Abstract
Developmental genetic studies in Drosophila unraveled the importance of Polycomb group (PcG) and Trithorax group (TrxG) genes in controlling cellular identity. PcG and TrxG proteins form histone modifying complexes that catalyze repressive or activating histone modifications, respectively, and thus maintaining the expression status of homeotic genes. Human orthologs of PcG and TrxG genes are implicated in tumorigenesis as well as in determining the prognosis of individual cancers. Recent whole genome analyses of cancers also highlighted the importance of histone modifying proteins in controlling tumorigenesis. Comprehensive understanding of the mechanistic relationship between histone regulation and tumorigenesis holds the promise of significantly advancing our understanding and management of cancer. It is anticipated that Drosophila melanogaster, the model organism that contributed significantly to our understanding of the functional role of histone regulation in development, could also provide unique insight for our understanding of how histone dysregulation can lead to cancer. In this review, we will discuss several recent advances in this regard.
Keywords: Polycomb, Epigenetics, Tumorigenesis, Drosophila
1. Introduction
1.1. Identification of PcG and TrxG proteins as chromatin regulators
The accurate placement of segmental structures along the anterior-posterior axis of animal body is defined by the highly conserved homeotic (Hox) gene family. Dysregulation of Hox gene leads to homeotic transformation—transformation of one body segment into the identity of another (Pearson et al., 2005). Therefore, in order to maintain cellular identity, the establishment and maintenance of Hox gene expression pattern has to be tightly controlled. In Drosophila, the Hox gene expression is initiated by transient transcriptional factors encoded by gap genes and pair-rule genes in early embryogenesis. However, the transient transcription factors disappear soon after turning on the Hox genes. Development of an organism requires a cellular system to “remember” the appropriate Hox gene expression status ascribed to individual cell.
Genetic analysis unraveled two groups of genes affecting cellular identity: Polycomb group (PcG) and Trithorax group (TrxG). The first two PcG genes—Extra sex combs (Esc) and Polycomb (Pc), were described in Drosophila by Lewis and colleagues in the 1940s. They were named after the mutation phenotype: male flies grew extra sex combs on the second and third legs, which were usually restricted to the first legs (Lewis, 1947, 1978). Following mechanistic studies revealed that the PcG mutant cells inappropriately reactivate specific Hox genes which should have been repressed in those cells, transforming one body segment into another (Struhl, 1983; Jurgens, 1985). This failure in the cellular memory system leads to the idea that PcG functions to maintain the repressed state of Hox gene (Pirrotta, 1997). Although originally identified in Drosophila, PcG function has been fairly well conserved along evolution: several Polycomb mutants in mice exhibit anterior-posterior transformations and other abnormalities of the axial skeleton (Akasaka et al., 1996; Core et al., 1997; del Mar Lorente et al., 2000).
TrxG proteins have been characterized as an antagonistic system of PcG proteins, which set up an active state for the Hox gene. In the absence of Trithorax (TRX), the best characterized member in TrxG, multiple homeotic genes become repressed in a PcG-dependent fashion from cells where they are expressed in early stage embryos. Consequently, flies show segmental transformations, similar to the phenotypes of Hox gene mutants (Breen and Harte, 1991; Orlando and Paro, 1995). Therefore, PcG and TrxG work together, through catalyzing either repressive or activating histone modifications, to achieve the appropriate temporal and spatial pattern of Hox gene expression. Although PcG and TrxG proteins were firstly discovered to regulate Hox genes, further studies have identified a variety of target genes involved in stem cell maintenance, cell cycle control, apoptosis, etc. Genetic lesions inducing inappropriate PcG and TrxG activity may perturb these fundamental biological processes, leading to pathogenesis such as tumor (Mills, 2010).
1.2. Chromatin structures and epigenetic regulations
The term “epigenetics” is used to define changes in gene expression that do not result from alternating primary DNA sequence and are mitotically heritable. Epigenetic inheritance can be produced by distinct mechanisms, such as DNA methylation, chromatin modifications, and non-coding RNA. Here we are focusing on the epigenetic regulation through the modification of chromatin structures.
In eukaryotic cells, DNA is wrapping around core histone octamers to form the basic chromosome structured—nucleosomes, which are further folded into higher order chromatin. Different chromatin conformations are usually associated with diverse DNA accessibilities and transcriptional potentials. In general, an open chromatin or “euchromatin” facilitates transcription whereas a compact chromatin or “heterochromatin” makes the underlying genes highly resistant to transcriptional activity. The impact of heterochromatin configuration on gene silencing was noticed decades ago through the study of position effect variegation (PEV), which reveals that gene activity is dependent on its position relative to a heterochromatin region on chromosome. When a Drosophila gene required for red eye pigmentation was placed in juxtapotition with the pericentric heterochromatin, it became silenced in a subset of cells and resulted in a mosaic eye color (Muller, 1932).
Changes in chromatin structures are effectively conducted through enzymatic modifications of the histone proteins. The N-terminal tails of core histones that protrude from nucleosomes are subject to a variety of post-translational covalent modifications. The histone modifications provide a scaffold for the recruitment of regulatory proteins or chromatin remodeling factors, which in turn define distinct chromatin states (Jenuwein and Allis, 2001). For example, trimethylation at lysine 9 or lysine 27 on histone H3 serves as repressive histone mark for the transcriptionally silent heterochromatin, whereas trimethylation at lysine 4 on histone H3 as well as acetylation on histone H3 have been closely linked to the transcriptionally active chromatin.
1.3. PcG/TrxG mediated chromatin modifications
In flies, there are at least two types of multi-protein complexes working together to conduct PcG-mediated silencing. They are referred to as the Polycomb Repressive Complex 1 and 2 (PRC1 and PRC2), which function to maintain and establish the silenced chromatin states, respectively. The later on characterized PhoRC mainly contains PHO/PHOL and SFMBT (Scmrelated gene containing four MBT domains), which may provide DNA-binding property (Schwartz and Pirrotta, 2007).
The biochemically purified Drosophila PRC2 core complex consists of Enhancer of zeste (E(Z)), Suppressor of zeste 12 (SU(Z)12), Extra sex comb (ESC) and NURF55. The SET domain in E(Z) has histone methyltransferase activity and is able to catalyze trimethylation at lysine27 of histone H3 (H3K27me3), a repressive histone mark associated with gene silencing (Cao et al., 2002; Czermin et al., 2002; Müller et al., 2002). The Drosophila PRC1 complex is more diverse and mainly contains Polycomb (PC), Posterior sex combs (PSC), Polyhomeotic (PH) and RING (Shao et al., 1999; Saurin et al., 2001). The chomodomain in PC specifically recognizes the H3K27me3 established by PRC2, to stabilize the repressive status of chromatin. The core components of mammalian PRC1 and PRC2 are very similar to those in Drosophila but containing more paralogs (Table 1), which may function as alternatives to target different genes or different tissues (Levine et al., 2002).
Table 1.
Many histone modifiers are evolutionary conserved and implicated in tumorigenesis.
| Complex | Drosophila protein | Human homologues | Functional domains | Biochemical activity |
|---|---|---|---|---|
| PcG proteins | ||||
| PRC1 | Polycomb (PC) | CBX2, CBX4, CBX6, CBX7, CBX8 | Chromodomain | Binding to H3K27me3 |
| Posterior sex comb (PSC) | PCGF1(NSPc1), PCGF2(MEL18), PCGF4(BMI1) |
Zinc finger | Cofactor for Ring | |
| Polyhomeotic (PH) | PHC1, PHC2, PHC3 | Zinc finger and SAM | Required for silencing | |
| Sex combs extra (SCE or RING) | RING1A, RING1B, RNF2 | RING zinc finger | H2AK119 ubiquitin ligase | |
| Sex comb on midleg (SCM) | SCMH1, SCML2 | SAM, MBT, Zinc finger | recruitment of the PcG protein |
|
| PRC2 | Enhancer of zeste (E(Z)) | EZH1 and EZH2 | SET | Histone methyltransferase, establish H3K27me3 |
| Extra sex combs (ESC) | EED | WD40 repeats | Co-factor for E(Z) | |
| Extra sex combs-like (ESCL) | EED | WD40 repeats | Co-factor for E(Z) | |
| Suppressor of zeste 12 (SU(Z)12) | SUZ12 | Zinc finger | Co-factor for E(Z) | |
| Polycomb-like (PCL) | PCL1(PHF1), PCL2(MTF2), PCL3(PHF19) |
PHD | ||
| NURF 55 | Nurf55 | WD40 repeats | Facilitating the nucleosome binding |
|
| PhoRC | Pleiohomeotic (PHO) | YY1, YY2 | Zinc finger | DNA binding |
| Pleiohomeotic-like (PHOL) | YY1, YY2 | Zinc finger | DNA binding | |
| SFMBT | L3MBTL2, MBTD1 | MBT, SAM | Binding to mono- and dimethyl H3K9, H4K20 |
|
| TrxG proteins | ||||
| TAC1 | Trithorax (TRX) | MLL, MLL2, MLL3, MLL5 | SET | Histone methyltransferase, establishes H3K4me3 |
| dCBP | CBP | KIX, IBiD, zinc finger | Histone acetyltransferase | |
| dUTX | UTX | JmjC | Di- and trimethylated H3K27 demethylase |
|
| ASH1 | ASH1L | SET | H3K4/H3K36 methylase | |
| ASH2 | ASH2L, WDR5 | WD40 repeats | Essential for H3K4me3 | |
| SWI-SNF nucleosome remodeling complex |
OSA | ARID1A, ARID1B | ||
| BRM | BRM, BRG1 | SWI-SNF-like helicase, Bromodomain |
ATPase activity, binds acetylated histones |
|
| SNR1 | SNF5, ARID4A, ARID4B | Non-catalytic core subunit | ||
| Other related histone modifiers |
SIR2 | SIRT1 | Zinc finger | NAD+-dependent class III histone deacetylase |
| LID | JARID1 | JmjC | Di- and trimethylated H3K4 demethylase |
|
| SU(VAR)3-3 | LSD1 | SWIRM, amine oxidase domain |
Mono- and dimethylated H3K4 demethylase |
PcG = polycomb group; TrxG = trithorax group; TAC = trithorax acetylation complex; NURF55 = nucleosome remodeling factor of 55 kDa; SFMBT = Scmrelated gene containing four MBT domains; CBP = CREB-binding protein; ASH = absent, small, or homeotic discs; SIR = silent information regulator; UTX = ubiquitously transcribed tetratricopeptide repeat, X chromosome; LID = little imaginal discs; CBX = chromobox homolog; PHC = polyhomeotic homologue; EZH = enhancer of zeste homologue; EED = embryonic ectoderm development; YY = Yin-Yang transcription factor; MLL = mixed lineage leukemia; LSD = lysine specific demethylase; PHD = plant homeodomain; WDR5 = WD repeat domain 5.
How PRCs modulate transcriptional silencing remains to be fully understood. The potential mechanisms include facilitating chromatin compaction, impeding RNA Pol II initiation and elongation, recruiting DNMTs (DNA methyltransferases) to target genes, as well as blocking the modulation of SWI/SNF chromatin remodeling complex (Sparmann and van Lohuizen, 2006). In addition, the RING finger domain of RING protein possesses E3 ubiquitin ligase activity and induces monoubiquitination of H2AK119, which is also associated with gene silencing (de Napoles et al., 2004; Wang et al., 2004; Cao et al., 2005).
Similar to PcGs, TrxG proteins also form multi-components complexes. They maintain an active chromatin state through either direct histone modification or ATP-dependent nucleosome remodeling (Strahl and Allis, 2000; Vignali et al., 2000). The founding member of Drosophila TrxG family, TRX, is a histone methyltransferase which catalyzes H3K4 trimethylation to favor transcriptional activation (Santos-Rosa et al., 2002). TrxG proteins also regulate chromatin dynamics through nucleosome remodeling: the SWI-SNF complex contains ATP-dependent chromatin remodeling proteins, which are able to alter the nucleosome structures to facilitate basal transcription machinery (Smith and Peterson, 2005).
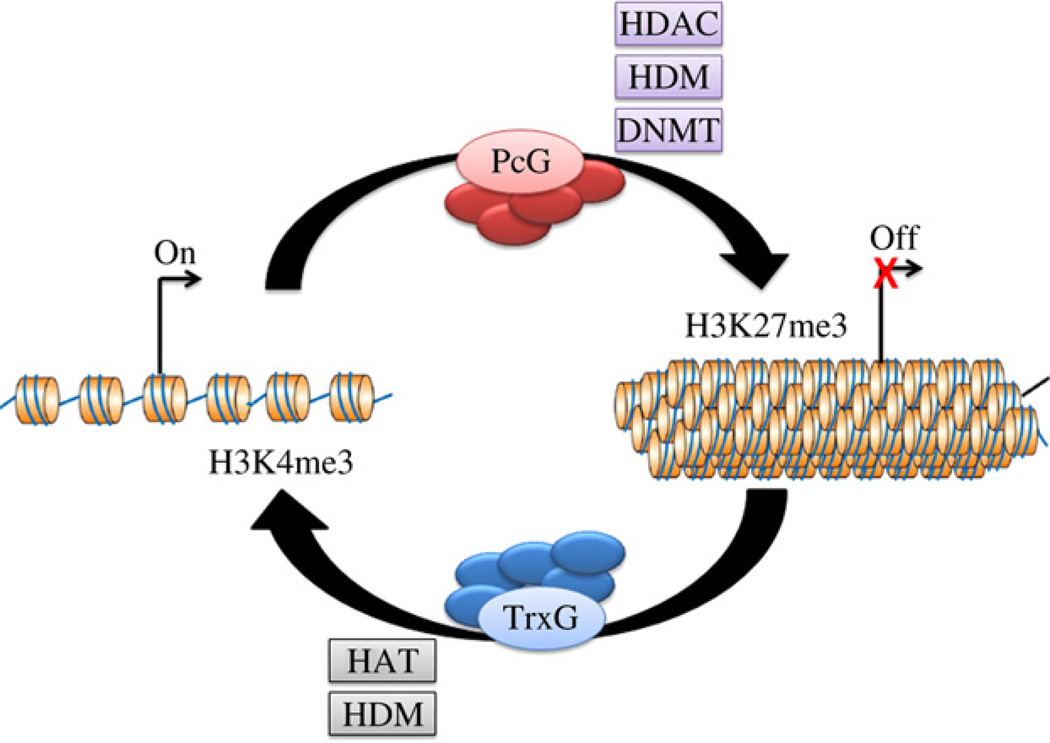
In addition to direct methylation, both PcG and TrxG complexes can recruit other histone modifiers to ensure transcriptional repression or activation. For example, they both can recruit histone demethylases (HDM): whereas TrxG proteins recruit HDMs (e.g., UTX) that specifically remove methyl groups from repressive histone mark of H3K27me3; a human HDM JARID1, which demethylates H3K4me3, has been found to associate with PcG proteins (Agger et al., 2007; Hong et al., 2007; Lee et al., 2007; Shi, 2007). Other histone modifiers such as histone acetyltransferase (HAT) and histone deacetylase (HDAC) are also recruited either directly or indirectly to modulate transcription (Sparmann and van Lohuizen, 2006; Mills, 2010) (Fig. 1). It is believed that the coordinated removal of repressive marks and deposition of positive marks (and vice versa) are important for chromatin dynamics and transcription.
Fig. 1.
PcG and TrxG proteins regulate gene expression by modulating chromatin structures. PcG and TrxG proteins directly methylate specific histone residues to establish repressive (H3K27me3) and active (H3K4me3) histone marks, respectively. In addition, they are able to recruit enzymes that modulate other histone modifications such as acetylation and demethylation as well as DNA methylation. PcG complexes can associate with HDACs, H3K4me3-specific HDMs and DNMTs to suppress gene expression, whereas TrxG complexes recruit HATs and H3K27me3-specific HDMs to activate gene expression.
For additional detailed information about the mechanism of PcG/TxG -mediated chromatin regulation, readers can refer to recent reviews (Muller and Verrijzer, 2009; Schuettengruber and Cavalli, 2009).
2. Chromatin regulation and cancer
Traditionally, cancer has been considered as a genetic disease, which can be dated back to the observation of aneuploidy associated with cancer cells. Over the last decade, it became increasingly clear that epigenetic regulation plays an important role in tumorigenesis (Jones and Baylin, 2002). Both abnormal DNA methylation and chromatin modifications are associated with cancer. Recent mechanistic studies have led to the hypothesis that DNA methylation functions to stabilize and maintain the silencing state initiated by histone methylation (Reik, 2007). The dysregualtion of chromatin modifications associated with tumorigenesis could manifest as global changes of particular modifications, such as the global reduction of H4K16 acetylation and H4K20 trimethylation observed in a skin carcinogenesis model (Fraga et al., 2005). However, the most common case is that epigenetic silencing is restrained to specific groups of genes, while the overall levels of suppressive histone modification are not significantly changed.
Several key PcG components were found dysregulated in cancers. For instance, EZH2 was found to be consistently upregulated in metastatic prostate cancer as compared to localized prostate cancer or normal tissues (Varambally et al., 2002). Subsequently, overexpression of EZH2 has been observed in a broad range of hematopoietic and solid human malignancies, such as multiple types of lymphoma, breast cancer, colon cancer etc. (van Kemenade et al., 2001; Visser et al., 2001; Varambally et al., 2002; Kleer et al., 2003; Mimori et al., 2005). The overloaded EZH2 activity results in ectopic repression of targeted genes, many of which are tumor-suppressor genes. In prostate cancer, the elevated EZH2 is responsible for silencing several important tumor-suppressor genes, including DAB2IP, MSMB (Chen et al., 2005; Beke et al., 2007). Thus, the oncogenic potential of PcG family members could potentially be exerted through transcriptional repression of tumor-suppressor genes.
In addition, PcG members might also contribute to tumorigenesis by “mis-specification” of cells to a stem cell fate. Recent studies demonstrate that PcG proteins paly important role in defining and maintaining pluripotency of stem cells, mainly through repressing developmental genes implicated in differentiation and lineage specification (Caretti et al., 2004; Bernstein et al., 2006; Lee et al., 2006; Ezhkova et al., 2009). It is well known that tumor cells share some common features with stem cells, such as extensive proliferation capacity and differentiation potential, which leads to the development of “cancer stem cell” hypothesis (Pardal et al., 2003; Sparmann and van Lohuizen, 2006). Although the existence of “cancer stem cells” or “tumor initiating cells” is still a subject of debate, the role of PcG in specifying stem cell fate likely contribute to their tumorigenic activity.
Recently, several large scale transcriptome/exome studies aimed at genomic analysis of tumor genetic abnormalities have again revealed the importance of chromatin regulation in tumorigenesis (van Haaften et al., 2009; Dalgliesh et al., 2010; Gui et al., 2011). Histone modifiers, such as the histone H3K27 demethylase UTX, were repeatedly identified to be mutated in a variety of cancers. Besides protein complexes that directly modify histones, other protein complexes, such as chromatin barrier, also play a role in affecting chromatin status and oncogenesis. It has been reported that the loss of CTCF (CCCTC-binding factor) binding, which is an insulator protein, will lead to the spreading of facultative heterochromatin into the promoters and/or transcribed regions of tumor-suppressor genes p16 (Witcher and Emerson, 2009) and p53 (Soto-Reyes and Recillas-Targa, 2010), thus resulting in the ectopic silencing of these tumor suppressors.
3. Unraveling epigenetic regulation and tumorigenesis in Drosophila
3.1. Tumor-like phenotype of PcG mutant clones
It has been noticed for some time that clones of cells mutated for PRC-1 components Psc-Su(z)2 or Polyhomeotic (Ph) in the developing wing disc display tumor-like hyperplasia phenotype (Beuchle et al., 2001). In addition to Hox genes de-repressed in these clones, CycB also appears up-regulated in cells lacking Psc-Su(z)2 or Ph (Oktaba et al., 2008). The proportion of cells stalled in G2/M was significantly increased for these mutant cells, which overall have larger nuclei compared to their wild-type sister cells. Notably, clones of cells mutated for Psc-Su(z)2 and/or Ph in the eye discs also display dramatic hyperplasia phenotype, accompanied by abnormal activation of the JAK-STAT pathway (Classen et al., 2009), or the Notch pathway (Martinez et al., 2009). A recent study demonstrated that Drosophila ovary follicle stem cells (FSCs) carrying the same Psc-Su(z)2 mutation exhibit sustained activation of Wnt signaling, and develop into neoplastic tumors (Li et al., 2010). The severity of the phenotype observed for mutant clones lacking various PcG genes appears to correlate with the timing and extend of the de-repression of silenced/repressed genes in the mutant cells. Suppression of the ectopically activated genes or signal transduction pathways can often alleviate the tumor-like phenotype.
At the superficial level these findings seem to contradict to the “simplified” notion that in mammalian systems PcG gain-of-function, instead of lose-of-function, is associated with cancer development. However, these studies pointed to the complexity and diversity of cellular consequences following dysregulation of key PcG proteins. First of all, losing the function of different PcG components has different impact on tissue homeostasis. Unlike Psc-Su(Z)2 and Ph, clones mutated for other PRC-1 components such as Pc and Scm in the wing disc did not display hyperplasia phenotype (Beuchle et al., 2001). Furthermore, the same study found that clones mutated for E(Z) were eliminated by cellular competition, indicating that similar to what was observed with EZH2 in prostate and breast cancer cells, E(Z) functions to increase the resistance to environmental stress induced cell death.
Secondly, the impact of compromised PcG function depends on the tissue/cell type and developmental stage. For instance, clones of cells mutated for Pc failed to show any tumor-like phenotype in the wing disc (Beuchle et al., 2001), yet, clones mutated for Pc (albeit a different allele) generated in the eye disc display massive hyperplasia phenotype (Classen et al., 2009). For individual PcG-repressed genes, the consequence of losing key PcG function varies significantly. In embryos lacking Ph or Psc-Su(Z)2, while some target genes are globally de-repressed, others are only de-repressed in a particular tissue or even a specific cell lineage (Oktaba et al., 2008). While there is some correspondence between PcG binding and de-repression of genes in mutants, only a small portion of targeted genes (i.e., bound by Ph) are de-repressed in cells mutated for Ph (Oktaba et al., 2008).
3.2. H3K27 demethylase dUTX and the Notch-dependent oncogenic pathway
The evidence that enhancement of PcG-mediated silencing may lead to tumor-like hyperplasia in Drosophila was revealed indirectly by experiments with dUTX. Clones of cells mutated for dUTX has increased level of H3K27me3, confirming its role as H3K27 demethylase (Herz et al., 2010). These clones significantly overgrow as compared to their sister clones (twin spots). This hyperplasia phenotype is significantly reduced or blocked in animals heterozygous to either Pc or E(Z) mutation, indicating that it is, at least partially, due to increased silencing of PcG-target genes (Herz et al., 2010). Interestingly, in addition to homeotic genes, several Notch pathway genes had increased H3K27me3 modifications and reduced mRNA levels in dUTX heterozygous animals. The interaction between dUTX and the Notch signaling pathway was also genetically verified. Intriguingly, several other histone demethylases, such as dLSD1 and Lid (litter imaginal discs), also interact with the Notch signaling pathway (Di Stefano et al., 2011; Mulligan et al., 2011), suggesting histone regulations play an important role in defining the pleiotropic Notch pathway.
The importance of UTX as a tumor-suppressor gene has been revealed by several independent large scale transcriptome/exome analyses (van Haaften et al., 2009; Dalgliesh et al., 2010; Gui et al., 2011). Loss-of-function UTX mutations (including mis/non- sense mutations, deletions, frame shift, etc) were found frequently in a variety of cancers, such as 59% of transitional cell carcinoma (Gui et al., 2011). These findings indicate that UTX has general tumor-suppression function and is a gate keeper for preventing hyperplasia in a variety of tissues.
3.3. Dysregulation of PcG targeting and epigenetic silencing of pro-apoptotic genes
Epigenetic regulation could be disturbed to promote tumorigenesis without significant changes of the global level of suppressive histone modifications. For instance, the long non-coding RNA HOTAIR can promote tumorigenesis through genome-wide re-targeting of PRC-2 to a pattern that resembles those typical of undifferentiated/lowly differentiated cells (Gupta et al., 2010). Such aberrant targeting of PcG proteins is known to promote the silencing of tumor-suppressor genes, which were in transcription-ready (bivalent) state in normal stem cells (Ohm et al., 2007).
Altered PcG targeting can be induced by oncogenic proteins. For instance, oncogenic Ras can lead to repression of Fas expression. Although at the end stage the silencing of Fas (and several other tumor-suppressor genes) is manifested as both DNA hypermethylation and increased suppressive histone modifications, a mechanistic analysis indicated that PcG proteins such as Bmi1 and EzH2 are required for Rasinduced silencing of Fas (Gazin et al., 2007). Interestingly, a small adenoviral protein, E4-ORF6, can activate a unknown mechanism that leads to the formation of facultative hetero-chromatins in P53-targeted stress-responsive genes (Soria et al., 2010). Since E4-ORF6 lacks any enzymatic domain that can modify histone tails, it must be acting through a cellular pathway that can specifically silence P53-targeted stress-responsive genes. Revealing this mechanism would certainly advance our understanding of how PcG-mediated epigenetic silencing can be targeted to specific groups of genes.
Epigenetic regulation of P53-targeted stress-responsive genes is also observed during Drosophila development. An about 30 kb intergenic region located in the pro-apoptotic gene cluster is responsible for mediating P53-dependent induction of reaper, hid, and sickle. This region, irradiation-responsive enhancer region (IRER), is open in early embryonic stages when most cells are proliferating, conveying high sensitivity to DNA damage and other stresses. However, at embryonic stage 12, when most cells enter post-mitotic differentiation, this region forms heterochromatin-like structure enriched for both H3K9me3 and H3K27me3. Consequently, the three pro-apoptotic genes can no longer be induced following irradiation (Zhang et al., 2008). This open-to-closed transition of IRER requires the function of PcG proteins such as Pc and Su(z)12, as well as HDAC and Su(var)3–9. Interestingly, the epigenetic suppression is strictly limited to the IRER without affecting the transcribed regions of the pro-apoptotic genes. This limitation is important since those pro-apoptotic genes are expressed in a cell lineage-specific pattern in late stage embryos, and are required for lineage specific cell death (Tan et al., 2011). The demarcation of epigenetic blocking is achieved by a chromatin barrier separating IRER from the promoter and transcribed regions of reaper. This barrier, located within a 294 bp DNA fragment, appears to be highly conserved as it can block heterochromatin spreading when tested in a vertebrate system (Lin et al., 2011).
The epigenetic blocking of IRER provided a nice system for elucidating the cellular mechanism that regulates targeted suppressive histone modification at tumor-suppressor/stress-responsive genes. Additional evidence indicated that an open IRER not only convey sensitivity to irradiation and DNA damage, but also increase the cellular sensitivity to competition-induced cell death. Clones of cells with IRER deletion (mimicking closed IRER) overgrow their sister clones that have wild-type IRER (Zhang et al., in preparation). When an ubi-DsRed reporter was inserted into IRER via homologous recombination to monitor the epigenetic status of this region, it appears that the accessibility of IRER varies in different tissues and cell types. Interestingly, even for the same cell type, there is significant variation of IRER accessibility, which likely reflects the stochastic nature of epigenetic regulation of stress-responsive genes in a given cell type.
4. Conclusion and perspectives
The utility of Drosophila as a model for unraveling the role of epigenetic regulation in tumorigenesis is emerging. There is no doubt that mechanistic studies in fruit fly will continually provide insights into the fundamental mechanisms of chromatin regulation. In addition, the fruit fly could also serve as valuable systems for addressing key questions related to tumorigenesis, such as how PcG-mediated suppression is targeted to specific genes and what pathway(s) controls epigenetic regulation of P53-targeted tumor-suppressor genes.
Acknowledgments
This work was supported by NIH CA095542 and AI079074.
References
- Agger K, Cloos PAC, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Akasaka T, Kanno M, Balling R, Mieza MA, Taniguchi M, Koseki H. A role for mel-18, a Polycomb group-related vertebrate gene, during theanteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. doi: 10.1242/dev.122.5.1513. [DOI] [PubMed] [Google Scholar]
- Beke L, Nuytten M, Van Eynde A, Beullens M, Bollen M. The gene encoding the prostatic tumor suppressor PSP94 is a target for repression by the Polycomb group protein EZH2. Oncogene. 2007;26:4590–4595. doi: 10.1038/sj.onc.1210248. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Beuchle D, Struhl G, Muller J. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development. 2001;128:993–1004. doi: 10.1242/dev.128.6.993. [DOI] [PubMed] [Google Scholar]
- Breen TR, Harte PJ. Molecular characterization of the trithorax gene, a positive regulator of homeotic gene expression in Drosophila. Mechanisms Dev. 1991;35:113–127. doi: 10.1016/0925-4773(91)90062-b. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y-i, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 Lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tu S-w, Hsieh J-T. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J. Biol. Chem. 2005;280:22437–22444. doi: 10.1074/jbc.M501379200. [DOI] [PubMed] [Google Scholar]
- Classen AK, Bunker BD, Harvey KF, Vaccari T, Bilder D. A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nat. Genet. 2009;41:1150–1155. doi: 10.1038/ng.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core N, Bel S, Gaunt SJ, Aurrand-Lions M, Pearce J, Fisher A, Djabali M. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development. 1997;124:721–729. doi: 10.1242/dev.124.3.721. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, Teague J, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Forbes S, Jia M, Jones D, Knott H, Kok CY, Lau KW, Leroy C, Lin ML, McBride DJ, Maddison M, Maguire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, O’Meara S, Pleasance E, Rajasingham A, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turrell K, Dykema KJ, Khoo SK, Petillo D, Wondergem B, Anema J, Kahnoski RJ, Teh BT, Stratton MR, Futreal PA. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquityl00ation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- del Mar Lorente M, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development. 2000;127:5093–5100. doi: 10.1242/dev.127.23.5093. [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Walker JA, Burgio G, Corona DFV, Mulligan P, Näär AM, Dyson NJ. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev. 2011;25:17–28. doi: 10.1101/gad.1983711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Pérez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Gazin C, Wajapeyee N, Gobeil S, Virbasius C-M, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, He M, Li Z, Sun X, Jia W, Chen J, Yang S, Zhou F, Zhao X, Wan S, Ye R, Liang C, Liu Z, Huang P, Liu C, Jiang H, Wang Y, Zheng H, Sun L, Liu X, Jiang Z, Feng D, Chen J, Wu S, Zou J, Zhang Z, Yang R, Zhao J, Xu C, Yin W, Guan Z, Ye J, Zhang H, Li J, Kristiansen K, Nickerson ML, Theodorescu D, Li Y, Zhang X, Li S, Wang J, Yang H, Wang J, Cai Z. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011;330:228–231. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, Davuluri R, Shilatifard A, Hariharan IK, Bergmann A. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol. Cell. Biol. 2010;30:2485–2497. doi: 10.1128/MCB.01633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Cho Y-W, Yu L-R, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jurgens G. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature. 1985;316:153–155. [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RGAB, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Norman J, Shilatifard A, Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a Polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K-i, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the Polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lewis PH. D. melanogaster new mutants: report of Pamela H. Lewis. Drosophila Inform. Ser. 1947;21:69. [Google Scholar]
- Li X, Han Y, Xi R. Polycomb group genes Psc and Su(z)2 restrict follicle stem cell self-renewal and extrusion by controlling canonical and noncanonical Wnt signaling. Genes Dev. 2010;24:933–946. doi: 10.1101/gad.1901510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin N, Li X, Cui K, Chepelev I, Tie F, Liu B, Li G, Harte P, Zhao K, Huang S, Zhou L. A barrier-only boundary element delimits the formation of facultative heterochromatin in Drosophila melanogaster and vertebrates. Mol. Cell. Biol. 2011;31:2729–2741. doi: 10.1128/MCB.05165-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez AM, Schuettengruber B, Sakr S, Janic A, Gonzalez C, Cavalli G. Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat. Genet. 2009;41:1076–1082. doi: 10.1038/ng.414. [DOI] [PubMed] [Google Scholar]
- Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat. Rev. Cancer. 2010;10:669–682. doi: 10.1038/nrc2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori K, Ogawa K, Okamoto M, Sudo T, Inoue H, Mori M. Clinical significance of enhancer of zeste homolog 2 expression in colorectal cancer cases. Eur. J. Surg. Oncol. 2005;31:376–380. doi: 10.1016/j.ejso.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Further studies on the nature and causes of gene mutations. Proc. 6th Int. Congr. Genet. 1932;1:213–255. [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Muller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr. Opin. Genet. Dev. 2009;19:150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Mulligan P, Yang F, Di Stefano L, Ji J-Y, Ouyang J, Nishikawa JL, Toiber D, Kulkarni M, Wang Q, Najafi-Shoushtari SH, Mostoslavsky R, Gygi SP, Gill G, Dyson NJ, Näär AM. A SIRT1-LSD1 corepressor complex regulates Notch target gene expression and development. Mol. Cell. 2011;42:689–699. doi: 10.1016/j.molcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, Berman DM, Jenuwein T, Pruitt K, Sharkis SJ, Watkins DN, Herman JG, Baylin SB. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, Muller J. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev. Cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Orlando V, Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr. Opin. Genet. Dev. 1995;5:174–179. doi: 10.1016/0959-437x(95)80005-0. [DOI] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat. Rev. Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 1997;13:314–318. doi: 10.1016/s0168-9525(97)01178-5. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu C-t, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat. Rev. Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr. Top Dev. Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- Soria C, Estermann FE, Espantman KC, O’Shea CC. Heterochromatin silencing of p53 target genes by a small viral protein. Nature. 2010;466:1076–1081. doi: 10.1038/nature09307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Reyes E, Recillas-Targa F. Epigenetic regulation of the human p53 gene promoter by the CTCF transcription factor in transformed cell lines. Oncogene. 2010;29:2217–2227. doi: 10.1038/onc.2009.509. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Struhl G. Role of the esc+ gene product in ensuring the selective expression of segment-specific homeotic genes in Drosophila. J. Embryol. Exp. Morphol. 1983;76:297–331. [PubMed] [Google Scholar]
- Tan Y, Yamada-Mabuchi M, Arya R, St Pierre S, Tang W, Tosa M, Brachmann C, White K. Coordinated expression of cell death genes regulates neuroblast apoptosis. Development. 2011;138:2197–2206. doi: 10.1242/dev.058826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O’Meara S, Teague J, Butler A, Hinton J, Latimer C, Andrews J, Barthorpe S, Beare D, Buck G, Campbell PJ, Cole J, Forbes S, Jia M, Jones D, Kok CY, Leroy C, Lin ML, McBride DJ, Maddison M, Maquire S, McLay K, Menzies A, Mironenko T, Mulderrig L, Mudie L, Pleasance E, Shepherd R, Smith R, Stebbings L, Stephens P, Tang G, Tarpey PS, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Collins VP, Ichimura K, Law S, Wong J, Yuen ST, Leung SY, Tonon G, DePinho RA, Tai YT, Anderson KC, Kahnoski RJ, Massie A, Khoo SK, Teh BT, Stratton MR, Futreal PA. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kemenade FJ, Raaphorst FM, Blokzijl T, Fieret E, Hamer KM, Satijn DPE, Otte AP, Meijer CJLM. Coexpression ofBMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood. 2001;97:3896–3901. doi: 10.1182/blood.v97.12.3896. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RGAB, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL. ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser HPJ, Gunster MJ, Kluin-Nelemans HC, Manders EMM, Raaphorst FM, Meijer CJLM, Willemze R, Otte AP. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Brit. J. Haematol. 2001;112:950–958. doi: 10.1046/j.1365-2141.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Witcher M, Emerson BM. Epigenetic silencing of the p16(INK4a) tumor suppressor is associated with loss of CTCF binding and a chromatin boundary. Mol. Cell. 2009;34:271–284. doi: 10.1016/j.molcel.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lin N, Carroll PM, Chan G, Guan B, Xiao H, Yao B, Wu SS, Zhou L. Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev. Cell. 2008;14:481–493. doi: 10.1016/j.devcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]