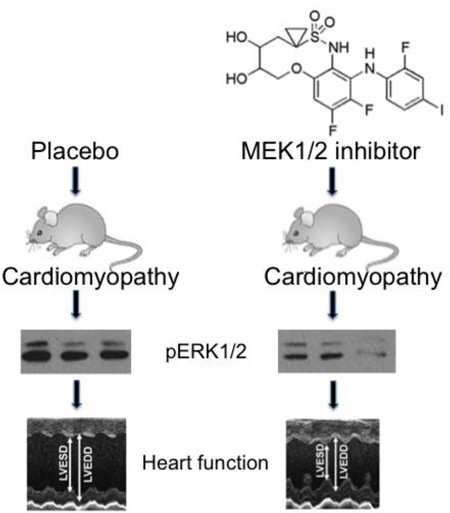
Abstract
Signaling mediated by extracellular signal-regulated kinases 1 and 2 (ERK1/2) is involved in numerous cellular processes. Mitogen-activated protein kinase kinases (MEK1/2) catalyze the phosphorylation of ERK1/2, converting it into an active kinase that regulates the expression of numerous genes and cellular processes. Inhibitors of MEK1/2 have demonstrated preclinical and clinical efficacy in certain cancers and types of cardiomyopathy. We report the synthesis of a novel, allosteric, macrocyclic MEK1/2 inhibitor that potently inhibits ERK1/2 activity in cultured cells and tissues of mice after systemic administration. Mice with dilated cardiomyopathy caused by a lamin A/C gene mutation have abnormally increased cardiac ERK1/2 activity. In these mice, this novel MEK1/2 inhibitor is well tolerated, improves left ventricular systolic function, decreases left ventricular fibrosis, has beneficial effects on skeletal muscle structure and pathology and prolongs survival. The novel MEK1/2 inhibitor described herein may therefore find clinical utility in the treatment of this rare cardiomyopathy, other types of cardiomyopathy and cancers in humans.
Keywords: cardiomyopathy, Emery-Dreifuss muscular dystrophy, extracellular signal-regulated kinase, lamin, MEK inhibitor, mitogen-activated protein kinase
Graphical abstract
1. Introduction
Extracellular signal-regulated kinases 1 and 2 (ERK1/2) are structurally similar protein-serine/threonine kinases that regulate a variety of cellular processes including adhesion, migration, survival, differentiation, metabolism, proliferation, transcription, cytoskeletal remodeling and cell cycle progression.1 Mitogen-activated protein kinase kinases 1 and 2 (MEK1/2) catalyze the phosphorylation of ERK1/2, which is required for enzyme activation.2 Activated ERK1/2 in turn catalyzes the phosphorylation of various cellular substrates, including other kinases and transcription factors that regulate various cellular processes and gene expression.1,2 Several non-ATP-competitive, allosteric MEK1/2 inhibitors have been developed and assessed in clinical studies, primarily for cancers in which ERK1/2 signaling is aberrantly activated.3–5
In addition to cancer, MEK1/2 inhibitors have potential to be useful in other diseases. RASopathies, a group of genetic syndromes that includes neurofibromatosis 1, Noonan syndrome, LEOPARD syndrome, Costello syndrome and cardiofaciocutaneous syndrome, are caused by germline mutations in genes that encode components or regulators of the Ras-ERK1/2 signaling pathway that may lead to abnormally increased ERK1/2 activity.6,7 Increased ERK1/2 signaling in heart in RASopathies can cause hypertrophic cardiomyopathy and MEK1/2 inhibitor treatment has been shown to be beneficial in a mouse model of Noonan syndrome.8.9 Data from transgenic mice with cardiomyocyte-specific overexpression of FasL also suggest that MEK1/2 inhibitors may be useful in the preventing the progression of dilated cardiomyopathy and heart failure.10
Mutations in the lamin A/C gene (LMNA) cause a variety of diseases called laminopathies.11 Most often, LMNA mutations cause dilated cardiomyopathy with variable skeletal myopathy, including autosomal Emery-Dreifuss muscular dystrophy, limb-girdle muscular dystrophy type 1B and congenital muscular dystrophy.12–17 ERK1/2 activity is increased in hearts of mice with dilated cardiomyopathy caused by Lmna mutation as well as in hearts of human subjects with LMNA mutations.18–20 ERK1/2 signaling is also increased in hearts of mice lacking emerin, which occurs in humans with X-linked Emery-Dreifuss muscular dystrophy that has dilated cardiomyopathy as a prominent feature.21,22 MEK1/2 inhibitors have beneficial effects in mice with an Lmna mutation that causes cardiomyopathy.20,23,24 Given the high incidence of heart failure in patients with LMNA mutation and poor efficacy of standard therapies,17 we synthesized a novel macrocyclic MEK1/2 inhibitor with improved pharmacological profile and evaluated its therapeutic potential in mice with dilated cardiomyopathy caused by Lmna mutation.
2. Results and discussion
2.1. Chemistry
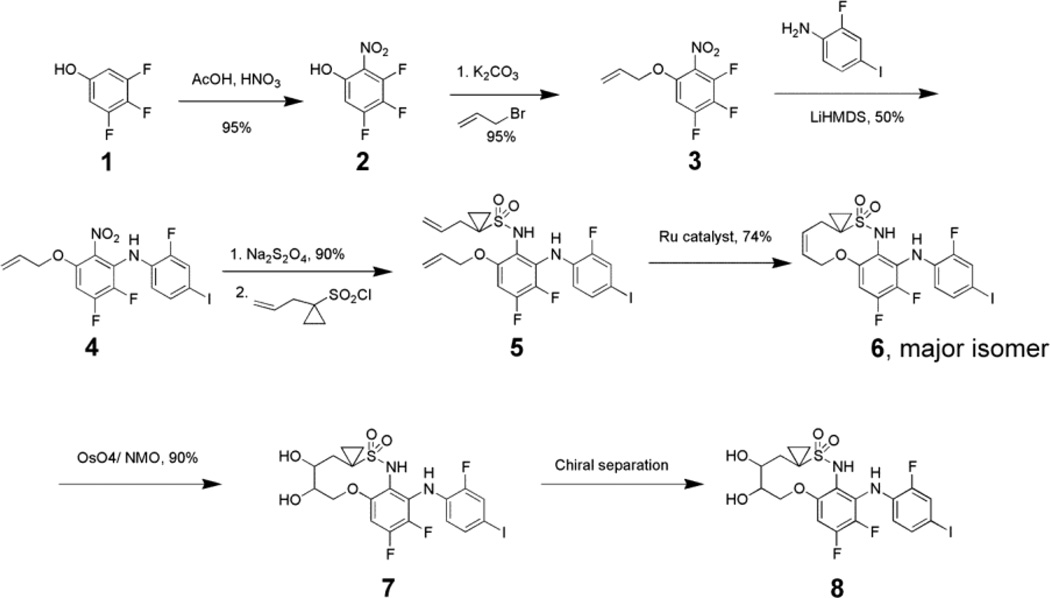
Several allosteric inhibitors of MEK1/2 are currently in clinical development and two of them are FDA approved drugs.3–5 These inhibitors are selective towards MEK1/2, as they bind to non-ATP-competitive allosteric sites. However, the poor pharmacokinetic profile of these MEK1/2 inhibitors, such as short half-life can be attributed to metabolically labile structural features. Our rational design of a novel class of MEK1/2 inhibitors sought to improve pharmacokinetic profile over existing molecules. We designed a macrocyclic inhibitor that maintains critical protein/ligand contacts through sulfonamide, iodine but does not possess a primary alcohol or hydroxamate functionality, a known metabolic liability.25 The structure of lead molecule 8 (Scheme 1) is distinct from other MEK1/2 inhibitors in that it is a macrocycle. It has been reported that macrocyclic scaffolds improve drug-like properties including target binding, selectivity and oral bioavailability.26
Scheme 1.
Synthesis of macrocyclic MEK1/2 inhibitor 8.
The synthesis of a lead molecule 8 is depicted in Scheme 1. Nitration of commercially available 3,4,5-trifluorophenol provided nitrophenol 2. Allylation of the phenol 2, followed by selective nucleophilic displacement with 2-fluoro, 4-iodoaniline provided advanced intermediate 4. Reduction of Nitro group afforded the aniline followed by coupling with sulfonyl chloride, yielded diene 5. Olefin metathesis reaction afforded the cis-olefin as a major product 6. Dihydroxylation catalyzed by osmium tetraoxide in the presence of N-methyl morpholine-N-oxide, of macrocyclic olefin 6 provided cis diol 7. The lead molecule, 8, was then obtained as a single stereoisomer from chiral chromatography.
We used an in vitro biochemical assay to test the racemic diol 7 for inhibition of MEK1 activity. The compound 7 was potent with IC50 = 21 nM. After chiral separation, we tested the individual enantiomeric diols for anti-proliferative activity on HT-29 cells, the proliferation of which is slowed by inhibiting MEK1/2. The fast moving isomer 8 on chiral HPLC was approximately 25-fold more potent as compared to the slow moving one (IC50 = 16 nM and 400 nM, respectively). We chose the more potent faster moving isomer for further evaluation.
2.2. Effects of 8 on heart and skeletal muscle of LmnaH222P/H222P mice
We evaluated the effects of 8 on heart and skeletal muscle in LmnaH222P/H222P mice, a validated model of cardiomyopathy caused by LMNA mutation in humans.27 We used male mice because female mice develop pathology at a significantly later age. Male LmnaH222P/H222P mice develop initial signs of cardiomyopathy at 8 to 10 weeks with progressive left ventricular dilatation and decreased left ventricular fractional shortening. By approximately 20 weeks of age, they also develop significant skeletal myopathy. Median survival is approximately 26 weeks. ERK1/2 signaling is abnormally increased of LmnaH222P/H222P mice, starting as early as 4 weeks of age.18,19 We have previously shown that treatment with MEK1/2 inhibitors such as PD98059 and selumetinib has a beneficial effect on left ventricular and skeletal muscle function and prolongs survivial.20,22–24,28
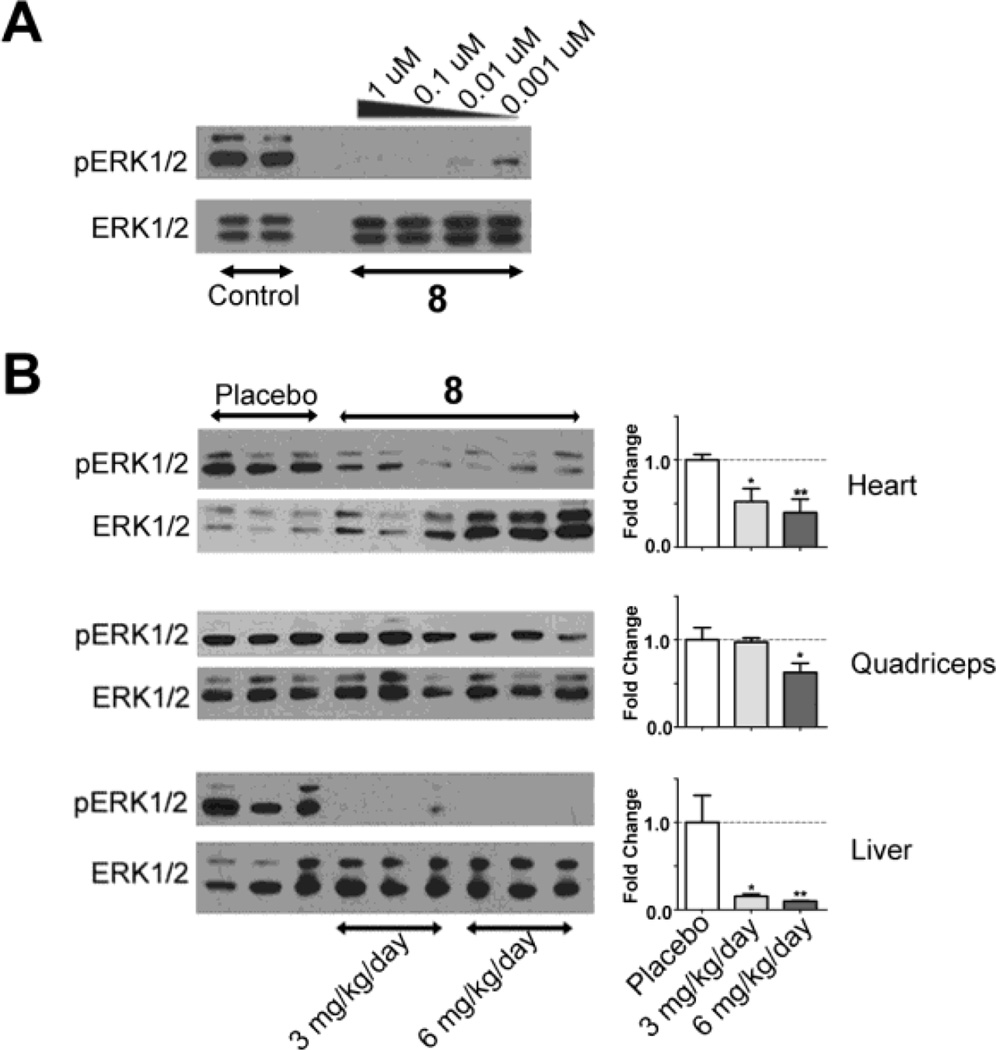
Prior to administering 8 to LmnaH222P/H222P mice, we tested its activity by examining phosphorylation (activation) of ERK1/2 in cultured HEK-293 cells. Addition of 1 µM to 0.001 µM of 8 to culture media essentially abolished ERK1/2 phosphorylation (Figure 1A). We next administered 3 mg/kg/day or 6 mg/kg/day of 8 to male LmnaH222P/H222P mice by oral gavage starting at 14 weeks of age, when there are symptoms of cardiomyopathy, up to 20 weeks of age. After six weeks of systemic administration, both doses led to significant decreases in phosphorylated ERK1/2 relative to total ERK1/2 in heart and liver, whereas only the higher dose produced a significant decrease in quadriceps muscle (Figure 1B). There were no differences in body mass between the three groups at the end of treatment [placebo = 23.19 ± 0.43 (n = 22); 3 mg/kg/day 8 = 24.06 ± 0.52 (n = 19); 6 mg/kg/day 8 = 22.93 ± 0.71 (n = 16); values are means ± standard errors].
Figure 1.
8 reduces ERK1/2 activity in cultured cells and mouse tissues. Representative immunoblots using antibodies against phosphorylated (activated) ERK1/2 (pERK1/2) and total ERK1/2 (ERK1/2). (A) Immonublot of proteins extracted from HEK-293 cells cultured in the presence of diluent (Control) or varying concentrations of 8 for 24 h. Concentrations of 8 used are indicated above each lane. (B) Immuoblots of proteins extracted from hearts, quadriceps and livers of LmnaH222P/H222P mice treated with diluent (placebo) or 8 at doses of 3 mg/kg/day or 6 mg/kg/day. The bar graphs at right show quantification of the pERK1/2 to total ERK1/2 ratio determined from scanned immunoblots (n = 3); values shown are means ± standard errors. *P<0.05, **P<0.01 compared to placebo.
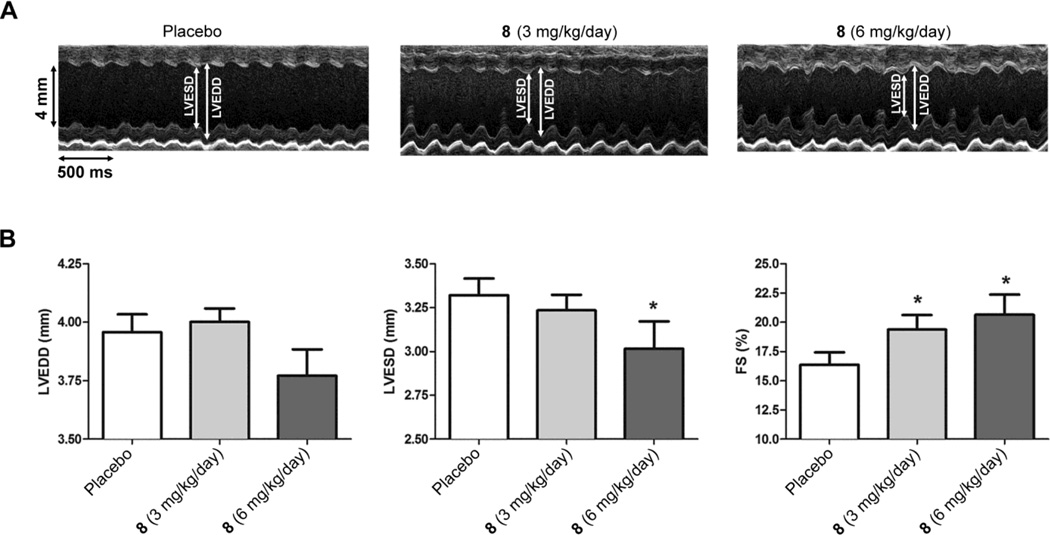
We examined the effects of six weeks of treatment with 8 on echocardiographic parameters that correlate with left ventricular function. Treatment was started at 14 weeks of age, when significant left ventricular dysfunction is already present, and mice were analyzed six weeks later at 20 weeks of age. Treatment with both 3 mg/kg/day and 6 mg/kg/day of 8 resulted in significant increases in left ventricular fractional shortening (the percentage the left ventricular diameter decreases with each contraction) and the higher dose also provided a significant decrease in left ventricular end diastolic diameter (Figure 2; Table 1). These echocardiographic changes were consistent with improved left ventricular systolic function.
Figure 2.
Effect of 8 on echocardiographic parameters in LmnaH222P/H222P mice. (A) Representative transthoracic M-mode echocardiographic tracings from LmnaH222P/H222P mice treated with placebo or 8 at 3 mg/kg/day or 6 mg/kg/day. Left ventricular end systolic diameter (LVESD), left ventricular end diastolic diameter (LVEDD) are indicated. (B) Bar graphs show means and standard errors of means for left ventricular end systolic diameter (LVESD), left ventricular end diastolic diameter (LVEDD) and left ventricular fractional shortening (FS) in LmnaH222P/H222P mice treated with placebo (n = 22), 8 3 mg/kg/day (n = 19) or 8 6 mg/kg/day (n = 16). *P<0.05 compared to placebo.
Table 1.
Echocardiographic data for LmnaH222P/H222P at age of 20 weeks of age treated with placebo or 8.
| Treatment | n | Heart rate (beats/min) |
LVEDD (mm) |
LVESD (mm) |
FS (%) |
|---|---|---|---|---|---|
| Placebo | 22 | 502.73 ± 0.46 | 3.96 ± 0.08 | 3.32 ± 0.10 | 16.36 ± 1.08 |
| 8 (3 mg/kg/day) | 19 | 503.63 ± 0.54 | 4.00 ± 0.06 | 3.24 ± 0.09 | 19.39 ± 1.25* |
| 8 (6 mg/kg/day) | 16 | 501.56 ± 0.61 | 3.77 ± 0.11 | 3.02 ± 0.16* | 20.65 ± 1.72* |
LVEDD, left ventricular end-diastolic diameter; LVESD, left ventricular end systolic diameter; FS, fractional shortening. Values are means ± standard errors.
P<0.05 compared to placebo.
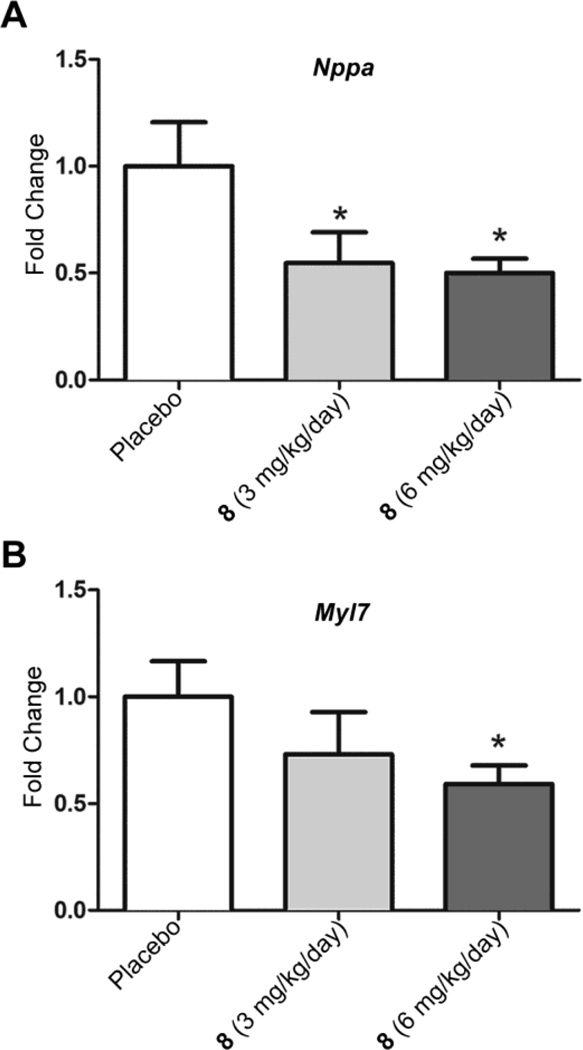
We previously reported pathological up-regulation of the gene encoding natriuretic peptide A (Nppa), the synthesis of which is increased in dilated hearts, and genes encoding proteins involved in sarcomere organization in the hearts of LmnaH222P/H222P mice.18,23,24 We therefore assayed expression of mRNAs encoded by Nppa and by the gene encoding cardiac myosin light chain 7 (Myl7) in mice treated with 8. Treatment with both 3 mg/kg/day and 6 mg/kg/day significantly reduced cardiac Nppa mRNA compared to placebo treatment (Figure 3A). Myl7 mRNA was significantly decreased after treatment with 6 mg/kg/day (Figure 3B).
Figure 3.
Effect of 8 on expression of natriuretic peptide A and myosin light chain 7 mRNAs in hearts of LmnaH222P/H222P mice. (A) Quantitative real-time RT–PCR analysis of mRNA encoding natriuretic peptide A (Nppa) in hearts from LmnaH222P/H222P mice treated with placebo or 8 at 3 mg/kg/day or 6 mg/kg/day. (B) Quantitative real-time RT–PCR analysis of mRNA encoding myosin light chain 7 (Myl7) in hearts from LmnaH222P/H222P mice treated with placebo or 8 at 3 mg/kg/day or 6 mg/kg/day. Results for placebo treatment (n = 5) are normalized to 1 and those for treatment with 3 mg/kg/day (n = 5) or 6 mg/kg/day (n = 6) of 8 represent as fold change compared to placebo treatment. Bar graphs show means and standard errors. *P<0.05 compared to placebo.
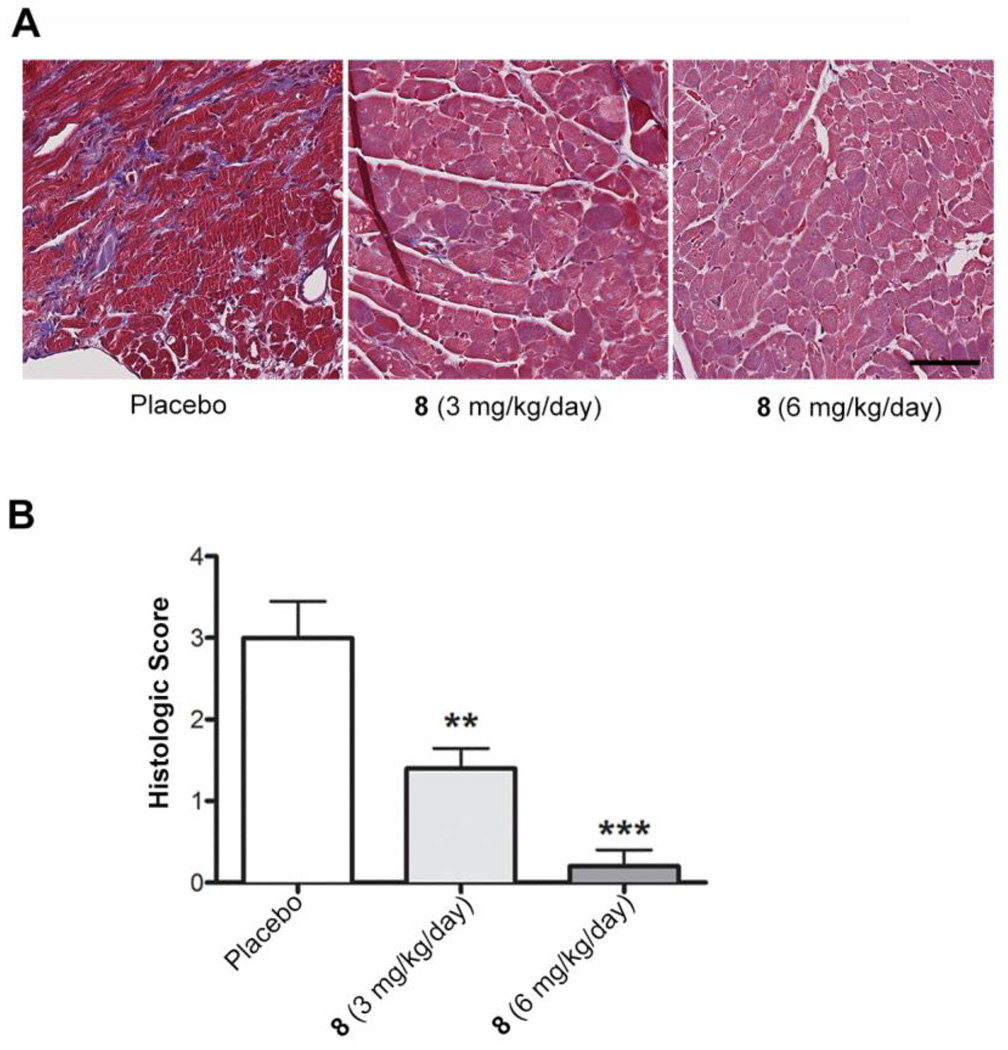
Cardiomyopathy in LmnaH222P/H222P mice is characterized by profound myocardial fibrosis, which is prominent in male mice by 20 weeks of age.20,23–24,27 This recapitulates the significant fibrosis that occurs in hearts of human subjects with cardiomyopathy caused by LMNA mutations.29–32 In humans with LMNA mutations, fibrosis in the interventricular septum may be the mechanism behind the early atrioventricular block and ventricular arrhythmias that frequently occur and can lead to sudden death.33 We therefore assessed the effects of 8 treatment on cardiac fibrosis in LmnaH222P/H222P mice. Staining with Masson trichrome of fixed sections of left ventricles showed that treatment of LmnaH222P/H222P mice with both 3 mg/kg/day and 6 mg/kg/day of 8 lead to decreased fibrosis (Figure 4A). A grading system was constructed based on the presence of two abnormalities that were found to be variably present between specimens: cytoplasmic vacuolation and cardiac fibrosis. Low-grade specimens had no fibrosis and either no vacuolation (grade 0), mild vacuolation (grade 1) or moderate vacuolation (grade 2). Higher-grade specimens possessed vacuolation and mild fibrosis (grade 3) or severe fibrosis (grade 4). This analysis showed that treatment 3 mg/kg/day and 6 mg/kg/day of 8 lead to dose-dependent statistically significant decreases in fibrosis (Figure 4B). Hence, treatment with 8 may prevent irreversible cardiac fibrosis in patients with LMNA mutations and possible the atrioventricular block and ventricular arrhythmias that can result from it.
Figure 4.
Effect of 8 on cardiac fibrosis in LmnaH222P/H222P mice. (A) Photomicrographs of representative left ventricle sections stained with Masson trichrome from LmnaH222P/H222P mice treated with placebo or 8 at 3 mg/kg/day or 6 mg/kg/day. Scale bar: 70 µm. The placebo image corresponds to grade 4 histology (severe fibrosis), the 3 mg/kg image corresponds to grade 2 histology (moderate vacuolation, no fibrosis) and the 6 mg/kg image corresponds to grade 1 histology (mild vacuolation, no fibrosis). (B) Cardiac fibrosis scores for LmnaH222P/H222P mice treated with placebo (n = 5) or 3 mg/kg/day (n = 5) or 6 mg/kg/day (n = 6) of 8. Bar graph shows means and standard errors. *P<0.05, **P<0.01 compared to placebo.
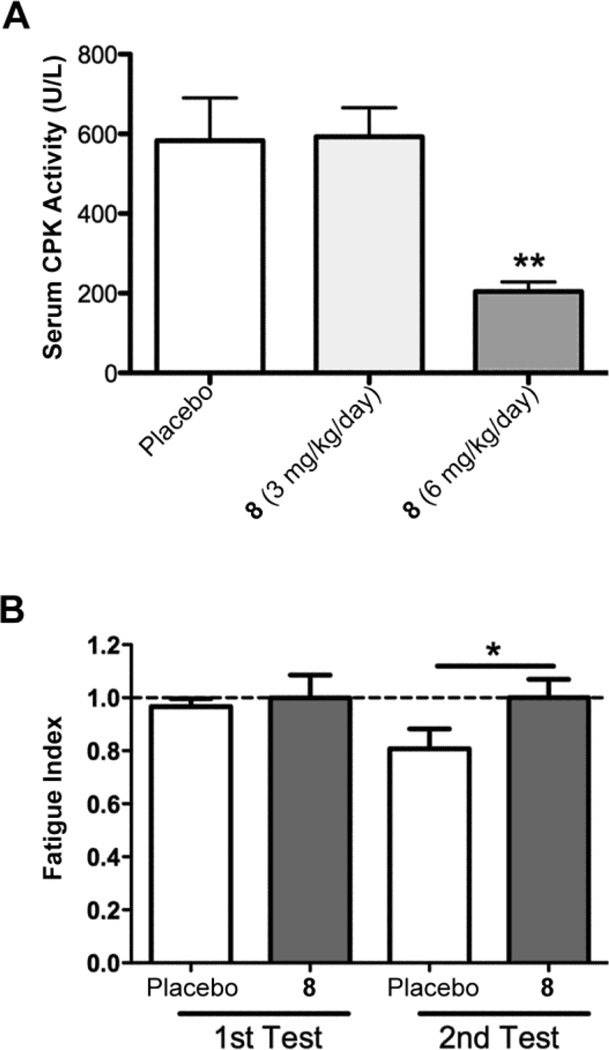
Human subjects with LMNA mutations that cause cardiomyopathy frequently have associated muscular dystrophy.12,14–17 Abnormally increased ERK1/2 activity appears to contribute to the development of skeletal muscle pathology in LmnaH222/H222P mice and MEK1/2 inhibition may have beneficial effects.28 To further assess this possibility, we examined the effects of 8 on a serum creatine phosphokinase, a biomarker of striated muscle breakdown, and skeletal muscle fatigue. Compared to placebo, treatment with 6 mg/kg/day but not 3 mg/kg/day of 8 significantly reduced serum creatine phosphokinase activity in LmnaH222P/H222P mice (Figure 5A). Treatment with 6 mg/kg/day of 8 also significantly reduced the forelimb grip fatigue index (Figure 5B). While the beneficial effects on skeletal muscle may have occurred secondary to improvement in heart function, the fact that only 6 mg/kg/day provided reduced serum creatine phosphokinase activity whereas both 3 mg/kg/day and 6 mg/kg/day had improved heart function suggests a direct effect. Histological examination of hematoxylin and eosin-stained sections of quadriceps muscle by a pathologist blind to treatment showed internally nucleated fibers in 3 of 6 samples from mice treated with placebo, in 1 of 5 samples from mice treated with 3 mg/kg/day or 8 and 0 of 5 samples from mice treated with 6 mg/kg/day of 8. Internally nucleated fibers are indicative of chronic myofiber injury and regeneration, as can be seen in a variety of muscular dystrophies and other disease states.
Figure 5.
Effect of 8 on skeletal muscle in LmnaH222P/H222P mice treated with placebo, 8 3mg/kg/day or 6 mg/kg/day. (A) Serum creatine phosphokinase (CPK) activities in LmnaH222P/H222P mice treated with placebo (n = 10), 8 3 mg/kg/day (n = 10) or 8 6 mg/kg/day (n = 10). Values are means ± standard errors. (B) Forelimb grip fatigue index of LmnaH222P/H222P mice treated with placebo (n = 7) or 8 6 mg/kg/day (n = 6). Values are means ± standard errors. *P<0.05, **P<0.01 compared to mice treated with placebo.
2.3. Toxicology analyses of 8 in LmnaH222P/H222P mice
We carried out a preliminary analysis of potential tissue toxicity of 8 in male LmnaH222P/H222P mice treated from 14 to 20 weeks of age. We measured serum alkaline phosphatase activity, alanine aminotransferase activity and bilirubin concentration to assess possible hepatic injury and liver function. We also measured serum creatinine and blood urea nitrogen concentrations as indicators of renal function and serum amylase activity as a marker of pancreatic injury. There were no statistically significant differences in any of these parameters between male LmnaH222P/H222P mice treated with 3 mg/kg/day or 6 mg/kg/day or placebo (Table 2). These results suggested that 6 weeks of treatment with 8 at a dose up to 6 mg/kg/day did not produce significant liver, kidney or pancreatic toxicity.
Table 2.
Selected serum chemistry values in LmnaH222P/H222P mice at 20 weeks of age treated for 6 weeks with placebo or 8.
| Treatment | Placebo | 8 (3 mg/kg/day) | 8 ( 6 mg/kg/day) |
|---|---|---|---|
| Alkaline Phosphatase (U/L) | 91.5 ± 6.99 | 104.5 ± 7.43 | 113.0 ± 6.55 |
| ALT (U/L) | 56 ± 10.55 | 69.6 ± 10.81 | 69.6 ± 14.31 |
| Total bilirubin (mg/L) | 0.36 ± 0.02 | 0.32 ± 0.02 | 0.37 ± 0.03 |
| Amylase (U/L) | 1,102 ± 53.63 | 1,372 ± 282.9 | 1,086 ± 40.18 |
| BUN (mg/dL) | 17.98 ± 0.94 | 18.04 ± 0.48 | 19.12 ± 1.83 |
| Creatinine (mg/dL) | 0.45 ± 0.01 | 0.46 ± 0.01 | 0.46 ± 0.01 |
| BUN/Creatinine | 20.14 ± 1.25 | 19.66 ± 0.61 | 20.77 ± 1.92 |
ALT, alanine aminotransferase; BUN, blood urea nitrogen. Values are means ± standard errors for n = 10 mice per group.
A pathologist who was blind to drug treatment and mouse genotype examined histological section of livers, kidneys and spleens from LmnaH222P/H222P mice after 6 weeks of receiving placebo or 8 (Table 3). Examination of the livers did not show hepatocyte necrosis, significant inflammation or cholestasis; only mild non-specific changes were observed in livers from mice treated with placebo, 3 mg/kg/day of 8 or 6 mg/kg/day of 8. The kidneys showed no evidence of glomeruli abnormalities or interstitial nephritis and were mostly normal except for some non-specific inflammation in mice treated with either placebo or 8. Spleens from some mice treated with either placebo or 8 contained hemosiderin-laden macrophages. These are normally present in rodent spleens34 but may also suggest old hemorrhage somewhere in the animals. One mouse receiving 8 had evidence of a splenic infarct, which is not typical for a drug reaction but can be a thromboembolic event in cardiomyopathy.35 Overall, there were no consistent or specific abnormalities in liver, kidney or spleen histopathology in mice receiving 8 and no alterations that typically occur with drug toxicity. The fact that minor abnormalities were present in both placebo-treated and drug-treated LmnaH222P/H222P mice suggested that they were a result of underlying disease and not drug toxicity.
Table 3.
Histological findings in livers, kidney and spleens from LmnaH222P/H222P mice at 20 weeks of age treated for 6 weeks with placebo or 8
| Livers | ||
|---|---|---|
| Treatment | Sample # | Histology |
| Placebo | 7603 | Normal |
| Placebo | 7604 | Normal |
| Placebo | 7605 | Normal |
| Placebo | 7606 | Greater vacuolation than in other specimens; uncertain significance; could be preparation artifact |
| Placebo | 7607 | Normal |
| Placebo | 7608 | Normal |
| 8 3 mg/kg/day | 7609 | Normal |
| 8 3 mg/kg/day | 7610 | Normal |
| 8 3 mg/kg/day | 7611 | Normal |
| 8 3 mg/kg/day | 7612 | Slightly greater vacuolation in some zones; similar to 7606 but milder; could be preparation artifact |
| 8 3 mg/kg/day | 7613 | Normal |
| 8 6 mg/kg/day | 7614 | Occasional hemosiderin laden macrophages. Some portal triads have mild lymphocytic inflammation; significance uncertain |
| 8 6 mg/kg/day | 7615 | Normal |
| 8 6 mg/kg/day | 7616 | Normal |
| 8 6 mg/kg/day | 7617 | Some intracellular vacuolation in some zones; similar to 7612; could be preparation artifact |
| 8 6 mg/kg/day | 7618 | Normal |
| Kidneys | ||
| Treatment | Sample # | Histology |
| Placebo | 7619 | Normal |
| Placebo | 7620 | Normal |
| Placebo | 7621 | Normal |
| Placebo | 7622 | Some lymphocytic inflammation around the renal pelvis (around the ureter); otherwise normal |
| Placebo | 7623 | Some lymphocytic inflammation around the renal pelvis (around the ureter); otherwise normal |
| Placebo | 7624 | Normal |
| 8 3 mg/kg/day | 7625 | Normal |
| 8 3 mg/kg/day | 7626 | Normal |
| 8 3 mg/kg/day | 7627 | Normal |
| 8 3 mg/kg/day | 7628 | Normal |
| 8 3 mg/kg/day | 7629 | Normal |
| 8 6 mg/kg/day | 7630 | Normal |
| 8 6 mg/kg/day | 7631 | Some lymphocytic inflammation around the renal pelvis (around the ureter) and occasional medium-sized arteries; more inflammation than 7623 and 7622 |
| 8 6 mg/kg/day | 7632 | Mostly normal but some mild lymphocytic inflammation around small arteries with occasional hemosiderin-laden macrophages |
| 8 6 mg/kg/day | 7633 | Normal |
| 8 6 mg/kg/day | 7634 | Normal |
| Spleens | ||
| Treatment | Sample # | Histology results |
| Placebo | 7651 | Normal |
| Placebo | 7652 | Normal |
| Placebo | 7653 | Normal |
| Placebo | 7654 | Normal |
| Placebo | 7655 | Normal |
| Placebo | 7656 | Moderately increased number of hemosiderin-laden macrophages suggestive of old hemorrhage Somewhere |
| 8 3 mg/kg/day | 7657 | Mildly increased number of hemosiderin-laden macrophages suggestive of old hemorrhage Somewhere |
| 8 3 mg/kg/day | 7658 | Normal (hemosiderin-laden macrophages present but at approximately normal levels) |
| 8 3 mg/kg/day | 7659 | Normal |
| 8 3 mg/kg/day | 7660 | Normal |
| 8 3 mg/kg/day | 7661 | Normal |
| 8 6 mg/kg/day | 7662 | Area of necrosis consistent with splenic infarct; moderately increased number of hemosiderin-laden macrophages suggestive of hemorrhage old somewhere |
| 8 6 mg/kg/day | 7663 | Normal |
| 8 6 mg/kg/day | 7664 | Normal |
| 8 6 mg/kg/day | 7665 | Normal |
| 8 6 mg/kg/day | 7666 | Moderately increased number of hemosiderin-laden macrophages suggestive of old hemorrhage Somewhere |
2.4. Effects of 8 on survival of LmnaH222P/H222P mice
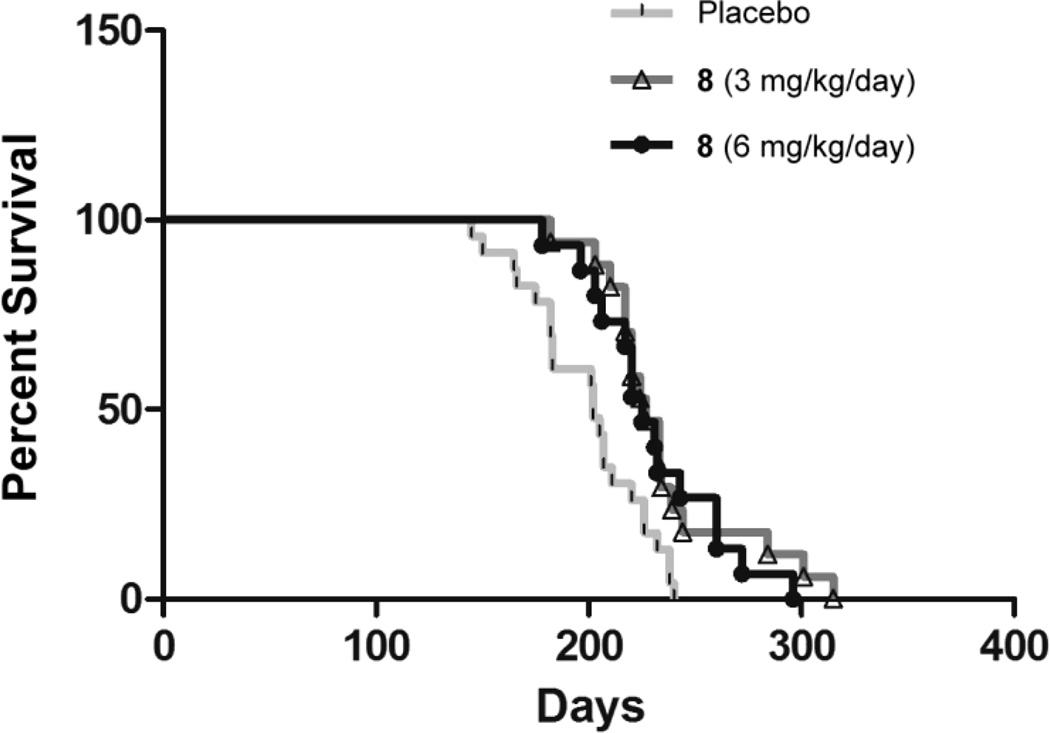
To assess the effects of 8 on survival, LmnaH222P/H222P mice started treatment with either placebo, 8 3 mg/kg/day or 8 6 mg/kg/day and were followed until death or euthanasia was recommended by a veterinarian blind to treatment group (Figure 6). Mice treated with placebo had a median survival of 202 days whereas median survival was 225 days for mice treated with 8 3 mg/kg/day and 225 days for mice treated with 8 6 mg/kg/day. The median survivals of mice treated with both doses of 8 were statistically significantly (P<0.05) longer than that for mice treated with placebo.
Figure 6.
Effect of 8 on survival of LmnaH222P/H222P mice. Kaplan-Meier plots for LmnaH222P/H222P mice treated with placebo (n = 23), 8 3 mg/kg/day (n = 17) or 8 6 mg/kg/day (n = 15) are shown.
3. Conclusions
We have synthesized a novel, allosteric, macrocyclic MEK1/2 inhibitor that potently inhibits ERK1/2 activity in cultured cells and in tissues of mice after systemic administration. Other MEK1/2 inhibitors are currently approved for or in late clinical development for various cancers.3–5 Our preclinical results suggest that this novel MEK1/2 inhibitor will be effective for treating cardiomyopathy caused by LMNA mutations.
There are several limitations to the present study that could be addressed in future research. We used only one mouse model, LmnaH222P/H222P mice, and the results could be generalized to additional models of cardiomyopathy caused by LMNA mutations.36–38 Mouse models also do not exactly recapitulate the corresponding cardiomyopathy caused by LMNA mutations, as the human disease is generally inherited in an autosomal dominant fashion whereas heterozygous mice are mostly asymptomatic or develop relatively less severe disease only at advanced ages.27,36–41 In addition, the current experiments were all performed in male LmnaH222P/H222P mice and studies in female mice, which develop pathology at older age,27 should be considered. Finally, future molecular docking studies and crystallization of the novel MEK1/2 inhibitor we describe will improve our understanding of its structure and mechanism of action.
Regardless of these limitations, the preclinical efficacy and lack of observed significant adverse events in male LmnaH222P/H222P mice indicates that clinical development of novel MEK1/2 inhibitor described in this paper is warranted. In addition to cardiomyopathy caused by LMNA mutations, this inhibitor may have utility in the treatment of cardiomyopathy in X-linked Emery-Dreifuss muscular dystrophy22 and in the treatment of RASopathies, including the hypertrophic cardiomyopathy in Noonan syndrome.8.9 A potent MEK1/2 may also find utility in the treatment of various cancers.3–5
4. Experimental
4.1. General chemistry information
NMR spectra were recorded on Agilent DD2 400 (400 MHz) instrument. Column chromatography was carried out using CombiFlash over RediSep column cartridges employing Merck silica gel (Kieselgel 60, 63–200 µm). Pre-coated silica gel plates F-254 were used for thin-layer analytical chromatography. Mass determinations were performed using electrospray ionization on Sciex API 365 LC/MS/MS system. Purity (>95%) was determined by using reverse phase using Gilson analytical HPLC, using Phenomenex Lune 5µ C18(2) 250 × 4.6 mm column. Samples were run at 1 mL/min using gradient mixtures of 5–100% water with 0.1% trifluoroacetic acid (A) and acetonitrile with 0.1% trifluoroacetic acid (B) for 10 min.
4.1.1. Phenol, 3,4,5-trifluoro-2-nitro- (2)
Nitric acid (16 mL, 247 mmol) was added dropwise to an ice-cold stirred solution of 3,4,5-trifluorophenol (20 g, 135 mmol) in glacial acetic acid (40 mL) in a 2 L flask. The reaction mixture was slowly brought to room temperature and stirred for 1 h. Thin-layer chromatography was used to monitor the progress of reaction. After completion, the reaction mixture was poured into ice water and the aqueous layer was extracted with 10% ethyl acetate in heptanes. The organic layer was washed with saturated NaHCO3 solution, water (X3), brine, dried over anhydrous Na2SO4 and concentrated to yield benzene, 1,2,3-trifluoro-4-nitro-5-(2-propen-1-yloxy)- 1, as yellow oil. Yield: 25.31 g (131 mmol, 97 %). Thin-layer chromatography (30% ethyl acetate:hexanes) Rf = 0.22 1H NMR (CDCl3): δ 6.84 (m, 1H), 10.28 (br s, 1H).
4.1.2 Benzene, 1,2,3-trifluoro-4-nitro-5-(2-propen-1-yloxy)- (3)
Potassium carbonate (51 g, 370 mmol) and allyl bromide (25.6 mL, 296 mmol) were added to a solution of phenol, 3,4,5-trifluoro-2-nitro- 1 (47.6 g, 247 mmol) in acetone (350 mL) at room temperature. The reaction mixture was heated at 70°C for 3 h, and cooled to room temperature and continued to stir overnight. Thin-layer chromatographic analysis indicated completion of reaction. Solvent was evaporated and the residue was dissolved on water and extracted with ethyl acetate. Organic layer was washed with brine and dried (Na2SO4). Organic solvents were evaporated and the residue was purified on silica gel column using 10–25% ethyl acetate/ heptanes as eluent. Yield: 49.2 g (211 mmol, 85%). Thin-layer chromatrograph (10% ethyl acetate:hexanes) Rf = 0.35 1H NMR (CDCl3): δ 4.65 (dt, 2H), 5.39 (d, 1H), 5.45 (d, 1H), 5.98 (m, 1H), 6.72 (m, 1H).
4.1.3 Benzenamine, 2,3-difluoro-N-(2-fluoro-4-iodophenyl)-6-nitro-5-(2-propen-1-yloxy)-(4)
A solution of 2-fluoro-4-iodoaniline (31.04 G, 131 mmol) in anhydrous tetrahydrofuran (600 mL) was cooled to −78°C and stirred for 1 h. Lithium bis(trimethylsilyl)amide (1 M in tetrahydrofuran solution) was added to the reaction mixture through addition funnel dropwise (over 30 min). To this mixture was then added solution of Benzene, 1,2,3-trifluoro-4-nitro-5-(2-propen-1-yloxy)- 3 (30.7 g, 132 mmol) in tetrahydrofuran (50 mL) was added dropwise, and the resulting purple mixture was stirred at −78°C for 4 h and then warmed to room temperature overnight. The mixture was quenched with water. Tetrahydrofuran was removed under vacuum and the resulting slurry was diluted with ethyl acetate and water, organic separated and washed with brine. After concentration, a solid residue was obtained. This residue was triturated with heptanes and a small amount of ether. The solid was filtered and washed with heptanes and dried. Yield: 53 g (89%). Thin-layer chromatography (10% ethyl acetate:hexanes) Rf = 0.23 1H NMR (CDCl3): δ 4.62 (dt, 2H), 5.33–5.36 (d, 1H), 5.48 (d, 1H), 5.98–6.02 (m, 1H), 6.22 (dd, 1H), 6.36 (dd, 1H), 7.04–7.08 (m, 1H), 7.45–7.52 (m, 2H), 7.79 (s, 1 H).
4.1.4 1,2-Benzenediamine, 3,4-difluoro-N2-(2-fluoro-4-iodophenyl)-6-(2-propen-1-yloxy)-
Compound 4 (12 g, 26.66 mmol) was dissolved in ethanol (330 mL) by heating at 90°C. The solution was refluxed for 30 min and then sodium dithionite (37.13 g, 213 mmol) in water (110 mL) was added and heating was continued for additional 1 h. The mixture was cooled to room temperature, concentrated to remove ethanol, extracted with mix of 20% heptanes in ethyl acetate, dried (Na2SO4), filtered and concentrated. The resulting solid was used in the next step without any further purification. Yield: quantitative. Thin-layer chromatography (10% ethyl acetate:hexanes) Rf = 0.18 MS: (M+H)+ 421.4.
4.1.5 (1-allyl-N-(6-(allyloxy)-3,4-difluoro-2-((2-fluoro-4-iodophenyl)amino)phenyl)cyclopropane-1-sulfonamide) (5)
1,2-Benzenediamine, 3,4-difluoro-N2-(2-fluoro-4-iodophenyl)-6-(2-propen-1-yloxy)-from the previous step (30.28 g, 72 mmol) was taken in a seal tube and 4-dimethylaminopyridine (500 mg), pyridine (9.1 mL), methylene chloride (15 mL) and 1-allylcyclopropane-1-sulfonyl chloride (13 g, 72 mmol) were added sequentially. The reaction mixture was then heated at 50°C in a sealed tube for 3 days. After cooling to room temperature, the solvent was evaporated and the residue was dissolved in ethyl acetate and filtered through a plug of silica gel to remove baseline impurities. The organic layer was evaporated and diethyl ether and hexanes added to the residue. The gray solid was filtered and dried to provide a clean product (1-allyl-N-(6-(allyloxy)-3,4-difluoro-2-((2-fluoro-4-iodophenyl)amino)phenyl)cyclopropane-1-sulfonamide). Yield: 37 g (92%). Thin-layer chromatography (10% ethyl acetate:hexanes) Rf = 0.35 1H NMR (CDCl3): δ 0.78 (m, 2H), 1,24 (m, 2H), 2.71 (d, J = 7.2 Hz, 2H), 4.59 (d, J = 5.6 Hz 2H), 5.06 (d, J = 18 Hz 1H), 5.11 (d, J = 10 Hz 1H), 5.41 (d, J = 10 Hz 1H), 5.47 (d, J = 14 Hz, 1H), 5.66–5.68 (m, 1H), 6.05–6.08 (m, 2H), 6.51–6.56 (m, 2H), 7.09 (d, J = 8.8 Hz, 1H), 7.21–7. 28 (m, 1H), 7.33 (s, 1H).
4.1.6 (Z)-10,11-difluoro-12-((2-fluoro-4-iodophenyl)amino)-4,7-dihydro-1H–spiro[benzo[b][1,5,4]oxathiazecine-3,1’-cyclopropane] 2,2-dioxide (6)
The catalyst (3-phenyl-1H-inden-1-ylidene[bis(i-butylphoban)]ruthenium(II) dichloride (671 mg, 5 mol%) was added to a solution of (1-allyl-N-(6-(allyloxy)-3,4-difluoro-2-((2-fluoro-4-iodophenyl)amino)phenyl)cyclopropane-1-sulfonamide) 5 (10 g) in dichloromethane (3 L) and the mixture was degassed for 10 min by bubbling nitrogen gas. The mixture was then stirred at 65°C for 15 h. The next day, the solvent was evaporated and the crude product purified by silica gel column (5%/50% ethyl acetate/heptanes). The desired cis isomer, (Z)-10,11-difluoro-12-((2-fluoro-4-iodophenyl)amino)-4,7-dihydro-1H-spiro[benzo[b][1,5,4]oxathiazecine-3,1’-cyclopropane] 2,2-dioxide was isolated (7g, 74%) and a minor trans product (E)-10,11-difluoro-12-((2-fluoro-4-iodophenyl)amino)-4,7-dihydro-1H-spiro[benzo[b][1,5,4]oxathiazecine-3,1’-cyclopropane] 2,2-dioxide (1.92 g, 20%) was also isolated. Rf for the desired product was 0.45 (thin-layer chromatography in 40% ethyl acetate/heptanes) and for the trans product was 0.2 under same conditions. MS: (M+H)+ 537, 1H NMR (CDCl3): δ 0.74 (br s, 2H), 1,14 (br s, 2H), 3.14 (m, 2H), 4.92 (s, 2H), 5.46 (dd, J = 12.0 Hz, 1 H), 5.72 (dd, J = 8.0, 12.0 Hz), 6.27 (s, 1H), 6.51 (m, 2H), 7.18 (s, 1H), 7.29 (d, J = 8.0 Hz, 1H). 7.41 (d, J = 12Hz, 1H).
4.1.7 10,11-difluoro-12-((2-fluoro-4-iodophenyl)amino)-5,6-dihydroxy-4,5,6,7-tetrahydro-1H–spiro[benzo[b][1,5,4]oxathiazecine-3,1’-cyclopropane] 2,2-dioxide (7)
N-methyl morpholine N-oxide (16.6 g, 3.3 equivalents) at room temperature followed by an aqueous solution of osmimum tetraoxide (6 mL of 5 mol% solution) were added to a 20-g solution of 2 ((Z)-10,11-difluoro-12-((2-fluoro-4-iodophenyl)amino)-4,7-dihydro-1H-spiro[benzo[b][1,5,4]oxathiazecine-3,1’-cyclopropane] 2,2-dioxide) in tetrahydrofuran (500 mL) and water (5 mL). The mixture was stirred at room temperature for 16 h and then quenched by adding 10 mL of 1M aqueous sodium sulfite solution and stirred for 1 h. The mixture was concentrated under vacuum and the residue was diluted with ethyl acetate and washed with 5 M sodium sulfite solution and brine. The organic layer was concentrated and the residue was purified on a small (plug) silica gel using 50%/90% ethyl acetate/hexanes to provide a quantitative yield of 10,11-difluoro-12-((2-fluoro-4-iodophenyl)amino)-5,6-dihydroxy-4,5,6,7-tetrahydro-1H–spiro[benzo[b][1,5,4]oxathiazecine-3,1’-cyclopropane] 2,2-dioxide. Thin-layer chromatography (neat ethyl acetate) Rf = 0.4; MS: (M+H)+ 570.9, 1H NMR (CDCl3): δ 0.68 −0.85 (br m, 3H), 1.17 (br m, 1H), 1,4 (s, 1H), 2.25 (d, 1H), 3.25–3.65 (m, 3H), 3.75 (m, 1H), 3.85 (d, 1H), 4.20 (br s, 1H), 4.42 (br t, 1H), 6.25 (m, 1H), 6.50–6.55 (m, 1H), 7.28 (s, 1H), 7.30–7.40 (d, J = 8.0 Hz, 1H), 7.60 (m, 1H).
4.1.8 10,11-difluoro-12-((2-fluoro-4-iodophenyl)amino)-5,6-dihydroxy-4,5,6,7-tetrahydro-1H–spiro[benzo[b][1,5,4]oxathiazecine-3,1’-cyclopropane] 2,2-dioxide (8)
The active enantiomer was obtained by chiral separation of the racemic diol (26.64 g) using Thar 350 preparative supercritical fluid chromatography (column: ChiralPak AD-10u, 300×50mmI.D., mobile phase: A for CO2 and B for Methanol(0.05%DEA), gradient B(45%), flow rate: 200 mL/min. The faster moving enantiomer, 8 (RT 7.00) was isolated (12.27 g), the slower moving isomer was also isolated (RT 10.92, 12.60 g).
4.2 In vitro biochemical assay of MEK1 activity
We used an in vitro biochemical assay to test 7 for inhibition of MEK1 activity. The reaction mixture contained 80 ng of a recombinant fusion protein of glutathione-S-transferase and full-length human MEK1 (Cell Signaling), 4 µg of ERK1/ERK2 peptide (Enzo Life Sciences), 100 µM or 1 mM ATP, 1 µM to 1.37 nM of 7 diluted in dimethyl sulfoxide and buffer containing 5 mM MOPS (pH 7.2), 2.5 mM β-glycerophosphate, 1 mM methylene glycol-bis(β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid, 0.4 mM ethylenediaminetetraacetic acid, 5 mM MgCl2 and 0.05 mM dithiothreitol. Reaction mixture was incubated at room temperature for 90 min. At the end of the reaction, 20 µL of ADP-Glo reagent (Promega) was added and incubated at room temperature for 40 min. Forty µL of Kinase Detection Reagent (Promega) was then added and incubated at room temperature for 1 h. Chemiluminescence was measured in a luminometer and IC50 calculated using SoftMax software.
4.3 Cell culture experiments
4.3.1 Anti-proliferative activity
HT-29 (American Type Culture Collection) were cultured at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with L-glutamine and 10% fetal bovine serum (Invitrogen). Proliferation was assessed by plating 2,000 cells/well in 100 µL of Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum or Roswell Park Memorial Institute medium supplemented with 10% fetal bovine serum in a 96-well plate and incubated overnight at 37°C with 5% CO2. After overnight incubation, the culture media were replaced with 100 µL of the same media containing various concentrations of the compounds. Compounds were added at 3-fold dilutions with concentrations ranging from 3.3 µM to 4.5 nM. After 72 h of incubation at 37°C with 5% CO2, cell viability was measured in a luminometer after adding 100 µM/well CellTiterGlo reagent (Promega). IC50s were calculated using SoftMax software.
4.3.2 Effects on phosphorylated ERK1/2
HEK-293 cells (American Type Culture Collection) were cultured at 37°C with 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. 8 was dissolved in 10% v/v Chremophor EL (BASF)/normal saline at a concentration of 100 µM. Cells were seeded in 6-well plate at a density of 5.0 × 105. After 24 h of incubation, the cells were rinsed twice with culture medium. Cells were then incubated for 24 h in culture medium with 100 µl 10% v/v Chremophor EL/normal saline as control or culture medium with serial dilutions of 8 at finial concentrations of 1 µM to 0.001 µM. Cells were then washed three times with phosphate-buffered saline and collected for protein extraction and immunoblotting as described below.
4.4 Mouse experiments
4.4.1. Mice
The Institutional Animal Care and Use Committee at Columbia University Medical Center approved the use of animals and the study protocol. LmnaH222P/H222P mice were bred and genotyped as described previously.23,25 Mice were fed a chow diet and housed in a barrier facility with 12 h/12 h light/dark cycles. Male LmnaH222P/H222P mice were used in all experiments because they develop signs and symptoms of cardiomyopathy at an earlier age and have a shorter lifespan than female LmnaH222P/H222P mice.25
4.4.2. Treatment of mice with 8
8 was dissolved in 10% v/v Chremophor EL/normal saline at concentrations of 0.75 mg/ml or 1.5 mg/ml and 0.1 ml was administered 6 days a week by oral gavage for doses of 3 mg/kg/day or 6 mg/kg/day. Placebo was the same volume of 10% v/v Chremophor EL/normal saline given by oral gavage. For biochemical analysis of tissue, echocardiographic analysis, analysis of fibrosis, analysis of skeletal muscle function and toxicology analyses, treatment was started at 14 weeks of age and continued until 20 weeks of age. For survival analyses, treatment was started at 10 weeks of age and continued either until death or a veterinarian blind to treatment group determined that euthanasia was necessary. Euthanasia was performed in a CO2 chamber followed by cervical dislocation, according to the protocol of the Institute of Comparative Medicine at Columbia University Medical Center. Euthanasia was confirmed by checking for lack of response to limb and tail pinch. Immediately after euthanasia, mouse organs were quickly excised and divided with one portion snap frozen in liquid nitrogen and the other portion placed in 4% formaldehyde solution.
4.4.3. Protein extraction and immunoblotting
Methods for protein extraction and immunoblotting have been described previously.18,23 Briefly, proteins were extracted from cultured cells and mouse tissues by homogenization in extraction buffer as described previously. Extracted proteins were separated by SDS-polyacrylamide slab gel electrophoresis, transferred to nitrocellulose membranes and blotted with primary antibodies against total ERK1/2 (no. Sc-94, Santa-Cruz) and phosphorylated ERK1/2 (no. 9101, Cell Signaling). Secondary antibodies were horseradish peroxidase-conjugated (GE Healthcare). SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) was used to visualize recognized proteins. For quantification of phosphorylated ERK1/2 compared to total ERK1/2, immunoblots were scanned and analyzed using ImageJ64 software.42
4.4.4. Echocardiography
At 20 weeks of age, mice were anaesthetized with 1.5% isoflurane by inhalation and placed on a heating pad (37°C). Echocardiography was performed using a Visualsonics Vevo 770 ultrasound with a 30 MHz transducer applied to the chest wall. Cardiac ventricular dimensions were measured in two-dimensional mode and M-mode three times for the number of animals indicated. Left ventricular fractional shortening (FS) was calculated from the left ventricular end diastolic diameter (LVEDD) and left ventricular end systolic diameter (LVESD) using the following formula: FS = [(LVEDD – LVESD)/LVEDD] × 100.
4.4.5. Quantitative real-time RT-PCR analysis
RNA was extracted from heart tissue using the RNeasy isolation kit (Qiagen). Complementary DNA was synthesized from total RNA using RevertAid RT Reverse Transcription Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. For each replicate in each experiment, RNA from tissue samples of different animals was used. Primers were designed corresponding to mouse RNA sequences using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi): for Nppa forward 5’-gcttccaggccatattggag-3’ and reverse 5’-ccctgcttcctcagtctgct-3’; for Myl7 forward 5’-tcaaggaagccttcagctgc-3’ and reverse 5’-cggaacacttaccctcccg-3’. Real-time RT-PCRs contained HotStart-IT SYBR green qPCR Master Mix (Affymetrix), 200 nM of each primer, and 0.2 µL of template in a 25 µL reaction volume. Amplification was carried out using the ABI 7300 Real-Time PCR System (Applied Biosystems) with an initial denaturation at 95°C for 2 min followed by 50 cycles at 95°C for 30 seconds and 62°C for 30 seconds. Relative levels of mRNA expression were calculated using the ΔΔCT method.43 Individual expression values were normalized by comparison with Gapdh mRNA (forward 5’-tgcaccaccaactgcttag-3’ and reverse 5’-ggatgcagggatgatgttc-3’).
4.4.6. Histological analysis
Tissue samples from LmnaH222P/H222P mice were fixed in 4% formaldehyde for 48 h, embedded in paraffin, sectioned at 5 µM and stained with hematoxylin and eosin or Masson trichrome. Representative stained sections were scanned using a Leica SCN400 scanner. Images were processed using Adobe Creative Cloud Photoshop CC (Adobe Systems). A pathologist blind to mouse genotype and treatment group analyzed pathology in heart sections stained with Masson trichrome. A grading system was constructed based on the presence of two abnormalities that were found to be variably present between specimens: cytoplasmic vacuolation and cardiac fibrosis. Specimens were scored as no fibrosis and no vacuolation (grade 0), no fibrosis and mild vacuolation (grade 1), no fibrosis and moderate vacuolation (grade 2), vacuolation and mild fibrosis (grade 3) or vacuolation and severe fibrosis (grade 4). The same blinded pathologist provided descriptive assessments of sections of other tissues stained with hematoxylin and eosin.
4.4.7. Serum biochemical analysis
Serum was separated from blood drawn from mice and stored at −80°C until analyzed. Routine clinical chemistry analysis was performed on an AutoAnalyzer at the Comparative Pathology Laboratory at Columbia University Medical Center.
4.4.8. Skeletal muscle fatigability analysis
Forelimb grip strength was assessed using a published protocol.44 An investigator blind to treatment performed the experiments. Each mouse was placed in front of a Chatillon DFIS-2 digital force gauge pull meter with angled mesh assembly and allowed to grasp the pull bar with forelimbs and then pulled backward by the tail until the grip was broken. Force applied to the bar the moment the grasp was released was automatically recorded. Each mouse pulled the grid three times in a row and then returned in the cage for a resting period of at least one minute. Each mouse performed a total of five series of pulls, each followed by a short resting period. In this way, each mouse had pulled a total of 15 times (3 pulls x 5 times = 15 pulls). The grip strength was normalized to body mass. The fatigue index was determined by calculating the decrement between the average of the first two and the last two series of pulls 1+2+3=A, 4+5+6=B, 10+11+12=C and 13+14+15=D. The formula (C+D)/(A+B) gives a value of 1 for mice that are not fatigued. The mice were tested for two consecutive days.
4.4.9. Survival Studies
LmnaH222P/H222P mice were treated with placebo, 3 mg/kg/day or 6 mg/kg/day of 8 as described above starting at 10 weeks of age and until time of death or signs of significant distress requiring euthanasia. Specific signs of significant distress included (i) difficulty with normal ambulatory movement, (ii) failure to eat or drink, (iii) weight loss of more than 20%, (iv) depression, (v) rough or unkempt hair coat, and (vi) significant respiratory distress. A staff veterinarian at the Institute of Comparative Medicine at Columbia University Medical Center, blind to treatment group, determined if euthanasia was required.
4.4.9 Statistics
Values for scanned immunoblots, echocardiographic parameters, real-time RT–PCR, blood chemistry results and cardiac fibrosis scores were compared using one-way ANOVA. To validate these results, a nonparametric (Mann-Whitney) test was performed and concordance checked. Mouse survival was analyzed using the Kaplan-Meier estimator.38 Differences in median survival were compared using a log-rank (Mantel-Cox) test with P<0.05 considered to be statistically significant.45 Statistical analyses were performed using GraphPad Prism software.
Acknowledgments
We thank Dr. Anne Rutkowski for help and advice. This work was supported by the National Institutes of Health [grant numbers R41TR001008 and K08AR059750].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
W.W. has no related interest to disclose. M.D.C. was an employee of Cheminpharma and is an inventor on patents related to this work. B.P. Hart has equity in AlloMek Therapeutics and is an inventor on patents related to the work. E.S. Kumarasinghe in an employee of Cheminpharma. M.K. Ji is an employee of Cheminpharma. A. Bhargava is an executive at Shakti Biorsearch and has equity in the company. M.W. Lawlor receives research support from Audentes Therapeutics, Solid GT and Demeter Therapeutics and is a member of the Scientific Advisory Board of Audentes Therapeutics. J.-Y. Shin has no related interest to disclose. F. Sera has no related interest to disclose. S. Homma has not related interest to disclose. A. Muchir is a member of the Scientific Advisory Board of AlloMek Therapeutics, has equity in AlloMek Therapeutics and is an inventor on a patent related to the work. U.R. Khire is an executive at AlloMek Therapeutics, an executive at Cheminpharma, has equity in both of these companies and is an inventor on patents related to the work. H.J. Worman previously received research support from AlloMek Therapeutics, is a member of the Scientific Advisory Board of AlloMek Therapeutics, has equity in AlloMek Therapeutics and is an inventor on a patent related to the work.
References and notes
- 1.Roskoski R., Jr Pharmacol. Res. 2012;66:105. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Roskoski R., Jr Biochem. Biophys. Res. Commun. 2012;417:5. doi: 10.1016/j.bbrc.2011.11.145. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y, Adjei AA. Nat. Rev. Clin. Oncol. 2014;11:385. doi: 10.1038/nrclinonc.2014.83. [DOI] [PubMed] [Google Scholar]
- 4.Caunt CJ, Sale MJ, Smith PD, Cook SJ. Nat. Rev. Cancer. 2015;15:577. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 5.Shang J, Lu S, Jiang Y, Zhang J. Chem. Biol. Drug Des. 2016;88:485. doi: 10.1111/cbdd.12780. [DOI] [PubMed] [Google Scholar]
- 6.Bentires-Alj M, Kontaridis MI, Neel BG. Nat. Med. 2006;12:283. doi: 10.1038/nm0306-283. [DOI] [PubMed] [Google Scholar]
- 7.Rauen KA. Annu. Rev. Genomics Hum Genet. 2013;14:355. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelb BD, Tartaglia M. J. Clin. Invest. 2011;121:844. doi: 10.1172/JCI46399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Simpson J, Hong JH, Kim KH, Thavarajah NK, Backx PH, Neel BG, Araki T. J. Clin. Invest. 2011;121:1009. doi: 10.1172/JCI44929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huby AC, Turdi S, James J, Towbin JA, Purevjav E. Clin. Sci. (Lond) 2016;130:289. doi: 10.1042/CS20150624. [DOI] [PubMed] [Google Scholar]
- 11.Worman HJ, Fong LG, Muchir A, Young SG. J. Clin. Invest. 2009;119:1825. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonne G, Di Barletta MR, Varnous S, Bécane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K. Nat. Genet. 1999;21:285. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 13.Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ, Jr, Spudich S, De Girolami U, Seidman JG, Seidman C, Muntoni F, Müehle G, Johnson W, McDonough B. N. Engl. J. Med. 1999;341:1715. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 14.Brodsky GL, Muntoni F, Miocic S, Sinagra G, Sewry C, Mestroni L. Circulation. 2000;101:473. doi: 10.1161/01.cir.101.5.473. [DOI] [PubMed] [Google Scholar]
- 15.Muchir A, Bonne G, van der Kooi AJ, van Meegen M, Baas F, Bolhuis PA, de Visser M, Schwartz K. Hum. Mol. Genet. 2000;9:1453. doi: 10.1093/hmg/9.9.1453. [DOI] [PubMed] [Google Scholar]
- 16.Quijano-Roy S, Mbieleu B, Bönnemann CG, Jeannet PY, Colomer J, Clarke NF, Cuisset JM, Roper H, De Meirleir L, D’Amico A, Ben Yaou R, Nascimento A, Barois A, Demay L, Bertini E, Ferreiro A, Sewry CA, Romero NB, Ryan M, Muntoni F, Guicheney P, Richard P, Bonne G, Estournet B. Ann. Neurol. 2008;64:177. doi: 10.1002/ana.21417. [DOI] [PubMed] [Google Scholar]
- 17.Lu JT, Muchir A, Nagy PL, Worman HJ. Dis. Model. Mech. 2011;4:562. doi: 10.1242/dmm.006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muchir A, Pavlidis P, Decostre V, Herron AJ, Arimura T, Bonne G, Worman HJ. J. Clin. Invest. 2007;117:1282. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi JC, Wu W, Muchir A, Iwata S, Homma S, Worman HJ. J. Biol. Chem. 2012;287:40513. doi: 10.1074/jbc.M112.404541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muchir A, Reilly SA, Wu W, Iwata S, Homma S, Bonne G, Worman HJ. Cardiovasc. Res. 2012;93:311. doi: 10.1093/cvr/cvr301. [DOI] [PubMed] [Google Scholar]
- 21.Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D. Nat. Genet. 1994;8:323. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- 22.Muchir A, Pavlidis P, Bonne G, Hayashi YK, Worman HJ. Hum. Mol. Genet. 2007;16:1884. doi: 10.1093/hmg/ddm137. [DOI] [PubMed] [Google Scholar]
- 23.Muchir A, Shan J, Bonne G, Lehnart SE, Worman HJ. Hum. Mol. Genet. 2009;18:241. doi: 10.1093/hmg/ddn343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu W, Muchir A, Shan J, Bonne G, Worman HJ. Circulation. 2011;123:53. doi: 10.1161/CIRCULATIONAHA.110.970673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isshiki Y, Kohchi Y, Iikura H, Matsubara Y, Asoh K, Murata T, Kohchi M, Mizuguchi E, Tsujii S, Hattori K, Miura T, Yoshimura Y, Aida S, Miwa M, Saitoh R, Murao N, Okabe H, Belunis C, Janson C, Lukacs Christine S, Verena Shimma N. Bioorg. Med. Chem. Lett. 2011;21(6):1795–1801. doi: 10.1016/j.bmcl.2011.01.062. [DOI] [PubMed] [Google Scholar]
- 26.Marsault E, Peterson MLJ. J. Med. Chem. 2011;54:1961. doi: 10.1021/jm1012374. [DOI] [PubMed] [Google Scholar]
- 27.Arimura T, Helbling-Leclerc A, Massart C, Varnous S, Niel F, Lacène E, Fromes Y, Toussaint M, Mura AM, Keller DI, Amthor H, Isnard R, Malissen M, Schwartz K, Bonne G. Hum. Mol. Genet. 2005;14:155. doi: 10.1093/hmg/ddi017. [DOI] [PubMed] [Google Scholar]
- 28.Muchir A, Kim YJ, Reilly SA, Wu W, Choi JC, Worman HJ. Skelet. Muscle. 2013;3:17. doi: 10.1186/2044-5040-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raman SV, Sparks EA, Baker PM, McCarthy B, Wooley CFJ. Cardiovasc. Magn. Reson. 2007;9:907. doi: 10.1080/10976640701693733. [DOI] [PubMed] [Google Scholar]
- 30.van Tintelen JP, Tio RA, Kerstjens-Frederikse WS, van Berlo JH, Boven LG, Suurmeijer AJ, White SJ, den Dunnen JT, te Meerman GJ, Vos YJ, van der Hout AH, Osinga J, van den Berg MP, van Veldhuisen DJ, Buys CH, Hofstra RM, Pinto YM. J. Am. Coll. Cardiol. 2007;49:2430. doi: 10.1016/j.jacc.2007.02.063. [DOI] [PubMed] [Google Scholar]
- 31.Holmström M, Kivistö S, Heliö T, Jurkko R, Kaartinen M, Antila M, Reissell E, Kuusisto J, Kärkkäinen S, Peuhkurinen K, Koikkalainen J, Lötjönen J, Lauerma K. J. Cardiovasc. Magn. Reson. 2011;13:30. doi: 10.1186/1532-429X-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontana M, Barison A, Botto N, Panchetti L, Ricci G, Milanesi M, Poletti R, Positano V, Siciliano G, Passino C, Lombardi M, Emdin M, Masci PG. JACC Cardiovasc. Imaging. 2013;6:124. doi: 10.1016/j.jcmg.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Hasselberg NE, Edvardsen T, Petri H, Berge KE, Leren TP, Bundgaard H, Haugaa KH. Europace. 2014;16:563. doi: 10.1093/europace/eut291. [DOI] [PubMed] [Google Scholar]
- 34.Masuda T, Satodate R, Tsuruga K, Kasai T. Tohoku J. Exp. Med. 1993;170:169. doi: 10.1620/tjem.170.169. [DOI] [PubMed] [Google Scholar]
- 35.Cosnett JE, Pudifin DJ. Br. Heart J. 1964;26:544. doi: 10.1136/hrt.26.4.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. J. Cell Biol. 1999;147:913. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mounkes LC, Kozlov SV, Rottman JN, Stewart CL. Hum. Mol. Genet. 2005;14:2167. doi: 10.1093/hmg/ddi221. [DOI] [PubMed] [Google Scholar]
- 38.Davies BS, Barnes RH, 2nd, Tu Y, Ren S, Andres DA, Spielmann HP, Lammerding J, Wang Y, Young SG, Fong LG. Hum. Mol. Genet. 2010;19:2682. doi: 10.1093/hmg/ddq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf CM, Wang L, Alcalai R, Pizard A, Burgon PG, Ahmad F, Sherwood M, Branco DM, Wakimoto H, Fishman GI, See V, Stewart CL, Conner DA, Berul CI, Seidman CE, Seidman JG. J. Mol. Cell. Cardiol. 2008;44:293. doi: 10.1016/j.yjmcc.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chandar S, Yeo LS, Leimena C, Tan JC, Xiao XH, Nikolova-Krstevski V, Yasuoka Y, Gardiner-Garden M, Wu J, Kesteven S, Karlsdotter L, Natarajan S, Carlton A, Rainer S, Feneley MP, Fatkin D. Circ. Res. 2010;106:573. doi: 10.1161/CIRCRESAHA.109.204388. [DOI] [PubMed] [Google Scholar]
- 41.Cattin ME, Bertrand AT, Schlossarek S, Le Bihan MC, Skov Jensen S, Neuber C, Crocini C, Maron S, Lainé J, Mougenot N, Varnous S, Fromes Y, Hansen A, Eschenhagen T, Decostre V, Carrier L, Bonne G. Hum. Mol. Genet. 2013;22:3152. doi: 10.1093/hmg/ddt172. [DOI] [PubMed] [Google Scholar]
- 42.Schneider CA, Rasband WS, Eliceiri KW. Nat. Methods. 2012;9:671. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ponchel F, Toomes C, Bransfield K, Leong FT, Douglas SH, Field SL, Bell SM, Combaret V, Puisieux A, Mighell AJ, Robinson PA, Inglehearn CF, Isaacs JD, Markham AF. BMC Biotechnol. 2003;3:18. doi: 10.1186/1472-6750-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aartsma-Rus A, van Putten M. J. Vis. Exp. 2014;85:e51303. doi: 10.3791/51303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaplan EL, Meier P. J. Amer. Statist. Assn. 1958;53:457. [Google Scholar]