Abstract
BACKGROUND. No laboratory test can predict the risk of nonrelapse mortality (NRM) or severe graft-versus-host disease (GVHD) after hematopoietic cellular transplantation (HCT) prior to the onset of GVHD symptoms.
METHODS. Patient blood samples on day 7 after HCT were obtained from a multicenter set of 1,287 patients, and 620 samples were assigned to a training set. We measured the concentrations of 4 GVHD biomarkers (ST2, REG3α, TNFR1, and IL-2Rα) and used them to model 6-month NRM using rigorous cross-validation strategies to identify the best algorithm that defined 2 distinct risk groups. We then applied the final algorithm in an independent test set (n = 309) and validation set (n = 358).
RESULTS. A 2-biomarker model using ST2 and REG3α concentrations identified patients with a cumulative incidence of 6-month NRM of 28% in the high-risk group and 7% in the low-risk group (P < 0.001). The algorithm performed equally well in the test set (33% vs. 7%, P < 0.001) and the multicenter validation set (26% vs. 10%, P < 0.001). Sixteen percent, 17%, and 20% of patients were at high risk in the training, test, and validation sets, respectively. GVHD-related mortality was greater in high-risk patients (18% vs. 4%, P < 0.001), as was severe gastrointestinal GVHD (17% vs. 8%, P < 0.001). The same algorithm can be successfully adapted to define 3 distinct risk groups at GVHD onset.
CONCLUSION. A biomarker algorithm based on a blood sample taken 7 days after HCT can consistently identify a group of patients at high risk for lethal GVHD and NRM.
FUNDING. The National Cancer Institute, American Cancer Society, and the Doris Duke Charitable Foundation.
Graft-versus-host-disease biomarkers measured one week after allogeneic transplant can risk stratify patients for lethal graft-versus-host disease and survival.
Introduction
Hematopoietic cellular transplantation (HCT) is an important treatment for high-risk hematologic malignancies whose curative potential depends on the graft-versus-leukemia (GVL) effect. Graft-versus-host disease (GVHD), the major cause of nonrelapse mortality (NRM) after HCT, is closely associated with GVL (1–4). Pretransplant clinical risk factors for GVHD include the degree of human leukocyte antigen (HLA) match between donor and recipient, recipient age, donor type, and conditioning regimen intensity (5, 6). Some centers use one or more of these risk factors to guide GVHD prophylaxis, such as the use of anti-thymocyte globulin when the donor is not an HLA-identical sibling (7), but such approaches are globally immunosuppressive and carry their own risks, in particular of opportunistic infections (8, 9).
Acute GVHD affects 40% to 60% of patients and targets the skin, liver, and gastrointestinal (GI) tract (6, 10). The median onset of acute GVHD is approximately 1 month after transplant (11, 12). Recently, a signature of 3 plasma biomarkers (TNFR1, ST2, and REG3α) at the onset of clinical symptoms has been shown to predict NRM and response to treatment (11). Our goal was to determine whether a biomarker signature early after HCT could predict NRM and GVHD before the development of overt clinical disease.
Results
Patients.
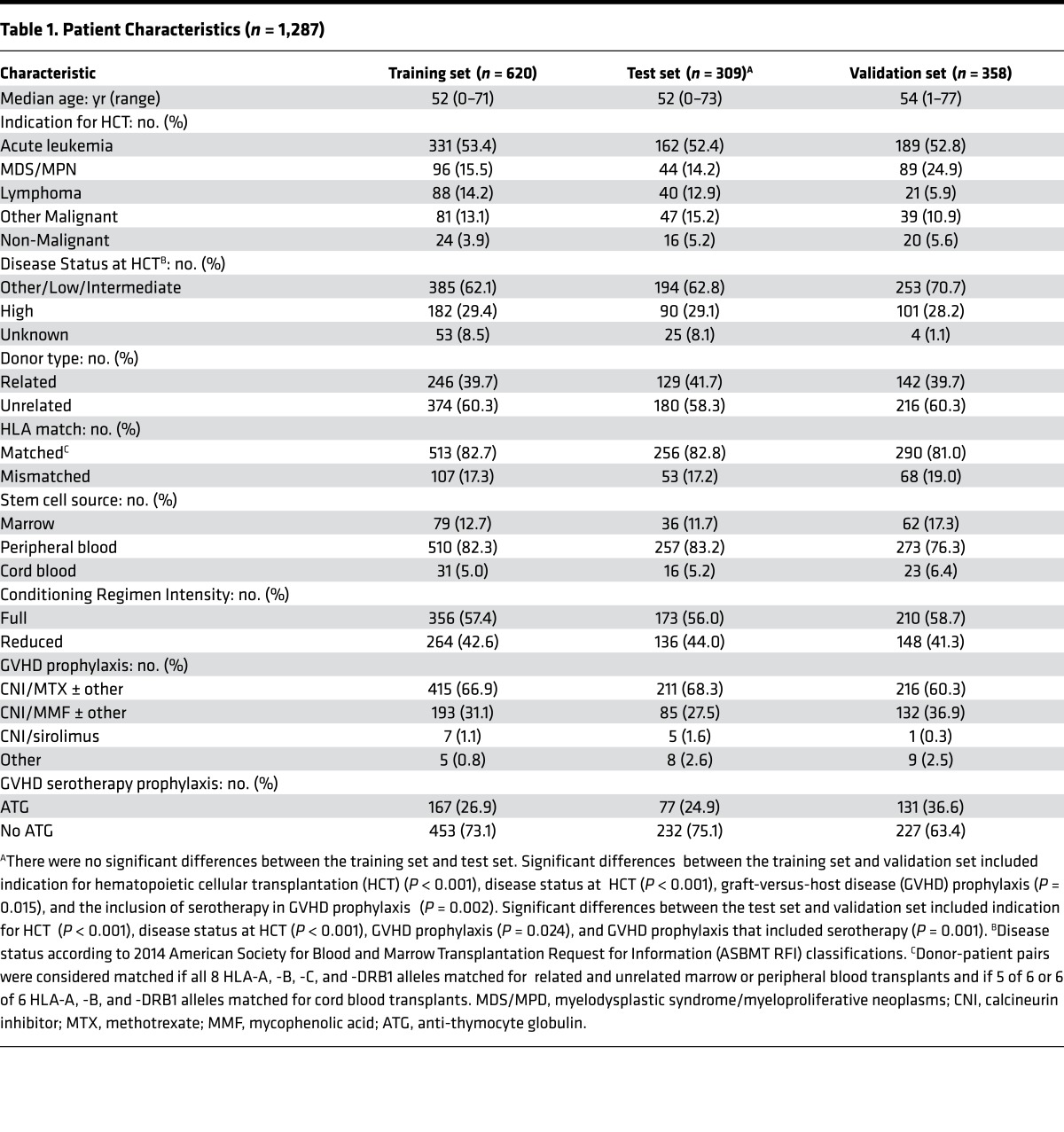
The clinical characteristics of all the patients are shown in Table 1. We observed no significant differences between training and test sets following randomization. An independent multicenter validation set (9 centers, 3 countries; n = 358) differed significantly from the training and test sets with myelodysplastic syndrome as a more frequent indication for HCT (25% vs. 14%, P < 0.001), fewer patients with unknown disease status at HCT (1% vs. 8%, P < 0.001), less use of methotrexate-containing GVHD prophylaxis (60% vs. 68%, P = 0.024) and more use of anti-thymocyte globulin (37% vs. 25%, P = 0.001). The overall incidence of 6-month NRM for the training, test, and validation sets was highly similar at 11%, 12%, and 13%, respectively. The median day of GVHD onset was 28 days in the training set and 29 days in the test and validation sets (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/jci.insight.89798DS1).
Table 1. Patient Characteristics (n = 1,287).
Algorithm development.
We developed a predictive model using biomarker combinations in samples from the training set through a rigorous strategy to maximize reproducibility (see Methods). The most accurate model included the concentrations of ST2 and REG3α and the area under the receiver operating characteristic curve is 0.68 (Supplemental Figure 1). A threshold of p̂ = 0.16 separated high-risk (HR) and low-risk (LR) groups to identify a maximum number of HR patients with a near-maximum difference in NRM. The median and range for all 4 biomarkers are shown in Supplemental Table 2.
Algorithm performance at day 7 after HCT.
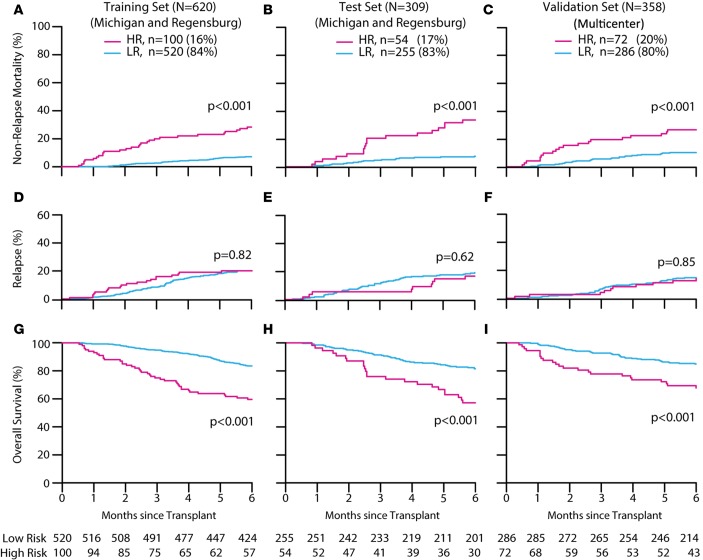
This final Mount Sinai Acute GVHD International Consortium (MAGIC) algorithm identified an HR group in the training set whose NRM (28%) was significantly greater (P < 0.001) than that of the LR group (7%) (Figure 1A). Application of this algorithm to the test set produced similar, highly statistically significant differences between HR and LR groups (Figure 1B). We performed a second validation in the multicenter set and again observed large differences between groups, with an HR 6-month NRM of 26% versus 10% in the LR group (P < 0.001) (Figure 1C). NRM remained largely the same through the first 12 months after transplant (Supplemental Table 3). The proportion of patients in the HR group was similar in all 3 patient sets (16% to 20%). Relapse rates were equivalent in both risk groups in all 3 sets (Figure 1, D–F), with the result that HR patients experienced significantly worse overall survival (P < 0.001) (Figure 1, G–I).
Figure 1. Outcomes according to MAGIC risk stratification.
Six-month cumulative incidences of nonrelapse mortality in high risk (HR) and low risk (LR) were defined by the MAGIC algorithm and compared using Gray’s test. Training set (A): HR 28% (95% CI, 20 to 37); LR 7% (95% CI, 5 to 10); test set (B): HR 33% (95% CI, 21 to 46); LR 7% (95% CI, 5 to 11); validation set (C): HR 26% (95% CI, 17 to 37); LR 10% (95% CI, 7 to 14). Six-month relapse rates were as follows: training set (D): HR 20% (95% CI, 13 to 29); LR 20% (95% CI, 17 to 24); test set (E): HR 17% (95% CI, 8 to 28); LR 19% (95% CI, 15 to 24); validation set (F): HR 14% (95% CI, 7 to 23); LR 15% (95% CI, 11 to 19). Six-month overall survival rates were calculated by the Kaplan-Meier method and compared by the log-rank test: training set (G): HR 60% (95% CI, 51 to 70); LR 84% (95% CI, 80 to 87); test set (H): HR 57% (95% CI, 45 to 72); LR 81% (95% CI, 77 to 86); validation set (I): HR 68% (95% CI, 58 to 80); LR 85% (95% CI, 81 to 89).
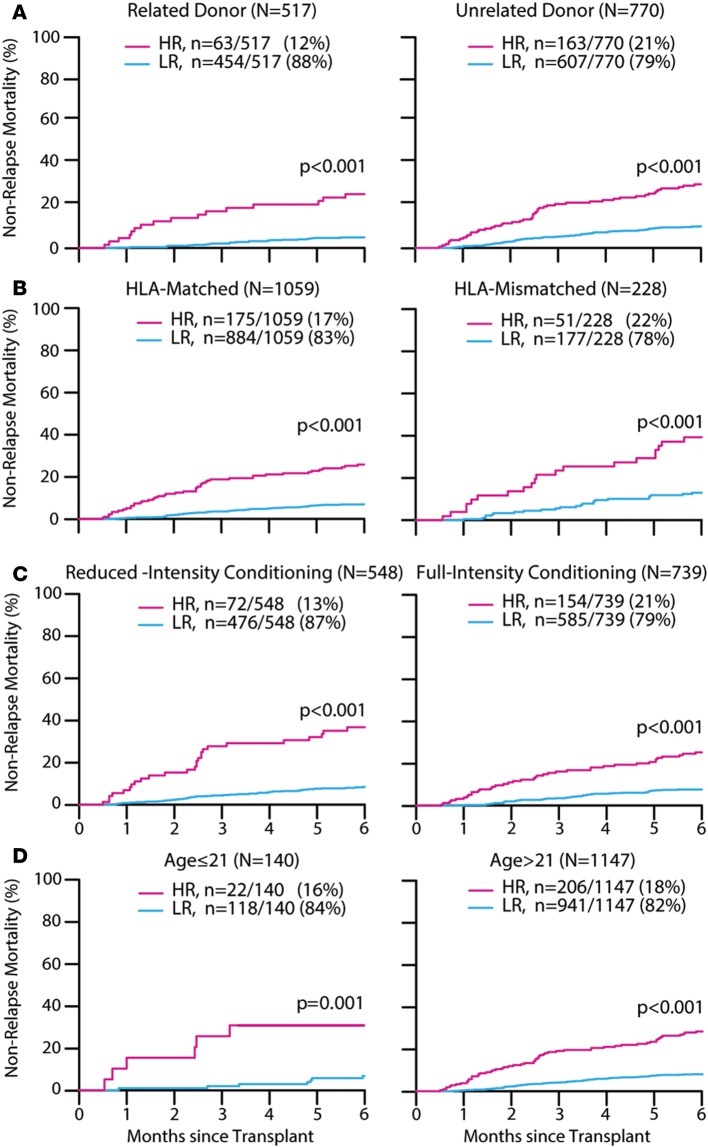
Several pre-HCT clinical risk factors predict a higher risk of NRM, such as HLA mismatch, non–family member donors, age of the recipient, and the intensity of the conditioning regimen (6, 13). Donor type and match were significant predictors of NRM in univariate analyses performed on the training set (Supplemental Table 4). Yet the MAGIC algorithm still stratified patients into 2 distinct risk groups independently of the degree of HLA match between donor and recipient, the genetic relationship of the donor to the recipient, the intensity of the conditioning regimen, and age (Figure 2, A–D). The differences between groups remained statistically significant in all 3 sets within each clinical risk factor except for pediatric patients in a few sets where the total number of patients was exceptionally small (Supplemental Table 5). Again, relapse rates were equivalent within all subgroups of clinical risk factors, resulting in a decrease of at least 20% in overall survival for HR patients (Supplemental Figures 2–4).
Figure 2. MAGIC risk groups.
Six-month cumulative incidence of nonrelapse mortality of all patients (n = 1,287) by (A) related donor: high risk (HR) 26% (95% CI, 15 to 37); low risk (LR) 5% (95% CI, 3 to 7); unrelated donor: HR 30% (95% CI, 23 to 37); LR 10% (95% CI, 8 to 13); (B) HLA matched: HR 26% (95% CI, 20 to 33); LR 7% (95% CI, 5 to 9); HLA mismatched: HR 39% (95% CI, 26 to 53); LR 13% (95% CI, 9 to 18); (C) reduced-intensity conditioning: HR 37% (95% CI, 26 to 48); LR 8% (95% CI, 6 to 11); full-intensity conditioning: HR 25% (95% CI, 19 to 32); LR 8% (95% CI, 6 to 10); (D) age ≤ 21: HR 27% (95% CI, 11 to 47); LR 6% (95% CI, 3 to 11); age > 21:HR 29% (95% CI, 23 to 36); LR 8% (95% CI, 7 to 10). Gray’s test was used for statistical comparisons between groups.
Causes of NRM.
We next analyzed the contribution of GVHD to NRM. HR patients were 3 times more likely to die from GVHD than LR patients when all 1,287 patients were considered (HR 19% vs. LR 6%, P < 0.001) and the difference was statistically significant within each set (Supplemental Figure 5). GVHD-related deaths reflected the efficacy of treatments that varied according to the standard of care at each center, but the majority of patients with grade II–IV acute GVHD received high-dose systemic steroids, and HR patients also experienced twice as much steroid-refractory GVHD as LR patients (HR 35% vs. 15%, P < 0.001). The GI tract is the GVHD target organ that is most resistant to treatment and represents a major cause of NRM (11, 14), and we observed twice as much severe GI GVHD (Supplemental Figure 6). Although severe skin GVHD is uncommon, affecting fewer than 5% of all patients, we found that HR patients experienced 4 times as much severe skin GVHD as LR patients (Supplemental Figure 6). All causes of NRM are shown in Supplemental Table 6.
Algorithm performance at onset of GVHD.
We have previously reported that an algorithm using 2 thresholds of 3 plasma biomarkers measured at the time of onset of GVHD symptoms is able to separate patients into 3 distinct risk strata (Ann Arbor [AA] scores 1, 2, and 3) regarding response to systemic treatment and NRM (11). We measured these 3 biomarkers (ST2, REG3α, and TNFRI) in 212 patients from this data set for whom samples were available at the onset of GVHD. Using thresholds to provide approximately the same NRM in each stratum as the 3-biomarker algorithm, the 2-biomarker algorithm successfully identified 3 distinct risk strata and assigned 45% of patients to the LR group, AA1 (Figure 3A). Thus, the same 2-biomarker algorithm that separated LR and HR on day 7 after transplant can be successfully adapted through the use of 2 thresholds to separate patients into 3 distinct risk strata at the onset of GVHD. When we applied the 3-biomarker algorithm to these same patient samples, it assigned substantially fewer patients to the LR group, with the result that NRM was lower for the intermediate risk group (AA2) (Figure 3B and Supplemental Table 7).
Figure 3. Graft-versus-host disease (GVHD)–related outcomes by MAGIC risk stratification and application of algorithm at GVHD onset.
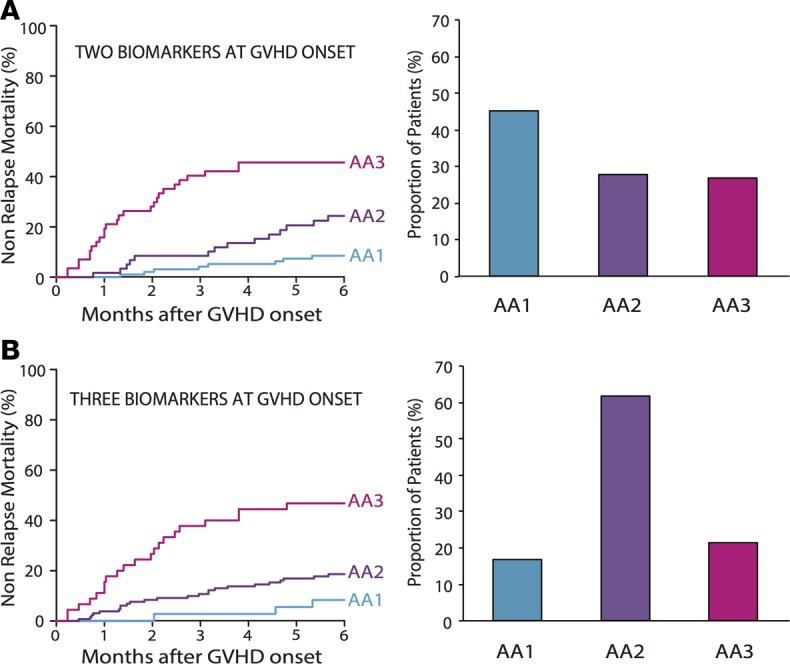
(A) Six-month cumulative incidences of nonrelapse mortality in Ann Arbor (AA) risk groups AA1, AA2, and AA3 were defined by the 2-biomarker-containing MAGIC algorithm applied at GVHD onset (n = 212): AA3 46% (95% CI, 32 to 58); AA2 24% (95% CI, 14 to 36); and AA1 8% (95% CI, 4 to 15). The proportion of patients in each risk group, as represented by the bar graph, were AA3 27% (n = 57), AA2 28% (n = 59), and AA1 45% (n = 96). (B) Six-month cumulative incidences of nonrelapse mortality in AA1, AA2, and AA3 were defined by the 3-biomarker-containing MAGIC algorithm applied at GVHD onset (n = 212): AA3 47% (95% CI, 32 to 61); AA2 19% (95% CI, 12 to 26); and AA1 8% (95% CI, 2 to 20). The proportion of patients in each risk group, as represented by the bar graph, were AA3 21% (n = 45), AA2 62% (n = 131), and AA1 17% (n = 36).
Discussion
A long-sought goal in HCT is the identification of individual patients at HR for severe GVHD. The day 7 MAGIC algorithm developed here identifies a significant number of such patients. The algorithm’s reproducibility among multiple transplant centers may be attributed to several elements of the study design. First, the acquisition rate of samples was very high (93%), ensuring a broad representation of patients. Second, the clinical data practices were standardized and monitored among all centers, thereby increasing the accuracy of the data. Third, the final algorithm was the result of a vigorous cross-validation strategy in a large number of patients that tested performance in 75 different combinations of the training set prior to the development of the final model and its validation in 2 independent sets.
The fidelity of risk assignment by the MAGIC algorithm transcends known clinical risk factors for GVHD, such as conditioning regimen, age, HLA mismatch, or relatedness of the donor. These latter 2 risk factors directly reflect the histocompatibility antigens in the host to which donor T cells respond within days of graft infusion and were significant predictors of NRM in univariate analysis in the training set, but their incorporation into the algorithm did not appreciably improve its performance (Supplemental Table 8). The GVH reaction is already in progress by day 7 and has led to increased biomarker concentrations even though clinical symptoms may not occur until days or weeks later. The same may be said for the conditioning regimen intensity, which correlates with the inflammation that amplifies donor T cell responses to host alloantigens (15). The MAGIC algorithm’s fidelity across these variables derives from assigning a greater percentage of patients with an adverse characteristic to the HR group. For example, 163 of 770 (21%) of unrelated donors are assigned to the HR group, compared with 63 of 517 (12%) of related donors (P < 0.001).
Importantly, when biomarkers are measured at the onset of GVHD symptoms, this 2-biomarker algorithm can be successfully adapted using 2 thresholds to define 3 distinct risk groups. Indeed, the 2-biomarker algorithm assigned more patients to both the HR and LR groups than the 3-biomarker algorithm. We speculate that the superior performance of the 2-biomarker algorithm is due, at least in part, to the increased sensitivity of the new ELISA assay for ST2 that was not available when the 3-biomarker algorithm was derived (Supplemental Table 7). We now appreciate that both ST2 and REG3α are closely associated with GI GVHD (16–18) and we speculate further that the levels of these biomarkers’ concentrations both on day 7 and at the onset of overall symptoms reflect GI pathology that is not yet clinically apparent. The overall incidence of severe GI GVHD in our study (9.6%) was similar to that of other reports (7.9%) (14), and the GI tract is key to overall GVHD severity because it is affected in 86% of severe cases (12). It is thus worth noting again that the algorithm allocated twice as many patients who would eventually develop severe GI GVHD to the HR group (Supplemental Figure 6).
This large study confirms earlier studies in which 50% of GVHD occurs after day 28 and 90% occurs after day 14 (Supplemental Table 1) (11, 12). Thus, the use of the MAGIC algorithm could facilitate preemptive intervention for GVHD prior to the onset of clinical disease in a substantial number of patients. One attractive strategy that avoids global immunosuppression and thus minimizes increased risk for relapse is to interrupt traffic of GVHD effector cells to the GI tract. Blockade of the α4β7 integrin expressed on donor T cells that home to the intestinal mucosa can abrogate experimental GVHD (19–21), and α4β7 is expressed on greater percentages of T cells in patients who later develop intestinal acute GVHD (22). The safety and efficacy of monoclonal antibodies such as vedolizumab (α4β7 antagonist), natalizumab (α4 antagonist), and etrolizumab (β7 antagonist) to treat inflammatory bowel disease is established (23–25), making them prime candidates for such intervention. Two clinical trials of such strategies in GVHD prophylaxis or treatment are currently ongoing (clinicaltrials.gov; NCT02133924 and NCT02728895).
The biomarkers for HR disease may identify additional pathways that could be therapeutically targeted. ST2, a decoy receptor for soluble IL-33, is shed from activated T cells as GVHD progresses, and soluble ST2 administration has been shown to reduce experimental GVHD (17, 18). Additional strategies may target IL-33 itself, which is released from dying GI epithelial cells during GVHD. REG3α is produced by GI epithelium, in particular Paneth cells, whose numbers decrease significantly during GVHD (26, 27). Thus, REG3α production decreases during GVHD even as its concentration increases in the bloodstream as a result of damaged epithelial mucosa (16). IL-22 induces REG3α, and lower numbers of circulating, IL-22–secreting innate lymphoid cells after transplant are associated with a higher risk for GVHD (28, 29). Administration of IL-22 restores REG3α homeostasis and accelerates repair of the epithelial mucosa, preventing GVHD in preclinical models (29, 30). This appealing approach avoids further immunosuppression altogether and in fact enhances the reconstitution of the innate immune system of the GI tract. A recombinant version of IL-22 has been approved for human use and is currently being tested in a clinical trial to treat GVHD (clinicaltrials.gov; NCT02406651).
Regardless of the nature of preemptive interventions, the MAGIC algorithm should prove a useful tool in clinical research of GVHD therapy because it identifies patients at HR for severe disease. The exact nature of the intervention, including its inherent risks as well as potential benefits, will largely determine the enthusiasm of patients and physicians for any particular approach. Future improvements to the algorithm might include the incorporation of additional biomarkers or repeating the test at a later time point to increase sensitivity. Nevertheless, the MAGIC algorithm represents an important advance toward precision medicine for HCT patients.
Methods
Study design and oversight.
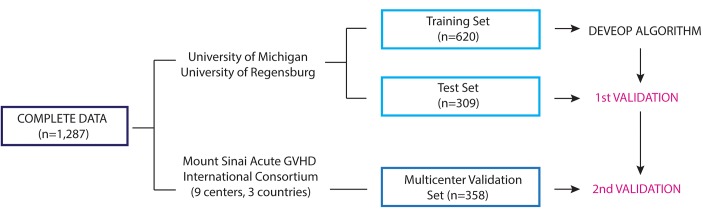
Patients from 11 centers in the Mount Sinai Acute GVHD International Consortium (MAGIC) underwent first allogeneic HCT from January 2005 to June 2015 and provided blood samples for a biorepository 7 days after HCT (Figure 4). Patient samples from the 2 largest contributing centers, the University of Michigan, Ann Arbor (n = 642) and the University of Regensburg, Germany (n = 287) were combined and were then randomly assigned to a training (n = 620) and test (n = 309) set, conditional to similar ratios of patients from each center, median HCT dates, and overall 6-month NRM. An independent group of 358 patients from the 9 other MAGIC centers constituted the validation set (Supplemental Table 9).
Figure 4. Study scheme of algorithm development and validation.
Clinical data and plasma samples from day 7 after hematopoietic cellular transplantation were available from 1,287 patients transplanted at 11 MAGIC centers. Patient samples from the 2 largest centers, the University of Michigan and the University of Regensburg, were randomly assigned to the training and test sets in a 2:1 proportion. The remaining 358 patients were assigned to the independent multicenter validation set. The training set alone (n = 620) was used to develop the algorithm. All possible combinations of 1 to 4 biomarkers were used to model 6-month nonrelapse mortality (NRM) by competing-risks regression. Rigorous comparison of models through a Monte Carlo cross validation of 75 different, randomly created training sets confirmed that the models using ST2 and REG3α were superior to all other biomarker combinations. We used this model to predict the probability of 6-month NRM in the patients from the training set, rank ordered them from lowest to highest, and chose a threshold to separate risk groups for the final algorithm (see Methods). We then applied the algorithm to the test set in a first validation and to the multicenter validation set in a second validation.
Clinical data, blood collection, and analysis.
GVHD clinical staging was standardized using published guidelines (31), and was prospectively reviewed during monthly data teleconferences starting in 2013 (n = 600). Blood samples were collected prospectively 7 ± 3 days after HCT. Nonrelapse deaths were considered related to GVHD if the patient died from either GVHD itself or from an infection that developed while receiving systemic steroids (at least 10 mg prednisone daily or equivalent) for the treatment of GVHD. Steroid-refractory GVHD was GVHD that either did not respond or required additional therapy within 28 days. Samples were shipped to a central laboratory where they were analyzed in batches for 4 GVHD biomarkers (ST2, REG3α, TNFR1, and IL-2Rα) by ELISA as previously described (16, 32, 33).
Statistics.
Biomarker concentrations were normalized by log-transformation and all 15 possible combinations of 1 to 4 biomarkers were used to model 6-month NRM by competing-risks regression, with relapse as the competing risk (34). We compared models using either Akaike’s information criterion (AIC) for non-nested comparisons or the Wald test for nested comparisons to determine the most accurate model (35). All models were sorted according to AIC and the lowest AIC was considered to have the best fit. Furthermore, the best 1-, 2-, and 3-biomarker models based on AIC and the 4-biomarker model were compared using the Wald test considering P values of less than 0.05 to indicate statistical significance. The combination of ST2 and REG3α best predicted 6-month NRM, which we confirmed with Monte Carlo cross validation by randomly creating 75 different training sets and repeating the modeling process (36). No combination of 1, 3, or 4 biomarkers was superior to the combination of these 2 biomarkers. Seventy-five of seventy-five (100%) of the 2-biomarker models included ST2 and 68 of 75 (91%) included REG3α. We then created a training set at random and repeated the entire process to generate a final model: log10[–log10(1 – p̂ )] = –11.263 + 1.844(log10ST2) + 0.577(log10REG3α), where p̂ = predicted probability of 6- month NRM. We determined the p̂ for each patient and rank ordered them from lowest to highest. We observed multiple thresholds that demarcated groups with a difference of at least 15% NRM, which we deemed clinically significant (Supplemental Table 10). We chose a threshold of p̂ = 0.16 to maximize the size of the HR patient group while maintaining a near-maximum difference in NRM. Differences in cumulative incidence of NRM and relapse between HR and LR groups were calculated by Gray’s test. Overall survival was estimated by the Kaplan-Meier method and differences between groups were calculated using the log-rank test.
Patient characteristics between training, test, and validation sets were compared using chi-squared or Wilcoxon rank-sum tests as appropriate. Differences in proportions for the cause of death analysis and GVHD incidence were calculated using chi-squared tests. Clinical risk factors that were statistically significant (P < 0.05) predictors for NRM were identified by univariate analysis in the training set (Supplemental Table 4). An algorithm to predict NRM that combined significant clinical risk factors and biomarkers was derived using the same training set used to derive the biomarkers-only algorithm. All analyses were performed using R statistical package version 3.2.3 (R Development Core Team 2015). Error bars represent the SEM in all figure parts where error bars are shown.
Study approval.
The institutional review boards at each of the 11 participating centers approved this study and written informed consent was received from participants prior to inclusion in the study. The participating centers are listed in Supplemental Table 9.
Author contributions
MJH, UO, JEL, and JLMF were involved in the conception and design of the study. MJH, UO, EH, ASR, HMM, MA, WJH, FA, YAE, EOH, UB, MQ, RO, MW, SM, AP, YBC, SD, MRL, KW, JEL, and JLMF acquired the data and/or analyzed and interpreted the data. All authors wrote the manuscript or were substantially involved in its revision before submission.
Supplementary Material
Acknowledgments
Supported by grants (R21CA173459, P01 CA03942, and P30 CA106521) from the National Cancer Institute, an American Cancer Society Clinical Research Professorship (to JLMF), and a Doris Duke Charitable Foundation Clinical Research Mentorship (to MH).
Footnotes
We thank Rachel Young, Project Manager for MAGIC, the many colleagues who contributed to data collection and phenotypic characterization of clinical samples, and all the patients who participated in this study.
Role of funding source: The funding sources played no role in the design of the study, data collection, analysis, or interpretation, writing the report, or the decision to submit the paper for publication.
Conflict of interest: J.E. Levine and J.L.M. Ferrara are coinventors of a patent (number 62/411,230) for GVHD biomarkers.
Reference information:JCI Insight. 2017;2(3):e89798. https://doi.org/10.1172/jci.insight.89798.
Contributor Information
Umut Özbek, Email: umut.ozbek@mountsinai.org.
Ernst Holler, Email: ernst.holler@klinik.uni-regensburg.de.
Anne S. Renteria, Email: anne.renteria@mssm.edu.
Hannah Major-Monfried, Email: hannah.major-monfried@icahn.mssm.edu.
Pavan Reddy, Email: reddypr@umich.edu.
Mina Aziz, Email: mina.aziz@icahn.mssm.edu.
Francis Ayuk, Email: ayuketang@uke.de.
Udomsak Bunworasate, Email: ubunworasate@gmail.com.
Muna Qayed, Email: Muna.Qayed@choa.org.
Rainer Ordemann, Email: Rainer.Ordemann@uniklinikum-dresden.de.
Matthias Wölfl, Email: Woelfl_M@ukw.de.
Stephan Mielke, Email: mielke_S@ukw.de.
Attaphol Pawarode, Email: pawarode@med.umich.edu.
Yi-Bin Chen, Email: YCHEN6@PARTNERS.ORG.
Steven Devine, Email: steven.devine@osumc.edu.
Andrew C. Harris, Email: Andrew.Harris@hsc.utah.edu.
Madan Jagasia, Email: madan.jagasia@Vanderbilt.Edu.
Nicolaus Kröger, Email: nkroeger@uke.de.
Franco Locatelli, Email: franco.locatelli@opbg.net.
George Morales, Email: george.morales@mssm.edu.
Ryotaro Nakamura, Email: RNakamura@coh.org.
Ran Reshef, Email: ran.reshef@columbia.edu.
Daniela Weber, Email: Daniela.Weber@klinik.uni-regensburg.de.
Kitsada Wudhikarn, Email: kwudhikarn@hotmail.com.
References
- 1.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anasetti C, et al. Peripheral-blood stem cells versus bone marrow from unrelated donors. N Engl J Med. 2012;367(16):1487–1496. doi: 10.1056/NEJMoa1203517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gooley TA, et al. Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363(22):2091–2101. doi: 10.1056/NEJMoa1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Socie G, Ritz J, Martin PJ. Current challenges in chronic graft-versus-host disease. Biol Blood Marrow Transplant. 2010;16(1 Suppl):S146–S151. doi: 10.1016/j.bbmt.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 5.Harris AC, Ferrara JL, Levine JE. Advances in predicting acute GVHD. Br J Haematol. 2013;160(3):288–302. doi: 10.1111/bjh.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jagasia M, et al. Risk factors for acute GVHD and survival after hematopoietic cell transplantation. Blood. 2012;119(1):296–307. doi: 10.1182/blood-2011-06-364265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finke J, et al. Standard graft-versus-host disease prophylaxis with or without anti-T-cell globulin in haematopoietic cell transplantation from matched unrelated donors: a randomised, open-label, multicentre phase 3 trial. Lancet Oncol. 2009;10(9):855–864. doi: 10.1016/S1470-2045(09)70225-6. [DOI] [PubMed] [Google Scholar]
- 8.Nucci M, et al. Infectious complications in patients randomized to receive allogeneic bone marrow or peripheral blood transplantation. Transpl Infect Dis. 2003;5(4):167–173. doi: 10.1111/j.1399-3062.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 9.Tomblyn M, et al. Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: a global perspective. Biol Blood Marrow Transplant. 2009;15(10):1143–1238. doi: 10.1016/j.bbmt.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majhail NS, et al. Significant improvement in survival after unrelated donor hematopoietic cell transplantation in the recent era. Biol Blood Marrow Transplant. 2015;21(1):142–150. doi: 10.1016/j.bbmt.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine JE, et al. A prognostic score for acute graft-versus-host disease based on biomarkers: a multicentre study. Lancet Haematol. 2015;2(1):e21–e29. doi: 10.1016/S2352-3026(14)00035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacMillan ML, et al. A refined risk score for acute graft-versus-host disease that predicts response to initial therapy, survival, and transplant-related mortality. Biol Blood Marrow Transplant. 2015;21(4):761–767. doi: 10.1016/j.bbmt.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kollman C, et al. The effect of donor characteristics on survival after unrelated donor transplantation for hematologic malignancy. Blood. 2016;127(2):260–267. doi: 10.1182/blood-2015-08-663823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castilla-Llorente C, et al. Prognostic factors and outcomes of severe gastrointestinal GVHD after allogeneic hematopoietic cell transplantation. Bone Marrow Transplant. 2014;49(7):966–971. doi: 10.1038/bmt.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill GR, Crawford JM, Cooke KR, Brinson YS, Pan L, Ferrara JL. Total body irradiation and acute graft-versus-host disease: the role of gastrointestinal damage and inflammatory cytokines. Blood. 1997;90(8):3204–3213. [PubMed] [Google Scholar]
- 16.Ferrara JL, et al. Regenerating islet-derived 3-alpha is a biomarker of gastrointestinal graft-versus-host disease. Blood. 2011;118(25):6702–6708. doi: 10.1182/blood-2011-08-375006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reichenbach DK, et al. The IL-33/ST2 axis augments effector T-cell responses during acute GVHD. Blood. 2015;125(20):3183–3192. doi: 10.1182/blood-2014-10-606830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, et al. ST2 blockade reduces sST2-producing T cells while maintaining protective mST2-expressing T cells during graft-versus-host disease. Sci Transl Med. 2015;7(308):308ra160. doi: 10.1126/scitranslmed.aab0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueha S, et al. Intervention of MAdCAM-1 or fractalkine alleviates graft-versus-host reaction associated intestinal injury while preserving graft-versus-tumor effects. J Leukoc Biol. 2007;81(1):176–185. doi: 10.1189/jlb.0306231. [DOI] [PubMed] [Google Scholar]
- 20.Waldman E, et al. Absence of beta7 integrin results in less graft-versus-host disease because of decreased homing of alloreactive T cells to intestine. Blood. 2006;107(4):1703–1711. doi: 10.1182/blood-2005-08-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murai M, et al. Peyer’s patch is the essential site in initiating murine acute and lethal graft-versus-host reaction. Nat Immunol. 2003;4(2):154–160. doi: 10.1038/ni879. [DOI] [PubMed] [Google Scholar]
- 22.Chen YB, et al. Up-regulation of alpha4beta7 integrin on peripheral T cell subsets correlates with the development of acute intestinal graft-versus-host disease following allogeneic stem cell transplantation. Biol Blood Marrow Transplant. 2009;15(9):1066–1076. doi: 10.1016/j.bbmt.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Targan SR, et al. Natalizumab for the treatment of active Crohn’s disease: results of the ENCORE Trial. Gastroenterology. 2007;132(5):1672–1683. doi: 10.1053/j.gastro.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 24.Sandborn WJ, et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med. 2013;369(8):711–721. doi: 10.1056/NEJMoa1215739. [DOI] [PubMed] [Google Scholar]
- 25.Vermeire S, et al. Etrolizumab as induction therapy for ulcerative colitis: a randomised, controlled, phase 2 trial. Lancet. 2014;384(9940):309–318. doi: 10.1016/S0140-6736(14)60661-9. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa H, et al. Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm Bowel Dis. 2003;9(3):162–170. doi: 10.1097/00054725-200305000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Levine JE, et al. Low Paneth cell numbers at onset of gastrointestinal graft-versus-host disease identify patients at high risk for nonrelapse mortality. Blood. 2013;122(8):1505–1509. doi: 10.1182/blood-2013-02-485813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munneke JM, et al. Activated innate lymphoid cells are associated with a reduced susceptibility to graft-versus-host disease. Blood. 2014;124(5):812–821. doi: 10.1182/blood-2013-11-536888. [DOI] [PubMed] [Google Scholar]
- 29.Sanos SL, Vonarbourg C, Mortha A, Diefenbach A. Control of epithelial cell function by interleukin-22-producing RORγt+ innate lymphoid cells. Immunology. 2011;132(4):453–465. doi: 10.1111/j.1365-2567.2011.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindemans CA, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528(7583):560–564. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris AC, et al. International, multicenter standardization of acute graft-versus-host disease clinical data collection: a report from the Mount Sinai Acute GVHD International Consortium. Biol Blood Marrow Transplant. 2016;22(1):4–10. doi: 10.1016/j.bbmt.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vander Lugt MT, et al. ST2 as a marker for risk of therapy-resistant graft-versus-host disease and death. N Engl J Med. 2013;369(6):529–539. doi: 10.1056/NEJMoa1213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paczesny S, et al. A biomarker panel for acute graft-versus-host disease. Blood. 2009;113(2):273–278. doi: 10.1182/blood-2008-07-167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94(446):496–509. doi: 10.1080/01621459.1999.10474144. [DOI] [Google Scholar]
- 35.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19(6):716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 36.Xu Q-S, Liang Y-Z. Monte Carlo cross validation. Chemometr Intell Lab. 2001;56(1):1–11. doi: 10.1016/S0169-7439(00)00122-2. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.