Abstract
Chromatin, consisting of deoxyribonucleic acid (DNA) wrapped around histone proteins, facilitates DNA compaction and allows identical DNA code to confer many different cellular phenotypes. This biological versatility is accomplished in large part by post-translational modifications to histones and chemical modifications to DNA. These modifications direct the cellular machinery to expand or compact specific chromatin regions, and mark regions of the DNA as important for cellular functions. While each of the four bases that make up DNA can be modified (Iyer et al. 2011), this chapter will focus on methylation of the 6th position on adenines (6mA), as this modification has been poorly characterized in recently evolved eukaryotes but shows promise as a new conserved layer of epigenetic regulation. 6mA was previously thought to be restricted to unicellular organisms, but recent work has revealed its presence in more recently evolved metazoa. Here, we will briefly describe the history of 6mA, examine its evolutionary conservation, and evaluate the current methods for detecting 6mA. We will discuss the enzymes that bind and regulate this mark and finally examine known and potential functions of 6mA in eukaryotes.
Introduction
DNA must faithfully transmit the blueprints of life from generation to generation. However, it is also necessary that different cell types have access to different portions of the genome, and that specific cell types can respond appropriately to changes in the environment. Such dynamic responses are mediated in part by transcription factor complexes, and by chemical modifications to chromatin. DNA is not as heavily modified as RNA, which has 141 different modifications identified to date (Machnicka et al. 2013; Grosjean 2015). The limited number of DNA modifications (relative to RNA) is presumably evolutionarily selected for to protect the DNA code from mutations, and to enable formation of the double helix. Nevertheless, several DNA modifications occur across the tree of life, and are important as both signals of DNA lesions and as epigenetic regulators of diverse biological processes. Importantly, DNA modifications increase the repertoire of cellular phenotypes that can be encoded by a single DNA sequence, without directly altering the integrity of the genetic code. Soon after DNA was discovered, variants of each base were identified. However, the role of DNA methylation in the context of normal biological processes and disease pathogenesis remains an active area of study.
Although 6mA was discovered soon after cytosine methylation (5mC), it was thought to exist predominantly in prokaryotes and was therefore not given the same amount of research attention in eukaryotes as 5mC. The discovery that 6mA exists in more recently evolved eukaryotes has revived interest in this DNA modification. To understand the dynamic regulation of and by adenine methylation, it is useful to view the role of 6mA across evolution. Here, we aim to provide a broad overview of the historical research on 6mA across the evolutionary spectrum and discuss the mechanisms by which N6-adenine methylation is established, reversed, and recognized. We examine 6mAs role in biology, discuss the possibility of 6mA maintaining epigenetic information across cell divisions and potentially across generations, and summarize exciting areas for future research.
Types of DNA modifications
Each DNA base is modified to varying degrees in different organisms. DNA methylation occurs either as non-enzymatic DNA damaging lesions or as directed modifications with signaling function, which are actively introduced by specific methyltransferase enzymes. DNA lesions include N1-methyladenine (1mA), N3-methyladenine (3mA), N7-methyladenine (7mA), N3-methylcytosine (3mC), N2-methylguanine (2mG), O6-methylguanine (6mG), N7-methylguanine (7mG), N3-methylthymine (3mT), and O4-methylthymine (4mT), while directed methylation includes N6-methyladenine (6mA), N4-methylcytosine (4mC), and C5-methylcytosine (5mC) (Sedgwick et al. 2007; Iyer et al. 2011; Grosjean 2009). Other DNA modifications include deaminated cytosines (Shapiro, Klein 1966; Lindahl, Nyberg 1974), oxidized derivatives of 5mC (5hmC, 5fC, and 5caC) (Wyatt, Cohen 1952; Privat, Sowers 1996; Shen et al. 2014) and the hypermodified thymine base J (Gommers-Ampt et al. 1993). These modifications are discussed in greater detail in other reviews; we will focus on 6mA, a relatively uncharacterized DNA modification in eukaryotes with potential epigenetic function.
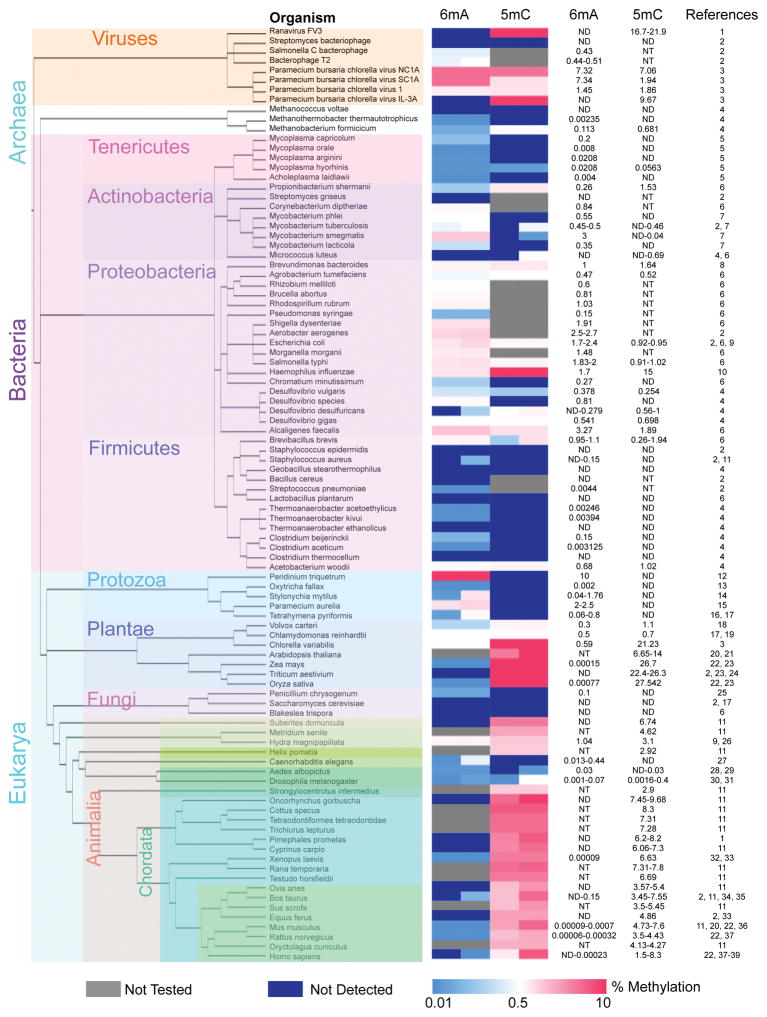
Of the directed DNA methylation events, 5mC is the most extensively studied. 5mC occurs at a higher frequency in more recently evolved organisms and its abundance in the genome ranges from 0.002% to 27% of cytosines, depending on the organism (Fig. 1). In mammals and plants, 5mC is the most abundant DNA modification (Iyer et al. 2011), and functions in the regulation of gene expression and maintenance of epigenetic memory (Bird 2002). 5mC in promoter regions typically leads to transcriptional gene silencing and therefore plays important roles in diverse cellular and developmental processes, including X-chromosome inactivation, genomic imprinting, stem cell pluripotency and differentiation (Bird 2002). Other directed DNA methylation events include 4mC and 6mA. 4mC has been identified mainly in thermophilic bacteria and archaea (Janulaitis et al. 1983; Ehrlich et al. 1985; Ehrlich et al. 1987; Grosjean 2009). Until recently, 6mA was also thought to be restricted to bacteria, archaea, and protists. However, its recent identification in several eukaryotes raises the possibility that 6mA serves as an epigenetic signaling modification within an organism and potentially across generations.
Figure 1. Abundance of 6mA and 5mC across the tree of life.
The relative abundance of 6mA and 5mC are displayed in a heat map. The first column of the heat map displays the percentage of adenines that are N6-methylated (%6mA/A) and the second column displays the percentage of cytosines that are C5-methylated (%5mC/C) for the organism indicated in each row. Blue color represents lower 6mA or 5mC abundance and red color represents higher 6mA or 5mC abundance. Grey color indicates that the methylation mark was not tested in that organism. Dark blue color indicates that the methylation mark was not detected in that organism, and therefore may or may not be present at levels below the limit of detection for the technique used. For some organisms the level of methylation has been shown to vary across multiple measurements, between different studies or between different cell types within the same organism. In such cases, a range is presented where the left half of the column reflects the lowest detected level (or not detected in some cases) and the right half of the column shows the highest detected level. Methylation values are presented on the right along with citations. The phylogenetic tree was generated using the PhyloT web server (http://phylot.biobyte.de/index.html) and visualized using the Interactive Tree Of Life web server (http://itol.embl.de/). The phylogenetic tree (‘rooted’ setting) displays the inferred evolutionary relationships between the indicated genera based on their genetic similarity (Letunic, Bork 2011). The tree was created using FigTree v1.4.2. The different organisms are subdivided into different colored boxes to represent different kingdoms and phyla. For some phyla only one organism has been examined. 1: (Willis, Granoff 1980), 2: (Dunn, Smith 1958), 3: (Van Etten et al. 1985), 4: (Ehrlich et al. 1985), 5: (Razin, Razin 1980), 6: (Vanyushin et al. 1968), 7: (Srivastava et al. 1981), 8: (Degnen, Morris 1973), 9: (Yuki et al. 1979), 10: (Drozdz et al. 2012), 11: (Vanyushin et al. 1970), 12: (Rae 1976), 13: (Rae, Spear 1978), 14: (Ammermann et al. 1981), 15: (Cummings et al. 1974), 16: (Gorovsky et al. 1973), 17: (Hattman et al. 1978), 18: (Babinger et al. 2001), 19: (Fu et al. 2015), 20: (Capuano et al. 2014), 21: (Kakutani et al. 1999), 22: (Huang et al. 2015), 23: (Wagner, Capesius 1981), 24: (Montero et al. 1992), 25: (Rogers et al. 1986), 26: (Hassel et al. 2010), 27: (Greer et al. 2015b), 28: (Adams et al. 1979), 29: (Proffitt et al. 1984), 30: (Zhang et al. 2015), 31: (Lyko et al. 2000), 32: (Koziol et al. 2016), 33: (Jabbari et al. 1997), 34: (Unger, Venner 1966), 35: (Romanov, Vanyushin 1981), 36: (Wu et al. 2016), 37: (Gama-Sosa et al. 1983), 38: (Tawa et al. 1992), 39: (Ehrlich et al. 1982).
Discovery of 6mA across eukaryotes
DNA N6-methyladenine (6mA) is a widespread modification in prokaryotes. Although 6mA is not necessary for viability in prokaryotes (Marinus, Morris 1973; Russell, Hirata 1989), it plays crucial roles in regulating DNA replication (Campbell, Kleckner 1990; Yamaki et al. 1988), repair (Pukkila et al. 1983), transposition (Roberts et al. 1985), transcription (Wallecha et al. 2002; Robbins-Manke et al. 2005), and cellular defense (Luria, Human 1952; Meselson, Yuan 1968; Linn, Arber 1968; Smith et al. 1972). For reviews on 6mA in prokaryotes, please see (Marinus, Lobner-Olesen 2014; Wion, Casadesus 2006; Murray 2002) and Chapter ???. An unknown base was initially identified in E. coli and, using several techniques, this base was compared to synthesized nucleotides to identify 6mA. Hydrolyzed bases were separated by two-dimensional paper chromatography in different solvents ultraviolet absorption spectrum maximums and minimums were measured, and electrophoretic mobility of this unknown base all confirmed the detection of 6mA (Dunn, Smith 1955, 1958). The existence of 6mA was subsequently confirmed in a variety of different bacterial species (Vanyushin et al. 1968). These initial detection techniques were capable of detecting 6mA at ~0.01% of total adenines (Vanyushin et al. 1970). This detection limit, combined with the confounding presence of commensal symbionts, technical variability, tissue-specific differences, development/stage-specific variability, or subtle environmental effects on 6mA levels initially led to contradictory reports of the identification of 6mA in eukaryotes. Indeed, 6mA was reported by one group to occur in bull and human sperm (Unger, Venner 1966), but other groups were unable to replicate this result or detect 6mA in other metazoa (Dunn, Smith 1958; Vanyushin et al. 1970). 6mA was reported to occur in some unicellular eukaryotes including Paramecium aurelia (Cummings et al. 1974), Stylonychia mytilus (Ammermann et al. 1981), Oxytricha fallax (Rae, Spear 1978), Chlorella variabilis (Van Etten et al. 1985), Tetrahymena pyriformis (Gorovsky et al. 1973) and Chlamydomonas reinhardi (Hattman et al. 1978). Two reports also identified 6mA in multicellular eukaryotes, including the mosquito Aedes albopictus (Adams et al. 1979) and the sponge Suberites domuncula (Vanyushin et al. 1970). However, detection of 6mA in mosquitos was not reproduced (Proffitt et al. 1984), and its detection in the sponge was dismissed as potentially coming from symbiotic prokaryotes or algae (Vanyushin et al. 1970). Therefore, until recently, 6mA was thought to be restricted to prokaryotes and unicellular eukaryotes (Casadesus, Low 2006).
With the advent of more sensitive detection techniques (discussed below), 6mA has recently been identified in multicellular eukaryotes including Caenorhabditis elegans and Drosophila melanogaster (Greer et al. 2015b; Zhang et al. 2015). Several other papers reported low levels of 6mA in more recently evolved eukaryotes, but each of these has caveats that we must acknowledge. 6mA was detected in Drosophila, calf thymus, and human placental samples by dot blots (Achwal et al. 1983). A recent paper detected 6mA by immunofluorescence in mouse heart tissues (Sun et al. 2015). Another group identified 6mA in the plants Oryza sativa and Zea mays, rat tissues, and human cells by high performance liquid chromatography coupled with mass spectrometry (HPLC-ms/ms) (Huang et al. 2015). More recently, 6mA was found by dot blots, HPLC, and methyl DNA immunoprecipitation followed by sequencing (MeDIPseq) in Xenopus laevis and mouse kidney (Koziol et al. 2016), and by dot blots, MeDIPseq, HPLC and SMRT-seq in mouse embryonic stem (ES) cells (Wu et al. 2016). While these papers raise the exciting possibility that 6mA may indeed be present across the tree of life, it is difficult to discount potential contaminating microbiota and to confirm that the detection of 6mA is real when the reported levels of 6mA are at the limit of detection. RNA m6A (discussed below) could also account for contaminating signal in dot blots and immunofluorescence if not properly removed. We must also recognize that the injection of N6-adenine methylated oligos into mice induces a greater immune response than unmethylated oligos, as measured by the production of IL-12 (Tsuchiya et al. 2005). But this does not necessarily confirm that 6mA is a foreign base in mice as unmethylated CpG motifs also induce a more substantial immune response (Tsuchiya et al. 2005). These results raise the possibility that 6mA is either not present in mammals, or present in sufficiently small quantities to keep it as an immunogenic species in the mammalian repertoire. To confirm the existence of 6mA across eukaryotes, it will be necessary to identify the enzymes that regulate 6mA and biological conditions under which the modification changes. These recent studies suggest that 6mA might be a conserved DNA modification, and raise several fundamental and largely unexplored questions about the evolutionary importance of 6mA across the tree of life. From an evolutionary perspective, why did higher eukaryotes shift from 6mA (the most pervasive DNA modification in prokaryotes), towards using 5mC as the more dominant DNA modification? To what extent are the ancient functions of 6mA and its modifying enzymes conserved from prokaryotes to more recent eukaryotes?
In contrast to DNA adenine methylation, RNA adenine methylation (m6A) has long been recognized as the most abundant post-transcriptional modification of prokaryotic and eukaryotic mRNAs (Niu et al. 2013). In humans, there are over 18,000 m6A sites representing approximately 7,000 unique mRNA transcripts (Jia et al. 2011; Meyer et al. 2012; Dominissini et al. 2012). Furthermore, m6A is enriched in 3′UTRs in highly conserved regions (Meyer et al. 2012; Dominissini et al. 2012; Deng et al. 2015), suggesting a shared function for m6A in evolutionarily distant species. N6-methyladenosine regulates multiple aspects of RNA metabolism, including mRNA stability/decay, translation, splicing and localization (Wang et al. 2014; Wang et al. 2015; Zhou et al. 2015; Niu et al. 2013), and participates in diverse cellular and biological processes including meiosis and embryonic stem cell differentiation (Yue et al. 2015; Batista et al. 2014; Hongay, Orr-Weaver 2011; Bodi et al. 2012). The prevalence of RNA m6A raises the possibility that DNA adenine methylation could be a consequence of methylated adenines in RNA recycled via the nucleotide salvage pathway. Another possibility is that DNA adenine methylation is catalyzed by RNA methyltransferases, either as an off-target effect of these enzymes or as a biologically regulated process. Unlike the better-characterized RNA m6A, relatively little is known about the functional importance of DNA 6mA in metazoan genomes, and whether 6mA plays a similarly conserved role in the dynamic regulation of biological processes. The phenotypic consequences of RNA m6A might provide clues to the roles of N6-adenine methylation on DNA.
Abundance of 6mA
The relative genomic abundance of 6mA can provide clues to its biological relevance across evolutionarily distinct organisms. 6mA and 5mC appear to have a large range of abundance in the genomes of different organisms across evolution (Gommers-Ampt, Borst 1995). 5mC is undetectable in many bacterial species, as well as the genome of S. cerevisiae, and ranges from 0.0016% of cytosines in D. melanogaster to as high as 10% in some mammals and 30% in certain plant species (Gommers-Ampt, Borst 1995; Capuano et al. 2014; Wagner, Capesius 1981). If we accept that published literature documenting the presence of 6mA in different organisms is in fact detecting 6mA in the reported organism (rather than in contaminating symbionts), the genomic abundance of 6mA varies by several orders of magnitude across the tree of life (Fig. 1). Generally, organisms with higher levels of 6mA such as bacteria and single-celled eukaryotes tend to have lower levels of 5mC, while organisms with higher levels of 5mC such as plants and mammals tend to have lower levels of 6mA. The detected level of 6mA ranges from ~0.0001–0.0003% of adenines in plants and mammals to as high as 3% of adenines in some species of bacteria, and up to 10% of adenines in the dinoflagellate Peridinium triquetrum (Rae 1976). Early studies of nucleic acid composition in the 1950’s examined the base composition of DNA in different strains of bacteria using 2D paper chromatography (Dunn, Smith 1958). It was found that 6mA comprised 1.75% of all adenines in E. coli and 2.5% of adenines in Aerobacter aerogenes (Dunn, Smith 1958). Subsequent studies examined the content of 6mA in the DNA of unicellular eukaryotes, such as the ciliate Tetrahymena pyriformis (0.65–0.8% of adenines) (Gorovsky et al. 1973), Paramecium aurelia (2.5%) (Cummings et al. 1974), and Stylonychia mytilus (0.176%) (Ammermann et al. 1981). The level of 6mA in these unicellular eukaryotes is comparable to the 6mA abundance in many species of bacteria. Interestingly Tetrahymena and Stylonychia mytilus have 4–13 fold lower 6mA levels in their micronucleus than their macronucleus (Gorovsky et al. 1973; Ammermann et al. 1981), suggesting that this modification plays an important role in determining the differences between the two nuclei in these species, which are separated by ~1159 million years of evolution (Parfrey et al. 2011).
Recently, 6mA was identified in the DNA of C. elegans, using both antibody-based approaches and antibody-independent methods of quantitation, including single molecule real time (SMRT) sequencing and ultra-high performance liquid chromatography followed by mass spectrometry (UHPLC-ms/ms) (Greer et al. 2015b). Based on the UHPLC-ms/ms data, the levels of 6mA ranged from 0.013% to 0.39% of adenines, representing a 30-fold variation in the global level of adenine methylation between different batches of wild-type C. elegans. The observation that 6mA abundance can vary by more than an order of magnitude within an isogenic population of animals is interesting, as it suggests that the levels of 6mA in these organisms might be particularly sensitive to subtle changes in the environment (e.g. stress stimuli).
A recent study quantified the genomic abundance of 6mA in plants, rat tissues and human cells using HPLC-ms/ms (Huang et al. 2015). These data must be viewed with caution, as there was no independent validation that the 6mA modification was occurring in the reported organisms, rather than contaminating symbionts. In that study, the abundance of 6mA in plant and mammalian genomes ranged from 0.00008% of adenines in rat lung DNA to as high as 0.0007% of adenines in plant DNA. The human cell lines had 0.0017% and 0.0023% 6mA (in Jurkat and 293T cells, respectively). Another group identified 6mA in 0.00009% of adenines in Xenopus laevis by HPLC and MeDIPseq (Koziol et al. 2016). More recently 6mA was identified in mouse ES cells at 0.0006–0.0007% of adenines (Wu et al. 2016). These finding suggests that 6mA in plants and mammalian genomes is ~1,000–40,000-fold lower than its abundance in some bacteria and single-celled eukaryotes. The large degree of variability in 6mA abundance between eukaryotes motivates further exploration into the environmental factors and evolutionary pressures that led to a decline in 6mA levels and an increase in 5mC levels during eukaryotic evolution. These differences could also indicate that at very low 6mA levels, 6mA is at the limit of detection. Therefore, quantitative differences between different samples could be attributed to technical errors, rather than true biological variability. Moreover, these modifications are typically detected under basal conditions. It is possible that 6mA levels are dramatically altered under specific environmental conditions. Finally, we should note that even if a relatively rare percentage of adenines are methylated, the presence of a single methylated adenine at a critical genomic location could have dramatic phenotypic consequences by affecting the binding of specific regulatory proteins (see cell cycle regulation below).
Methods of detecting 6mA
Detection of DNA methylation has evolved over the years to become increasingly sensitive and accurate. Detecting different DNA modifications started with a technique of combining the cytosine fraction with picric acid to form crystalline picrate. After purification by crystallization, salt crystals were compared to synthetic pyrimidines of known structure. By this method, the authors reported the identification of 5mC in Mycobacterium tuberculosis in 1925 (Johnson, Coghill 1925). Detection techniques shifted to paper chromatography (Hotchkiss 1948), which had a limit of detection of 1%, and was used to compare synthetically generated 5mC to the content of 5mC in animal, plant, viral, and bacterial DNA (Wyatt 1950). By the time, 6mA was first identified in 1955, its presence was confirmed by a combination of ultraviolet absorption spectrum (Mason 1954), electrophoretic mobility, and its paper chromatographic movement in different solvents (Dunn, Smith 1955). Because these early methods were relatively insensitive, the presence of 6mA in a number of animal species was undetectable. Researchers quickly realized that they could take advantage of restriction enzymes to identify methylated residues (Bird, Southern 1978; Geier, Modrich 1979). A limitation of this approach is that detection of methylation sites is dependent on the methylated residue occurring in the appropriate restriction enzyme target motif, and whether the restriction enzyme preferentially recognizes un-, hemi- or fully-methylated substrates. Therefore, not all sequence contexts can be addressed with this method.
High-performance liquid chromatography was subsequently used to determine that E. coli has 1.4% 6mA (Yuki et al. 1979). Liquid chromatography has become increasingly sensitive and, recently, ultra high performance liquid chromatography coupled with mass spectrometry (UHPLC-ms/ms) has been used to detect concentrations of 6mA on the order of 0.00001% (Huang et al. 2015). An alternative technique, called capillary electrophoresis and laser-induced fluorescence (CE-LIF), uses the fluorescent dye boron-dipyrromethene (BODIPY), to specifically bind to 6mA, followed by capillary electrophoresis combined with laser-induced fluorescence to detect 6mA levels (Krais et al. 2010). This technique has a lower limit of detection of 0.01% 6mA and was used to confirm the presence of 6mA in Bacteriophage λ, E. coli, and to identify 6mA’s presence in Hydra magnipapillata (1.04% of adenines) (Krais et al. 2010). At this limit of detection, the authors could not detect 6mA in calf thymus or human kidney samples.
While the aforementioned techniques have proven useful for detecting whether 6mA is present in a particular organism, they do not provide information on the genomic location of this modification. To determine the genomic locations of 6mA, several methylation-sensitive sequencing techniques have been developed. Methylated DNA immunoprecipitation (MeDIP) coupled with microarray analysis (Weber et al. 2005) has evolved into MeDIP sequencing (MeDIP-seq) (Pomraning et al. 2009). MeDIP-seq has been optimized by a combination of photo-crosslinking, exonuclease digestion, and restriction enzyme digestion to achieve near single-nucleotide resolution of 6mA (Chen et al. 2015; Fu et al. 2015). MeDIP-seq, however, is dependent on the antibody specifically recognizing 6mA. Alternative techniques have also been developed to identify where throughout the genome 6mA occurs. One such technique consists of radioactive methylation of DNA followed by restriction digest, electrophoresis, and sequencing (Posfai, Szybalski 1988). Single-molecule real-time sequencing (SMRT-seq) is a next-generation sequencing technique which provides accurate sequence reads and measures the kinetic rate of nucleotide incorporation during sequencing (Flusberg et al. 2010). Since different DNA modifications result in different kinetic signatures, SMRT-seq can identify every DNA modification at single-base resolution. This technology, however, does have troubles distinguishing several closely related modifications from each other, including 1mA from 6mA. However, when coupled with UHPLC-ms/ms (which can distinguish 1mA from 6mA), this technique can give rather unambiguous confirmation of both the presence and genomic location of 6mA in a specific organism (Greer et al. 2015b). Methylated residues can be confirmed by restriction digest coupled with real-time RT PCR to determine the methylation at a specific locus (Fu et al. 2015). Alternatively, sequence-specific probes have been developed that can selectively bind to 6mA or unmodified adenines in specific sequence contexts (Dohno et al. 2010).
To convincingly identify rare modifications, such as 6mA, a combination of multiple complimentary techniques is ideal since each technique has its own set of limitations (Table 1). UHPLC-ms/ms can be complemented by restriction enzyme digestion confirmation (as long as 6mA occurs in the appropriate motif), dot blots and MeDIP with a 6mA-specific antibody, and SMRT-seq. For a complementary discussion of the methods for detection of 5mC see chapter ???.
Table 1. Recent methods for detecting and quantifying 6mA.
Recent methods for detecting and quantifying 6mA are summarized in this table. Relative limitations and sensitivity of each method are discussed and references to the primary papers are cited.
| Detection/Quantitation Method | Description/ Limitations | 6mA Specificity | Sensitivity (lower limit of detection) | Genomic sequence information | Reference(s) |
|---|---|---|---|---|---|
| 6mA-sensitive restriction enzymes | Restriction endonuclease cleavage of methylated motifs. Cannot detect 6mA outside of restriction recognition sites. |
High (enzyme-dependent) | Can identify single methylated adenine so long as it occurs within specific recognition motifs (e.g. GATC) Preference for hemi-methylated or dually methylated depending on the enzyme |
None normally Can be used, in combination with real-time RT PCR, to validate sites identifed by sequencing methods |
Bird AP et al. 1978, Geier GE et al. 1979 |
| 6mA Dot Blotting | Antibody-dependent semi-quantitative detection of 6mA levels in genomic DNA samples | High (antibody-dependent) If DNA is not single stranded 6mA antibodies generally recognize 1mA as well | Moderate (can not distinguish lowly methylated samples from each other) | None | Achwal CW et al. 1983 |
| Immunofluorescence | Antibody-dependent method for detecting 6mA in whole animals or tissues at cell-level resolution. Very difficult to validate that signal is coming from 6mA rather than background (ideally need to manipulate the 6mA regulating enzymes) |
High (antibody-dependent) | Moderate. Immunofluorescence is not good for assessing relative changes in 6mA. (antibody-dependent) | None normally Could be used in combination with DNA probes to confirm 6mA localization in specific genomic regions |
Greer EL et al. 2015, Sun Q et al 2015 |
| MeDIP-seq | Antibody-dependent method for identifying genomic regions harboring 6mA | High (antibody-dependent) | High (antibody-and organism-dependent (C. elegans and D. melanogaster gave very low coverage depth)) | Genome-wide 6mA localization at near base pair resolution |
Pomraning KR et al. 2009 Chen K et al. 2015 |
| SMRT-seq | Provides base modifications at single-nucleotide resolution 6mA and 1mA are indistinguishible and quite expensive. |
Medium (1mA and 6mA are indistinguishible) | Can identify single methylated adenine with sufficient coverage | Single base resolution 6mA detection genome-wide | Flusberg BA et al 2010 |
| UHPLC-MS/MS | Chemical separation and detection by mass spectrometry. | Highest | High (As low as 0.0001% 6mA/A) | None |
Yuki H et al 1979 Zhang G et al 2015 Fu Y et al 2015 Greer EL et al 2015 Huang W et al. 2015 |
| 6mA-specific probes | DNA probe containing a formyl group on the O6 position of a G base discriminates between adenine and 6mA via formation of an interstrand cross-link (ICL). 6mA can not form ICL. ICLs detected by PAGE or HPLC | Untested for other modifications (such as 1mA) | Detection by electrophoresis (6mA has no ICLs and will be single-stranded on the gel) | Can confirm 6mA in specific genomic locations not a discovery tool | Dohno C et al. 2010 |
| CE-LIF | BODIPY FL EDA binds covalently to the phosphate group of deoxyribonucleotide after activated by carbodiimide reagent. Run by CE-LIF which distinguishes different bases from each other and methylated bases based on migration time. | Can distinguish from 5mC or other bases but untested with 1mA | Moderate (0.01% limit) | None | Krais AM et al. 2010 |
Abbreviations: MeDIP-seq: Methylated DNA immunoprecipitation followed by high throughput DNA-sequencing; SMRT-seq: Single-Molecule Real Time sequencing; UHPLC-MS/MS: Ultra high performance liquid chromatography coupled to tandem mass spectrometry. BODIPY FL EDA: fluorescent dye 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-propionyl ethylene diamine hydrochloride; CE-LIF: Capillary electrophoresis with laser-induced fluorescence
6mA regulating enzymes
DNA methyltransferases
An important step in the confirmation of 6mA as a regulated mark of biological significance has been the identification of enzymes that deposit and remove this mark. It was previously thought that methylated adenines were incorporated premade into genomic DNA. This assumption likely hampered initial efforts to identify 6mA in eukaryotes. A study in the early 1970s concluded that 6mA did not exist in eukaryotes, because radioactively labeled adenines, but not methylated adenines were incorporated into DNA when added exogenously (Vanyushin et al. 1970). However, several groups demonstrated that DNA could be glycosylated and RNA could be methylated at the N6 position of adenines after incorporation into polynucleotides, rather than pre-methylated nucleotides being incorporated during the biosynthesis of polynucleotide (Kornberg et al. 1959; Kornberg et al. 1961; Fleissner, Borek 1962). These findings led to the hypothesis that methylation occurs after DNA synthesis (Theil, Zamenhof 1963), rather than on unincorporated nucleotides, and spurred attempts to identify the DNA methylating enzymes. The first studies were conducted in E. coli by fractionation of total protein lysates followed by methylation assays with each fraction. Early studies identified a single fraction that methylated DNA at the C5 position of cytosines and the N6 position of adenines, but this fraction was only efficient at methylating foreign DNA (Gold et al. 1963; Gold, Hurwitz 1964). Subsequent studies using increasingly subdivided fractions were able to identify multiple adenine and cytosine methyltransferases in E. coli (Nikolskaya et al. 1976; Nikolskaya et al. 1981).
Additional evidence for the widespread presence and functional importance of 6mA in eukaryotic genomes comes from the observation that members of the MT-A70 family of known or putative N6-adenine methyltransferases exist in most organisms, ranging from bacteria to humans (Luo et al. 2015). Based on structural orthology to other members of the MT-A70 family of methyltransferases, the candidate DNA adenine methyltransferase enzymes in multicellular organisms likely evolved from the bacterial M.MunI-like 6mA methyltransferase, which functions in the host restriction modification system (Iyer et al. 2011). The MT-A70 family includes both RNA and DNA methyltransferases, including IME4 (also called SPO8) in S. cerevisiae (Clancy et al. 2002), DAMT-1 in C. elegans (Greer et al. 2015b), and members of the methyltransferase-like (METTL) family in mammals, including METTL3 (an N6-adenosine RNA methyltransferase) (Liu et al. 2014), and METTL4 (a homolog of DAMT-1) (Greer et al. 2015b). Whether the same enzymes catalyze both RNA and DNA adenine methylation in different organisms remains an open question. Notably, biochemical in vitro studies have suggested that the mammalian RNA methyltransferase METTL3 also methylates DNA (personal communications C. He), suggesting that the same enzymes are capable of methylating both RNA and DNA in certain contexts, but the substrate specificity (i.e. RNA, DNA or both) for each member of the different MT-A70 family members remains incompletely characterized. At the structural level, all of these enzymes are characterized by a 7-β-strand methyltransferase domain at their C-terminus, fused to a predicted alpha-helical domain at their N-terminus and require S-adenosyl-L-methionine (SAM) as a methyl donor (Iyer et al. 2011). The high degree of amino acid sequence conservation among the predicted N6-adenine methyltransferases motivates further exploration into their potential functional conservation.
How adenine methyltransferases of recently evolved eukaryotes recognize their substrates still remains to be determined. The utilization of adenine methylation by the restriction-modification system suggests that bacterial 6mA methyltransferases evolved to recognize specific sequences for methylation. In bacteria and the unicellular eukaryote Tetrahymena DNA adenine methylation occurs in a palindromic sequence-specific manner in vitro and in vivo (Geier, Modrich 1979; Zelinkova et al. 1990; Bromberg et al. 1982). However, sequence-specific adenine methylation is not observed in all organisms and some bacterial DNA adenine methyltransferases show no sequence specificity (Drozdz et al. 2012). Similarly, 6mA sites in C. elegans only appeared modestly enriched in specific sequence contexts (Greer et al. 2015b), suggesting that targeted adenines might be selected by more complicated metrics than simply sequence codes. It remains to be seen whether other multicellular eukaryotes, which possess 6mA, show a sequence-specific pattern of adenine methylation (similar to bacteria and unicellular eukaryotes), or whether these organisms show little to no sequence-specificity in their adenine methylation pattern, as observed for C. elegans. It remains to be seen whether methyltransferases that do not recognize specific DNA sequences are recruited to specific locations of the genome by other DNA-binding proteins or other epigenetic chromatin features.
Mechanism of 6mA methyltransferases
Substantial work in prokaryotes has identified the mechanism of action, the preferred methyl donor, and the kinetics of 6mA methyltransferases. Whether these regulatory principles are conserved in eukaryotes remains to be seen. There was an initial debate as to whether N6 was directly methylated, or if adenines were first methylated on the N1 position and then, following a Dimroth rearrangement, the methyl group would be transferred to the N6 position. However, the enzyme EcoRI had been shown to methylate N6 directly rather than through an initial N1 methylation (Pogolotti et al. 1988). This result, combined with the slow rate of Dimroth reactions at endogenous pH (Macon, Wolfenden 1968), suggests that N6 is the direct target of methyltransferases. This conclusion has been confirmed by the structures of different adenine-N6 methyltransferases in complex with DNA, showing a direct approximation of the N6 atom towards the methyl-donor (Goedecke et al. 2001; Horton et al. 2005; Horton et al. 2006).
Early reports identifying that DNA was methylated suggested that S-adenosyl-L-methionine (SAM) was the primary methyl donor (Gold et al. 1963), and future work has shown that SAM is the predominant methyl donor for not only DNA and RNA methylation, but also for proteins and lipids (Chiang et al. 1996). However, 5,10-methylene tetrahydrofolate has been identified as the methyl donor for tRNAs in Streptococcus faecalis and Bacillus subtilis (Delk, Rabinowitz 1975; Delk et al. 1976; Urbonavicius et al. 2005). While the enzyme that utilizes 5,10-methylene tetrahydrofolate in B. subtilis, GidA, is absent in eukaryotes (Urbonavicius et al. 2005), this finding raises the possibility that some DNA methyltransferases might use alternative methyl donors.
Kinetic rates have been measured for the T4 bacteriophage DNA adenine methyltransferase, Dam (Malygin et al. 2000) and the EcoRI adenine methyltransferase (Reich, Mashhoon 1991). For Dam the methylation rate constant (kmeth) was significantly faster than the overall reaction rate constant (kcat) (0.56 and 0.47 s−1 vs 0.023 s−1) suggesting that product dissociation is the rate-limiting step. Similar, but faster results were observed with EcoRI (Reich, Mashhoon 1991). These enzymes function by binding, flipping out the adenine, methylating, and restacking of the modified base (Allan et al. 1998). Whether these hold true for M.MunI-like methyltransferases remains to be determined. Reducing the double strand duplex stability did not alter the kmeth, suggesting that base-flipping is not a rate limiting step in the methylation reaction (Malygin et al. 2000). Additionally, EcoRI enzyme-DNA complexes were less efficient compared to enzyme-SAM complexes, suggesting that the enzyme first binds to SAM before methylating its substrates (Reich, Mashhoon 1991). This is opposite to what has been observed with Dam and the bacterial 5mC methyltransferase HhaI, where the methyltransferase first binds to DNA, followed by SAM (Urig et al. 2002; Wu, Santi 1987), suggesting that the sequence of binding events in the DNA methylation reaction is enzyme-dependent.
An important step for the confirmation of the presence and role of 6mA in more recently evolved eukaryotes will be the identification of genuine 6mA methyltransferases. The conservation of MT-A70 domain containing proteins in conjunction with the identification of 6mA in many eukaryotes suggests that this modification is conserved. Whether eukaryotic DNA methyltransferases function in a similar manner to prokaryote methyltransferases remains to be seen. Interestingly, the RNA m6A methyltransferase, METTL3, functions in complex with METTL14 (Liu et al. 2014), raising the possibility that DNA methyltransferase enzymes, like many other chromatin regulating enzymes, function in multi-protein complexes. These multi-protein complexes could help the enzymes achieve their specificity.
DNA adenine demethylation
The identification of the enzymes that catalyze the removal of 6mA from DNA strongly suggests that 6mA is a regulated and dynamic epigenetic mark. Examination of the enzymes responsible for the removal of DNA base damage fostered the identification and characterization of the DNA demethylation processes. DNA base damage, in the form of 1mA and 3mC, was shown to be removed by the Fe(II)- and α-ketoglutarate-dependent dioxygenase AlkB in E. coli (Trewick et al. 2002). The AlkB family of dealkylating enzymes is highly conserved from bacteria to humans (Fedeles et al. 2015; Wei et al. 1996). AlkB enzymes can demethylate many DNA substrates, including the DNA lesions 1mA, 3mC and 3mT (80, 122). Notably, humans have nine AlkB family members (ALKBH1–8 and FTO). Like E. coli AlkB enzymes, the mammalian enzymes ALKBH2 and ALKBH3 function in the repair of DNA alkylation damage (Duncan et al. 2002). In addition to their DNA demethylase activity, AlkB members catalyze oxidative demethylation of RNA (Aas et al. 2003). Interestingly, AlkB enzymes in RNA viruses preferentially demethylate RNA substrates, suggesting these AlkBs are necessary for maintaining the integrity of the viral RNA genome (van den Born et al. 2008). More recently, it was found that AlkB family members function in the oxidative demethylation of N6-methyladenosine in RNA, catalyzed by ALKBH5 and FTO in mammals (Jia et al. 2011; Zheng et al. 2013), and that the AlkB family member NMAD-1 in C. elegans demethylates 6mA in DNA (28). FTO was also shown to demethylate 6mA in single-stranded DNA in vitro (Jia et al. 2011), raising the possibility that these enzymes might regulate both DNA and RNA 6mA. Whether NMAD-1 can also demethylate m6A on RNA remains to be tested. Recently ALKBH1 was also shown to demethylate 6mA in single-stranded DNA in vitro (Wu et al. 2016). Additionally ALKBH1 knockout causes an increase in global 6mA levels in mouse embryonic stem cells and this increase can be rescued by a wildtype, but not a catalytic domain mutant of ALKBH1 (Wu et al. 2016), suggesting that ALKBH1 functions as a 6mA demethylase in mammals.
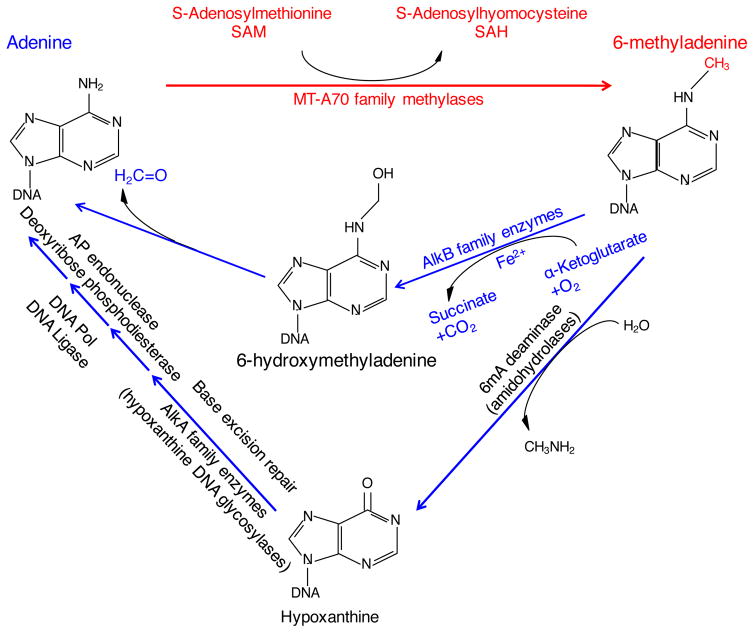
Several studies have begun to dissect the mechanism of action of AlkB demethylases. In the presence of their essential cofactors α-ketoglutarate and Fe(II), AlkB demethylases use molecular oxygen to oxidize the methyl group of 6mA, forming the unstable intermediate 6-hydroxymethyladenine (6hmA), which spontaneously releases its aldehyde group, regenerating the unmodified adenine base (Figure 2) (Fedeles et al. 2015). Whether the same mechanism occurs for the demethylation of 6mA in eukaryotes and if so, whether 6hmA has any additional function remains to be seen. In mammals, FTO was recently shown to oxidize m6A on RNA to N6-hydroxymethyladenosine (hm6A) and N6-formyladenosine (f6A) (Fu et al. 2013). These mRNA derivatives have half-lives of ~3 hours (Fu et al. 2013), suggesting that if 6hmA does have additional functions they would require a 6hmA specific binding protein which could stabilize the intermediate. The same oxidation reaction mechanism is used by AlkB enzymes to demethylate 1mA and 3mC during the cellular response to DNA alkylation damage (Falnes et al. 2002; Trewick et al. 2002).
Figure 2. Mechanisms of N6-adenine methylation and demethylation.
MT-A70 family methylases catalyze the methylation of adenine at the sixth position of the purine ring. MT-A70 methylases use S-adenosylmethionine (SAM) as their methyl donor to generate 6-methyladenine and S-adenosylhomocysteine (SAH).
Adenine could be regenerated from 6mA by several different enzymatic mechanisms: AlkB family enzymes catalyze the oxidative demethylation of 6mA. AlkB enzymes require α-ketoglutarate and Fe2+ and use oxygen to oxidize the methyl group. This oxidative demethylation reaction first generates 6-hydroxymethyladenine, which releases its formaldehyde group to generate adenine. Alternatively, 6mA can be deaminated and subsequently removed via the base excision repair pathway. First, 6mA deaminase hydrolyzes the methylamine to generate hypoxanthine. Hypoxanthine is recognized as a damaged base by AlkA family enzymes, which cleave the glycosyl bond to remove the base. Apurinic (AP) endonuclease cleaves the phosphodiester backbone at the abasic site, exposing the residual 5′ deoxyribose phosphate group, which is then removed by deoxyribose phosphodiesterase. Finally, DNA polymerase I incorporates the unmodified adenine and DNA ligase catalyzes the formation of the phosphodiester bond.
In addition to demethylation of 6mA by the AlkB demethylase family, 6mA can also be converted to hypoxanthine by a 6mA deaminase (Kamat et al. 2011). This modified base can then undergo base excision repair by hypoxanthine DNA glycosylases of the AlkA family (Saparbaev, Laval 1994) (Figure 2). If hypoxanthine is not removed, it can cause a transition mutation (AT pairs would be converted to GC pairs), since hypoxanthine pairs with cytosine instead of thymine. Recently, 6mA was found to be correlated with increased point mutations in Neisseria meningitidis (Sater et al. 2015), suggesting that this modified base might be mutagenic, potentially as a consequence of unrepaired 6mA deamination. However, 6mA deaminases in Neisseria meningiditis have not yet been identified. In contrast to 6mA deamination, which is only mutagenic if not removed, 5mC is converted to thymine when deaminated, which leads to a transition mutation in a single step (Lindahl, Nyberg 1974; Heindell et al. 1978). Deamination of adenine, 6mA, or cytosine all lead to non-natural bases, which can easily be identified by specific glycosylases. Deamination of 5mC, on the other hand, leads to thymine which requires a more complicated repair process. This more direct mutational path might explain why 5mC is more prone to mutation than 6mA. This divergence begs the question as to why evolution has selected for a higher prevalence of the more mutagenic DNA modification in more recently evolved species.
In E. coli, AlkB expression is induced by DNA damage and the enzyme functions in DNA repair via direct removal of base alkylation damage (Trewick et al. 2002). AlkB mutant E. coli are sensitized to cell death induced by the alkylating agent methyl methanesulfonate (MMS), and the predicted human ortholog of AlkB is sufficient to partially rescue the MMS-induced cytotoxicity seen in AlkB mutants (Wei et al. 1996). Interestingly, MMS treatment of human skin fibroblasts did not result in the same induction of AlkB seen in E. coli, suggesting that the regulation of AlkB expression may have diverged during the evolution of more recent eukaryotes (Wei et al. 1996), or that the induction by different alkylating agents is cell-type specific, and may only occur in certain cell types. In C. elegans, deletion of NMAD-1, a member of the AlkB family, causes a global increase in 6mA and purified NMAD-1 is capable of demethylating N6-adenine methylated oligonucleotides in vitro (Greer et al. 2015b). Importantly, mutation of the NMAD-1 catalytic domain abolished the in vitro demethylase activity of NMAD-1, identifying NMAD-1 as a 6mA demethylase in C. elegans and highlighting the mechanistic conservation of AlkB enzymes from bacteria to metazoa (Greer et al. 2015b).
Interestingly, a different family of enzymes, ten-eleven translocation (Tet) proteins, has been shown to demethylate 5mC in many organisms (Tahiliani et al. 2009; Ito et al. 2010; Ito et al. 2011). Unlike AlkB proteins, whose crystal structures have revealed that the enzymes flip out the base to facilitate demethylation (Yang et al. 2008; Sundheim et al. 2008), crystal structure of the TET family catalytic domains are not suitable for accommodating flipped out purines (Aravind et al. 2015) suggesting that they cannot act on mA. The TET family has a good phyletic correlation with DNA cytosine methylases, but not with DAMT-1 or other Dam family methylases (Aravind et al. 2015). Additionally in bacteria, there is little evidence that TET is capable of demethylating purines (Aravind et al. 2015). Given these findings, it is surprising that the D. melanogaster ortholog of Tet (named DMAD) was reported to function as a 6mA demethylase on DNA (Zhang et al. 2015). Nuclear extracts from DMAD mutant flies showed reduced in vitro demethylation activity compared to nuclear extracts from wild-type flies, while addition of purified DMAD was sufficient to increase adenine demethylation in these assays (Zhang et al. 2015). It remains to be seen whether this 6mA demethylase activity can be biochemically confirmed using purified DMAD, and whether Tet proteins play a conserved role as 6mA demethylases.
6mA binding proteins
Beyond the machinery that catalyzes addition and removal of 6mA, cells have evolved mechanisms to recognize 6mA as a regulatory signal that can be translated into different biological consequences (see Biological functions of 6mA). We will discuss later in this chapter the direct chemical consequences of adenine methylation, but 6mA can be recognized by specific effector molecules that alter chromatin architecture and/or transcriptional states directly, or indirectly via recruitment of other DNA-binding proteins. Alternatively, methylation could function by preventing binding. Methyladenine-binding proteins have evolved to recognize and transduce 6mA signals into specific biological outcomes. For example, in E. coli the MutS enzyme binds to mismatch base pairs as a homodimer, facilitating recruitment of the MutL protein, which binds MutS. The MutS-MutL-DNA complex then loops out until it finds the nearest hemimethylated GATC site, which is bound by the endonuclease MutH. Upon binding of MutL-MutS to the MutH-DNA complex, MutH is activated and nicks the unmethylated daughter strand, allowing helicase and exonucleases to excise the single-stranded mismatch region (Su, Modrich 1986). Thus, hemimethylated of GATC sites are used to specifically direct mismatch repair of the daughter strand (Lahue et al. 1987) Similarly, the oriC region of E. coli is hemimethylated to prevent premature replication before the cell has divided. These hemimethylated adenine sites are recognized and bound by the SeqA protein (Brendler et al. 1995; Slater et al. 1995), which prevents assembly of the DNA replication machinery at this region (von Freiesleben et al. 1994; Wold et al. 1998). The crystal structure for SeqA has revealed why SeqA binds preferentially to hemimethylated over fully methylated DNA (Guarne et al. 2002; Fujikawa et al. 2004), highlighting the importance of determining the crystal structure of 6mA binding proteins for deciphering the chemical and biological consequences of their binding. Thus far, these binding proteins have only been identified in prokaryotes, but an important next step to fully understand the possible biological roles of 6mA will be to identify eukaryotic 6mA binding proteins.
Biological Functions of 6mA
The direct effects of adenine methylation on the structure of DNA and its roles in prokaryote biology have been well characterized (see also chapter ???). Whether 6mA plays a conserved functional role in eukaryotes remains to be seen, but discussing its functional effects in prokaryotes raises several interesting potential functions which will need to be further explored in eukaryotes.
Effects of adenine methylation on DNA structure
One possible role for adenine methylation, beyond providing a binding site for effector proteins, is to directly alter the overall structure of DNA. An early crystal structure suggested that 6mA might alter the secondary structure of DNA (Sternglanz, Bugg 1973). Adenine methylation is thought to affect DNA double helix formation through altering both base pair stability and base stacking. Ultraviolet photoelectron studies suggested that adenine methylation would lower the ionization potentials and cause the destabilization of valence electrons to increase base stacking in methylated adenines (Peng et al. 1976). This increased base stacking would be offset by a slight destabilization of base pairing ranging from ~0.35–0.95 kcal/mol (Engel, von Hippel 1978b). Interestingly, 5mC behaves oppositely to 6mA in these regards. 5mC causes an increase in helix stability, while adenine methylation destabilizes the DNA, as measured by denaturing gradient gel electrophoresis (Collins, Myers 1987). 6mA within GATC sequences causes slight DNA unwinding of 0.5°/methyl group (Cheng et al. 1985). Consistent with these observations, two-dimensional NMR studies suggest that, in almost all cases, 6mA has only minor effects on helix conformation, as it retains the canonical B-form (Fazakerley et al. 1985; Quignard et al. 1985; Fazakerley et al. 1987). 6mA occurring directly after thymines, on the other hand, causes severe unwinding and bending of the DNA helix relative to the canonical B conformation (Fazakerley et al. 1989). However, 6mA lowers melting temperatures and slows the rate of helix formation, as demonstrated by enthalpy of dissociation studies (Quignard et al. 1985; Fazakerley et al. 1985). These studies suggest that methylated adenines are associated with DNA regions that spend prolonged periods in the open state. These effects were confirmed by cruciform extrusion assays where 5mC inhibits extrusion and 6mA facilitates initial opening of DNA (Murchie, Lilley 1989). These consequences seem to be in line with the reported effects of 5mC and 6mA on gene transcription; 5mC is generally believed to be a repressor of gene transcription when it occurs at promoters, while 6mA is hypothesized to be an activator. However, the correlation between 5mC and gene transcription is dependent on the genomic context in which it occurs. When 5mC occurs within gene bodies, rather than promoters, it is correlated with gene transcription (Reviewed in (Jones 2012)). Thus, the effects of 6mA on gene transcription may depend on its location in the genome.
The effects of 6mA on the thermodynamic stability and folding of DNA appear to be sequence-specific (Fazakerley et al. 1987). Indeed, when 6mA occurs directly after a T this can cause a highly altered structure that is overwound and bent (Fazakerley et al. 1989). While 6mA does not dramatically alter helix rigidity (Hagerman, Hagerman 1996; Mills, Hagerman 2004), it can increase DNA curvature to variable degrees, depending on sequence context (Diekmann 1987).
Restriction-modification systems
In prokaryotes, DNA N6-adenine methylation is used to discriminate self from foreign DNA, as part of restriction modification systems; a bacterial immune system by which pathogenic DNA from bacteriophages is recognized by endonucleases that selectively cleave unmethylated DNA at specific restriction sites that are methylated in the host’s genome, and thus protected from endonuclease digestion (Low et al. 2001; Iyer et al. 2011). Interestingly, enterobacteriophages appear to have evolved to contain fewer GATCs to avoid the GATC R-M system of their hosts (McClelland 1984). This system does not appear to be conserved in eukaryotes that have evolved more complex immune systems.
DNA damage control
Early reports suggested that dam mutant E. coli had higher mutation rates and were more sensitive to UV and mitomycin C, suggesting that 6mA could protect against DNA damage (Marinus, Morris 1974). It was subsequently suggested that 6mA could help to distinguish the parental DNA strand from the mutated daughter strand (Glickman et al. 1978; Glickman 1979). Similarly, Penicillium chrysogenum mutants deficient in 6mA had higher sensitivity to mutagenic agents without changes in the number of mutations (Rogers et al. 1986).
In E. coli and other gram-negative bacteria, DNA adenine methylation plays an important role in the DNA mismatch repair pathway, a strand-specific repair pathway that relies on the transient post-replicative hemimethylation of DNA. The DNA adenine methylase, Dam, binds selectively to hemimethylated DNA substrates and methylates GATC sites after DNA replication. The delay between DNA synthesis and methylation of the newly synthesized daughter strand is crucial for the fidelity of DNA mismatch repair (Pukkila et al. 1983). When DNA replication errors lead to base pair mismatches, the DNA repair machinery uses adenine methylation to distinguish the already methylated template strand from the newly synthesized unmethylated daughter strand. As described above (6mA binding proteins) hemimethylated DNA allows MutL, MutS, and MutH to identify and specifically cleave the daughter strand, allowing helicase and exonucleases to excise the single-stranded mismatch region. Subsequently, DNA polymerase III re-synthesizes the mismatch region of single-stranded DNA using the methylated parental strand as a template (Pukkila et al. 1983).
Effect on transcription
Several studies have suggested that N6-adenine methylation correlates with increased gene expression. Whether this is due to the direct effect on relaxing DNA structure (as discussed above), recruitment of 6mA-specific binding proteins, or both, remains unknown. It is still also unclear as to whether this phenomena is conserved across all organisms that contain 6mA. While 5mC CpG methylation had little effect on transcription in barley, 6mA methylation increased transcription two to five-fold (Rogers, Rogers 1995). Similarly, 6mA but not 5mC methylation increased gene expression by 3–50 fold of reporter constructs in tobacco or wheat protoplast, or intact wheat tissues (Graham, Larkin 1995). Luciferase reporter constructs purified from dam+dcm+ bacteria (with 5mC and 6mA methylation) had 2–6 fold increased luciferase production compared to constructs purified from dam-dcm- bacteria in rat or mouse cell lines, or when electroporated into mice (Allamane et al. 2000). Together, these results suggest that 6mA promotes gene expression.
6mA can also directly affect binding of transcription factors. Methylation of a HNF1 binding site reduces HNF1 binding affinity, but this only causes a minor reduction in gene transcription (Tronche et al. 1989; Lichtsteiner, Schibler 1989). Conversely, 6mA increases binding affinity for the transcription factor AGP1 in tobacco (Sugimoto et al. 2003). These results suggest that the effects of adenine methylation on transcription will be sequence- and transcription factor specific.
Similar to DNA cytosine methylation in metazoa, bacterial DNA adenine methylation regulates gene expression programs, including those related to virulence and phase variation (Low et al. 2001; Wallecha et al. 2002; Zaleski et al. 2005; Sarnacki et al. 2013), suggesting that 6mA levels might be sensitive to changes in environmental conditions. Similarly, recent data suggest that 6mA may play a role in transcriptional regulation in the single-celled eukaryote Chlamydomonas reinhardti, where 6mA occurs preferentially near actively transcribed genes (Fu et al. 2015). As preliminary evidence that 6mA levels might be relevant to human physiology and disease, it was reported that human patients with type 2 diabetes have reduced levels of m6A on RNA and 6mA on DNA, as measured by HPLC-ms/ms and it was proposed that these differences might be regulated by the cellular fat mass and obesity associated protein (FTO) (Huang et al. 2015), which was shown to function as an RNA m6A and single-stranded DNA 6mA demethylase (Jia et al. 2011) and DNA 3mT demethylase (Gerken et al. 2007). Future studies will be required to confirm the existence of 6mA in human DNA using independent detection methods. Despite recent progress in defining the potential functions of 6mA in different organisms, the roles of 6mA in more recently evolved eukaryotes, including its possible roles in human health and disease remain unknown. Given the high degree of evolutionary conservation of MT-A70 family methyltransferases and Alkb family demethylases, along with the recent discovery of 6mA in eukaryotes, we propose that 6mA is likely to play an important role in the regulation of diverse biological processes in metazoa.
Nucleosome positioning
In the protists Tetrahymena and Chlamydomonas 6mA is preferentially located in the linker regions between nucleosomes (Karrer, VanNuland 2002; Fu et al. 2015; Pratt, Hattman 1983), raising the possibility that 6mA could help to direct nucleosome positioning. Alternatively, enrichment of 6mA in linker regions may reflect increased accessibility, or recruitment of the methyltransferase at regions of open chromatin. Enrichment for 6mA in specific genomic regions was not observed in C. elegans (Greer et al. 2015b), but the analysis was performed on mixed tissue samples, which could have obscured any positional bias that may exist in specific cell types. In future studies, it will be interesting to examine whether 6mA directs nucleosome positioning and whether it does so in a conserved manner, or whether other open chromatin modifications can direct N6-adenine methylation at those sites.
Cell cycle regulation
N6-adenine methylation marks regions for DNA replication initiation in prokaryotes and has been shown to alter the rate of cell cycle progression (see chapter ???). In E. coli, the Dam methyltransferase is necessary for precise timing between DNA replication events (Bakker, Smith 1989; Boye, Lobner-Olesen 1990). The hemimethylation of DNA plays an important role in modulating the initiation of DNA replication; the SeqA protein binds to hemimethylated DNA adjacent to the origin of replication OriC, preventing its methylation by Dam, and leading to a delay in DNA replication before the cell has divided, which is only initiated from a fully methylated promoter (Low et al. 2001; Lu et al. 1994). When DNA replication is desired, adenine methylation at the oriC region lowers the thermal melting temperature which could facilitate the unwinding at the origin of replication (Yamaki et al. 1988). Interestingly, 6mA also slows the rate of DNA polymerase I catalysis, presumably due to the effects of 6mA on base pairing (discussed above) (Engel, von Hippel 1978a).
In Caulobacter crescentus, the cell cycle regulated DNA adenine methylase (CcrM) controls the timing of DNA replication and progression through the cell cycle (Collier et al. 2007). In contrast to E. Coli Dam methylase, which does not have a preference for hemimethylated sites, C. crescentus CcrM preferentially methylates hemimethylated DNA after replication (Berdis et al. 1998), and is essential for cell viability (Stephens et al. 1996). In C. crescentus, 6mA levels change throughout the cell cycle from fully to hemimethylated as the replication forks progress (Kozdon et al. 2013). The promoter of the replication initiation factor DnaA is preferentially activated when its promoter is fully methylated, leading to DnaA accumulation and progression through the cell cycle (Collier et al. 2007). Whether 6mA plays a similar role in controlling the cell cycle in eukaryotes remains to be seen.
Transgenerational inheritance
DNA methylation provides the most parsimonious method by which epigenetic information could be transmitted across generations. Because of the semi-conservative nature of DNA replication, methylation events on the parental strand can be replicated on the newly synthesized daughter strand. In mammals, 5mC methylation patterns are established by the de novo methyltransferases Dnmt3a and Dnmt3b during early embryonic development (Okano et al. 1999). Inheritance of cytosine methylation patterns through cell division is mediated by the maintenance methyltransferase Dnmt1 (Bestor et al. 1988). Dnmt1 preferentially binds hemimethylated DNA at the replication fork and copies parental-strand methylation patterns onto the unmethylated daughter strand (Stein et al. 1982; Yoder et al. 1997; Bestor 2000; Martin, Zhang 2007). Whether adenine methylation propagates non-genetic information through cell divisions, or from parents to their offspring remains to be seen. However, there are some hints that 6mA could transmit non-genetic information. Labeling experiments showed that newly synthesized E. coli DNA in Okazaki fragments were quickly N6-adenine methylated (Marinus 1976), consistent with the idea that parental methylation patterns might be passed on to their descendants during DNA replication. In some bacteria, DNA adenine methylation is tightly coordinated with cell division (Casadesus, Low 2006)(see cell cycle regulation above), enabling inheritance of parental methylation patterns. Thus, a key unanswered question is whether there exists a mode of inheritance of adenine methylation in eukaryotes, or whether different organisms have evolved different mechanisms for the inheritance of parental DNA methylation through somatic nuclear divisions and across generations. In the ciliate Tetrahymena thermophila macronucleus, analysis of methylation patterns using methylation-sensitive restriction enzymes showed that both actively replicating and non-replicating DNA contained hemimethylated sites, and that the vegetatively growing macronucleus contained a combination of partially methylated sites and fully methylated sites (Capowski et al. 1989). These findings are inconsistent with a simple semi-conservative 6mA inheritance mechanism, and suggest that inheritance of 6mA in some organisms may rely on hemi-methylation-independent mechanisms of 6mA maintenance through cell division (Capowski et al. 1989).
In C. elegans, loss of the histone H3 lysine 4 dimethyl (H3K4me2) demethylase spr-5 causes a progressive transgenerational loss of fertility (Katz et al. 2009) and a transgenerational extension in lifespan (Greer et al. 2015a). This is accompanied by a progressive decline in H3K9me3 and accumulation of H3K4me2 and 6mA (Greer et al. 2014; Greer et al. 2015b). Deletion of the 6mA demethylase, nmad-1, accelerates the progressive fertility decline, while deletion of the putative 6mA methyltransferase, damt-1, suppresses the transgenerational H3K4me2 accumulation, fertility, and longevity phenotypes (Greer et al. ; Greer et al. 2015a), raising the possibility that 6mA might transmit epigenetic information across generations. It remains to be seen whether methylated adenines themselves are transmitted across generations as they are transmitted across cell divisions, or whether 6mA is erased in the germline and established de novo during somatic development (see cell cycle discussion above), or if 6mA is more indirectly involved in these processes. Future studies will also reveal whether 6mA can regulate transgenerational inheritance in other species.
Many years of research have shown that chromatin modifications do not occur in isolation, but rather actively communicate with each other. For example, 5mC and H3K9me3 are coordinately regulated in plants (see chapter ???). The H3K9 methyltransferase binds to 5mC methylated DNA (Jackson et al. 2002; Johnson et al. 2007; Malagnac et al. 2002) and the DNA methyltransferase binds to H3K9me-containing nucleosomes (Du et al. 2012). It is possible that a similar reciprocal cross-talk occurs between 6mA and H3K4 methylation in C. elegans, as described in the previous paragraph (Greer et al. 2015b). It remains to be seen whether this reciprocal cross-talk is real and whether other species show a similar co-association between 6mA levels and H3Kme2 levels. Future work should reveal whether 6mA methyltransferases can bind to specific methylated histones to direct DNA methylation to particular loci.
Conclusions and future directions
As detection techniques are becoming increasingly sensitive 6mA has begun to be convincingly observed in several metazoa. The conservation of 6mA methyltransferases and demethylases along with the initial detection of 6mA in several metazoa suggest that N6-adenine methylation might be a conserved signaling modification. However, it will be important to rigorously examine whether 6mA is present across the tree of life using a combination of rapidly evolving detection techniques (discussed in this review). For metazoa that are confirmed to have 6mA in their DNA, it will be important to define the biological functions of 6mA and its genomic localization patterns in different cell types. A fundamental question is whether the biological functions of 6mA in bacteria are conserved in higher eukaryotes or whether 6mA has evolved new biological functions in these organisms. As 6mA occurs less frequently in more recently evolved organisms, this might reflect a more specialized functional role.
A growing body of work has revealed an important role for m6A on mRNAs in the regulation of gene expression and cellular differentiation in eukaryotes (Niu et al. 2013; Meyer et al. 2012; Dominissini et al. 2012; Deng et al. 2015; Wang et al. 2014; Wang et al. 2015; Zhou et al. 2015; Yue et al. 2015; Batista et al. 2014). Therefore, another open question is whether N6-adenine methylation of DNA is coordinately regulated with N6-adenine methylation on RNA. Given that substrates of the AlkB family of demethylases and MT-A70 family of methyltransferases can include both RNA and DNA, it will be of interest to better characterize the substrate specificity of these enzymes in different organisms and to examine whether the same enzymes regulate both RNA and DNA N6-adenine methylation in different organisms. Moreover, it will be relevant to find out if in cases of overlapping substrate specificities, whether methylation of DNA or RNA (or both) is the biologically relevant signal under different physiological conditions.
The inheritance of 6mA methylation during bacterial cell division (Wion, Casadesus 2006) raises the question of whether 6mA can be inherited in eukaryotes. Is 6mA passed on through successive generations, or erased in the germline? Recently described paradigms of transgenerational inheritance in C. elegans have raised the possibility that 6mA itself might carry epigenetic information across generations (Greer et al. 2015b). Alternatively, 6mA might communicate with other heritable epigenetic marks that reciprocally regulate the levels of 6mA. Future studies should reveal whether 6mA is incompletely erased in the germline and inherited in subsequent generations. In mice, 5mC is mostly erased by passive demethylation during the expansion of primordial germ cells preceding the formation of gametes (Seisenberger et al. 2012); methylation patterns are then re-established during early embryonic development by the de novo methyltransferases Dnmt3a and Dnmt3b (Okano et al. 1999) (see chapter ???). However, some regions of 5mC, such as those near imprinted genes, escape the typical erasure and can therefore carry non-genetic information across generations (Breiling, Lyko 2015). Whether a similar situation exists for 6mA remains to be seen.
Given the dynamic nature of 5mC in mammalian development and cell differentiation (Okano et al. 1999), it will be of interest to define the dynamics and potential functions of 6mA during mammalian development, if its presence in mammals can be rigorously confirmed. Future studies should also reveal the environmental factors that regulate the levels of 6mA and its modifying enzymes in metazoa, which should provide clues to its evolutionary conservation and biological relevance. The diversity of methods for detection of 6mA in DNA will allow for comprehensive and detailed examination of 6mA’s presence, localization patterns and potential functions in the genomes of diverse organisms. All in all, the newly developed and more sensitive tools for detection, along with the recent discovery of 6mA in metazoa open an exciting new chapter of discovery in the field of adenine methylation.
Acknowledgments
We thank S. Burger, N. O’Brown, and E. Pollina for critical reading of the manuscript. We thank C. He for helpful discussions. The work from the Greer laboratory is supported by a grant from the NIH (AG043550). Z.K.O. is supported by 5T32HD7466-19. We apologize for literature omitted owing to space limitations.
References
- Aas PA, Otterlei M, Falnes PO, Vagbo CB, Skorpen F, Akbari M, et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421(6925):859–63. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- Achwal CW, Iyer CA, Chandra HS. Immunochemical evidence for the presence of 5mC, 6mA and 7mG in human, Drosophila and mealybug DNA. FEBS Lett. 1983;158(2):353–8. doi: 10.1016/0014-5793(83)80612-7. [DOI] [PubMed] [Google Scholar]
- Adams RL, McKay EL, Craig LM, Burdon RH. Methylation of mosquito DNA. Biochim Biophys Acta. 1979;563(1):72–81. doi: 10.1016/0005-2787(79)90008-x. [DOI] [PubMed] [Google Scholar]
- Allamane S, Jourdes P, Ratel D, Vicat JM, Dupre I, Laine M, et al. Bacterial DNA methylation and gene transfer efficiency. Biochem Biophys Res Commun. 2000;276(3):1261–4. doi: 10.1006/bbrc.2000.3603. [DOI] [PubMed] [Google Scholar]
- Allan BW, Beechem JM, Lindstrom WM, Reich NO. Direct real time observation of base flipping by the EcoRI DNA methyltransferase. The Journal of biological chemistry. 1998;273(4):2368–73. doi: 10.1074/jbc.273.4.2368. [DOI] [PubMed] [Google Scholar]
- Ammermann D, Steinbruck G, Baur R, Wohlert H. Methylated bases in the DNA of the ciliate Stylonychia mytilus. Eur J Cell Biol. 1981;24(1):154–6. [PubMed] [Google Scholar]
- Aravind L, Zhang D, Iyer LM. The TET/JBP Family of Nucleic Acid Base-Modifying 2-Oxoglutarate and Iron-Dependent Dioxygenases. In: Hausinger R, Schofield C, editors. 2-Oxoglutarate-Dependent Oxygenases. Royal Society of Chemistry; 2015. [Google Scholar]
- Babinger P, Kobl I, Mages W, Schmitt R. A link between DNA methylation and epigenetic silencing in transgenic Volvox carteri. Nucleic Acids Res. 2001;29(6):1261–71. doi: 10.1093/nar/29.6.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker A, Smith DW. Methylation of GATC sites is required for precise timing between rounds of DNA replication in Escherichia coli. J Bacteriol. 1989;171(10):5738–42. doi: 10.1128/jb.171.10.5738-5742.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15(6):707–19. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdis AJ, Lee I, Coward JK, Stephens C, Wright R, Shapiro L, et al. A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(6):2874–9. doi: 10.1073/pnas.95.6.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988;203(4):971–83. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9(16):2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird AP, Southern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, et al. Adenosine Methylation in Arabidopsis mRNA is Associated with the 3′ End and Reduced Levels Cause Developmental Defects. Front Plant Sci. 2012;3:48. doi: 10.3389/fpls.2012.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E, Lobner-Olesen A. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell. 1990;62(5):981–9. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- Breiling A, Lyko F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin. 2015;8:24. doi: 10.1186/s13072-015-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendler T, Abeles A, Austin S. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. The EMBO journal. 1995;14(16):4083–9. doi: 10.1002/j.1460-2075.1995.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg S, Pratt K, Hattman S. Sequence specificity of DNA adenine methylase in the protozoan Tetrahymena thermophila. J Bacteriol. 1982;150(2):993–6. doi: 10.1128/jb.150.2.993-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JL, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62(5):967–79. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- Capowski EE, Wells JM, Harrison GS, Karrer KM. Molecular analysis of N6-methyladenine patterns in Tetrahymena thermophila nuclear DNA. Mol Cell Biol. 1989;9(6):2598–605. doi: 10.1128/mcb.9.6.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuano F, Mulleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem. 2014;86(8):3697–702. doi: 10.1021/ac500447w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadesus J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70(3):830–56. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Luo GZ, He C. High-Resolution Mapping of N(6)-Methyladenosine in Transcriptome and Genome Using a Photo-Crosslinking-Assisted Strategy. Methods Enzymol. 2015;560:161–85. doi: 10.1016/bs.mie.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Cheng SC, Herman G, Modrich P. Extent of equilibrium perturbation of the DNA helix upon enzymatic methylation of adenine residues. The Journal of biological chemistry. 1985;260(1):191–4. [PubMed] [Google Scholar]
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, et al. S-Adenosylmethionine and methylation. FASEB J. 1996;10(4):471–80. [PubMed] [Google Scholar]
- Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene. Nucleic Acids Res. 2002;30(20):4509–18. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, McAdams HH, Shapiro L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(43):17111–6. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M, Myers RM. Alterations in DNA helix stability due to base modifications can be evaluated using denaturing gradient gel electrophoresis. J Mol Biol. 1987;198(4):737–44. doi: 10.1016/0022-2836(87)90214-2. [DOI] [PubMed] [Google Scholar]
- Cummings DJ, Tait A, Goddard JM. Methylated bases in DNA from Paramecium aurelia. Biochim Biophys Acta. 1974;374(1):1–11. doi: 10.1016/0005-2787(74)90194-4. [DOI] [PubMed] [Google Scholar]
- Degnen ST, Morris NR. Deoxyribonucleic acid methylation and development in Caulobacter bacteroides. J Bacteriol. 1973;116(1):48–53. doi: 10.1128/jb.116.1.48-53.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delk AS, Rabinowitz JC. Biosynthesis of ribosylthymine in the transfer RNA of Streptococcus faecalis: a folate-dependent methylation not involving S-adenosylmethionine. Proceedings of the National Academy of Sciences of the United States of America. 1975;72(2):528–30. doi: 10.1073/pnas.72.2.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delk AS, Romeo JM, Nagle DP, Jr, Rabinowitz JC. Biosynthesis of ribothymidine in the transfer RNA of Streptococcus faecalis and Bacillus subtilis. A methylation of RNA involving 5,10-methylenetetrahydrofolate. The Journal of biological chemistry. 1976;251(23):7649–56. [PubMed] [Google Scholar]
- Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, et al. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 2015;43(13):6557–67. doi: 10.1093/nar/gkv596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S. DNA methylation can enhance or induce DNA curvature. The EMBO journal. 1987;6(13):4213–7. doi: 10.1002/j.1460-2075.1987.tb02769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohno C, Shibata T, Nakatani K. Discrimination of N6-methyl adenine in a specific DNA sequence. Chem Commun (Camb) 2010;46(30):5530–2. doi: 10.1039/c0cc00172d. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–6. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Drozdz M, Piekarowicz A, Bujnicki JM, Radlinska M. Novel non-specific DNA adenine methyltransferases. Nucleic Acids Res. 2012;40(5):2119–30. doi: 10.1093/nar/gkr1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151(1):167–80. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(26):16660–5. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn DB, Smith JD. Occurrence of a new base in the deoxyribonucleic acid of a strain of Bacterium coli. Nature. 1955;175(4451):336–7. doi: 10.1038/175336a0. [DOI] [PubMed] [Google Scholar]
- Dunn DB, Smith JD. The occurrence of 6-methylaminopurine in deoxyribonucleic acids. Biochem J. 1958;68(4):627–36. doi: 10.1042/bj0680627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Carreira LH, Ljungdahl LG, Kuo KC, Gehrke CW. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 1985;13(4):1399–412. doi: 10.1093/nar/13.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10(8):2709–21. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Wilson GG, Kuo KC, Gehrke CW. N4-methylcytosine as a minor base in bacterial DNA. J Bacteriol. 1987;169(3):939–43. doi: 10.1128/jb.169.3.939-943.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel JD, von Hippel PH. D(M6ATP) as a probe of the fidelity of base incorporation into polynucleotides by Escherichia coli DNA polymerase I. The Journal of biological chemistry. 1978a;253(3):935–9. [PubMed] [Google Scholar]
- Engel JD, von Hippel PH. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. The Journal of biological chemistry. 1978b;253(3):927–34. [PubMed] [Google Scholar]
- Falnes PO, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419(6903):178–82. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- Fazakerley GV, Gabarro-Arpa J, Lebret M, Guy A, Guschlbauer W. The GTm6AC sequence is overwound and bent. Nucleic Acids Res. 1989;17(7):2541–56. doi: 10.1093/nar/17.7.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazakerley GV, Guy A, Teoule R, Quignard E, Guschlbauer W. A proton 2D-NMR study of an oligodeoxyribonucleotide containing N6-methyladenine:d(GGm6ATATCC) Biochimie. 1985;67(7–8):819–22. doi: 10.1016/s0300-9084(85)80173-5. [DOI] [PubMed] [Google Scholar]
- Fazakerley GV, Quignard E, Teoule R, Guy A, Guschlbauer W. A two-dimensional 1H-NMR study of the dam methylase site: comparison between the hemimethylated GATC sequence, its unmethylated analogue and a hemimethylated CATG sequence. The sequence dependence of methylation upon base-pair lifetimes. Eur J Biochem. 1987;167(3):397–404. doi: 10.1111/j.1432-1033.1987.tb13351.x. [DOI] [PubMed] [Google Scholar]
- Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB Family of Fe(II)/alpha-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and Beyond. The Journal of biological chemistry. 2015;290(34):20734–42. doi: 10.1074/jbc.R115.656462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleissner E, Borek E. A new enzyme of RNA synthesis: RNA methylase. Proceedings of the National Academy of Sciences of the United States of America. 1962;48:1199–203. doi: 10.1073/pnas.48.7.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7(6):461–5. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798. doi: 10.1038/ncomms2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Luo GZ, Chen K, Deng X, Yu M, Han D, et al. N(6)-methyldeoxyadenosine marks active transcription start sites in chlamydomonas. Cell. 2015;161(4):879–92. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa N, Kurumizaka H, Nureki O, Tanaka Y, Yamazoe M, Hiraga S, et al. Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein. Nucleic Acids Res. 2004;32(1):82–92. doi: 10.1093/nar/gkh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Sosa MA, Midgett RM, Slagel VA, Githens S, Kuo KC, Gehrke CW, et al. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta. 1983;740(2):212–9. doi: 10.1016/0167-4781(83)90079-9. [DOI] [PubMed] [Google Scholar]
- Geier GE, Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. The Journal of biological chemistry. 1979;254(4):1408–13. [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science (New York, NY. 2007;318(5855):1469–72. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B, van den Elsen P, Radman M. Induced mutagenesis in dam- mutants of Escherichia coli: a role for 6-methyladenine residues in mutation avoidance. Mol Gen Genet. 1978;163(3):307–12. doi: 10.1007/BF00271960. [DOI] [PubMed] [Google Scholar]
- Glickman BW. Spontaneous mutagenesis in Escherichia coli strains lacking 6-methyladenine residues in their DNA: an altered mutational spectrum in dam- mutants. Mutat Res. 1979;61(2):153–62. doi: 10.1016/0027-5107(79)90122-2. [DOI] [PubMed] [Google Scholar]
- Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat Struct Biol. 2001;8(2):121–5. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- Gold M, Hurwitz J. The Enzymatic Methylation of Ribonucleic Acid and Deoxyribonucleic Acid. V. Purification and Properties of the Deoxyribonucleic Acid-Methylating Activity of Escherichia Coli. The Journal of biological chemistry. 1964;239:3858–65. [PubMed] [Google Scholar]
- Gold M, Hurwitz J, Anders M. The enzymatic methylation of RNA and DNA. Biochem Biophys Res Commun. 1963;11:107–14. doi: 10.1016/0006-291x(63)90075-5. [DOI] [PubMed] [Google Scholar]
- Gommers-Ampt JH, Borst P. Hypermodified bases in DNA. FASEB J. 1995;9(11):1034–42. doi: 10.1096/fasebj.9.11.7649402. [DOI] [PubMed] [Google Scholar]
- Gommers-Ampt JH, Van Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, et al. beta-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75(6):1129–36. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- Gorovsky MA, Hattman S, Pleger GL. (6 N)methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J Cell Biol. 1973;56(3):697–701. doi: 10.1083/jcb.56.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MW, Larkin PJ. Adenine methylation at dam sites increases transient gene expression in plant cells. Transgenic Res. 1995;4(5):324–31. doi: 10.1007/BF01972529. [DOI] [PubMed] [Google Scholar]
- Greer EL, Becker B, Latza C, Antebi A, Shi Y. Mutation of C. elegans demethylase spr-5 extends transgenerational longevity. Cell Res. 2015a doi: 10.1038/cr.2015.148.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Beese-Sims SE, Brookes E, Spadafora R, Zhu Y, Rothbart SB, et al. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Reports. 2014;7(1):113–26. doi: 10.1016/j.celrep.2014.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizabal-Corrales D, et al. DNA Methylation on N(6)-Adenine in C. elegans. Cell. 2015b;161(4):868–78. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H. Nucleic Acids Are Not Boring Long Polymers of Only Four Types of Nucleotides: A Guided Tour. In: Grosjean H, editor. DNA and RNA Modi cation Enzymes: Structure, Mechanism, Function and Evolution. CRC Press; 2009. pp. 1–18. [Google Scholar]
- Grosjean H. RNA modification: the Golden Period 1995–2015. RNA. 2015;21(4):625–6. doi: 10.1261/rna.049866.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarne A, Zhao Q, Ghirlando R, Yang W. Insights into negative modulation of E. coli replication initiation from the structure of SeqA-hemimethylated DNA complex. Nat Struct Biol. 2002;9(11):839–43. doi: 10.1038/nsb857. [DOI] [PubMed] [Google Scholar]
- Hagerman KR, Hagerman PJ. Helix rigidity of DNA: the meroduplex as an experimental paradigm. J Mol Biol. 1996;260(2):207–23. doi: 10.1006/jmbi.1996.0393. [DOI] [PubMed] [Google Scholar]
- Hassel M, Cornelius MG, Vom Brocke J, Schmeiser HH. Total nucleotide analysis of Hydra DNA and RNA by MEKC with LIF detection and 32P-postlabeling. Electrophoresis. 2010;31(2):299–302. doi: 10.1002/elps.200900458. [DOI] [PubMed] [Google Scholar]
- Hattman S, Kenny C, Berger L, Pratt K. Comparative study of DNA methylation in three unicellular eucaryotes. J Bacteriol. 1978;135(3):1156–7. doi: 10.1128/jb.135.3.1156-1157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindell HC, Liu A, Paddock GV, Studnicka GM, Salser WA. The primary sequence of rabbit alpha-globin mRNA. Cell. 1978;15(1):43–54. doi: 10.1016/0092-8674(78)90081-8. [DOI] [PubMed] [Google Scholar]
- Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(36):14855–60. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Liebert K, Bekes M, Jeltsch A, Cheng X. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J Mol Biol. 2006;358(2):559–70. doi: 10.1016/j.jmb.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng X. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of dam methyltransferase. Cell. 2005;121(3):349–61. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. The Journal of biological chemistry. 1948;175(1):315–32. [PubMed] [Google Scholar]
- Huang W, Xiong J, Yang Y, Liu SM, Yuan BF, Feng YQ. Determination of DNA adenine methylation in genomes of mammals and plants by liquid chromatography/mass spectrometry. Royal Society of Chemistry Advances. 2015;(5):64046–54. [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–33. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science (New York, NY. 2011;333(6047):1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Abhiman S, Aravind L. Natural history of eukaryotic DNA methylation systems. Prog Mol Biol Transl Sci. 2011;101:25–104. doi: 10.1016/B978-0-12-387685-0.00002-0. [DOI] [PubMed] [Google Scholar]
- Jabbari K, Caccio S, Pais de Barros JP, Desgres J, Bernardi G. Evolutionary changes in CpG and methylation levels in the genome of vertebrates. Gene. 1997;205(1–2):109–18. doi: 10.1016/s0378-1119(97)00475-7. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416(6880):556–60. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Janulaitis A, Klimasauskas S, Petrusyte M, Butkus V. Cytosine modification in DNA by BcnI methylase yields N4-methylcytosine. FEBS Lett. 1983;161(1):131–4. doi: 10.1016/0014-5793(83)80745-5. [DOI] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–7. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, et al. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Current biology : CB. 2007;17(4):379–84. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TB, Coghill RD. The discovery of 5-methyl-cytosine in tuberculinic acid, the nucleic acid of the Tubercle bacillus. Journal of the American Chemical Society. 1925;47:2838–44. [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Kakutani T, Munakata K, Richards EJ, Hirochika H. Meiotically and mitotically stable inheritance of DNA hypomethylation induced by ddm1 mutation of Arabidopsis thaliana. Genetics. 1999;151(2):831–8. doi: 10.1093/genetics/151.2.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat SS, Fan H, Sauder JM, Burley SK, Shoichet BK, Sali A, et al. Enzymatic deamination of the epigenetic base N-6-methyladenine. J Am Chem Soc. 2011;133(7):2080–3. doi: 10.1021/ja110157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer KM, VanNuland TA. Methylation of adenine in the nuclear DNA of Tetrahymena is internucleosomal and independent of histone H1. Nucleic Acids Res. 2002;30(6):1364–70. doi: 10.1093/nar/30.6.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137(2):308–20. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Zimmerman SB, Kornberg SR, Josse J. Enzymatic Synthesis of Deoxyribonucleic Acid. Influence of Bacteriophage T2 on the Synthetic Pathway in Host Cells. Proceedings of the National Academy of Sciences of the United States of America. 1959;45(6):772–85. doi: 10.1073/pnas.45.6.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg SR, Zimmerman SB, Kornberg A. Glucosylation of deoxyribonucleic acid by enzymes from bacteriophage-infected Escherichia coli. The Journal of biological chemistry. 1961;236:1487–93. [PubMed] [Google Scholar]
- Kozdon JB, Melfi MD, Luong K, Clark TA, Boitano M, Wang S, et al. Global methylation state at base-pair resolution of the Caulobacter genome throughout the cell cycle. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(48):E4658–67. doi: 10.1073/pnas.1319315110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol MJ, Bradshaw CR, Allen GE, Costa AS, Frezza C, Gurdon JB. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nature structural & molecular biology. 2016;23(1):24–30. doi: 10.1038/nsmb.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krais AM, Cornelius MG, Schmeiser HH. Genomic N(6)-methyladenine determination by MEKC with LIF. Electrophoresis. 2010;31(21):3548–51. doi: 10.1002/elps.201000357. [DOI] [PubMed] [Google Scholar]
- Lahue RS, Su SS, Modrich P. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(6):1482–6. doi: 10.1073/pnas.84.6.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39(Web Server issue):W475–8. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtsteiner S, Schibler U. A glycosylated liver-specific transcription factor stimulates transcription of the albumin gene. Cell. 1989;57(7):1179–87. doi: 10.1016/0092-8674(89)90055-x. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13(16):3405–10. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- Linn S, Arber W. Host specificity of DNA produced by Escherichia coli, X. In vitro restriction of phage fd replicative form. Proceedings of the National Academy of Sciences of the United States of America. 1968;59(4):1300–6. doi: 10.1073/pnas.59.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–5. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun. 2001;69(12):7197–204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Campbell JL, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77(3):413–26. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- Luo GZ, Blanco MA, Greer EL, He C, Shi Y. DNA N-methyladenine: a new epigenetic mark in eukaryotes? Nature reviews. 2015;16(12):705–10. doi: 10.1038/nrm4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, Human ML. A nonhereditary, host-induced variation of bacterial viruses. J Bacteriol. 1952;64(4):557–69. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in Drosophila melanogaster. Nature. 2000;408(6812):538–40. doi: 10.1038/35046205. [DOI] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Res. 2013;41(Database issue):D262–7. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macon JB, Wolfenden R. 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry. 1968;7(10):3453–8. doi: 10.1021/bi00850a021. [DOI] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. The EMBO journal. 2002;21(24):6842–52. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malygin EG, Lindstrom WM, Jr, Schlagman SL, Hattman S, Reich NO. Pre-steady state kinetics of bacteriophage T4 dam DNA-[N(6)-adenine] methyltransferase: interaction with native (GATC) or modified sites. Nucleic Acids Res. 2000;28(21):4207–11. doi: 10.1093/nar/28.21.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus MG. Adenine methylation of Okazaki fragments in Escherichia coli. J Bacteriol. 1976;128(3):853–4. doi: 10.1128/jb.128.3.853-854.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus MG, Lobner-Olesen A. DNA Methylation. Ecosal Plus. 2014;6(1) doi: 10.1128/ecosalplus.ESP-0003-2013.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus MG, Morris NR. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973;114(3):1143–50. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus MG, Morris NR. Biological function for 6-methyladenine residues in the DNA of Escherichia coli K12. J Mol Biol. 1974;85(2):309–22. doi: 10.1016/0022-2836(74)90366-0. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19(3):266–72. doi: 10.1016/j.ceb.2007.04.002. S0955-0674(07)00054-3 [pii] [DOI] [PubMed] [Google Scholar]
- Mason SF. Purine Studies. Part II. The Ultra-violet absorption spectra of some mono- and poly-substituted purines. Journal of the Chemical Society. 1954:2071–81. [Google Scholar]
- McClelland M. Selection against dam methylation sites in the genomes of DNA of enterobacteriophages. J Mol Evol. 1984;21(4):317–22. doi: 10.1007/BF02115649. [DOI] [PubMed] [Google Scholar]
- Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217(5134):1110–4. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149(7):1635–46. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JB, Hagerman PJ. Origin of the intrinsic rigidity of DNA. Nucleic Acids Res. 2004;32(13):4055–9. doi: 10.1093/nar/gkh740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero LM, Filipski J, Gil P, Capel J, Martinez-Zapater JM, Salinas J. The distribution of 5-methylcytosine in the nuclear genome of plants. Nucleic Acids Res. 1992;20(12):3207–10. doi: 10.1093/nar/20.12.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie AI, Lilley DM. Base methylation and local DNA helix stability. Effect on the kinetics of cruciform extrusion. J Mol Biol. 1989;205(3):593–602. doi: 10.1016/0022-2836(89)90228-3. [DOI] [PubMed] [Google Scholar]
- Murray NE. 2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self. Microbiology. 2002;148(Pt 1):3–20. doi: 10.1099/00221287-148-1-3. [DOI] [PubMed] [Google Scholar]
- Nikolskaya II, Lopatina NG, Chaplygina NM, Debov SS. The host specificity system in Escherichia coli SK. Mol Cell Biochem. 1976;13(2):79–87. doi: 10.1007/BF01837057. [DOI] [PubMed] [Google Scholar]
- Nikolskaya II, Lopatina NG, Debov SS. On heterogeneity of DNA methylases from Escherichia coli SK cells. Mol Cell Biochem. 1981;35(1):3–10. doi: 10.1007/BF02358182. [DOI] [PubMed] [Google Scholar]
- Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteomics Bioinformatics. 2013;11(1):8–17. doi: 10.1016/j.gpb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(33):13624–9. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Padva A, LeBreton PR. Ultraviolet photoelectron studies of biological purines: the valence electronic structure of adenine. Proceedings of the National Academy of Sciences of the United States of America. 1976;73(9):2966–8. doi: 10.1073/pnas.73.9.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogolotti AL, Jr, Ono A, Subramaniam R, Santi DV. On the mechanism of DNA-adenine methylase. The Journal of biological chemistry. 1988;263(16):7461–4. [PubMed] [Google Scholar]
- Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47(3):142–50. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Posfai G, Szybalski W. A simple method for locating methylated bases in DNA using class-IIS restriction enzymes. Gene. 1988;74(1):179–81. doi: 10.1016/0378-1119(88)90280-6. [DOI] [PubMed] [Google Scholar]
- Pratt K, Hattman S. Nucleosome phasing in Tetrahymena macronuclei. J Protozool. 1983;30(3):592–8. doi: 10.1111/j.1550-7408.1983.tb01428.x. [DOI] [PubMed] [Google Scholar]
- Privat E, Sowers LC. Photochemical deamination and demethylation of 5-methylcytosine. Chem Res Toxicol. 1996;9(4):745–50. doi: 10.1021/tx950182o. [DOI] [PubMed] [Google Scholar]
- Proffitt JH, Davie JR, Swinton D, Hattman S. 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol Cell Biol. 1984;4(5):985–8. doi: 10.1128/mcb.4.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila PJ, Peterson J, Herman G, Modrich P, Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983;104(4):571–82. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignard E, Fazakerley GV, Teoule R, Guy A, Guschlbauer W. Consequences of methylation on the amino group of adenine. A proton two-dimensional NMR study of d(GGATATCC) and d(GGm6ATATCC) Eur J Biochem. 1985;152(1):99–105. doi: 10.1111/j.1432-1033.1985.tb09168.x. [DOI] [PubMed] [Google Scholar]
- Rae PM. Hydroxymethyluracil in eukaryote DNA: a natural feature of the pyrrophyta (dinoflagellates) Science (New York, NY. 1976;194(4269):1062–4. doi: 10.1126/science.988637. [DOI] [PubMed] [Google Scholar]
- Rae PM, Spear BB. Macronuclear DNA of the hypotrichous ciliate Oxytricha fallax. Proceedings of the National Academy of Sciences of the United States of America. 1978;75(10):4992–6. doi: 10.1073/pnas.75.10.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, Razin S. Methylated bases in mycoplasmal DNA. Nucleic Acids Res. 1980;8(6):1383–90. doi: 10.1093/nar/8.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich NO, Mashhoon N. Kinetic mechanism of the EcoRI DNA methyltransferase. Biochemistry. 1991;30(11):2933–9. doi: 10.1021/bi00225a029. [DOI] [PubMed] [Google Scholar]
- Robbins-Manke JL, Zdraveski ZZ, Marinus M, Essigmann JM. Analysis of global gene expression and double-strand-break formation in DNA adenine methyltransferase- and mismatch repair-deficient Escherichia coli. J Bacteriol. 2005;187(20):7027–37. doi: 10.1128/JB.187.20.7027-7037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D, Hoopes BC, McClure WR, Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43(1):117–30. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Rogers JC, Rogers SW. Comparison of the effects of N6-methyldeoxyadenosine and N5-methyldeoxycytosine on transcription from nuclear gene promoters in barley. Plant J. 1995;7(2):221–33. doi: 10.1046/j.1365-313x.1995.7020221.x. [DOI] [PubMed] [Google Scholar]
- Rogers SD, Rogers ME, Saunders G, Holt G. Isolation of mutants sensitive to 2-aminopurine and alkylating agents and evidence for the role of DNA methylation in Penicillium chrysogenum. Curr Genet. 1986;10(7):557–60. doi: 10.1007/BF00447390. [DOI] [PubMed] [Google Scholar]
- Romanov GA, Vanyushin BF. Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction. Biochim Biophys Acta. 1981;653(2):204–18. doi: 10.1016/0005-2787(81)90156-8. [DOI] [PubMed] [Google Scholar]
- Russell DW, Hirata RK. The detection of extremely rare DNA modifications. Methylation in dam- and hsd- Escherichia coli strains. The Journal of biological chemistry. 1989;264(18):10787–94. [PubMed] [Google Scholar]
- Saparbaev M, Laval J. Excision of hypoxanthine from DNA containing dIMP residues by the Escherichia coli, yeast, rat, and human alkylpurine DNA glycosylases. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(13):5873–7. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnacki SH, Castaneda Mdel R, Noto Llana M, Giacomodonato MN, Valvano MA, Cerquetti MC. Dam methylation participates in the regulation of PmrA/PmrB and RcsC/RcsD/RcsB two component regulatory systems in Salmonella enterica serovar Enteritidis. PLoS One. 2013;8(2):e56474. doi: 10.1371/journal.pone.0056474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sater MR, Lamelas A, Wang G, Clark TA, Roltgen K, Mane S, et al. DNA Methylation Assessed by SMRT Sequencing Is Linked to Mutations in Neisseria meningitidis Isolates. PLoS One. 2015;10(12):e0144612. doi: 10.1371/journal.pone.0144612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T. Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007;6(4):429–42. doi: 10.1016/j.dnarep.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular cell. 2012;48(6):849–62. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R, Klein RS. The deamination of cytidine and cytosine by acidic buffer solutions. Mutagenic implications. Biochemistry. 1966;5(7):2358–62. doi: 10.1021/bi00871a026. [DOI] [PubMed] [Google Scholar]
- Shen L, Song CX, He C, Zhang Y. Mechanism and function of oxidative reversal of DNA and RNA methylation. Annu Rev Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82(6):927–36. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- Smith JD, Arber W, Kuhnlein U. Host specificity of DNA produced by Escherichia coli. XIV. The role of nucleotide methylation in in vivo B-specific modification. J Mol Biol. 1972;63(1):1–8. doi: 10.1016/0022-2836(72)90517-7. [DOI] [PubMed] [Google Scholar]
- Srivastava R, Gopinathan KP, Ramakrishnan T. Deoxyribonucleic acid methylation in mycobacteria. J Bacteriol. 1981;148(2):716–9. doi: 10.1128/jb.148.2.716-719.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R, Gruenbaum Y, Pollack Y, Razin A, Cedar H. Clonal inheritance of the pattern of DNA methylation in mouse cells. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(1):61–5. doi: 10.1073/pnas.79.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens C, Reisenauer A, Wright R, Shapiro L. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(3):1210–4. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternglanz H, Bugg CE. Conformation of N6-methyladenine, a base involved in DNA modification: restriction processes. Science (New York, NY. 1973;182(4114):833–4. doi: 10.1126/science.182.4114.833. [DOI] [PubMed] [Google Scholar]
- Su SS, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(14):5057–61. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Takeda S, Hirochika H. Transcriptional activation mediated by binding of a plant GATA-type zinc finger protein AGP1 to the AG-motif (AGATCCAA) of the wound-inducible Myb gene NtMyb2. Plant J. 2003;36(4):550–64. doi: 10.1046/j.1365-313x.2003.01899.x. [DOI] [PubMed] [Google Scholar]
- Sun Q, Huang S, Wang X, Zhu Y, Chen Z, Chen D. N(6) -methyladenine functions as a potential epigenetic mark in eukaryotes. Bioessays. 2015;37(11):1155–62. doi: 10.1002/bies.201500076. [DOI] [PubMed] [Google Scholar]
- Sundheim O, Talstad VA, Vagbo CB, Slupphaug G, Krokan HE. AlkB demethylases flip out in different ways. DNA Repair (Amst) 2008;7(11):1916–23. doi: 10.1016/j.dnarep.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (New York, NY. 2009;324(5929):930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawa R, Ueno S, Yamamoto K, Yamamoto Y, Sagisaka K, Katakura R, et al. Methylated cytosine level in human liver DNA does not decline in aging process. Mechanisms of ageing and development. 1992;62(3):255–61. doi: 10.1016/0047-6374(92)90111-p. [DOI] [PubMed] [Google Scholar]
- Theil EC, Zamenhof S. Studies on 6-Methylaminopurine (6-Methyladenine) in Bacterial Deoxyribonucleic Acid. The Journal of biological chemistry. 1963;238:3058–64. [PubMed] [Google Scholar]
- Trewick SC, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419(6903):174–8. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- Tronche F, Rollier A, Bach I, Weiss MC, Yaniv M. The rat albumin promoter: cooperation with upstream elements is required when binding of APF/HNF1 to the proximal element is partially impaired by mutation or bacterial methylation. Mol Cell Biol. 1989;9(11):4759–66. doi: 10.1128/mcb.9.11.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya H, Matsuda T, Harashima H, Kamiya H. Cytokine induction by a bacterial DNA-specific modified base. Biochem Biophys Res Commun. 2005;326(4):777–81. doi: 10.1016/j.bbrc.2004.11.115. [DOI] [PubMed] [Google Scholar]
- Unger G, Venner H. Remarks on minor bases in spermatic desoxyribonucleic acid. Hoppe Seylers Z Physiol Chem. 1966;344(4):280–3. [PubMed] [Google Scholar]
- Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria--evolutionary implications. Nucleic Acids Res. 2005;33(13):3955–64. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urig S, Gowher H, Hermann A, Beck C, Fatemi M, Humeny A, et al. The Escherichia coli dam DNA methyltransferase modifies DNA in a highly processive reaction. J Mol Biol. 2002;319(5):1085–96. doi: 10.1016/S0022-2836(02)00371-6. [DOI] [PubMed] [Google Scholar]
- van den Born E, Omelchenko MV, Bekkelund A, Leihne V, Koonin EV, Dolja VV, et al. Viral AlkB proteins repair RNA damage by oxidative demethylation. Nucleic Acids Res. 2008;36(17):5451–61. doi: 10.1093/nar/gkn519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten JL, Schuster AM, Girton L, Burbank DE, Swinton D, Hattman S. DNA methylation of viruses infecting a eukaryotic Chlorella-like green alga. Nucleic Acids Res. 1985;13(10):3471–8. doi: 10.1093/nar/13.10.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin BF, Belozersky AN, Kokurina NA, Kadirova DX. 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature. 1968;218(5146):1066–7. doi: 10.1038/2181066a0. [DOI] [PubMed] [Google Scholar]
- Vanyushin BF, Tkacheva SG, Belozersky AN. Rare bases in animal DNA. Nature. 1970;225(5236):948–9. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- von Freiesleben U, Rasmussen KV, Schaechter M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol. 1994;14(4):763–72. doi: 10.1111/j.1365-2958.1994.tb01313.x. [DOI] [PubMed] [Google Scholar]
- Wagner I, Capesius I. Determination of 5-methylcytosine from plant DNA by high-performance liquid chromatography. Biochim Biophys Acta. 1981;654(1):52–6. doi: 10.1016/0005-2787(81)90135-0. [DOI] [PubMed] [Google Scholar]
- Wallecha A, Munster V, Correnti J, Chan T, van der Woude M. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J Bacteriol. 2002;184(12):3338–47. doi: 10.1128/JB.184.12.3338-3347.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–20. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161(6):1388–99. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature genetics. 2005;37(8):853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wei YF, Carter KC, Wang RP, Shell BK. Molecular cloning and functional analysis of a human cDNA encoding an Escherichia coli AlkB homolog, a protein involved in DNA alkylation damage repair. Nucleic Acids Res. 1996;24(5):931–37. doi: 10.1093/nar/24.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis DB, Granoff A. Frog virus 3 DNA is heavily methylated at CpG sequences. Virology. 1980;107(1):250–7. doi: 10.1016/0042-6822(80)90290-1. [DOI] [PubMed] [Google Scholar]
- Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol. 2006;4(3):183–92. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Boye E, Slater S, Kleckner N, Skarstad K. Effects of purified SeqA protein on oriC-dependent DNA replication in vitro. The EMBO journal. 1998;17(14):4158–65. doi: 10.1093/emboj/17.14.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JC, Santi DV. Kinetic and catalytic mechanism of HhaI methyltransferase. The Journal of biological chemistry. 1987;262(10):4778–86. [PubMed] [Google Scholar]
- Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, et al. DNA methylation on N-adenine in mammalian embryonic stem cells. Nature. 2016 doi: 10.1038/nature17640.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt GR. Occurrence of 5-methylcytosine in nucleic acids. Nature. 1950;166(4214):237–8. doi: 10.1038/166237b0. [DOI] [PubMed] [Google Scholar]
- Wyatt GR, Cohen SS. A new pyrimidine base from bacteriophage nucleic acids. Nature. 1952;170(4338):1072–3. doi: 10.1038/1701072a0. [DOI] [PubMed] [Google Scholar]
- Yamaki H, Ohtsubo E, Nagai K, Maeda Y. The oriC unwinding by dam methylation in Escherichia coli. Nucleic Acids Res. 1988;16(11):5067–73. doi: 10.1093/nar/16.11.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature. 2008;452(7190):961–5. doi: 10.1038/nature06889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Soman NS, Verdine GL, Bestor TH. DNA (cytosine-5)-methyltransferases in mouse cells and tissues. Studies with a mechanism-based probe. J Mol Biol. 1997;270(3):385–95. doi: 10.1006/jmbi.1997.1125. [DOI] [PubMed] [Google Scholar]
- Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes & development. 2015;29(13):1343–55. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki H, Kawasaki H, Imayuki A, Yajima T. Determination of 6-methyladenine in DNA by high-performance liquid chromatography. J Chromatogr. 1979;168(2):489–94. doi: 10.1016/0021-9673(79)80020-5. [DOI] [PubMed] [Google Scholar]
- Zaleski P, Wojciechowski M, Piekarowicz A. The role of Dam methylation in phase variation of Haemophilus influenzae genes involved in defence against phage infection. Microbiology. 2005;151(Pt 10):3361–9. doi: 10.1099/mic.0.28184-0. [DOI] [PubMed] [Google Scholar]
- Zelinkova E, Paulicek M, Zelinka J. Modification methylase M.Sau3239I from Streptomyces aureofaciens 3239. FEBS Lett. 1990;271(1–2):147–8. doi: 10.1016/0014-5793(90)80393-w. [DOI] [PubMed] [Google Scholar]
- Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, et al. N(6)-methyladenine DNA modification in Drosophila. Cell. 2015;161(4):893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526(7574):591–4. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]