Abstract
It has been shown that streptococcal pyrogenic exotoxin B (SPE B) can induce cells to undergo apoptosis. The present study is to dissect the role of SPE B protease and SPE B protein in the apoptotic process of A549 cells and to elucidate the SPE B-induced apoptotic pathway. Recombinant SPE B (rSPE B) and C192S, a mutant of SPE B without protease activity, were expressed in Escherichia coli and purified by using an affinity column. The apoptosis of A549 cells was assayed by propidium iodide staining, followed by flow cytometry analysis. Our results showed that SPE B induced apoptosis in a dose-dependent manner, whereas C192S did not. When cells were pretreated with rSPE B (2 μg/ml) for as briefly as 5 min and then incubated with C192S of 28 kDa, an apoptosis that is proportional to the period of pretreatment was observed but not with C192S of 42 kDa. These results suggest that the extracellular protease activity of rSPE B is required for the initiation of apoptosis and that the size of SPE B is important for an effective induction of apoptosis. The time course analysis revealed that molecules activated in apoptosis were in the following order: caspase-8 (1.5 h), t-Bid (2.5 h), Bax (3 h), cytochrome c release (6 h), caspase-9 (7 h), and caspase-3 (8 h). The overexpression of Bcl-2 inhibited depolarization of mitochondrial membrane, cytochrome c release, and apoptosis. The results of the present study suggest that SPE B-induced apoptosis is mediated through a receptor-like mechanism and a mitochondrion-dependent pathway.
Group A streptococcus (GAS) is an important human pathogen that causes various severe infections, including pharyngitis and cellulitis, and severe invasive diseases such as rheumatic fever, necrotizing fasciitis, and streptococcal toxic shock syndrome (27, 32, 34). Streptococcal pyrogenic exotoxin B (SPE B), an exotoxin produced by all GAS, is an important factor in streptococcal infections (12, 13, 23, 42). Patients with low levels of SPE B antibody were more susceptible to the streptococcal infection than were individuals with high antibody titers (7, 25). GAS with a disrupted speB gene caused less mortality and tissue damage in mice than wild-type strains (18, 24).
SPE B is also a cysteine protease. It is synthesized as a 40-kDa zymogen and can be cleaved to a 28-kDa active enzyme by autolysis. The enzyme has been shown to cleave human fibronectin and degrade vitronectin that may facilitate bacterial dissemination, colonization, and invasion (15). It was also shown to cleave interleukin-1β precursor to produce biologically active IL-1β, a major mediator of inflammation (14). Treatment of U937 monocytic cells with SPE B decreased specific-125I-labeled single-chain u-PA binding by up to 85%. The urokinase receptor was cleaved by SPE B, and an active fragment of the receptor was released from the cell surface, suggesting that the cleavage of uPAR by SPE B may potentiate streptococcal virulence by reducing the capacity of phagocytic leukocytes to bind u-PA (43). In an in vitro study conducted on A549 cells, a human respiratory epithelial cell line, the mutant strains without protease had a decrease in invasion activity two- to threefold that of wild-type strains. The inhibition of cysteine protease with E64, a specific cysteine protease inhibitor, also decreased the invasion activity of GAS (40). These results suggested that protease activity makes a great contribution to the function of SPE B.
Apoptosis in many tissues and cells is associated with infection. Several pathogens have been identified as mediators of apoptosis both in vitro and in vivo (3, 4, 21, 22, 26, 44, 45). Studies from our group showed that the SPE B-producing strains of Streptococcus pyogenes caused a greater extent of apoptosis in U937 cells than did its isogenic SPE B-negative mutants and that SPE B alone could induce U937 cells to undergo apoptosis (19). SPE B also induced cell death in A549 cells, but its isogenic mutants were less effective at inducing apoptosis than wild-type strains were (41). Despite these reports suggesting the important role that SPE B may play at the cellular level, its molecular mechanism remains unclear. The present study was designed to examine how SPE B interacts with cells to initiate the apoptotic pathway in A549 cells, with a special emphasis on dissecting the role of “SPE B protein” and “SPE protease” in the process, and to determine what components in the apoptotic pathway are activated by SPE B. We used two forms of SPE B for the present study: wild-type recombinant SPE B, which has protease activity, and the mutant form, C192S, which has no protease activity due to the conversion of cysteine residue at the 192 position to serine.
Our results suggest that SPE B-induced apoptosis in A549 cells may be mediated by a receptor-like mechanism and that the protease activity of SPE B is required to initiate the apoptotic process by working on the cell surface to possibly expose the binding site for SPE B. An array of molecules in the apoptotic pathway was activated by SPE B, which includes caspase activation, mitochondrial membrane potential change, and cytochrome c release.
MATERIALS AND METHODS
Expression and purification of rSPE B and C192S.
The expression and purification of rSPE B and its mutant C192S have been described elsewhere (2). The DNA of Streptococcus pyogenes was extracted from strain A-20. The gene of ProSPE B was amplified by PCR with a His6 tag and BamHI recognition sites. The PCR product was purified and cloned into the BamHI site of pET21a vector, which was then transformed into Escherichia coli BL21(DE3) pLys strain. Stock cells in 250 μl were grown in 250 ml of Luria-Bertani medium containing 100 μg of ampicillin/ml until the optical density reached 0.5 to 1.0. An aliquot of 250 μl of IPTG (isopropyl-β-d-thiogalactopyranoside; 100 mg/ml) was added to induce protein expression. For the expression of rSPE B, cells were grown at 35°C overnight, while for the expression of C192S cells were grown at 28°C overnight. After induction, cells were centrifuged at 5,000 rpm for 15 min to pellet the cells, which were resuspended in 80 ml of buffer A (20 mM Tris, 200 mM NaCl [pH 7.0]). The resuspended pellet was broken three times by using a French press at 1,500 kg/cm2 for 30 s. The whole contents were centrifuged at 12,000 rpm for 10 min at 4°C to collect supernatant, which was then separated on an Ni-chelated column (Amersham Biosciences) and eluted by a 0 to 200 mM imidazole gradient. The collected fractions were concentrated by Amicon ultrafiltration with a 3-kDa cutoff membrane and exchanged with phosphate-buffered saline (PBS). The purity was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). rSPE B was expressed as a 42-kDa zymogen and converted to a 28-kDa active enzyme during the course of purification. The purified rSPE B and C192S were passed through Detoxi-Gel (Pierce) to remove possible lipopolysaccharide contamination.
Cell culture.
A549 cells are routinely maintained in the laboratory in complete medium (Dulbecco modified Eagle medium [DMEM] supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamate, and 50 μg of gentamicin/ml). Cells were incubated in a CO2 incubator at 37°C and 5% CO2 in a humidified atmosphere. For the experiment, cells at a density of 8 × 104 cells/well were seeded into a 24-well plate for 24 h, washed with PBS, and incubated in the complete medium containing the specific agents for indicated period of time as described for each experiment.
Analysis of apoptosis.
After incubation with SPE B or other agents, cells were fixed at −20°C with 70% ethanol and then treated with RNase (100 μg/ml). After they had been washed once with PBS, cells were stained with propidium iodide (40 μg/ml in PBS) for 30 min at room temperature in the dark. Apoptotic cells were quantified by FACScan with CELLQuest software (Becton Dickinson) and presented as the percentage of hypodiploid cells (40).
Western blotting.
Cells at a density of 2 × 105/well were seeded into a six-well plate for 24 h. The cells were washed with cold PBS and incubated with various agents for various times as described for each experiment. After incubation, the medium was removed, and the cells were washed twice with cold PBS. The cells were lysed in Laemmli sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 5% 2-mercaptoethanol, 0.001% bromophenol) and boiled for 5 min. After SDS-PAGE, proteins were transferred to polyvinylidene difluoride membrane, blocked with PBS containing 5% low-fat milk, and probed with different primary antibodies. After three washes with PBST (PBS containing 0.05% Tween 20), membranes were incubated with peroxidase-conjugated goat anti-mouse or goat anti-rabbit antibody (Calbiochem). Membranes were washed three times with PBST. The protein bands were visualized with a Liquid DAB-black substrate kit or by enhanced chemiluminescence (Amersham Biosciences). Primary antibodies used in the present study were rabbit polyclonal anti-caspase-8 antibody and anti-caspase-9 antibody (Oncogene), mouse monoclonal anti-caspase-3 antibody and anti-Bax antibody (Oncogene), rabbit polyclonal anti-t-Bid antibody (Biosource), mouse monoclonal anti-cytochrome c antibody (Pharmingen), and mouse monoclonal anti-β-actin antibody (Sigma).
Isolation of cytosolic and mitochondrial fractions.
After incubation, the cells were harvested by scraping on ice, washed with ice-cold PBS, and resuspended in 500 μl of buffer A (20 mM HEPES [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 40 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride). After incubation on ice for 1 h, cells were lysed by passing through a 27-gauge needle 20 to 30 times. The lysates were centrifuged at 750 × g for 5 min at 4°C, and the supernatant was centrifuged at 10,000 × g for 15 min at 4°C. The mitochondrial pellet was washed once in buffer A and lysed in Laemmli sample buffer. The supernatant was centrifuged at 100,000 × g for 30 min at 4°C to generate the cytosolic fraction.
Iodination of SPE B and C192S and binding assay.
An aliquot of 100 μl of rSPE B or C192S at a concentration of 2 mg/ml was added to an Eppendorf tube coated with 100 μl of iodogen (80 μg/ml) and incubated with 500 μCi of Na125I (Amersham Biosciences) at room temperature for 10 min. Whole reaction mixtures were then applied to a PD-10 column (Amersham Biosciences) and eluted with 0.01 M phosphate buffer (pH 7.2). Fractions after void volume that contained labeled protein were collected. Specific activity was determined from a trichloroacetic acid precipitates. Specific activities of 2.36 μCi/μg for SPE B and 2.23 μCi/μg for C192S were obtained. For a binding assay, cells at a density of 8 × 104 cells/well were plated in a 24-well plate and incubated in the CO2 incubator for 24 to 36 h. The culture medium was replaced with fresh medium that contained 2 × 106 cpm-labeled rSPE B (or C192S) with or without 100-fold excess unlabeled rSPE B (or C192S) in a total volume of 1 ml. The cells were reincubated at 37 or 4°C for 2 h. At the end of incubation, the medium was removed, and the cells were washed three times with PBS. The cells were finally dissolved in 100 μl of 1 N NaOH, and the radioactivity was determined in a gamma counter.
Transfection experiments.
A549 cells were plated at a density of 5 × 104 cells per well in a six-well plate. When the cell density reached 30 to 50% confluence, the cells were transfected with 2 μg of pUSE vector containing bcl-2 cDNA or empty vector (Upstate) by using Lipofectamine transfection reagent (Invitrogen). Stable cell lines were selected in the presence of 3 mg of G418/ml. Bcl-2 expression by the cells was evaluated by anti-Bcl-2 (Dako) immunoblotting.
Measurement of mitochondrial membrane potential change.
Fluorescent rhodamine dye was used to monitor the membrane potential change of mitochondria in A549 cells that were treated with or without SPE B (30). Cells were plated at a density of 3 × 105 cells per well in a six-well plate overnight. The cells were treated with rSPE B (20 μg/ml) for 4 or 8 h and then incubated with 5 μM Rhodamine-123 (Sigma) for 30 min at 37°C in the dark. The cells were removed from the plate by treatment with trypsin and centrifuged at 750 × g at 4°C for 10 min, and the cell pellets were resuspended and washed with PBS. The intensity of green fluorescence was then determined by FACScalibur flow cytometry (Becton Dickinson).
Caspase-8 short interfering RNA (siRNA) preparation.
Plasmids expressing short hairpin RNA were constructed by standard techniques. The pSUPER/enhanced green fluorescent protein (EGFP) vector was kindly provided by R. Agami (The Netherlands Cancer Institute, Amsterdam, The Netherlands). To generate pSUPER-Casp8/EGFP, the pSUPER/EGFP was digested with BglII and HindIII, and the annealed oligonucleotides (5′-gatccccGTTCCTGAGCCTGGACTACttcaagagaGTAGTCCAGGCTCAGGAACtttttggaaa-3′ and 5′-agcttttccaaaaaGTTCCTGAGCCTGGACTACtctcttgaaGTAGTCCAGGCTCAGGAACggg-3′; lowercase letters at 3′ and 5′ ends represent the restriction enzyme cutting sites for construction, and lowercase letters in the middles of the sequences represent spacers for siRNA to form a loop structure) (37) were ligated into the vector. A549 cells were cultured in 10-cm dish in DMEM supplemented with 10% fetal calf serum. Before transfection, 5 × 106 cells were washed with serum-free DMEM and mixed with 5 μl of Lipofectamine 2000 (Invitrogen) and 2.5 μg of DNA. After 24 h of incubation, cells were maintained in DMEM containing 10% fetal calf serum for an additional 24 h before the experiments. The FACSAria (Becton Dickinson) was used to sort EGFP-positive cells with purities ranging from 72 to 76% in different experiments.
Statistical analysis.
Fisher protected least significant difference was used for the statistical analysis. Differences were considered significant at a P value of <0.05.
RESULTS
Preparations of rSPE B and its mutant C192S.
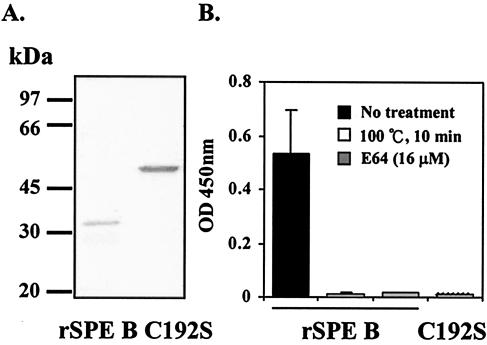
As described elsewhere (2), an E. coli expression system was used to express rSPE B and its mutant C192S. After IPTG induction, cells expressing rSPE B and C192S were collected and broken to release the expressed proteins. Pure proteins of rSPE B and rC192S could be obtained from an Ni-chelated column separation and eluted by an imidazole gradient. The rSPE B was expressed as a 42-kDa zymogen and converted to a 28-kDa form by an autolysis during the course of purification. The purified protein of C192S is 42 kDa and has no protease activity, whereas the rSPE B is 28 kDa and is an active enzyme (Fig. 1A). The protease activity of rSPE B can be completely inhibited either by heat or the cysteine protease inhibitor E64 (Fig. 1B). The purified rSPE B and rC192S were passed through Detoxi-Gel to remove possible lipopolysaccharide contamination.
FIG. 1.
The purity and protease activity of rSPE B and C192S. Purified rSPE B and C192S were separated on a SDS-12% PAGE. (A) Single bands of 28-kDa rSPE B and 42-kDa C192S were observed. Each protein was incubated with azocasein, and their protease activities were determined from the absorption at 450 nm of the supernatant of the reaction mixture. (B) The protease activity of rSPE B was also determined in the presence of E64 or in the heat-denatured form.
SPE B-induced apoptosis in A549 cells.
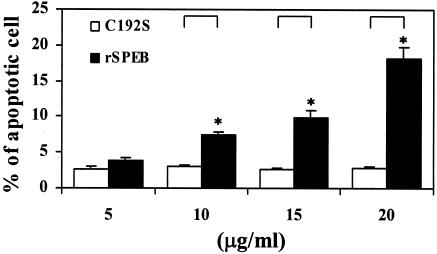
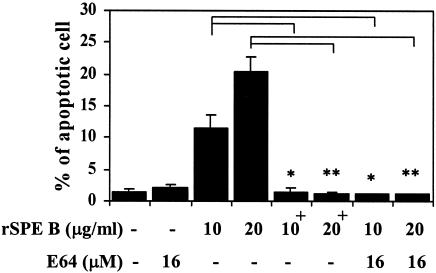
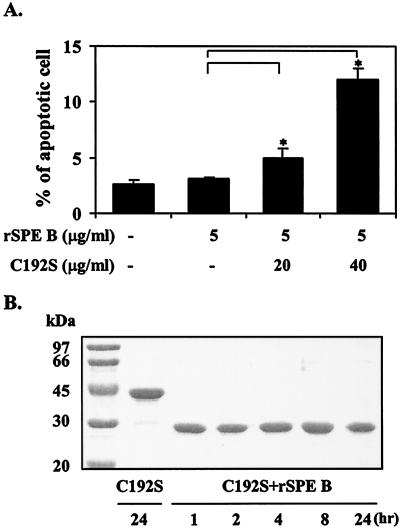
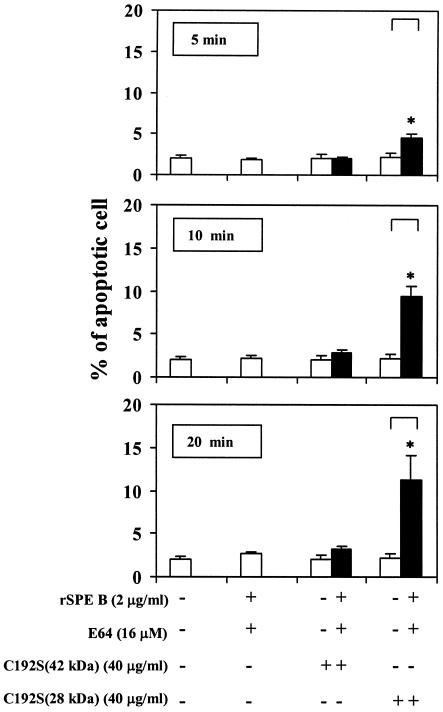
Because SPE B is also a cysteine protease, the role of protease in SPE B-induced apoptosis was first studied. When purified rSPE B and C192S were incubated with A549 cells, typical apoptosis, as analyzed by flow cytometry, could be induced only by rSPE B but not by C192S. The extent of apoptosis induced by rSPE B was also proportional to concentrations of rSPE B used (Fig. 2). To further confirm the role of protease in apoptosis, the protease activity of rSPE B was either inactivated by heat or inhibited by the addition of E64, an impermeable inhibitor of protease, to the culture. Under these conditions the apoptosis induced by rSPE B was completely abolished (Fig. 3). These results suggest that an extracellular protease is essential for SPE B-induced apoptosis. The C192S has no protease activity; however, it is a good substrate of rSPE B (2). An experiment was carried out to test whether C192S can be used as a competitive inhibitor of SPE B for SPE B-induced apoptosis. When cells were incubated with various concentrations of C192S in the presence of a low concentration of rSPE B (5 μg/ml), which did not induce apoptosis on its own, the percentage of apoptotic cells was proportional to the concentrations of C192S used (Fig. 4A). This observation was very interesting. Since neither rSPE B at 5 μg/ml nor C192S alone could induce apoptosis, the observed apoptosis is very likely attributable to the presence of the modified C192S. An electrophoretic analysis of the products, generated from the incubation of rSPE B and C192S in a ratio of 1/20, showed that C192S was cleaved to 28 kDa by rSPE B during the incubation period (Fig. 4B). This result indicated that the apoptosis induced by C192S was due to the presence of 28-kDa C192S. However, 28-kDa C192S alone, prepared by the trypsin digestion or by the rSPE B modification, did not induce apoptosis (data not shown). The likely explanation for these findings is that the cell surface must be also modified by the low concentration of rSPE B. To test this possibility, cells were preincubated with a low concentration of rSPE B (2 μg/ml) for 5, 10, or 20 min, added with E64 to block the protease activity of rSPE B, and then incubated with 28-kDa C192S or 42-kDa C192S for 24 h. The apoptosis was induced by 28-kDa C192S but not by 42-kDa C192S under the same conditions (Fig. 5). These results indicate that in addition to the required protease activity of SPE B, the size of the SPE B is also important for the induction of apoptosis.
FIG. 2.
Apoptosis in A549 cells induced by rSPE B and C192S. A549 cells were incubated with rSPE B or C192S at various concentrations for 24 h. Cells were fixed at −20°C with 70% ethanol in PBS, washed once with PBS, and stained with propidium iodide for 30 min at room temperature in the dark. Apoptotic cells were quantified by flow cytometry and are presented as the percentage of hypodiploid cells. The data are shown as the means ± the standard deviations (SD) for triplicate cultures. ∗, P < 0.05 (comparing groups treated with rSPE B or C192S).
FIG. 3.
Effect of protease activity of rSPE B on apoptosis of A549 cells. A549 cells were incubated with rSPE B or heat-inactivated rSPE B+ at various concentrations or in the presence of protease inhibitor E64. After a 24-h incubation, apoptosis was determined as described in Fig. 2. The data are shown as the means ± the SD for triplicate cultures. ∗, P < 0.05; ∗∗, P < 0.01 (comparing groups with or without treatment).
FIG. 4.
Apoptosis induced by a low concentration of rSPE B in combination with various concentrations of C192S. (A) A549 cells were incubated with a low concentration of rSPE B (5 μg/ml) and various concentrations of C192S, and the percentages of apoptotic cells were determined after 24 h. The data are shown as the means ± the SD for triplicate cultures. ∗, P < 0.05 (comparing groups that are with or without C192S treatment). (B) SDS-PAGE analysis of the products from the incubation of rSPE B and C192S at a ratio of 1 to 20.
FIG. 5.
Apoptosis induced by C192S. A549 cells were pretreated with rSPE B at 2 μg/ml for the time intervals indicated. E64 (to block the protease activity of rSPE B) and C192S (42 or 28 kDa) were then added to cultures and incubated for 24 h. The percentage of apoptotic cells was determined as described in Fig. 2. The data are shown as the means ± the SD for triplicate cultures. ∗, P < 0.05 (comparing groups with or without C192S 28 kDa).
Binding and internalization of SPE B.
To study how SPE B initiates the signal for apoptosis, the binding of iodinated rSPE B or C192S to A549 cells was evaluated at 4 and 37°C. As indicated in Table 1, there was a specific binding for rSPE B at both 4 and 37°C but not for C192S. The specific binding at 37°C was much higher than that at 4°C. In this type of experiment, the values of specific binding at 37°C represent the total radioactivity of [125I]rSPE B bound to the cell surface and internalized, whereas the value at 4°C represents the binding to the cell surface only, since internalization requires energy.
TABLE 1.
Binding of SPE B to A549 cells
| Ligand | Temp (°C) | Binding (cpm)
|
||
|---|---|---|---|---|
| Totala | Nonspecificb | Specificc | ||
| rSPE B | 4 | 6,939 | 5,871 | 1,068 |
| 37 | 12,641 | 7,402 | 5,239 | |
| C192S | 4 | 3,124 | 3,134 | 0 |
| 37 | 4,795 | 4,899 | 0 | |
Values of total binding were obtained from the incubation of cells with labeled rSPE B or labeled C192S.
Values of nonspecific binding were obtained from the incubation of cells with labeled rSPE B (or labeled C192S) plus 100-fold excess unlabeled rSPE B (or C192S).
Specific binding were calculated from the subtraction of nonspecific binding from total binding.
Apoptotic pathway induced by SPE B.
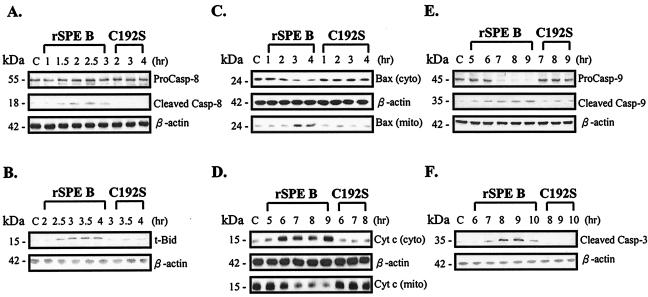
After the study of how the apoptosis in A549 cells was initiated by SPE B, we then investigated what molecules were activated by SPE B during apoptosis. As shown in Fig. 6, caspase-8 was first activated 1.5 h after the cells had been exposed to SPE B; caspase-8 was not activated by C192S, however (Fig. 6A). In a similar pattern, the formation of active t-Bid (Fig. 6B), the translocation of Bax to mitochondria (Fig. 6C), the release of cytochrome c from mitochondria to cytosol (Fig. 6D), the activation of caspase-9 (Fig. 6E), and the activation of caspase-3 (Fig. 6F) were sequentially activated at 2.5, 3, 6, 7, and 8 h, respectively, by SPE B.
FIG. 6.
Molecules activated by SPE B during apoptosis. Caspase-8, -9, and -3 and t-Bid were assayed from cell lysates (A, B, E, and F), and Bax and cytochrome c were assayed in both mitochondria and cytosol (C and D).
Effect of Bcl-2 overexpression on SPE B-induced mitochondrial membrane potential change and cytochrome c release.
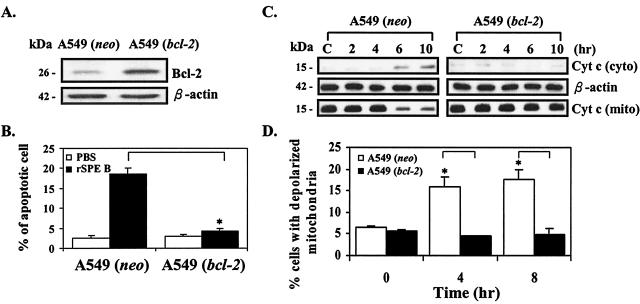
Since cytochrome c release was also activated by SPE B, we then investigated whether Bcl-2 could protect cells from the apoptosis caused by SPE B. A549 cells were transfected with bcl-2 carrying vector, and a stable clone was selected (Fig. 7A). When Bcl-2 was overexpressed, there was no apoptosis, whereas without Bcl-2 overexpression cells still underwent apoptosis in the presence of SPE B (Fig. 7B). Under the same conditions, cytochrome c release was inhibited by Bcl-2 overexpression (Fig. 7C). Changes in mitochondrial membrane potential are believed to be associated with the release of cytochrome c from mitochondria (6, 36). A mitochondrial potential change and cytochrome c release were further compared in cells that did or did not overexpress Bcl-2. In the presence of SPE B, the percentage of depolarized mitochondria was increased in cells that did not overexpress Bcl-2. On the other hand, there was no change in the percentage of depolarized mitochondria in cells that did overexpress Bcl-2 (Fig. 7D).
FIG. 7.
Overexpression of Bcl-2 protects cells from apoptosis. (A) The expression of Bcl-2 in A549 cells with bcl-2 vector (A549 [bcl-2]) or without bcl-2 vector (A549 [neo]). (B) Effect of Bcl-2 overexpression on SPE B-induced apoptosis. (C) Effect of Bcl-2 overexpression on cytochrome c release. (D) Effect of Bcl-2 overexpression on mitochondrial membrane potential changes. ∗, P < 0.05 (comparing groups with or without Bcl-2).
Effect of caspase-8 inhibitor on SPE B-induced mitochondrial membrane potential change.
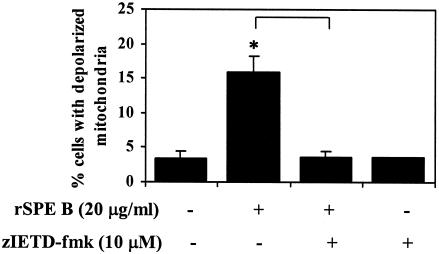
When A549 cells were incubated with rSPE B and zIETD-fmk, a caspase-8 inhibitor, SPE B induction of depolarized mitochondria was completely abolished (Fig. 8).
FIG. 8.
Effect of caspase-8 inhibitor on mitochondrial membrane potential change. A549 cells were incubated with rSPE B (20 μg/ml) in the presence or absence of caspase-8 inhibitor at 37°C for 4 h, and the percentage of depolarized mitochondria was determined as described in Materials and Methods. ∗, P < 0.05 (comparing groups with or without inhibitor).
Inhibition of caspase-8 prevented SPE B-induced apoptosis.
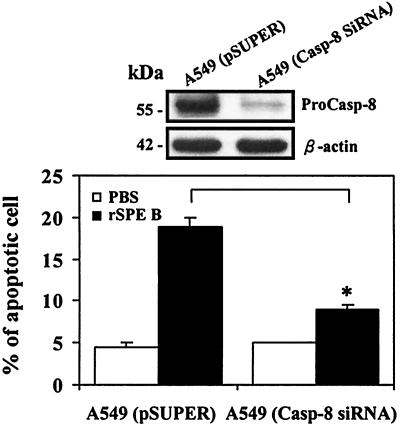
Since zIETD-fmk may also inhibit other caspases, other strategy was used to assess the role of caspase-8 in the SPE B-induce cell apoptosis. A549 cells were transfected with caspase-8 siRNA and subsequently treated with rSPE B. When caspase-8 was silenced by siRNA, the procaspase-8 was significantly reduced and rSPE B-induced apoptosis was effectively inhibited (Fig. 9).
FIG. 9.
Effect of caspase-8 siRNA on rSPE B-induced apoptosis. After transfection and sorting of EGFP-positive cells, the expression of procaspase-8 in A549 cells (A549-Casp-8 siRNA and A549-pSUPER) was measured (upper panel). Cells were incubated with rSPE B (20 μg/ml) and the percentages of apoptotic cells were determined after 24 h. The data are shown as the means ± the SD for triplicate cultures. ∗, P < 0.05 (comparing groups with or without caspase-8 siRNA [lower panel]).
DISCUSSION
Our previous studies demonstrated that SPE B induced apoptosis in A549 cells and U937 cells and that the protease of SPE B was essential for this function (19, 41). In the present study, our results show that SPE B induced apoptosis in A549 cells in a dose-dependent manner but that C192S did not induce apoptosis. When the protease activity of SPE B was inactivated either by heat or by the addition of extracellular protease inhibitor, its ability to induce apoptosis was abolished (Fig. 2 and 3). These results indicate that extracellular proteolytic hydrolysis is crucial for SPE B-induced apoptosis. The observation that the pretreatment of cells with a low concentration of rSPE B for as short as 5 min would enable 28-kDa C192S, but not 42-kDa C192S, to induce apoptosis is very interesting (Fig. 5). Since SPE B at a concentration lower than 5 μg/ml could not induce apoptosis (Fig. 2), the pretreatment of cells with 2 μg of SPE B/ml was obviously not enough to induce apoptosis. The observed effect of this pretreatment must be due to the proteolytic action of SPE B on the cell surface, presumably to expose the binding site for C192S. That 28-kDa C192S rather than 42-kDa C192S induced apoptosis after the pretreatment implies that a receptor-like mechanism is involved in the initial reaction of SPE B-induced apoptosis. In addition, the binding assay indicated that there was a low specific binding and an internalization of SPE B but not of C192S (Table 1). The low specific binding of SPE B to A549 cells may explain why a concentration threshold for SPE B was required to induce apoptosis (Fig. 2). Furthermore, it was also observed that the internalization of SPE B into A549 cells was lysosome and clathrin dependent, suggesting that it may be a process of receptor-mediated endocytosis (C. W. Chang, W. H. Tsai, W. J. Chuang, Y. S. Lin, J. J. Wu, and M. T. Lin, submitted for publication). Many bacterial protein toxins are internalized via receptor-mediated endocytosis (1, 11, 29, 38). These results therefore suggest that SPE B initiates the apoptotic pathway through a receptor-like mechanism. The process may be as follows: the truncation of SPE B precursor to active form by autolysis, the exposure of the receptor binding site by the proteolytic action of SPE B, the binding of SPE B to the receptor, and the initiation of the apoptotic pathway. A study of neutrophil apoptosis induced by proteolytic enzymes also showed that the stimulation of neutrophil apoptosis by protease depended on a proteolytic activity exerted on the cell surface structure (39). The protease-activated receptors belong to the large superfamily of G-protein-coupled receptors. The activation of receptors is achieved by the proteolytic unmasking of the N-terminal receptor-activating fragment that then binds to the same receptor molecule (16). Thrombin is a multifunctional serine protease that has an important role in hemostasis and wound healing. Its cellular actions are mostly attributed to the activation of thrombin receptor, a member of this receptor family. Thrombin also induces apoptosis in cultured neurons and astrocytes via the activation of its receptor and the initiation of a downstream cell death pathway that includes tyrosine kinase and RhoA activity (5).
There are two well-characterized caspase-activating cascades known to regulate apoptosis (10, 31, 35). One pathway that leads to caspase activation is initiated by the engagement of cell surface receptors. In this pathway, caspase-8 is first activated by the ligand-receptor complex, followed by activation of caspase-3 or other effector caspases. Another pathway involves the changes of mitochondrial membrane integrity, the release of cytochrome c from mitochondria to cytosol, and activation of caspase-3 via caspase-9 activation. In the present study, we have demonstrated that in A549 cells caspase-8, -9, and -3 are sequentially activated by SPE B at 1.5, 7, and 8 h, respectively, after incubation, which is consistent with the general pattern of receptor-mediated apoptosis, that is, that caspase-8 is involved in the initial phase of apoptosis and caspase-9 and -3 participate in the later phase (Fig. 6). The Helicobacter pylori infection also induced the activation of caspase-8, -9, and -3 (33).
We have also demonstrated in the present study that following the activation of caspase-8, t-Bid, Bax, and cytochrome c release were sequentially activated by SPE B at 2.5, 3, and 6 h before the activation of caspase-9 (Fig. 6). Apparently, the SPE B-induced cell death signal is passed from caspase-8 to the mitochondrial pathway. This is also supported by the experiment showing that in the presence of caspase-8 inhibitor, the mitochondrial membrane potential changes induced by SPE B were completely abolished (Fig. 8) and inhibition of caspase-8 by siRNA prevented rSPE B-induced apoptosis (Fig. 9). The mitochondrial pathway of apoptosis can be blocked by the antiapoptotic protein Bcl-2, which is localized to the mitochondrial membrane (17). The overexpression of Bcl-2 becomes a good tool for evaluating the involvement of the mitochondrial pathway in a particular apoptotic pathway (9). When A549 cells were transfected with vector carrying bcl-2, the overexpression of Bcl-2 inhibited the depolarization of mitochondria, cytochrome c release, and SPE B-induced apoptosis. These results further supported the importance of the mitochondrial pathway in SPE B-induced apoptosis. Whether the mitochondrial pathway is in the center stage of cell death has been debated (8). An elegant experiment from Kuwana et al. (20) suggested that the mitochondrial pathway may play a role in the amplification of the cell death signal from the activation of caspase-8; these authors found that, in the absence of mitochondria, high concentrations of caspase-8 were required to activate downstream caspases and that when mitochondria were present the effects of low concentrations of caspase-8 were amplified through cytochrome c-dependent caspase activation. In the case of SPE B, its binding to the cell surface was low and so was its induction of caspase-8; therefore, a mitochondrial pathway to amplify its death signal was necessary. A study of human pharyngeal epithelial cells (Hep-2) infected by Streptococcus pyogenes also showed the involvement of mitochondria in apoptosis and that the activation of caspase-9 was mediated by cytochrome c. Their confocal microscopy analysis revealed that Bax translocation to mitochondria and cytochrome c release occurred after 4 h of infection (28).
It was demonstrated previously by our group that YVAD-CMK, an irreversible caspase-1 inhibitor, could effectively inhibit the SPE B-induced apoptosis in U937 cells. Our recent studies suggested that in A549 cells caspase-1 also played an important role in apoptosis. When YVAD was added to cells with SPE B, the cytochrome c release from mitochondria was inhibited (data not shown). These results suggested that caspase-1 might act as upstream caspase in the SPE B-induced apoptosis in A549 cells.
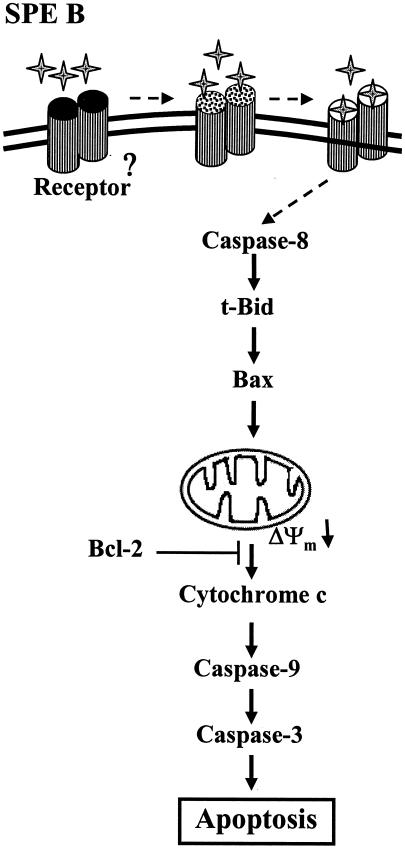
We have shown that SPE B-induced apoptosis in A549 cells is mediated through a receptor- and mitochondrion-dependent pathway. In this pathway, we have identified the earliest event to be the activation of caspase-8, which occurred 1.5 h after exposure to SPE B. The initial signal for the activation of caspase-8 has not been elucidated, however (Fig. 10).
FIG. 10.
SPE B-induced apoptosis in A549 cells. The binding site is exposed by the protease activity of SPE B, followed by the binding of SPE B to its receptor to initiate apoptosis. The death signal is passed through caspase-8 to mitochondria. The mitochondrial membrane potential (ΔΨm) is decreased, and the cytochrome c is released from mitochondria to cytosol. Finally, caspase-9 and caspase-3 are activated and death occurs.
Acknowledgments
This study was supported by grant NHRI-EX91-9027SP from the National Health Research Institutes of Taiwan.
We thank Bill Franke for editing the manuscript.
Editor: V. J. DiRita
REFERENCES
- 1.Abrami, L., S. Liu, P. Cosson, S. H. Leppla, and F. G. Van Der Goot. 2003. Anthrax toxin triggers endocytosis of its receptor via a lipid raft-mediated clathrin-dependent process. J. Cell Biol. 3:321-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, C. Y., C. F. Kuo, Y. S. Lin, J. J. Wu, M. T. Lin, C. C. Liu, and W. J. Chuang. 2003. Maturation processing and characterization of streptopain. J. Biol. Chem. 278:17336-17343. [DOI] [PubMed] [Google Scholar]
- 3.Chen, Y., and A. Zychlinsky. 1994. Apoptosis induced by bacterial pathogens. Microb. Pathog. 17:203-212. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Y. L., C. K. Yu, and H. Y. Lei. 1999. Propionibacterium acnes induces acute TNF alpha-mediated apoptosis of hepatocytes followed by inflammatory T-cell-mediated granulomatous hepatitis in mice. J. Biomed. Sci. 6:349-356. [DOI] [PubMed] [Google Scholar]
- 5.Donovan, F. M., C. J. Pike, C. W. Cotman, and D. D. Cunningham. 1997. Thrombin induces apoptosis in cultured neurons and astrocytes via a pathway requiring tyrosine kinase and RhoA activities. J. Neurosci. 17:5316-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earnshaw, W. C. 1999. A cellular cupboard. Nature 396:119.. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson, B. K. G., J. Andersson, S. E. Holm, and M. Norgren. 1999. Invasive group A streptococcal infections: T1M1 isolates expressing pyrogenic exotoxins A and B in combination with selective lack of toxin-neutralizing antibodies are associated with increased risk of streptococcal toxic shock syndrome. J. Infect. Dis. 180:410-418. [DOI] [PubMed] [Google Scholar]
- 8.Finkel, E. 2001. The mitochondrion: is it central to apoptosis? Science 292:624-629. [DOI] [PubMed] [Google Scholar]
- 9.Fulda, S., E. Meyer, and K. M. Debatin. 2002. Inhibition of TRAIL-induced apoptosis by Bcl-2 overexpression. Oncogene 21:2283-2294. [DOI] [PubMed] [Google Scholar]
- 10.Green, D. R. 2000. Apoptotic pathways: paper wraps stone blunts scissors. Cell 102:1-4. [DOI] [PubMed] [Google Scholar]
- 11.Herreros, J., T. Ng, and G. Schiavo. 2001. Lipid rafts act as specialized domains for tetanus toxin binding and internalization into neurons. Mol. Biol. Cell 12:2947-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holm, S. E., A. A. Norrby, M. Bergholm, and M. Norgren. 1992. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988-1989. J. Infect. Dis. 166:31-37. [DOI] [PubMed] [Google Scholar]
- 13.Hsueh, P. R., J. J. Wu, P. J. Tsai, J. W. Liu, Y. C. Chuang, and K. T. Luh. 1998. Invasive group A streptococcal pyrogenic exotoxin genes. Clin. Infect. Dis. 26:584-589. [DOI] [PubMed] [Google Scholar]
- 14.Kapur, V., M. W. Majesky, L. L. Li, R. A. Black, and J. M. Musser. 1993. Cleavage of interleukin 1β (IL-1β) precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 90:7676-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapur, V., S. Topouzis, M. W. Majesky, L. L. Li, M. R. Hamrick, R. J. Hamill, J. M. Patti, and J. M. Musser. 1993. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb. Pathog. 15:327-346. [DOI] [PubMed] [Google Scholar]
- 16.Kawabata, A., and R. Kuroda. 2000. Protease-activated receptor (PAR), a novel family of G protein-coupled seven trans-membrane domain receptors: activation mechanisms and physiological roles. Jpn. J. Pharmacol. 82:171-174. [DOI] [PubMed] [Google Scholar]
- 17.Kroemer, G. 1997. The proto-oncogene Bcl-2 and its role in regulating apoptosis. Nat. Med. 3:614-620. [DOI] [PubMed] [Google Scholar]
- 18.Kuo, C. F., J. J. Wu, K. Y. Lin, P. J. Tsai, S. C. Lee, Y. T. Jin, H. Y. Lei, and Y. S. Lin. 1998. Role of streptococcal pyrogenic exotoxin B in the mouse model of group A streptococcal infection. Infect. Immun. 66:3931-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo, C. F., J. J. Wu, P. J. Tsai, F. J. Kao, H. Y. Lei, M. T. Lin, and Y. S. Lin. 1999. Streptococcal pyrogenic exotoxin B induces apoptosis and reduces phagocytic activity in U937 cells. Infect. Immun. 67:126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuwana, T., J. J. Smith, M. Muzio, V. Dixit, D. D. Newmeyer, and S. Kornbluth. 1998. Apoptosis induction by caspase-8 is amplified through the mitochondrial release of cytochrome c. J. Biol. Chem. 273:16589-16594. [DOI] [PubMed] [Google Scholar]
- 21.Leite, F., C. Malazdrewich, H. S. Yoo, S. K. Maheswaran, and C. J. Czuprynski. 1999. Use of TUNEL staining to detect apoptotic cells in the lungs of cattle experimentally infected with Pasteurella haemolytica. Microb. Pathog. 27:179-185. [DOI] [PubMed] [Google Scholar]
- 22.Liles, W. C. 1997. Apoptosis: role in infection and inflammation. Curr. Opin. Infect. Dis. 10:165-170. [Google Scholar]
- 23.Lukomski, S., S. Sreevatsan, C. Amberg, W. Reichardt, M. Woischnik, A. Podvielski, and J. M. Musser. 1997. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J. Clin. Investig. 99:2574-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukomski, S., C. A. Montgomery, J. Rurangirwa, R. S. Geske, J. P. Barrish, G. J. Adams, and J. M. Musser. 1999. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect. Immun. 67:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mascini, E. M., M. Jansze, J. F. Schellekens, J. M. Musser, J. A. Faber, L. A. Verhoef-Verhage, L. Schouls, W. J. van Leeuwen, J. Verhoef, and H. van Dijk. 2000. Invasive group A streptococcal disease in The Netherlands: evidence for a protective role of anti-exotoxin A antibodies. J. Infect. Dis. 181:631-638. [DOI] [PubMed] [Google Scholar]
- 26.Merien, F., J. Truccolo, Y. Rougier, G. Baranton, and P. Perolat. 1998. In vivo apoptosis of hepatocytes in guinea pigs infected with Leptospira interrogans serovar icterohaemorrhgiae. FEMS Microbiol. Lett. 169:95-102. [DOI] [PubMed] [Google Scholar]
- 27.Musser, J. M., A. R. Hauser, M. H. Kim, P. M. Schlievert, K. Nelson, and R. K. Selander. 1991. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases; clonal diversity and pyrogenic exotoxin expression. Proc. Natl. Acad. Sci. USA 88:2668-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa, I., M. Nakata, S. Kawabata, and S. Hamada. 2001. Cytochrome c-mediated caspase-9 activation triggers apoptosis in Streptococcus pyogenes-infected epithelial cells. Cell. Microbiol. 3:395-405. [DOI] [PubMed] [Google Scholar]
- 29.Sandvig, K., and S. Grimmer. 2002. Pathways followed by ricin and Shiga toxin into cells. Histochem. Cell Biol. 117:131-141. [DOI] [PubMed] [Google Scholar]
- 30.Scaduto, R. C., and L. W. Grotyohann. 1999. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys. J. 76:469-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scaffidi, C., S. Fulda, A. Srinivasan, C. Friesen, F. Li, K. J. Tomaselli, K. M. Debatin, P. H. Krammer, and M. E. Peter. 1998. Two CD95 (APO-1/FAS) signaling pathways. EMBO J. 17:1675-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlievrt, P. M., A. P. Assimacopoulos, and P. P. Cleary. 1996. Severe invasive group A streptococcal disease: clinical description and mechanisms of pathogenesis. J. Lab. Clin. Med. 127:13-22. [DOI] [PubMed] [Google Scholar]
- 33.Shibayama, K., Y. Doi, N. Shibata, T. Yagi, T. Nada, Y. Iinuma, and Y. Arakawa. 2001. Apoptotic signaling pathway activated by Helicobacter pylori infection and increase of apoptosis-inducing activity under serum-starved conditions. Infect. Immun. 69:3181-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens, D. L., M. H. Tanner, J. Winship, R. Swarts, K. M. Ries, P. M. Schlivert, and E. Kaplan. 1989. Severe group A streptococcal infection associated with a toxic-like syndrome and scarlet fever toxin A. N. Engl. J. Med. 321:1-7. [DOI] [PubMed] [Google Scholar]
- 35.Strasser, A., L. O'Connor, and V. M. Dixit. 2000. Apoptosis signaling. Annu. Rev. Biochem. 69:217-245. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, W. D., X. D. Zhang, A. V. Franco, T. Nguyen, and P. Hersey. 2000. TNF-related apoptosis-inducing ligand-induced apoptosis of melanoma is associated with changes in mitochondrial membrane potential and perinuclear clustering of mitochondria. J. Immunol. 165:5612-5620. [DOI] [PubMed] [Google Scholar]
- 37.Tomar, R. S., H. Matta, and P. M. Chaudhary. 2003. Use of adeno-associated viral vector of delivery of small interfering RNA. Oncogene 22:5712-5715. [DOI] [PubMed] [Google Scholar]
- 38.Torgersen, M. L., G. Skretting, B. van Deurs, and K. Sandvig. 2001. Internalization of cholera toxin by different endocytic mechanisms. J. Cell Sci. 114:3737-3747. [DOI] [PubMed] [Google Scholar]
- 39.Trevani, A. S., G. Andonegui, M. Giordano, M. Nociari, P. Fontain, G. Dran, and J. R. Geffner. 1996. Neutrophil apoptosis induced by proteolytic enzymes. Lab. Investig. 74:711-721. [PubMed] [Google Scholar]
- 40.Tsai, P. J., C. F. Kuo, K. Y. Lin, Y. S. Lin, H. Y. Lei, F. F. Chen, J. R. Wang, and J. J. Wu. 1998. Effect of group A streptococcal cysteine protease on invasion of epithelial cells. Infect. Immun. 66:1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai, P. J., Y. S. Lin, C. F. Kuo, H. Y. Lei, and J. J. Wu. 1999. Group A streptococcus induces apoptosis in human epithelial cells. Infect. Immun. 67:4334-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wheeler, M. C., M. H. Roe, E. L. Kaplan, P. M. Schlievert, and J. K. Todd. 1991. Outbreak of group A streptococcus septicemia in children: clinical, epidemiologic, and microbiological correlates. JAMA 266:533-537. [PubMed] [Google Scholar]
- 43.Wolf, B. D., C. A. Gibson, V. Kapur, I. M. Hussain, J. M. Musser, and S. L. Gonias. 1994. Proteolytically active streptococcal pyrogenic exotoxin B cleaves monocytic cell urokinase receptor and release an active fragment of the receptor from the cells surface. J. Biol. Chem. 269:30682-30687. [PubMed] [Google Scholar]
- 44.Zychlinsky, A., K. Thirumalai, J. Arondel, J. R. Cantey, A. O. Aliprantis, and P. J. Sansonetti. 1996. In vivo apoptosis in Shigella flexneri infections, Infect. Immun. 64:5357-5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zychlinsky, A., and P. J. Sansonetti. 1997. Apoptosis in bacterial pathogenesis. J. Clin. Investig. 100:S63-S65. [DOI] [PMC free article] [PubMed] [Google Scholar]