Mendelian frequency and were indistinguishable in appearance from control HO-1fl/fl littermates or WT/Myh6MerCreMer (WT/Cre). To generate the Hmox1-deficient cardiomyocyte strain (HO-1(CM)–/–), 8- to 10-week-old mice were given tamoxifen (i.p.) for 3 days, and 8 days later, the hearts were harvested and examined (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.89676DS1)
Eight days after tamoxifen, HO-1(CM)–/– and WT/Cre mice were exposed to 100% O2 for 2 days (48 hours) and then transferred to filtered air cages for 8 days (Supplemental Figure 1). Immediately after hyperoxia, the hearts appeared normal, although microscopically, the hearts of the HO-1(CM)–/– mice showed a few foci of neutrophilic inflammation (Supplemental Figure 2A). Seven days after tamoxifen, HO-1(CM)–/– mice appeared healthy. Forty-eight–hour exposures to 100% O2 are not lethal to WT mice or to HO-1(CM)–/– mice. At day 8 after hyperoxia, 2/46 WT mice had died, and 8/46 of the HO-1(CM)–/– mice had died (17.4%; P = 0.09).Those mice had elevated NLRP3 inflammasome protein levels (Supplemental Figure 2B), failed to upregulate cardiac HO-1 mRNA (Supplemental Figure 2C), and showed little response in the content of mitochondrial TCA cycle enzyme citrate synthase (CS; Supplemental Figure 2D).
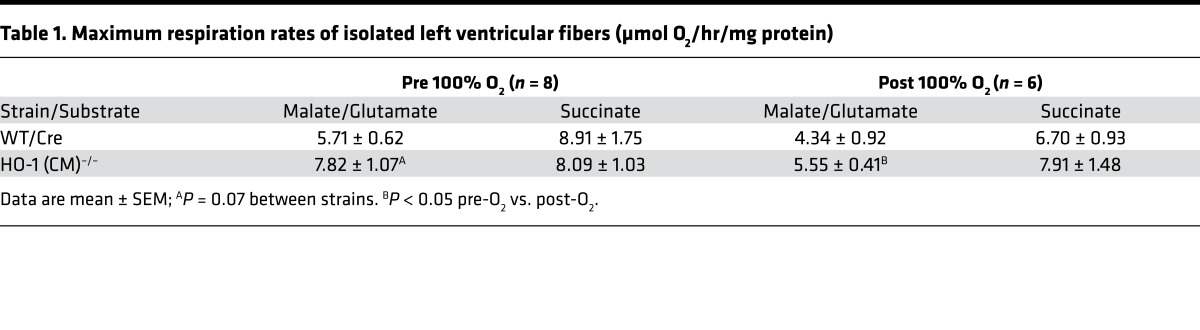
The cardiomyocyte respiratory capacity evaluated by maximum ADP respiration rates of freshly isolated left ventricular cells were the same in both strains of mice before 100% O2 exposure; however, respiration rates for Complex I substrates after hyperoxia were reduced in both strains (Table 1). This may be related in part to low mRNA levels of NADH dehydrogenase 1β complex subunit 8 (NDUFB8), a nuclear DNA–encoded Complex I protein integral to Complex I assembly (47), and NDUFAF1, a protein necessary for the assembly and stability of Complex I in the HO-1–deficient heart (48) (Supplemental Figure 3).
Table 1. Maximum respiration rates of isolated left ventricular fibers (μmol O2/hr/mg protein).
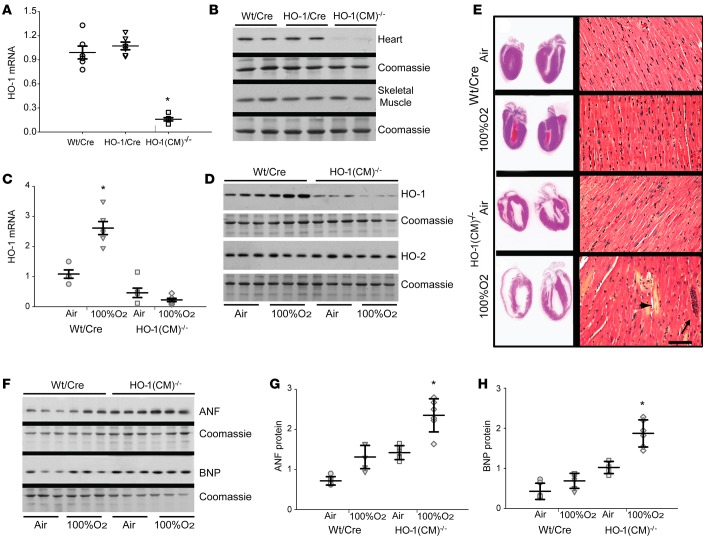
Even 8 days after O2, low HO-1 transcript and protein by quantitative PCR (qPCR) and immunoblotting were still apparent in the hearts of HO-1(CM)–/– mice (Figure 1, A and B; note stable HO-1 protein in skeletal muscle). Also at 8 days, cardiac HO-1 expression remained 2.5-fold higher in WT/Cre than in HO-1(CM)–/– mice, but the HO-2 constitutive isoform was stable in both strains (Figure 1, C and D). HO-1(CM)–/– hearts had enlarged by 8 days after O2 (Figure 1E, left), and ventricle/body weight ratios (3.2 ± 0.27 before to 5.6 ± 0.24 after O2) had increased significantly compared with WT/Cre hearts (2.9 ± 0.14 before to 3.3 ± 0.17 after O2; mean ± SEM mg/g body weight).
Figure 1. Cardiomyocyte-specific ablation of HO-1 and responses to transient hyperoxia.
(A) qPCR analysis of cardiac HO-1 in WT/Cre, HO-1/Cre, and HO-1(CM)–/– mice 8d after tamoxifen administration. Results are expressed mean ± SEM; horizontal bars represent mean values.*P < 0.05 for pre- vs. posthyperoxia; n = 6/group). (B) Western blot shows HO-1 protein analysis. (C) HO-1 mRNA expression in WT/Cre and HO-1(CM)–/– after tamoxifen before and after 100% O2 (hyperoxia). Results are mean ± SEM; horizontal bars represent mean values.*P < 0.05 for pre- vs. posthyperoxia; n = 6). (D) Representative Western blot shows HO-1 and HO-2 protein levels in WT/Cre and HO-1(CM)–/– from hearts pre- and posthyperoxia. (E) Images of mid-sagittal cardiac sections from WT/Cre and HO-1(CM)–/– mice after tamoxifen pre- and posthyperoxia, demonstrate thinning of ventricular walls in HO-1(CM)–/– hearts after O2. Right panels are photomicrographs of LV sections stained with H&E of control WT/Cre mice after tamoxifen in air and exposed to hyperoxia for 48h and air for 8d; control HO-1(CM)–/– mice after tamoxifen in air and hyperoxia for 48h and air for 8d. Deletion of HO-1 followed by O2 leads to foci of acute inflammation (long arrow). There is myofibrillar loss, cardiomyocyte swelling, paleness, and cell death (short arrow) compared with cardiomyocytes in WT/Cre. Tamoxifen alone did not perturb cytoarchitecture, but after HO-1 deletion, hyperoxia causes persistent cardiac damage (original magnification, ×250; scale bar = 10 μm). (F) Western analysis for myocardial ANF and BNP in WT/Cre and HO-1(CM)–/– mice. Coomassie staining was used as a loading control. Densitometry for (G) ANF and (H) BNP (mean ± SEM; *P < 0.05 for pre- vs. posthyperoxia; n = 6/group; 2-way ANOVA).
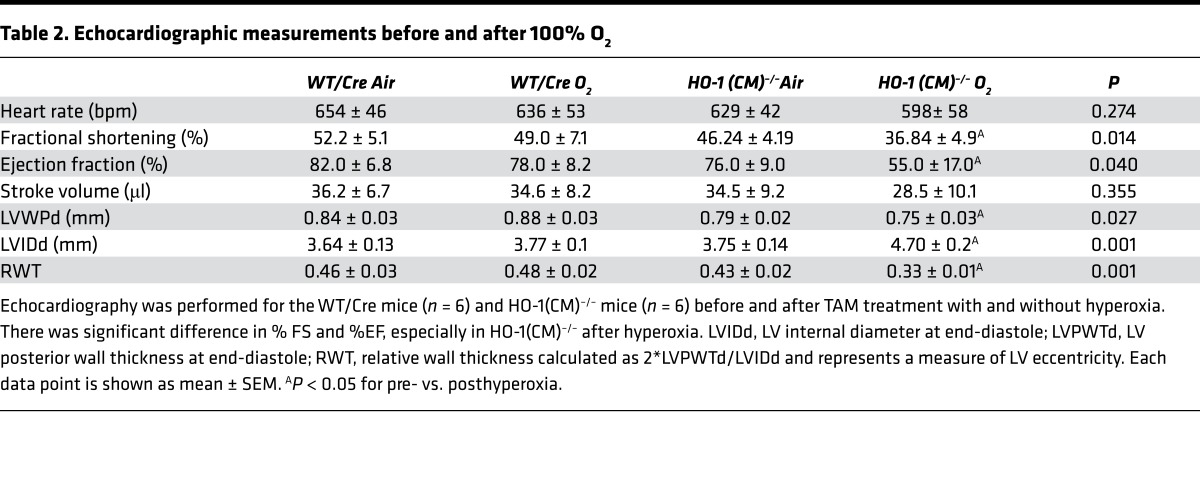
The increase in ventricular size and eccentric hypertrophy in HO-1(CM)–/– hearts was consistent with dilated cardiomyopathy. H&E-stained cross sections of HO-1(CM)–/– hearts at 8 days after 100% O2 revealed myofibrillar loss, myocyte degeneration, paleness, swelling, and leukocyte infiltration that were not seen in WT/Cre mice (Figure 1E, right). Echocardiographic data from HO-1(CM)–/– mice revealed a minimal decrease in fractional shortening (%FS) compared with WT/Cre. However, %FS and ejection fraction (%EF) in HO-1(CM)–/– mice had decreased significantly while the wall thickness changed significantly, suggesting eccentric hypertrophy in the HO-1(CM)–/– group (Table 2). Ventricular function was not impaired in tamoxifen-treated WT/Cre mice; thus, the effect was not due to the presence of Cre recombinase. The ventricular hypertrophy was reflected by increased tissue expression of hormonal hypertrophy markers, atrial natriuretic factor (ANF), and brain natriuretic peptide (BNP) in HO-1(CM)–/– hearts (Figure 1, F–H).
Table 2. Echocardiographic measurements before and after 100% O2.
Cardiac oxidative stress, inflammation, and apoptosis.
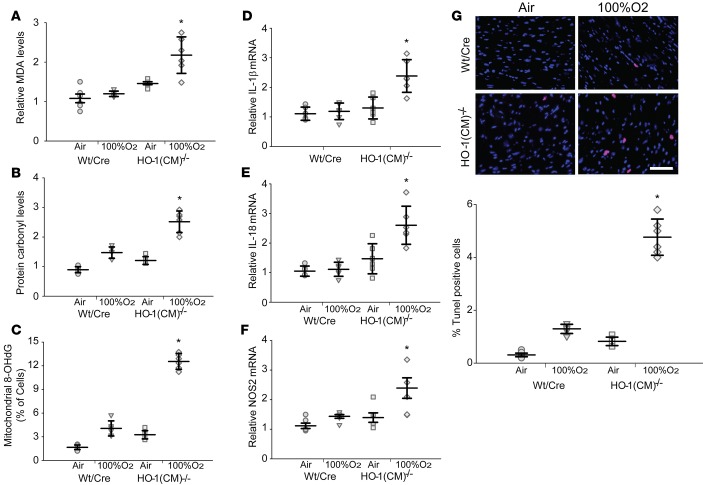
The extent of oxidative tissue damage was assessed by the quantification of total levels of protein carbonylation and malondialdehyde (MDA) adduct formation, and the level of mitochondrial DNA (mtDNA) damage was by mitochondrial 8-hydroxy-2’ -deoxyguanosine (8-OHdG) content (Figure 2, A–C). The data from 8 days after 100% O2 show that cardiac MDA and protein carbonyl levels were about twice as high in HO-1(CM)–/– mice compared with WT/Cre mice. However, mitochondrial 8-OHdG levels were increased by 4-fold in HO-1(CM)–/– mice compared with WT/Cre mice. These findings indicate the importance of HO-1 induction not only in antioxidant protection, but also in mtDNA integrity during oxidative stress. HO-1(CM)–/– mouse hearts showed inducible NOS (iNOS) levels and inflammasome output cytokines IL-1β and IL-18 that were twice as high as WT/Cre mice at 8 days after 100% O2 (Figure 2, D–F). We compared the number of TUNEL-positive cells in the hearts of HO-1(CM)–/– and WT/Cre mice before and after 100% O2 and found about a 4-fold increase in positive cells in HO-1(CM)–/– mice compared with WT/Cre mice, indicating significant increases in cell death rates after Hmox1 ablation (Figure 2G).
Figure 2. Posthyperoxia oxidative stress, inflammation, and cell death after ablation of cardiac HO-1 in mice.
(A) Cardiac levels of malonaldehyde, (B) cardiac level of carbonylated proteins, and (C) 8- OHdG levels in the 4 groups of mice (mean ± SEM; *P < 0.05 for pre- vs. posthyperoxia; n = 6/ group). (D–F) Inflammatory marker analysis by qPCR for IL-1β, IL-18, and iNOS (NOS2) (mean ± SEM; *P < 0.05 for pre- vs. posthyperoxia; n = 6/group). (G) TUNEL-positive cells in cardiac sections of the 4 groups of mice. Images are overlays of TUNEL-positive nuclei (red) with DAPI stained nuclei (blue). The magenta florescence indicates TUNEL positive cells. Excessive TUNEL staining is observed after tamoxifen in HO-1(CM)–/– mice after hyperoxia compared with the other groups (scale bar = 20 μm). (H) Quantitation of TUNEL positive nuclei in the heart sections (mean ± SEM; horizontal bars represent mean values.*P < 0.05 for pre- vs. posthyperoxia; n = 6/group; 2-way ANOVA).
HO-1 and mitochondrial biogenesis.
We evaluated mitochondrial biogenesis and its relationship to tissue damage recovery after oxidative stress because HO-1 is a known activator of the program (5, 49). Formalin-fixed heart sections were first stained for CS, a marker of mitochondrial density and distribution. The hearts of HO-1(CM)–/–, but not WT/Cre mice, demonstrated a patchy loss of CS that had not improved by 8 days after hyperoxia (Figure 3A). In contrast, CS levels increased in WT/Cre mice, consistent with an increase in mitochondrial mass by 8 days. This was corroborated by increased transcription factor A, mitocondrial (Tfam) protein levels in WT/Cre but not in HO-1(CM)–/– hearts 8 days after hyperoxia (Figure 3B). Using transmission electron microscopy (TEM) to examine cardiac ultrastructure, we found mitochondrial swelling and evidence of dilated intramembranous spaces at 8 days after tamoxifen administration in HO-1(CM)–/– mice (Figure 3C). There was more extensive mitochondrial damage 8 days after hyperoxia in HO-1(CM)–/– hearts, which showed many swollen mitochondria containing large electron-lucent areas (Figure 3C). Normal mitochondrial architecture was disrupted, and the distribution of sarcolemma appeared clustered instead of regularized (Figure 3C). HO-1–deficient cardiomyocytes showed widespread disorganization of myofibrils and disruption of contractile elements. In some areas, myofibrils were replaced by irregular mitochondria. At 8 days after O2, WT/Cre hearts maintained a structurally normal appearance, with regular arrays of myofibrils within the sarcomeres and normal size and number of mitochondria (Figure 3C).
Figure 3. Mitochondrial density and structure in mouse heart after hyperoxia.
(A) Photomicrographs of immunofluorescence staining of citrate synthase (CS, green) in the hearts. Control WT/Cre heart shows normal mitochondrial labeling with CS in the cardiomyocytes; WT/Cre heart 8d after hyperoxia shows increased CS labeling and mitochondrial fluorescence intensity; HO-1(CM)–/– mouse heart kept on air only shows modest decreases in mitochondrial density; HO-1(CM)–/– mouse hearts 8d after hyperoxia show decreased CS labeling and loss of intensity of mitochondrial fluorescence, suggesting that hyperoxia had caused persistent mitochondrial damage and cell degeneration in HO-1 deficiency. Original magnification, 200×; scale bar = 100 μm. (B) By Western blotting, mitochondrial CS and Tfam expression were significantly increased 8d after hyperoxia in WT/Cre mice but not in HO-1(CM)–/– mice (mean ± SEM; horizontal bars represent mean values.*P < 0.05 for pre- vs. posthyperoxia; n = 6 per group). (C) Electron microscopic findings of myocardial ultrastructure. Electron photomicrographs of cardiomyocytes of control WT/Cre mice showing linear myofibril arrangement and normal mitochondria; WT/Cre mouse exposed to hyperoxia showing enhanced autophagy, and arrows indicating mitophagosomes; electron micrograph from HO-1(CM)–/– mouse showing dilated intramembranous spaces (long arrows); HO-1(CM)–/– after hyperoxia have severe injury, including disoriented myofilaments and aggregates of enlarged mitochondria (M). The images reveal swollen and disorganized mitochondria with abnormal cristae structure (short arrow) and fragmented mitochondria (long arrow), suggesting poor respiratory capacity (n = 6/group). Note lack of mitophagosomes. Original magnification, 12,000×; scale bar = 1 μm. (D) Drp1 mRNA expression in hearts determined by qPCR. (E) Opa1 mRNA expression in hearts determined by qPCR. Values normalized to 18S levels (mean ± SEM; horizontal bars represent mean values.*P < 0.05 for pre- vs. posthyperoxia; n = 6/group; 2-way ANOVA).
The mitochondrial dynamics genes Drp1 and Opa1 mRNA increased significantly 8 days hyperoxia in WT/Cre hearts, but not in HO-1(CM)–/– hearts (Figure 3, D and E). The total mtDNA content and mitochondrial DNA transcription assessed by mitochondrial mRNA levels for ND1, as well as nuclear-encoded ND5, were all decreased in HO-1(CM)–/– hearts, suggesting the failure of mitochondrial biogenesis induction in the HO-1–deficient state (Figure 4, A–C). To understand the mechanism, we evaluated the expression levels of two transcriptional regulators of mitochondrial biogenesis, NRF-1 and PGC-1α. WT/Cre hearts showed a 2-fold increase in protein levels for NRF-1 and its PGC-1α partner, whereas NRF-1 and PGC-1α protein levels were markedly decreased in HO-1(CM)–/– hearts (Figure 4, D–F). This implicated HO-1 as a key mediator of the mitochondrial content differences in the two strains of mice after the oxidative stress of hyperoxia.
Figure 4. HO-1 and regulation of cardiac mitochondrial biogenesis.
(A) Deletion of HO-1 decreases cardiac mRNA expression of mitochondrial-encoded gene ND1. (B) Deletion of HO-1 decreases cardiac mRNA expression of nuclear-encoded gene ND5. (C) Mitochondrial mtDNA copy number by qPCR shows significant decline in mtDNA copies after hyperoxia in HO-1–deficient hearts. (D) Representative Western blots for nuclear-encoded NRF-1 and PGC-1α proteins that regulate the mitochondrial genome. (E and F) The quantification of NRF-1 and PGC-1α is, by densitometry, relative to loading control Lamin B. HO-1 ablation led to decreases in NRF-1 transcription factor and PGC-1α coactivator protein expression in the heart (mean ± SEM; horizontal bars represent mean values.*P < 0.05 for pre- vs. posthyperoxia; n = 6/group; 2-way ANOVA).
Autophagy and mitophagy.
Surprisingly few mitophagosomes were identified in tamoxifen-treated HO-1(CM)–/– hearts by electron microscopy (Figure 3C). The relative lack of autophagic vacuoles was striking, given the extent of mitochondrial disruption and myocardial damage in HO-1–deficient myocytes. To explore this apparent defect in autophagy/mitophagy, we examined LC3II/LC3I and p62 levels in WT/Cre and HO-1(CM)–/– hearts before and after O2. The conversion of LC3I to LC3II is indicative of autophagosome formation (50), whereas p62 decorating damaged mitochondria is degraded by autophagy and only accumulates when autophagy is reduced or impaired (51). LC3II/LC3I ratios were significantly reduced, and p62 levels were higher in HO-1(CM)–/– hearts after hyperoxia (Figure 5, A and B). Total LC3 levels were unaffected, indicating that loss of HO-1 somehow impairs LC3 processing rather than LC3 expression (Figure 5A). Also, Atg4B level, an autophagy protein that cleaves pro-LC3 to LC3-I, was decreased in HO-1(CM)–/– hearts compared with WT/Cre (Figure 5C). HO-1(CM)–/– hearts also exhibited lower levels of Beclin-1 protein, which is essential for autophagosomal membrane formation (Figure 5D). These data collectively imply that mitophagy in the HO-1(CM)–/– hearts has been interrupted after hyperoxia.
Figure 5. Loss of HO-1 impairs autophagy/mitophagy in the murine heart.
(A–D) Heart total protein from WT/Cre and HO-1(CM)–/– mice pre- and posthyperoxia were quantified for LC3II/I, p62, Atg4B, and Beclin1 protein expression. Coomassie staining was used as loading controls. The ratio of LC3-II/LC3-I was significantly decreased in the HO-1(CM)–/– mice at 8d after hyperoxia. In contrast, in the WT/Cre mice, LC3-I expression was increased, and its conversion to LC3-II was significantly enhanced in the hearts after hyperoxia (mean ± SEM; horizontal bars represent mean values. *P < 0.05 for pre- vs. posthyperoxia; †P < 0.05 for WT/Cre vs. HO-1(CM)–/–; n = 6 per group; 2-way ANOVA).
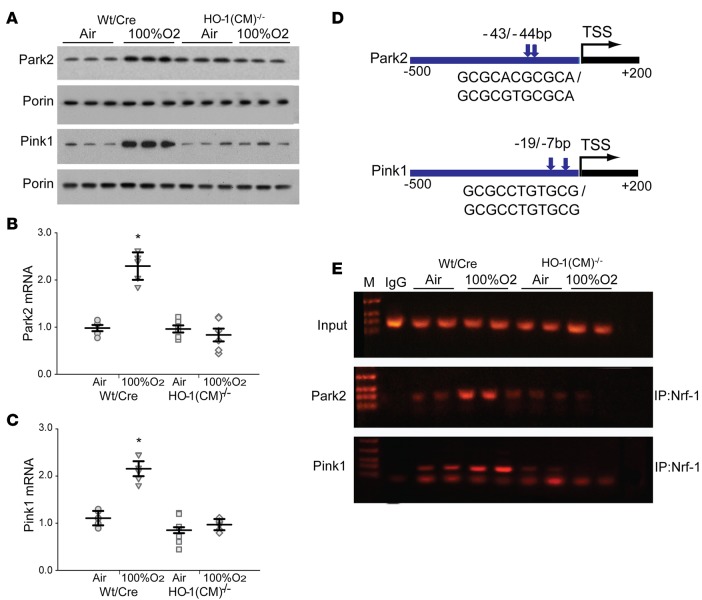
Mitophagy is mediated by the PINK1-PARK2 signaling pathway or by mitophagy receptor Bnip3l (Nix), and depolarized mitochondria are recognized by PINK1 and labeled through PARK2-mediated protein ubiquitination, which subsequently activates autophagy-related genes (ATGs) to engage autophagy (35). We compared activation levels of myocardial PINK1/PARK2 before and after hyperoxia and found that HO-1 deficiency drastically blocked the cellular enrichment of mitochondrial PINK1 and PARK2 after hyperoxia (Figure 6A), suggesting defective PINK1/PARK2 signaling. We realized that PARK2 total protein levels tracked positively with HO-1 levels (Figure 1, C and D), which suggested that HO-1 activity sets the transcriptional regulation of the Park2 gene. Accordingly, HO-1–dependent differences in Park2 and PINK1 mRNA levels were detected in the hearts (Figure 6, B and C). To determine whether Park2 and PINK1 are direct transcriptional targets of the NRF-1 axis downstream of HO-1, we used comprehensive in silico analysis of transcription factors binding to the promoter regions of mouse Park2 and PINK1 (5). The analysis of transcription factor binding sites in mouse Park2 and Pink1 (–500 to +200 Bp) promoter regions showed multiple NRF-1 transcription factor binding sites with nearby Sp1 sites within the immediate promoter sites of both genes. The Sp1 transcription factor activates or represses transcription in response to a range of pathological stimuli.
Figure 6. HO-1 is required for Pink1 and Park2 gene expression through NRF-1 activation after hyperoxia.
(A) Representative Western blots of Parkin (Park2) and PINK1 in mitochondrial fractions of the heart from WT/Cre and HO-1(CM)–/– mice prepared before and after hyperoxia. Porin was used as a loading control. (B) Deletion of HO-1 in HO-1(CM)–/– mice decreases cardiac expression of Park2 mRNA. (C) Lack of HO-1 in the heart of HO-1(CM)–/– mice decreased expression of PINK1 mRNA. (D) Schematic diagrams (–500 to +200 bp) of the Pink1 (ENSMUST00000030536) and Park2 (ENSMUST00000191124) promoter regions. Sequences were aligned between human and mouse using rVISTA 2.0. A search for NRF-1 binding sites upstream of the transcription start site (TSS) identified multiple consensus motifs for NRF-1 in human and mouse Pink1 and Park2 gene promoters by Genomatix and DNAsis software. (E) NRF-1 occupancy of promoters in vivo was investigated by ChIP analysis. Input lanes show the PCR product derived from chromatin prior to immunoprecipitation. Antibodies against NRF-1 used for immunoprecipitation, while IgG was used as negative control. Precipitated DNA was analyzed by PCR with primer sets specific for the 3 promoters (mean ± SEM; horizontal bars represent mean values. *P < 0.05 for pre- vs. posthyperoxia; n = 6 per group; 2-way ANOVA).
To assess transcriptional regulation of the Park2 and Pink1 promoter motifs in vivo, ChIP analysis was performed on nuclear DNA recovered from the hearts under control and 8 days after hyperoxic conditions using primers for highly proximal locations, as shown in Figure 6D. NRF-1 occupancy of Park2 and Pink1 promoters in vivo was identified (Figure 6E). Hyperoxia increased NRF-1 recruitment to Park2 and Pink1 promoters in WT/Cre heart (Figure 6E). However, NRF-1 was not recruited to the Park2 or Pink1 sites in HO-1(CM)–/– mice. WT/Cre NRF-1 recruitment corresponded to increased HO-1 and NRF-1 expression after hyperoxia. This demonstrated that HO-1 mediates mitophagy through transcriptional regulation of Park2 and Pink1 by NRF-1. An impairment of mitophagy in HO-1–deficient hearts after oxidative stress may not only jeopardize ATP production, but also induce myocardial inflammation, as revealed by a persistent inflammatory signature (Figure 2, D–F). Thus, the data support that HO-1 ablation impairs mitophagy, leading to accumulation of damaged mitochondria, oxidized macromolecules, and myocardial inflammation.
We used cultured beating HL-1 cells (5) to demonstrate that NRF-1 overexpression increased NRF-1 protein and downstream mitophagy genes Park2 and Pink1. In contrast, NRF-1 silencing significantly reduced Park2 and Pink1 gene expression (Supplemental Figure 4). These findings imply that HO-1–dependent induction of NRF-1 enhances mitochondrial quality control (QC) pathway function, allowing the beating rate of the cells to improve, while the silencing of HO-1 and NRF-1 decreases QC pathway function and reduces the rate of beating and the survival of these cells. Furthermore, HO-1 overexpression will increase the accumulation of the reduced form of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) in mitochondria, which reflects overall oxidative metabolic activity of the cells (Supplemental Figure 5). MTT is used as a surrogate for total respiration under standardized conditions and does not define the behavior of individual respiratory complexes. Meanwhile HO-1 silencing decreased the formazan accumulation in HL-1 cells, implying that HO-1 overexpression leads to augmented respiratory activity, while HO-1 silencing leads to a decline in respiration. Moreover, 72 hours after HO-1 transfection, the transduced HL-1 cells had increased their beating frequency, while HO-1 silencing significantly reduced beating rate (Supplemental Figure 5).
Cardiac fibrosis.
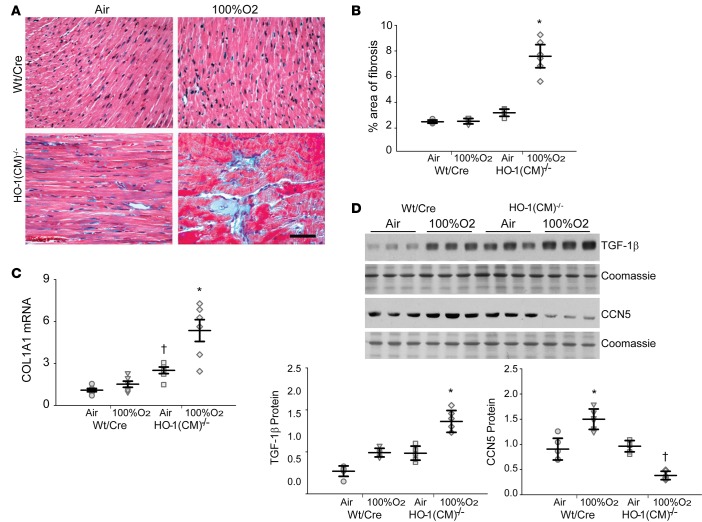
Studies with Masson’s trichrome–stained heart sections demonstrated extensive areas of interstitial fibrosis in the hearts 8 days after O2 in HO-1(CM)–/– relative to WT/Cre littermates (Figure 7, A and B). Fibrotic gene activation was evaluated by qPCR and showed greatly increased collagen mRNA levels in HO-1(CM)–/– hearts (Figure 7C) that was not present in WT/Cre mouse hearts. By immunoblotting, it was apparent that TGF-β1 levels were increased in the HO-1(CM)–/– mice, while levels of the antifibrotic protein CCN5 were decreased relative to WT/Cre littermates (Figure 7D). Thus, the inability to upregulate HO-1 resulted in extensive oxidative tissue damage and inflammation that led to significant fibrosis for many days after an O2 exposure that was normally be well-tolerated by the heart.
Figure 7. HO-1 deletion enhanced cardiac fibrosis after hyperoxia.
(A) Heart sections were stained with Masson’s trichrome to locate collagen (blue, fibrous collagen), which revealed extensive fibrosis of the left ventricle of HO-1(CM)–/– mice, mainly after hyperoxia (Scale bar = 50 μm). (B) The graph shows quantitation of fibrosis by densitometry. (C) Analysis of Col1A1 mRNA levels in the two strains of mice before and after hyperoxia. (D). Western blots of TGF-β and CCN5 proteins expression in heart lysates prepared before and after hyperoxia. The graph shows TGF-β and CCN5 protein quantification by densitometry relative to coomassie loading control (mean ± SEM; horizontal bars represent mean values; *P < 0.05 for pre- vs. posthyperoxia; †P < 0.05 for WT/Cre vs. HO-1(CM)–/–; n = 6 per group; 2-way ANOVA.).
Discussion
Each of the products of heme catabolism provides a different aspect of cell protection to avert serious oxidative tissue damage (52). Thus, the HO-1 system is far more than an antioxidant enzyme; for example, physiological CO production activates an essential mitochondrial defense by signaling the transcription of nuclear genes for mitochondrial biogenesis (49). But because free heme is a potent oxidant, it has been difficult to distinguish the cytoprotective effects of heme elimination by HO-1 from its mitochondrial role. (4, 5, 49). Here we demonstrate that HO-1(CM)–/– mice exposed to 100% O2 develop an evolving postexposure oxidative cardiac injury and HF due to the failure of mitochondrial quality control. In doing so, we confirm the induction of mitochondrial biogenesis by HO-1 but, more importantly, uncover the HO-1 system as an activator of transcriptional genes for selective mitophagy of cardiac mitochondria through nuclear respiratory factor–dependent (NRF-1–dependent) expression of the PINK1/PARK2 mitophagy gene pathway.
Normally, 48 hours of O2 exposure would produce mild self-limited cardiac injury and minimal mortality in mice, and the hearts would recover over a week or two. However, HO-1(CM)–/– mice showed a severely maladaptive cardiac response to this moderate oxidative stress compared with WT/Cre littermates. Although we had anticipated acute cellular damage due to loss of HO-1’s heme-scavenging function, these hearts displayed persistent inflammation and mitochondrial damage, resulting in cell death and rapid development of ventricular dilation and fibrosis (53, 54). A striking finding was impairment of autophagy/mitophagy, both under basal conditions and impressively in response to the oxidative stress. These results collectively support HO-1 as a critical regulator of mitochondrial quality control in the heart following oxidative tissue damage.
Prolonged O2 breathing causes minor hemodynamic changes, such as bradycardia, and slight decreases in stroke volume and cardiac output (55). After O2 exposure, we observed nothing outside of the physiological range in terms of heart rate and cardiac output in HO-1(CM)–/– mice. However, within a few days after hyperoxia, there were reductions in cardiac %EF and %FS in HO-1(CM)–/– mice (Table 2) compared with WT/Cre mice. Inspection of these hearts disclosed an increase in ventricular size, with dilation of both ventricles suggesting functional decompensation after hyperoxia. The supporting molecular evidence indicated that the heart is undergoing hypertrophy and remodeling related to the loss of the enzyme and damage to mitochondrial quality control, although secondary molecular responses related to the progressive decompensation cannot be excluded.
The induction of HO-1 by O2 is well known to be found in WT/Cre mice (56) but was not present in HO-1(CM)–/– mice. This is consistent with the upregulation of WT HO-1 by CO in the amelioration of cardiac cell death and fibrosis after doxorubicin, as well as with HO-1’s inhibitory effect on inflammatory and fibrotic responses in the injured myocardium in mice (6, 25). The requirement for HO-1 in the activation of mitochondrial quality control also means that CO administration or other HO-1 products alone could not provide complete protection, thereby excluding several protective possibilities attributed to the products of heme breakdown (7, 8).
HO-1 also has a critical protective role in the development of inflammation in the cardiovascular system (25, 57), and our results confirm previous studies demonstrating that the expression of certain inflammatory genes is increased by HO-1 inhibition (58). In addition, persistent inflammation in the O2-exposed HO-1(CM)–/– mouse heart is reflected by the increased levels of inflammatory mediators IL-1β, IL-18, and NOS2. These data confirm that the antiinflammatory effects of HO-1 are part of its known cytoprotective role.
In the failing heart, HO-1 induction allows upregulation of an important adaptation that opposes pathological remodeling and is mediated in part by product-dependent inhibition of apoptosis (59). Moreover, apoptosis from severe oxidative stress and inflammation intersect in cardiac injury (60, 61). Here, the numbers of TUNEL-positive cells increased in HO-1(CM)–/– mice compared with WT/Cre mice, supporting apoptosis as an important mechanism of cardiomyocyte death, although in some areas of the injured HO-1(CM)–/– hearts, cardiomyocyte death appeared to be due to necrosis. HO-1 overexpression prevents oxidative damage in cells (53, 54); hence, its absence would allow oxidant damage in the form of protein carbonylation, lipid peroxidation, and mtDNA oxidation to accumulate after hyperoxia and contribute to cardiomyocyte death.
The cell’s need for continuous OXPHOS makes mitochondria both generators and targets of ROS (62); damaged mitochondria elaborate excessive superoxide anion and H2O2 that can cause oxidative cell damage unless those organelles are removed from the cell (63, 64). Mitochondrial dysfunction and impaired mitochondrial biogenesis has repeatedly been observed in the failing heart in animal models and in patients, but the molecular mechanisms are poorly understood (65). Among the pathways for HO-1–induced improvement in cardiac function is transcriptional signaling for the maintenance of mitochondria (5, 49). That mitochondrial biogenesis is impaired in the HO-1(CM)–/– heart was anticipated and supported by immunochemistry, mtDNA depletion, and significant loss of mtDNA-encoded genes. WT/Cre mice avoided this mitochondrial destruction by significantly increased transcriptional levels of genes required for mitochondrial function (CS, ND1, Opa1, and Tfam) compared with HO-1(CM)–/– mice, which were not protected. Accordingly, the expression levels of mtDNA regulatory proteins (NRF-1, PGC-1α) were significantly upregulated in WT/Cre but not in HO-1(CM)–/– mice.
Oxidative mitochondrial damage in cardiac I/R injury has been implicated as a cause of cell death (66), but the prevention of heme accumulation and nonfunctional protein aggregates that lead to a cycle of oxidative stress and deterioration of mitochondrial function is clearly protective (67). There is further evidence that energy depletion together with ROS and protein aggregation induces autophagy (68, 69). One study showed that activation of autophagy clearly reduced myocardial infarct size in a mouse model of I/R injury (70), and the regulation of autophagy via HO-1 may play a protective role in other tissues (71). In the heart, autophagy is essential for homeostasis both under basal conditions and in response to stress (70, 72). For example, autophagy is important for protecting cardiomyocytes from hyperglycemic damage (73), and suppression of autophagy contributes to the development of cardiomyopathy in diabetes (74).
Healthy mitophagy is designed to clear damaged mitochondria effectively before they cause cellular harm (31). Damaged mitochondria release factors into the intermembrane space that signal not only macroautophagy, but also selective degradation of individual mitochondria (75, 76). Impaired mitophagy in the heart leads to accumulation of aberrant mitochondria and, subsequently, to contractile dysfunction and worsening of overall cardiac function (72, 77). Here, the lack of HO-1 decreased the number of autophagosomes; decreased LC3 II, Atg4B, and Beclin1 expression; and increased levels of p62 in the HO-1(CM)–/– hearts, consistent with impairment of autophagy/mitophagy after hyperoxia. We observed by electron microscopy (EM) that defective mitochondria after hyperoxia were engulfed by autophagosomes in WT/Cre mice but not in HO-1(CM)–/– mice. In WT/Cre mice, mitophagy is associated with upregulation of Drp1 and PARK2 protein levels. In HO-1(CM)–/– mice, however, 100% O2 significantly decreases Opa1, reflecting cristae destruction and a lack of autophagosomes accompanied by depressed Complex I respiration. This likely reflects low NDUFB8 and NDUFAF1 assembly protein expression, low mtDNA levels, and increased oxidant production (78). The implication is that HO-1 is necessary for the early activation of mitophagy that prevents the evolution to persistent cardiac injury following hyperoxia.
PARK2 is also recruited to damaged mitochondrial outer membranes in order to allow signaling for mitochondrial degradation (33, 79). Thus, mitochondrial dysfunction during HO-1 deficiency leading to the inability to clear damaged mitochondria by mitophagy would be detrimental to cardiomyocyte survival. Since PARK2 and Pink1 operate together, and the Pink1/PARK2 pathway is altered in HO-1 deficiency, we searched for Pink1/PARK2 regulation by transcription factors downstream of HO-1 by comprehensive in silico analysis. We uncovered the probable regulation of Pink1/PARK2 by NRF-1 by the presence of tandem Sp1 sites. This was tested experimentally in vivo by occupation of NRF-1 at consensus sites in the PINK1/PARK2 proximal gene promoters in WT/Cre mice and HO-1(CM)–/– mice. NRF-1 occupancy of PINK1/PARK2 promoter sites was significantly enhanced after hyperoxia in WT/Cre but was strikingly absent on NRF-1 binding in HO-1(CM)–/– mice. Moreover, NRF-1 silencing or transfection in beating HL-1 cardiomyocytes led to low vs. high gene expression, respectively, for Pink1 and Park2. These results are consistent with data on mitochondrial biogenesis, demonstrating the HO-1–dependent nuclear accumulation of NRF-1 (80). Thus, HO-1–dependent NRF-1 induction permits the genetic upregulation of both mitochondrial biogenesis and mitophagy in the heart.
The remodeling of ventricular dilation involves individual cardiomyocytes, which undergo eccentric hypertrophy, together with interstitial cell proliferation by fibroblasts, which adversely affects cardiac structure and function, ultimately leading to HF (81). A key effector of pathological remodeling operates through connective tissue growth factor regulated by the TGF-β1 pathway, which promotes fibroblast proliferation and extracellular matrix production in connective tissues (82, 83). We found increased fibrosis in HO-1(CM)–/– mice, accompanied by increases in TGF-β1 expression, which stimulates the accumulation of extracellular matrix proteins and fibrosis (84, 85). In WT/Cre mice, TGF-β1 was not significantly induced after hyperoxia, and there was a significant increase in CCN5, a negative regulator of α-SMA and collagen I expression and a potent repressor of TGF-β1 (86, 87). CCN5 expression, however, was reduced in HO-1(CM)–/– mice. It is intriguing to speculate that HO-1 upregulation opposes the myocardial fibrotic response through this pathway.
In summary, we report a new mechanism for HO-1 protection of the heart from ongoing oxidative injury through the maintenance of mitochondrial homeostasis by the transcriptional regulation of key mitophagy proteins. This mechanism extends far beyond heme clearance by encompassing the well-known role of HO-1 in mitochondrial biogenesis (49), as well as the facilitation of mitophagosome clearance of damaged mitochondria leading to mitigation of cellular oxidant production. In this context, HO-1–induced cardioprotection is driven by sentinel transcriptional events involving activation of redox-sensitive transcription factors, such as NRF-1, that prevent the arrest of stress-induced mitophagy from consumption of tagged organelles labeled with essential decorator proteins such as Pink1 and PARK2. In summary, HO-1–dependent induction of NRF-1 is a critical regulator of the cardiac transcriptional circuitry for the overall mitochondrial quality control cycle that arrests late cell death and appears to prevent fibrosis after oxidative stress.
Methods
Mice.
Mice were provided access to food and water ad libitum and maintained on a 12-hour light/dark cycle. Hmox1 genomic fragment of 9 kb, encompassing exons 1–5 and surrounding sequences, was obtained by high fidelity PCR from the Hmox1 locus and cloned into a gene targeting vector (pMSCV-loxp-dsRed). The targeting vector was modified by inserting the proximal loxP site into intron 2 and the distal loxP site, including an FRT-flanked neomycin-resistance gene subsequently inserted downstream of exon 5. After the neo gene was excised from the targeted locus by Flp treatment, neo-minus cells were microinjected into C57BL/6J mouse blastocysts, then implanted into pseudo pregnant recipients. High-chimera males were mated with C57BL/6J females to produce F1 heterozygotes Hmox1fl/+ (HO-1fl/+). Mice were genotyped using a multiplex PCR reaction with 3 primers: Hmox1F, 5′ tcactatgcaactctgttggagg 3′; Hmox1R, 5′ gtctgtaatcctagcactcgaa 3′; and Dsred-R, 5′ ggaaggacagcttcttgtagtcg 3′. Mice harboring a homozygous conditional null mutation in HO-1fl/fl were crossed to transgenic Myh6MerCreMer mice (The Jackson Laboratory) that express Cre recombinase under the control of the cardiomyocyte-specific Myh6 promoter to generate cardiomyocyte-specific Hmox1 KO mice (Supplemental Figure 1). All mice were on a C57BL/6J background and Cre littermates were used as controls (WT/Cre). To selectively delete cardiomyocyte Hmox1, 8- to 10-week-old mice were given 40 mg/kg tamoxifen for 3 days. This resulted in rapid and effective knockdown of the Hmox1 gene without cardiotoxicity.
Respiration measurements.
Hearts were quickly harvested from euthanized mice, and left ventricular muscle fibers were separated in cold, calcium-free isolation buffer (88) and exposed with gentle mixing to 1 mg/ml collagenase (Sigma-Aldrich, C0130) for 10 minutes followed by saponin (50 g/ml, Sigma-Aldrich, S4521) for 30 minutes to permeabilize the cardiomyocytes. The fibers were thoroughly washed 3 times in respiration buffer to arrest permeabilization. The O2 uptake of the fibers was measured in a temperature-controlled respirometer (Mitocell MT200; Strathkelvin Instruments) using malate (5 mM) plus glutamate (10 mM) or succinate (10 mM) (State 4). Maximum respiration rates (State 3) were obtained by adding ADP (2.5 mM). The values are reported as μmol O2/hr/mg of fiber protein. No increase in respiration was observed by addition of 10 mM cytochrome c after ADP.
Oxygen exposures.
Mice, aged 8–10 weeks, were placed in cages in airtight chambers (50 × 50 × 30 cm) with access to food and water ad libitum and exposed to 100% O2 for 48 hours, and then moved to filtered room air for 8 days. The chamber oxygen concentration was monitored continuously with an oxygen analyzer (Vascular Technology) as described (89). Mouse hearts from these exposures were harvested immediately or at day 19, snap frozen in liquid N2, and stored at −80°C. Fresh hearts were fixed in 10% formalin for histology or 0.4% gluteraldehyde for electron microscopy (Supplemental Figure 2).
Oxidative stress assessment.
To evaluate tissue lipid peroxidation, levels of MDA were determined in fresh ventricle using assay kits (Abcam) with the manufacturer’s instructions. To detect protein oxidation, we used the OxyBlot Oxidized Protein Detection kit (Chemicon). Protein homogenates from normoxic and hyperoxic heart were derivatized with 1,3-dinitrophenylhydrazine for 15 minutes according to the manufacturer’s instructions, and dot blot analysis was performed and the data densitized.
Histology and EM.
Hearts were isolated and perfusion-fixed at a constant pressure (80 mmHg) with 10% formalin, dehydrated in 70% ethanol, and embedded in paraffin. Sections were stained with H&E, and gross morphology was viewed under a low-power field (×5). Masson’s trichrome stain was used to evaluate the degree of fibrosis in hearts. The extent of fibrosis in these photomicrographs was quantified by a blinded observer using the Nikon image software (NIS-Elements Basic Research 3.2). For immunofluorescence, sections were stained with CS primary antibodies (1:200; GeneTex Inc.). Sections were then extensively washed in PBS and incubated in secondary anti-rabbit IgG, Alexa Fluor 488 (Thermo Fisher Scientific, catalog A-11034). Conventional fluorescence images were obtained using a Nikon Microphot-FXA fluorescence microscope. Small pieces of left heart were fixed in 2% glutaraldehyde and 2% paraformaldehyde at room temperature for 2 hours, followed by 24 hours at 4°C. Specimens were rinsed in 0.1 mol/l phosphate buffer (PB), pH 7.4, and fixed in 1% osmium tetroxide in PB (pH 7.4) at 4°C for 1 hour, and then dehydrated in graded ethanol, followed by propylene oxide, and embedded in Eponate 812 (Ted Pella Inc.). Ultrathin sections (around 60–70 nm) from longitudinal parts were prepared and contrasted with uranyl acetate, followed by lead citrate, and examined in Philips CM12 electron microscope.
TUNEL assays.
Thin paraffin-embedded heart sections were used for the TUNEL assay commercial kit (In Situ Cell Death Detection Kit, Roche Diagnostics) following the manufacturer’s instructions. Negative and positive control sections were prepared with label solution only or by preinduction of strand breakage with recombinant DNase I. Nuclei were counterstained with DAPI. Sections from 4–5 hearts in each group were used in the staining, and at least 5 unique fields/section were quantified in each experiment by a blinded observer using Nikon image software (NIS lements BR 3.2).
Echocardiography.
Cardiac function in WT/Cre and HO-1(CM)–/– normoxic or hyperoxic mice was compared at day 19 by transthoracic echocardiography performed using a Vevo770 Ultrasonograph (VisualSonics) equipped with a 45-MHz imaging transducer. The mice were lightly anesthetized with 1%–1.5% isoflurane, and body temperature was maintained at 37°C. Two-dimensional (2-D) mode in the parasternal long axis and parasternal short axis at the mid-papillary muscle level were imaged. From this parasternal short axis view, the 2-D–guided M-mode across the anterior wall and posterior wall were recorded. Left ventricular anterior wall (LVAW), left ventricular interior diameter (LVID), and left ventricular posterior wall thickness (LVPW) at systole (s) and diastole (d) were measured. Relative wall thickness (RWT) was calculated from the LIVDd and LVPWd measurements (2 × LVPWTd/LVIDd). %EF was calculated as (EDV − ESV)/EDV × 100%, where EDV and ESV are end-diastolic and end-systolic volume, respectively. %FS was calculated by (LVIDd − LVIDs)/LVIDd × 100%.
qPCR.
Total RNA was isolated from hearts using the RNAqueous-4 PCR kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Samples were treated with DNase I (4 units) for 2 hours at 37°C that was then inactivated, and the cDNA was prepared with a high-capacity cDNA archive kit (Applied Biosystems), according to the manufacturer’s protocol. qPCR primers for HO-1, Tfam, ND1, ND5, IL-1β, IL-18, NOS2, Drp1, Opa1, Park2, Pink1, Myh6, Myh7, NDUFB8, NDUFAF1, and 18S from Applied Biosystems were used with the manufacturer’s gene expression assay protocol. All reactions were conducted on StepOnePlus for 40 cycles. The results were analyzed using the 2−ΔΔCt method after normalized to the level 18S in each samples.
mtDNA copy number.
Total cellular DNA was extracted using the Sigma-Aldrich DNA isolation kit. mtDNA copy number was quantified with real-time PCR on StepOnePlus Sequence Detector System (Applied Biosystems). Primers were designed for cytochrome b (cyt b) with ABI Probe Design software (Applied Biosystem), and amplifications were performed on 10 ng total mtDNA using PCR primers (cyt b-sense [cyt b-s] and cyt b-antisense [cyt b-as]). One copy of linearized pGEMT-cyt b vector was used for standard mtDNA quantification (90). The cyt b probe, 5′ FAM-ttcctccacgaaacaggatcaaa-TAMRA 3′, contained FAM (6-carboxy-fluorescein) at the 5′ end as a fluorescent reporter dye and TAMRA (6-carboxy-tetramethyl-rhodamine) at the 3′ end as a quencher dye selected from a highly conserved region of mouse cyt b gene. Serial dilutions of 1 × 105 to 1 × 1010 copies of standard cyt b plasmid were prepared for a standard curve. Samples were tested for mtDNA at 1:100 and 1:1,000 dilutions. Samples were analyzed in triplicate; mtDNA copy number/ng DNA was determined relative to the standards of known mtDNA copies per dilution.
Immunoblot analysis.
Protein extracts from mouse hearts (n = 5–6 per group) were analyzed by Western blotting as described (90). Total heart protein, mitochondrial protein, or nuclear protein were resolved after fractionation by SDS-PAGE on gradient or 14% gels (Bio-Rad), transferred to polyvinylidene difluoride membranes (Millipore), blocked with 4% nonfat dry milk in Tris-buffered saline with Tween 20, and probed overnight at 4°C with antibodies against HO-1 or HO-2 (1:1,000; Enzo Life Sciences), mitochondrial transcription factor A (Tfam; 1:1,000; developed in our laboratory), CS (1:1,000; GeneTex, catalog GTX110624), NRF-1 (1:2,000, in house), PGC-1 (1:1,000; Abcam, catalog ab54481), Pink1 (1:500; Abcam, catalog ab23707), PARK2 (1:500; Santa Cruz Biotechnology Inc., catalog sc-32282), ANF (1:500; Santa Cruz Biotechnology Inc., catalog sc-18811), BNP (1:1,000; Santa Cruz Biotechnology Inc., catalog sc-18813), LC3I/II (1:1,000; Cell Signaling Technology, catalog 41085), p62 (1:1,000; Cell Signaling Technology, catalog 8025), Atg4B (1:500; Santa Cruz Biotechnology Inc., catalogsc130968), Beclin1 (1:1,000; Cell Signaling Technology, catalog 37385), TGF-β (1:500; Santa Cruz Biotechnology Inc., catalog sc-8829), or CCN5 (1:500; Santa Cruz Biotechnology Inc., catalog sc-25442). Protein loading was confirmed by nuclear protein Lamin B (Santa Cruz Biotechnology Inc, sc-374015), tubulin (1:1000; Sigma-Aldrich, catalog T-5168), or the mitochondrial porin (1:500; Santa Cruz Biotechnology, catalog sc-8829) and/or Coomassie blue staining. After incubation with primary antibody, the membranes were treated with horseradish peroxidase–conjugated secondary antibody (1:2,000, Santa Cruz Biotechnology Inc., catalog sc-516102) and developed by enhanced chemiluminescence (Western Blotting Luminol Reagent, Santa Cruz Biotechnology Inc.). The protein bands were quantified on digitized images in the mid-dynamic range using Quantity One (Bio-Rad). Densitometry measurements were normalized to tubulin, Porin, Lamin B, or Coomassie in the same samples.
ChIP analysis.
ChIP assays were performed in cardiac cell nuclei that were sonicated in shearing buffer. Shearing effectiveness was confirmed by electrophoresis on ethidium bromide–stained agarose gels. Samples were processed for immunoprecipitation using ChIP IT assay kit (Active Motif) and NRF-1 antibody (5) at a concentration of 2–5 μg/ml. After precipitation, cross-linking was reversed, and PCR was carried out in the precipitated and input samples using 1 μl of each sample (input DNA dilution 1:10, immunoprecipitated fractions undiluted) in PCR buffer (Qiagen) containing dNTP (Invitrogen) and Taq DNA polymerase (Qiagen) using primers –98 5′ ctgccggaggcgaatcttac 3′ and +165 5′ gcgcggagagattgtacct 3′ to amplify the selected NRF-1 site on the murine Park2 promoter. The primers –149 5′ ttgttcacaacccctcgacc 3′ and +6 5′ caacaacaaacttcgggggc 3′ were used to amplify the selected NRF-1 site on the Pink1 promoter. DNA concentration was measured and standardized across samples prior to PCR. PCR products were analyzed by agarose gel electrophoresis.
Cell culture assay.
HL-1 cardiomyocytes (1 × 104 per treatment) were cultured in flasks coated in fibronectin (12.5 mg/l) gelatin (0.02%) containing Claycomb medium (Sigma-Aldrich) supplemented with 10% FBS (PAN Biotech GmbH), 2 mM L-glutamine (PAN Biotech GmbH), 0.1 mM norepinephrine (Sigma-Aldrich), ascorbic acid 0.3 mM (Sigma-Aldrich), 100 U/ml penicillin (Sigma-Aldrich), and 100 μg/ml streptomycin (Sigma-Aldrich) in a humid atmosphere of 5% CO2/95% air at 37°C (5). Cells were transfected in 6-well plates at 70%–80% confluence with pGFPCRS vector containing HO-1 oligonucleotide sequence for optimal suppression of HO-1 (Origene, TR30018), HO-1 TrueORF (Origene, MR203944), or NRF-1 TrueORF (Origene, MG208550) using FuGENE-HD (Promega, E2311). After 48–72 hours, the cells were harvested near confluence for further measurements. To confirm HO-1 or NRF-1 overexpression or knockdown, qPCR was used. We used the MTT assay to estimate total cellular metabolic activity based on the reduction of the compound to the formazan by mitochondrial dehydrogenases (91). Beating rate was estimated by counting the number of beats per minute in 5 different cell clusters in 4 blinded experiments. Each experiment was conducted in triplicate and at least repeated 3 times.
Statistics.
Analysis was performed after a normality test, and results were expressed as mean ± SEM. Data from multiple groups were compared by two-way ANOVA. Fisher’s protected least significant difference tests were used for post-hoc corrections. Student’s t test assuming unequal variance was used for comparisons involving two groups at any single time. The Wilcoxon rank-sum test was performed only when data were not normally distributed. P < 0.05 was considered significant.
Study approval.
All animal experiments were approved by the Institutional Animal Care and Use Committee at the Duke University Medical Center, Durham, North Carolina, USA.
Author contributions
HBS conceptualized and designed experiments, collected and assembled data, analyzed and interpreted data, wrote the manuscript, and gave final approval of the manuscript. CAP conceptualized and designed experiments, assembled data, analyzed and interpreted data, wrote the manuscript, gave final approval of the manuscript, and received financial support. JEK collected and interpreted the data, offered technical assistance, and approved of the manuscript.
Supplementary Material
Acknowledgments
We appreciate the technical assistance of Craig Marshall, Martha Salinas, Linhua Song, and Lynn Tatro. We thank Randy Threasure for his help with the generation of the floxed HO-1 mice. This work was supported by NIH grants R01-AI095424 and VA Merit Review grant to CAP.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2017;2(3):e89676. https://doi.org/10.1172/jci.insight.89676.
References
- 1.Maines MD. The heme oxygenase system: a regulator of second messenger gases. Annu Rev Pharmacol Toxicol. 1997;37:517–554. doi: 10.1146/annurev.pharmtox.37.1.517. [DOI] [PubMed] [Google Scholar]
- 2.Wagener FA, et al. Heme is a potent inducer of inflammation in mice and is counteracted by heme oxygenase. Blood. 2001;98(6):1802–1811. doi: 10.1182/blood.V98.6.1802. [DOI] [PubMed] [Google Scholar]
- 3.Wagener FA, et al. Different faces of the heme-heme oxygenase system in inflammation. Pharmacol Rev. 2003;55(3):551–571. doi: 10.1124/pr.55.3.5. [DOI] [PubMed] [Google Scholar]
- 4.Maines MD. The heme oxygenase system: update 2005. Antioxid Redox Signal. 2005;7(11-12):1761–1766. doi: 10.1089/ars.2005.7.1761. [DOI] [PubMed] [Google Scholar]
- 5.Piantadosi CA, Carraway MS, Babiker A, Suliman HB. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ Res. 2008;103(11):1232–1240. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suliman HB, Carraway MS, Ali AS, Reynolds CM, Welty-Wolf KE, Piantadosi CA. The CO/HO system reverses inhibition of mitochondrial biogenesis and prevents murine doxorubicin cardiomyopathy. J Clin Invest. 2007;117(12):3730–3741. doi: 10.1172/JCI32967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lancel S, Hassoun SM, Favory R, Decoster B, Motterlini R, Neviere R. Carbon monoxide rescues mice from lethal sepsis by supporting mitochondrial energetic metabolism and activating mitochondrial biogenesis. J Pharmacol Exp Ther. 2009;329(2):641–648. doi: 10.1124/jpet.108.148049. [DOI] [PubMed] [Google Scholar]
- 8.Morse D, Lin L, Choi AM, Ryter SW. Heme oxygenase-1, a critical arbitrator of cell death pathways in lung injury and disease. Free Radic Biol Med. 2009;47(1):1–12. doi: 10.1016/j.freeradbiomed.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kapturczak MH, et al. Heme oxygenase-1 modulates early inflammatory responses: evidence from the heme oxygenase-1-deficient mouse. Am J Pathol. 2004;165(3):1045–1053. doi: 10.1016/S0002-9440(10)63365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida T, Maulik N, Ho YS, Alam J, Das DK. H(mox-1) constitutes an adaptive response to effect antioxidant cardioprotection: A study with transgenic mice heterozygous for targeted disruption of the Heme oxygenase-1 gene. Circulation. 2001;103(12):1695–1701. doi: 10.1161/01.CIR.103.12.1695. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Wei J, Peng DH, Layne MD, Yet SF. Absence of heme oxygenase-1 exacerbates myocardial ischemia/reperfusion injury in diabetic mice. Diabetes. 2005;54(3):778–784. doi: 10.2337/diabetes.54.3.778. [DOI] [PubMed] [Google Scholar]
- 12.Shioi T, Inuzuka Y. Aging as a substrate of heart failure. J Cardiol. 2012;60(6):423–428. doi: 10.1016/j.jjcc.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Shih H, Lee B, Lee RJ, Boyle AJ. The aging heart and post-infarction left ventricular remodeling. J Am Coll Cardiol. 2011;57(1):9–17. doi: 10.1016/j.jacc.2010.08.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abel ED, Litwin SE, Sweeney G. Cardiac remodeling in obesity. Physiol Rev. 2008;88(2):389–419. doi: 10.1152/physrev.00017.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehnart SE. Understanding the physiology of heart failure through cellular and in vivo models-towards targeting of complex mechanisms. Exp Physiol. 2013;98(3):622–628. doi: 10.1113/expphysiol.2012.068262. [DOI] [PubMed] [Google Scholar]
- 16.Chen YH, et al. Microsatellite polymorphism in promoter of heme oxygenase-1 gene is associated with susceptibility to coronary artery disease in type 2 diabetic patients. Hum Genet. 2002;111(1):1–8. doi: 10.1007/s00439-002-0769-4. [DOI] [PubMed] [Google Scholar]
- 17.Exner M, et al. Heme oxygenase-1 gene promoter microsatellite polymorphism is associated with restenosis after percutaneous transluminal angioplasty. J Endovasc Ther. 2001;8(5):433–440. doi: 10.1177/152660280100800501. [DOI] [PubMed] [Google Scholar]
- 18.Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiol Rev. 2010;90(1):207–258. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- 19.Barth E, Stämmler G, Speiser B, Schaper J. Ultrastructural quantitation of mitochondria and myofilaments in cardiac muscle from 10 different animal species including man. J Mol Cell Cardiol. 1992;24(7):669–681. doi: 10.1016/0022-2828(92)93381-S. [DOI] [PubMed] [Google Scholar]
- 20.Lesnefsky EJ, Moghaddas S, Tandler B, Kerner J, Hoppel CL. Mitochondrial dysfunction in cardiac disease: ischemia--reperfusion, aging, and heart failure. J Mol Cell Cardiol. 2001;33(6):1065–1089. doi: 10.1006/jmcc.2001.1378. [DOI] [PubMed] [Google Scholar]
- 21.Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345(6203):1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kluge MA, Fetterman JL, Vita JA. Mitochondria and endothelial function. Circ Res. 2013;112(8):1171–1188. doi: 10.1161/CIRCRESAHA.111.300233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piantadosi CA, Suliman HB. Redox regulation of mitochondrial biogenesis. Free Radic Biol Med. 2012;53(11):2043–2053. doi: 10.1016/j.freeradbiomed.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zukor H, et al. HO-1-mediated macroautophagy: a mechanism for unregulated iron deposition in aging and degenerating neural tissues. J Neurochem. 2009;109(3):776–791. doi: 10.1111/j.1471-4159.2009.06007.x. [DOI] [PubMed] [Google Scholar]
- 25.Hull TD, et al. Heme oxygenase-1 regulates mitochondrial quality control in the heart. JCI Insight. 2016;1(2):e85817. doi: 10.1172/jci.insight.85817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong C, et al. Heme oxygenase-1 enhances autophagy in podocytes as a protective mechanism against high glucose-induced apoptosis. Exp Cell Res. 2015;337(2):146–159. doi: 10.1016/j.yexcr.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Wu W, et al. PINK1-Parkin-Mediated Mitophagy Protects Mitochondrial Integrity and Prevents Metabolic Stress-Induced Endothelial Injury. PLoS ONE. 2015;10(7):e0132499. doi: 10.1371/journal.pone.0132499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song M, et al. Interdependence of Parkin-Mediated Mitophagy and Mitochondrial Fission in Adult Mouse Hearts. Circ Res. 2015;117(4):346–351. doi: 10.1161/CIRCRESAHA.117.306859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupfer C, et al. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat Immunol. 2013;14(5):480–488. doi: 10.1038/ni.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marzetti E, Csiszar A, Dutta D, Balagopal G, Calvani R, Leeuwenburgh C. Role of mitochondrial dysfunction and altered autophagy in cardiovascular aging and disease: from mechanisms to therapeutics. Am J Physiol Heart Circ Physiol. 2013;305(4):H459–H476. doi: 10.1152/ajpheart.00936.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubli DA, et al. Parkin protein deficiency exacerbates cardiac injury and reduces survival following myocardial infarction. J Biol Chem. 2013;288(2):915–926. doi: 10.1074/jbc.M112.411363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siddall HK, et al. Loss of PINK1 increases the heart’s vulnerability to ischemia-reperfusion injury. PLoS ONE. 2013;8(4):e62400. doi: 10.1371/journal.pone.0062400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191(5):933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Dorn GW. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340(6131):471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205(2):143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koyano F, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510(7503):162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- 38.Galan JM, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16(19):5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamark T, Kirkin V, Dikic I, Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8(13):1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 41.Piquereau J, et al. Protective role of PARK2/Parkin in sepsis-induced cardiac contractile and mitochondrial dysfunction. Autophagy. 2013;9(11):1837–1851. doi: 10.4161/auto.26502. [DOI] [PubMed] [Google Scholar]
- 42.Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci USA. 2011;108(23):9572–9577. doi: 10.1073/pnas.1106291108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labruto F, Yang J, Vaage J, Valen G. Role of tumor necrosis factor alpha and its receptor I in preconditioning by hyperoxia. Basic Res Cardiol. 2005;100(3):198–207. doi: 10.1007/s00395-004-0509-0. [DOI] [PubMed] [Google Scholar]
- 44.Pourkhalili K, et al. Ischemia and reperfusion-induced arrhythmias: role of hyperoxic preconditioning. J Cardiovasc Med (Hagerstown) 2009;10(8):635–642. doi: 10.2459/JCM.0b013e32832997f3. [DOI] [PubMed] [Google Scholar]
- 45.Tähepõld P, Ruusalepp A, Li G, Vaage J, Starkopf J, Valen G. Cardioprotection by breathing hyperoxic gas-relation to oxygen concentration and exposure time in rats and mice. Eur J Cardiothorac Surg. 2002;21(6):987–994. doi: 10.1016/S1010-7940(02)00125-2. [DOI] [PubMed] [Google Scholar]
- 46.Ryter SW, Choi AM. Regulation of autophagy in oxygen-dependent cellular stress. Curr Pharm Des. 2013;19(15):2747–2756. doi: 10.2174/1381612811319150010. [DOI] [PubMed] [Google Scholar]
- 47.Francis BM, et al. Reduced levels of mitochondrial complex I subunit NDUFB8 and linked complex I + III oxidoreductase activity in the TgCRND8 mouse model of Alzheimer’s disease. J Alzheimers Dis. 2014;39(2):347–355. doi: 10.3233/JAD-131499. [DOI] [PubMed] [Google Scholar]
- 48.Vogel RO, et al. Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J. 2005;272(20):5317–5326. doi: 10.1111/j.1742-4658.2005.04928.x. [DOI] [PubMed] [Google Scholar]
- 49.Suliman HB, Carraway MS, Tatro LG, Piantadosi CA. A new activating role for CO in cardiac mitochondrial biogenesis. J Cell Sci. 2007;120(Pt 2):299–308. doi: 10.1242/jcs.03318. [DOI] [PubMed] [Google Scholar]
- 50.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komatsu M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 52.Immenschuh S, Schröder H. Heme oxygenase-1 and cardiovascular disease. Histol Histopathol. 2006;21(6):679–685. doi: 10.14670/HH-21.679. [DOI] [PubMed] [Google Scholar]
- 53.Lee PJ, Alam J, Wiegand GW, Choi AM. Overexpression of heme oxygenase-1 in human pulmonary epithelial cells results in cell growth arrest and increased resistance to hyperoxia. Proc Natl Acad Sci USA. 1996;93(19):10393–10398. doi: 10.1073/pnas.93.19.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Motterlini R, Foresti R, Intaglietta M, Winslow RM. NO-mediated activation of heme oxygenase: endogenous cytoprotection against oxidative stress to endothelium. Am J Physiol. 1996;270(1 Pt 2):H107–H114. doi: 10.1152/ajpheart.1996.270.1.H107. [DOI] [PubMed] [Google Scholar]
- 55.Bak Z, Sjöberg F, Rousseau A, Steinvall I, Janerot-Sjoberg B. Human cardiovascular dose-response to supplemental oxygen. Acta Physiol (Oxf) 2007;191(1):15–24. doi: 10.1111/j.1748-1716.2007.01710.x. [DOI] [PubMed] [Google Scholar]
- 56.Bach FH. Heme oxygenase-1 as a protective gene. Wien Klin Wochenschr. 2002;114 Suppl 4:1–3. [PubMed] [Google Scholar]
- 57.Wu ML, Ho YC, Yet SF. A central role of heme oxygenase-1 in cardiovascular protection. Antioxid Redox Signal. 2011;15(7):1835–1846. doi: 10.1089/ars.2010.3726. [DOI] [PubMed] [Google Scholar]
- 58.Piantadosi CA, et al. Heme oxygenase-1 couples activation of mitochondrial biogenesis to anti-inflammatory cytokine expression. J Biol Chem. 2011;286(18):16374–16385. doi: 10.1074/jbc.M110.207738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang G, et al. Cardioprotective and antiapoptotic effects of heme oxygenase-1 in the failing heart. Circulation. 2010;121(17):1912–1925. doi: 10.1161/CIRCULATIONAHA.109.905471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewis P, et al. Lack of the antioxidant enzyme glutathione peroxidase-1 accelerates atherosclerosis in diabetic apolipoprotein E-deficient mice. Circulation. 2007;115(16):2178–2187. doi: 10.1161/CIRCULATIONAHA.106.664250. [DOI] [PubMed] [Google Scholar]
- 61.Lorenzo O, Picatoste B, Ares-Carrasco S, Ramírez E, Egido J, Tuñón J. Potential role of nuclear factor κB in diabetic cardiomyopathy. Mediators Inflamm. 2011;2011:652097. doi: 10.1155/2011/652097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown GC. Control of respiration and ATP synthesis in mammalian mitochondria and cells. Biochem J. 1992;284(Pt 1):1–13. doi: 10.1042/bj2840001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Becker LB, vanden Hoek TL, Shao ZH, Li CQ, Schumacker PT. Generation of superoxide in cardiomyocytes during ischemia before reperfusion. Am J Physiol. 1999;277(6 Pt 2):H2240–H2246. doi: 10.1152/ajpheart.1999.277.6.H2240. [DOI] [PubMed] [Google Scholar]
- 64.Vanden Hoek TL, Li C, Shao Z, Schumacker PT, Becker LB. Significant levels of oxidants are generated by isolated cardiomyocytes during ischemia prior to reperfusion. J Mol Cell Cardiol. 1997;29(9):2571–2583. doi: 10.1006/jmcc.1997.0497. [DOI] [PubMed] [Google Scholar]
- 65.Arany Z, Novikov M, Chin S, Ma Y, Rosenzweig A, Spiegelman BM. Transverse aortic constriction leads to accelerated heart failure in mice lacking PPAR-gamma coactivator 1alpha. Proc Natl Acad Sci USA. 2006;103(26):10086–10091. doi: 10.1073/pnas.0603615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Melo LG, et al. Gene therapy strategy for long-term myocardial protection using adeno-associated virus-mediated delivery of heme oxygenase gene. Circulation. 2002;105(5):602–607. doi: 10.1161/hc0502.103363. [DOI] [PubMed] [Google Scholar]
- 67.Sadagurski M, et al. IRS2 increases mitochondrial dysfunction and oxidative stress in a mouse model of Huntington disease. J Clin Invest. 2011;121(10):4070–4081. doi: 10.1172/JCI46305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joubert PE, et al. Chikungunya-induced cell death is limited by ER and oxidative stress-induced autophagy. Autophagy. 2012;8(8):1261–1263. doi: 10.4161/auto.20751. [DOI] [PubMed] [Google Scholar]
- 69.Tannous P, et al. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117(24):3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Matsui Y, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100(6):914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 71.Carchman EH, Rao J, Loughran PA, Rosengart MR, Zuckerbraun BS. Heme oxygenase-1-mediated autophagy protects against hepatocyte cell death and hepatic injury from infection/sepsis in mice. Hepatology. 2011;53(6):2053–2062. doi: 10.1002/hep.24324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakai A, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13(5):619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 73.Xie Z, He C, Zou MH. AMP-activated protein kinase modulates cardiac autophagy in diabetic cardiomyopathy. Autophagy. 2011;7(10):1254–1255. doi: 10.4161/auto.7.10.16740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khullar M, Al-Shudiefat AA, Ludke A, Binepal G, Singal PK. Oxidative stress: a key contributor to diabetic cardiomyopathy. Can J Physiol Pharmacol. 2010;88(3):233–240. doi: 10.1139/Y10-016. [DOI] [PubMed] [Google Scholar]
- 75.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462(2):245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15(12):2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- 77.Oka T, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485(7397):251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen L, et al. OPA1 mutation and late-onset cardiomyopathy: mitochondrial dysfunction and mtDNA instability. J Am Heart Assoc. 2012;1(5):e003012. doi: 10.1161/JAHA.112.003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci USA. 2008;105(5):1638–1643. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Piantadosi CA, Suliman HB. Transcriptional control of mitochondrial biogenesis and its interface with inflammatory processes. Biochim Biophys Acta. 2012;1820(4):532–541. doi: 10.1016/j.bbagen.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khan R, Sheppard R. Fibrosis in heart disease: understanding the role of transforming growth factor-beta in cardiomyopathy, valvular disease and arrhythmia. Immunology. 2006;118(1):10–24. doi: 10.1111/j.1365-2567.2006.02336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abreu JG, Ketpura NI, Reversade B, De Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol. 2002;4(8):599–604. doi: 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blom IE, Goldschmeding R, Leask A. Gene regulation of connective tissue growth factor: new targets for antifibrotic therapy? Matrix Biol. 2002;21(6):473–482. doi: 10.1016/S0945-053X(02)00055-0. [DOI] [PubMed] [Google Scholar]
- 84.Border WA, Noble NA. Transforming growth factor beta in tissue fibrosis. N Engl J Med. 1994;331(19):1286–1292. doi: 10.1056/NEJM199411103311907. [DOI] [PubMed] [Google Scholar]
- 85.MacLellan WR, Brand T, Schneider MD. Transforming growth factor-beta in cardiac ontogeny and adaptation. Circ Res. 1993;73(5):783–791. doi: 10.1161/01.RES.73.5.783. [DOI] [PubMed] [Google Scholar]
- 86.Jeong D, et al. Matricellular Protein CCN5 Reverses Established Cardiac Fibrosis. J Am Coll Cardiol. 2016;67(13):1556–1568. doi: 10.1016/j.jacc.2016.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoon PO, et al. The opposing effects of CCN2 and CCN5 on the development of cardiac hypertrophy and fibrosis. J Mol Cell Cardiol. 2010;49(2):294–303. doi: 10.1016/j.yjmcc.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 88.Cronje FJ, Carraway MS, Freiberger JJ, Suliman HB, Piantadosi CA. Carbon monoxide actuates O(2)-limited heme degradation in the rat brain. Free Radic Biol Med. 2004;37(11):1802–1812. doi: 10.1016/j.freeradbiomed.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 89.Carraway MS, Suliman HB, Kliment C, Welty-Wolf KE, Oury TD, Piantadosi CA. Mitochondrial biogenesis in the pulmonary vasculature during inhalational lung injury and fibrosis. Antioxid Redox Signal. 2008;10(2):269–275. doi: 10.1089/ars.2007.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Suliman HB, Carraway MS, Welty-Wolf KE, Whorton AR, Piantadosi CA. Lipopolysaccharide stimulates mitochondrial biogenesis via activation of nuclear respiratory factor-1. J Biol Chem. 2003;278(42):41510–41518. doi: 10.1074/jbc.M304719200. [DOI] [PubMed] [Google Scholar]
- 91.Suliman HB, Zobi F, Piantadosi CA. Heme Oxygenase-1/Carbon Monoxide System and Embryonic Stem Cell Differentiation and Maturation into Cardiomyocytes. Antioxid Redox Signal. 2016;24(7):345–360. doi: 10.1089/ars.2015.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.