Abstract
Single cell analysis provides a new framework for understanding biology and disease, however, an absolute quantification of single cell gene expression still faces many challenges. Microfluidic digital polymerase chain reaction (PCR) provides a unique method to absolutely quantify the single cell gene expression, but only limited devices are developed to analyze a single cell with detection variation. This paper describes a self-priming compartmentalization (SPC) microfluidic digital polymerase chain reaction chip being capable of performing single molecule amplification from single cell. The chip can be used to detect four single cells simultaneously with 85% of sample digitization. With the optimized protocol for the SPC chip, we first tested the ability, precision, and sensitivity of our SPC digital PCR chip by assessing β-actin DNA gene expression in 1, 10, 100, and 1000 cells. And the reproducibility of the SPC chip is evaluated by testing 18S rRNA of single cells with 1.6%–4.6% of coefficient of variation. At last, by detecting the lung cancer related genes, PLAU gene expression of A549 cells at the single cell level, the single cell heterogeneity was demonstrated. So, with the power-free, valve-free SPC chip, the gene copy number of single cells can be quantified absolutely with higher sensitivity, reduced labor time, and reagent. We expect that this chip will enable new studies for biology and disease.
I. INTRODUCTION
Cells are the fundamental unit of biology and the heterogeneity of cells resulting from stochastic expression of genes, proteins, and metabolites plays an important role in cell differentiation, origins of disease and cellar reaction.1 However, an individual cell in general weighs a few picograms and its volume is about 1 pl, which makes it difficult to analyze at single-cell level. The exiting approaches are mostly based on studying the bulk cell population rather than single cell, which would unavoidably lead to an average result and possibly disguise the significant heterogeneity of functional cells.2 Moreover, some theoretical and practical problems can be solved only at single-cell level.3,4 In order to understand the functions of individual cell in the context of its microenvironment, distinguish the heterogeneity of single cells, and analyze the rare single cell gene expression, such as cancer stem cell, circulating tumour cell (CTC), and so on, single-cell analysis is necessary.
In fact, a single-cell analysis has recently reached a new stage since the vital advances in technologies have provided more sensitive and accurate methods to analyze a single cell.5,6 Although a single cell RNA-seq and genome-seq has been developed, it is too expensive for specific gene markers.7–9 Microfluidic technology by the accurate control can realize low-cost and efficient single cell analysis.10,11 The scale of microfluidic channels is generally 1–100 μm, the volume of reaction chambers is in picoliter or nanoliter level. The small internal dimensions of microfluidic devices are appropriate for individual cells. Meanwhile, small reaction volume increasing the relative concentration of sample, thus improves sensitivity and precision, lowers cost of reagent and reduces reaction time.12–15 Many microfluidic devices for single-cell gene expression analysis have been demonstrated including single cell isolation, lysis, complementary DNA (cDNA) synthesis,3,16 and real-time quantitative polymerase chain reaction (qPCR)17 following off-chip protocol.18,19 Especially, digital polymerase chain reaction (PCR) devices have supplied an efficient and precise platform for single-cell analysis because of their well-fit scale and higher precision.20–23 Digital PCR chip with microvalves have been used to map the cellular subpopulations in normal tissues and tumors.24 An integrated device was developed to accomplish the necessary steps for single-cell analysis including cell capture, cell lysis, reverse transcription, and digital PCR.25 However, these devices still need microvalves which are complicated in manufacture and control.
To realize self-digitization in microfluidic device, a self-digitization chip was developed for single cell digital reverse transcription polymerase chain reaction (RT-PCR), which can perform reverse transcription in the digitized volumes.26 Compared with this method, we have developed two kinds of simpler and more efficient chips called self-priming compartmentalization (SPC) digital LAMP chip and integrated self-priming compartmentalization (SPC) digital PCR chip.27,28 Both two kinds of chips have taken the advantage of the high gas solubility of poly(dimethylsiloxane) (PDMS).29,30 Degassed by a vacuum, the chip has much lower air pressure of its inside than the atmosphere. Thus the air pressure difference provides a built-in power so that the sample solution and oil can sequentially be sucked into the channels and microwells to realize “divide and conquer” for single molecule amplification without microvalves or control system.23 However, the above-mentioned two kinds of microfluidic chips are still for gene expression quantification of large populations of cells.
In this work, we applied the self-priming compartmentalization digital PCR chip to the determination of the gene expression in single cells based on our previous work, making single cell gene expression quantification easier to manipulate. We applied this absolute quantification method to different gene expressions in single cells and different population of cells. We then compared the result to study the heterogeneity of cells. We also used qPCR to show the limitation of it in single cell study. We provided an alternative method to quantify the absolute gene expressions within a single cell.
II. MATERIALS AND METHODS
A. Device design and fabrication
The self-priming compartmentalization (SPC) chip microfluidic chips for single-cell digital PCR were fabricated by multilayer soft lithography techniques. The patterns of the SPC chip were designed with Corel DRAW X4 and printed on a transparency films by a high resolution printer. The flow mold was fabricated with three lithographic steps according to manufacturer specifications. Every chip was composed of a five-layer structure. From the bottom to the top, the first layer was a glass coverslip as the base. The second layer was a 250 μm-thick thin layer of PDMS called the flow layer. This layer was created by spin-coating 5A:1B (excess Si–H groups) mixture of PDMS on the flow mold at 1000 rpm for 30 s. The flow layer contained four panels to demonstrate the gene expression measurement, and each panel contained 8 flow channels and 1280 independent microwells. The flow channel was 50 μm in width and 25 μm in height. The microwell was both 150 μm in width and length while the height was 250 μm. The layer above the flow layer was nano water-proof layer preventing water vapor transport and evaporation of the sample solution in the microwells during thermocycling. After the second layer (microwell array) was baked for 5 min on 85 °C, a 10 nm-thick water-proof layer was created by spinning a low-permeability fluorosilane polymer (EGC-1720, 3M) on the cured PDMS layer. This coating can dry immediately, usually, this layer was coated two times. After coated, the mold was baked for 3 min on 85 °C, and then, 5A:1B PDMS was poured on the coating layer. The water-proof layer does not need to be activated, which can bind with uncured PDMS to form a PDMS-fluorosilane polymer-PDMS hybrid after being baked. On the top of the water-proof layer was the inlet and outlet layer made of a 4 mm-thick PDMS. The diameter of the sample inlet is 0.5 mm. Another 4 mm-thick PDMS layer was punched with holes (2.5 mm in diameter) and was aligned to the inlet and outlet layer as the suction chambers which could provide additional power to drive the flow (Fig. 1). After the sample solution was sucked into the microwells and individually separated by oil, the suction chambers layer would be removed, the inlet and outlet holes would be sealed by PDMS and silicone oil mix, and another glass coverslip as the seal would be pressed on the upper surface of the chip.
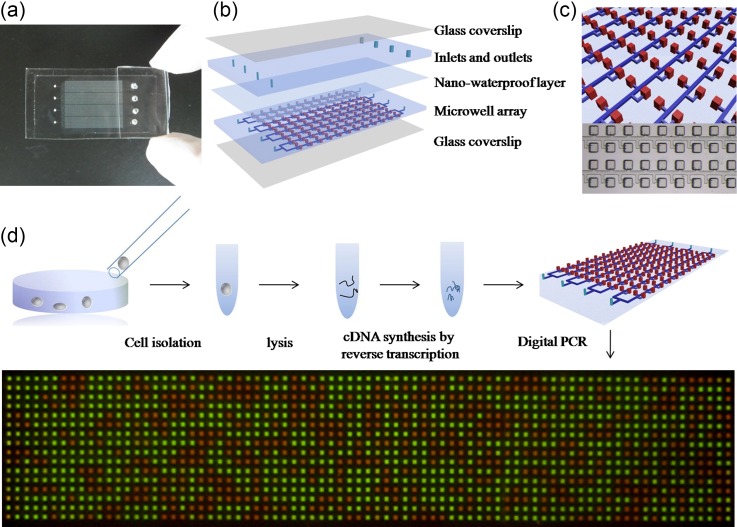
FIG. 1.
Schematic drawing of the self-priming compartmentalization (SPC) digital PCR chip and procedure of single cell digital PCR. (a) Photograph of the prototype of the SPC chip. It contains four individual panels, and suction chambers are on the top of the outlets. (b) Schematic diagram of the layered device structure of the chip, which is composed of two layers of PDMS (PDMS layer and Microarray layer), a nano-waterproof layer and two glass coverslip layers. (c) Diagram of the detail of the chip design. Thousands of microwells are connected with the main channel, green line represents the main channels and the red boxes represent the reaction microwells, the size of the microwells is 150 μm × 150 μm × 250 μm, the size of the main channel is 50 μm × 25 μm. (d) The process of the single cell gene expression digital PCR.
B. Device operation
First, the SPC digital PCR chip was attached a transparent adhesive tape to its top surface and placed in the vacuum pump to perform the procedure of degassing (1 kPa for 20 min). After the degassing procedure finished, the chip was taken out and put in a normal atmosphere. The transparent adhesive tape was punctured with a syringe needle. An 8.5 μl of reaction mix was then dispensed via the sample inlet with a conventional micropipette and was sequentially sucked into the channels and microwells because of the air pressure difference. When the reaction mix was totally sucked into the channels and microwells, silicone oil was added in the sample inlet and followed the reaction mix into the channels. When the reaction mix was individually separated by oil into the microwells, the suction chamber layers were removed. Then a mixture of PDMS (1.1 g of uncured PDMS was mixed at the ratio of 10A:1B) and silicone oil (4 g) prepared before was dispensed into the inlets and outlets. At last, the upper surface of the chip was bonded by a glass coverslip without bubbles. Thus, the chip was encapsulated into a whole set and ready to perform digital PCR reaction. Mineral oil was applied between the chip and the flatbed block of the PCR machine to improve the thermal contact during thermocycling.
C. Cell preparation
The reaction mix was prepared from the TaqMan® Gene Expression Cells-to-CT™ Kit (Life Technologies). Before the cell preparation, the stop solution was thawed at the temperature of 4 °C and placed on ice. Phosphate buffered saline (PBS) buffer was stored at 4 °C. Lysis solution was added into a new tube. A549 cells were digested by trypsin and washed in cold PBS prepared before to remove the culture medium and re-suspended in cold PBS buffer. The cell suspension was then serially diluted to get 100 cells and 1000 cells sample by pipetting under microscope. 1 cell, 10 cells were picked up by a capillary tube from single-cell suspension and put into a tube which contains the 6 μl lysis solutions incubating for 5 min at room temperature. The 100 cells and 1000 cells samples were centrifuged to remove the PBS buffer and added with 6 μl lysis solutions, respectively. After incubated for 5 min at room temperature, each lysis reaction was added to 0.6 μl stop solution and incubated for 2 min at room temperature. Each lysate was mixed with 10 μl 2× RT Buffer, 1 μl 20× RT Enzyme and 2.4 μl nuclease-free water, then the mix was placed on the PCR machine (MGL96G, Long Gene) to run the RT thermal cycler program which included a 60 min incubating step at 37 °C and 5 min RT enzyme inactivating step at 95 °C. After transcripts were reverse transcribed to cDNA, the cDNA sample may be stored at −20 °C for following digital PCR and quantitative real-time PCR.
D. Single-cell digital PCR
We first performed the measurements of β-actin gene expression to establish the capability of single-cell digital PCR analysis on the SPC digital PCR chip and test the sensitivity, precision, and uniformity of the chip. All reaction components, including the PCR master mix, primers, probe, and template were assembled and mixed off-chip before analysis. The reaction mix was supplemented with 0.1% TWEEN-20 to prevent reaction components from being absorbed by PDMS during the reaction. For housekeeping gene β-actin DNA from 1, 10, 100, and 1000 A549 cells, the reaction mix (20 μl) includes 10 μl of 2× TaqMan Gene Expression Master Mix, 2 μl of downstream-primer, 2 μl of upstream-primer, 1 μl of probe, 1 μl of TWEEN-20 (0.1%), and 4 μl of template (cDNA). The reaction mix was then performed on the digital PCR reaction using the SPC digital PCR chip. The thermocycling procedure included a 2 min heating step at 50 °C, 10 min hot start at 95 °C; and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min.
We then demonstrated the measurements of single cell 18S rRNA gene using the SPC digital PCR chip. Due to the high abundance of 18S rRNA gene expression in single cells, the total number of target DNA template was much higher than the theoretical device capacity. cDNA sample of single A549 cell was serially diluted 10-fold and 100-fold. Then 4 μl of the dilution series of cDNA sample was mixed with 10 μl of 2× TaqMan Gene Expression Master Mix, 1 μl of 2× TaqMan Gene Expression Assay, 4 μl of nuclease-free water, and 1 μl of TWEEN-20 (0.1%). 8.5 μl of the reaction mix was loaded onto the chip to perform the digital PCR reaction. The thermocycling procedure included a 2 min heating step at 50 °C, 10 min hot start at 95 °C; and then 40 cycles of 95 °C for 15 s and 60 °C for 1 min.
Finally, we applied the SPC digital PCR chip to absolutely measure the lung cancer related genes PLAU expression. The reaction mix (20 μl) includes 10 μl of 2× TaqMan Gene Expression Master Mix, 1 μl of 2× TaqMan Gene Expression Assay, 4 μl of nuclease-free water, 1 μl of TWEEN-20 (0.1%) and 4 μl of template (cDNA). 8.5 μl of the reaction mix was performed on the digital PCR reaction using the microchip. Each experiment mentioned above was repeated three times. The thermocycling procedure included a 2 min heating step at 50 °C, 10 min hot start at 95 °C; and then 40 cycles of 95 °C for 15 s and 61 °C for 1 min.
E. Cell qPCR
Real-time qPCR is inadequate for single-cell gene expression measurement because of the limitation in sensitivity and precision. In order to demonstrate, that, as well as the capability of single-cell digital PCR analysis on the SPC digital PCR chip, the Applied Biosystems 7900HT system was used for all real-time PCR measurements of β-actin DNA from single, 10, 100, and 1000 A549 cells. The result was then compared to the digital PCR result.
F. Data acquisition and analysis
The chip after amplification was detected using Maestro Ex INVIVO Imaging System (CRI Maestro). Fluorescence images were acquired by using a large area CCD system. An enlarged image can be observed by using the fluorescence microscope (OLYMPUS). The fluorescence was excited at 455 nm and the emitted light was accepted by the CCD through a 520 nm long-pass filter. The ROX as the reference fluorescence was detected at the same time.
III. RESULTS AND DISCUSSION
The self-priming compartmentalization (SPC) chip which is capable of performing single cell digital PCR is shown in Fig. 1(a). The chip contains 4 separate panels, and each panel contains 1280 independent 5.625 nl microchambers, which is capable of detecting 4 cells simultaneously. The structure of the SPC chip is shown in Fig. 1(b), the chip is composed of two layers of the Poly(dimethylsiloxane) (PDMS) bonded to a glass coverslip and one nano-waterproof layer preventing the water evaporation. It is important to prevent the water evaporation during digital PCR reaction in PDMS devices, the nano-waterproof layer made of a low-permeability fluorosilane polymer was embedded ∼50 μm above the microwell array to create an effective permeation barrier, preventing the water from evaporation. The amount of lost water is about 2.88%, which can ensure the performance of digital PCR successfully. The details of the structures of the chip are shown in Fig. 1(c). Each panel of the chip consists of 8 branching rectangular main channel with rectangular microwells. To avoid the cross-reaction among them, microwells are evenly distributed along both sides of the channel in a staggered manner; Figure S1 in the supplementary material shows the principle of the SPC chip. For the single cell gene expression analysis, the SPC chip has its advantages. First, the self-priming ability of the chip can ensure a sample introduction without any loss. After the solution was aspirated to the tip of pipette, the tip was inserted into the inlet of the chip, and the solution can be sucked into the chip totally without any loss. Second, the viscous oil phase can slow down the speed of the solution introduction, which can provide enough time to realize the compartmentalization of the sample. Third, the diameter of the suction chamber of the chip was decreased compared to the previous chip, reducing the volume of waste. This chip was used to realize 85% of the sample digitization. For each panel of the chip, a 7.2 μl of 8.5 μl of reaction mixture was introduced into the chip to realize compartmentalization, and only 1.3 μl was sucked into the suction chamber.
The procedure of the single cell gene expression digital PCR analysis is shown in Fig. 1(d). Single cell can be picked up by a capillary tube from single-cell suspension and put into a tube which contains the lysis solution. After cell lysis, stop solution was added and then the reverse transcription reagent was added. The tube was then put on a PCR machine for reverse transcription. From cell lysis to reverse transcription, only one tube was used for each single cell by optimizing the protocol, which not only ensured the efficiency of the reverse transcription, but also avoided the DNA loss. When the reverse transcription was accomplished, the reaction product was mixed with the PCR reagent, and the mixture was then dispensed into the inlet of the SPC chip. After the sample was totally sucked into the chip, the oil was added in the injection port and was sucked into the chip following the sample solution. After the sample was isolated by the oil, the chip was sealed by the previously prepared PDMS, and then the chip was placed on the PCR machine to carry out PCR; Figure S2 in the supplementary material shows the operation procedure of the SPC chip. The result was shown in Fig. 1(d), green dots represent the positive reaction, and red dots represent the negative reaction, by counting the number of the positive dots, the copies number of gene can be calculated.
The theoretical dynamic range of the SPC digital PCR chip is related with the number of the chambers of the chip, which is given by the concentration of sample beyond which the array becomes saturated completely. The number of the SPC chip for each panel is 1280, and the total number of the whole chip with four panels is 5120. So, for one panel, the dynamic range can cover more than 3 orders of magnitude, whereas the theoretical upper limit is approximately 9100 target copies. If you use the whole chip with 4 panels to detect one sample, the dynamic range can cover more than 4 orders of magnitude, whereas the theoretical upper limit is approximately 43 700 target copies (Figure S3 in the supplementary material). In order to increase the measurement precision, sensitivity, and dynamic range of the SPC chip, the higher density chip should be developed. As we all know, soft lithography was capable of reproducing smaller scale features, but the microwells having minimum dimensions of less than ∼2 μm are prone to collapse, likely due to elastic structure effects. When the devices are degassed in a vacuum, the microwells are even more prone to collapse. Thus, it is estimated that the chamber which is smaller than the size of 10 × 10 × 10 μm is not appropriate to be fabricated using PDMS. Based on our protocol, a chip with an array of dimensions of 50 × 50 × 50 μm was easy to develop, corresponding to 0.125 nl with a density of 10 000 cm−2, and a scale of approximately 100 000 reactions per device. We think that the number of microwells is enough for the most applications.
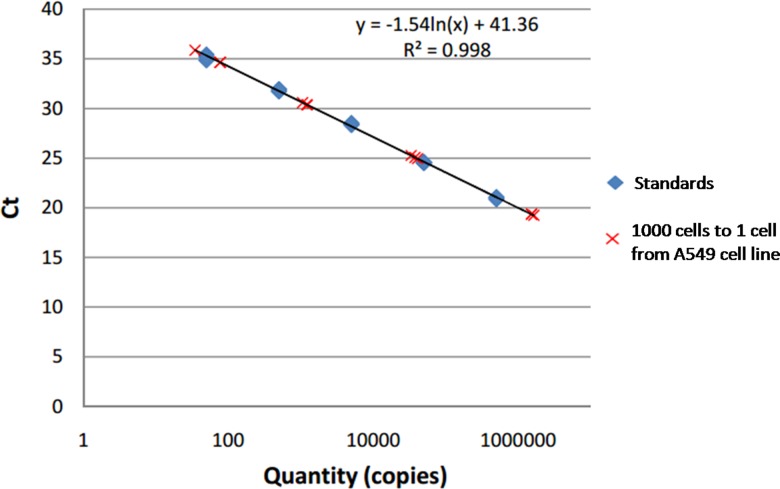
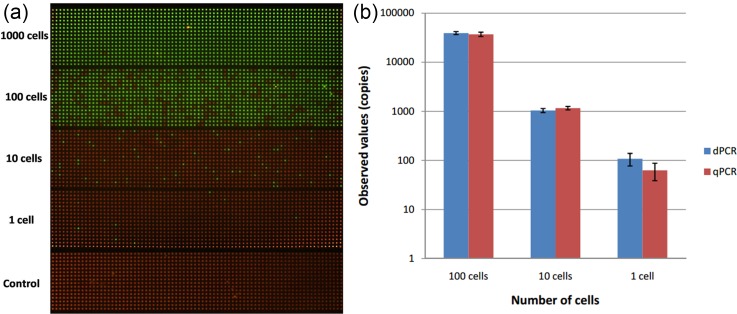
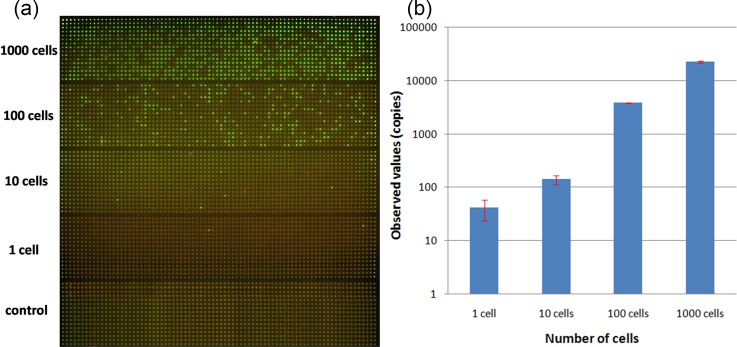
We first tested the ability, precision, and sensitivity of our SPC digital PCR chip by assessing β-actin gene expression in 1, 10, 100, and 1000 cells. Before we directly carried out the on-chip digital PCR measurement, we performed a real-time qPCR of the same reaction components using the 7900HT system (Applied Biosystems). According to the result shown in Fig. 2, cycle threshold (CT) was linearly proportional to ln(copies), which meant that β-actin DNA had a stable expression in A549 cells. So as the housekeeping gene with stable expression in a single cell, β-actin DNA is suitable for the validation of the on-chip single-cell digital PCR. We then carried out the digital PCR on the SPC chip by detecting different numbers of cells. For the same cell concentration, the experiment was repeated three times. The result was taken by the Maestro Ex IN-VIVO Imaging System and showed in Fig. 3(a). We calculated the number of positive microwells of each cell concentration and applied the mathematical correction of the Poisson distribution according to our previously reported SPC digital PCR chip. The 1000 cells sample has so high target gene expression that exceeds the maximum theoretical dynamic range of the SPC digital PCR chip, these results are not used to calculate.
FIG. 2.
Real-time PCR measurement results of β-actin gene with a serial of A549 cells ranging from 1000 cells to 1 cell.
FIG. 3.
Digital PCR and qPCR measurement results of β-actin gene on the different numbers of A549 cells. (a) Digital PCR fluorescence images of β-actin gene of different numbers of cells ranging from 1000 cells to 1 cell and no-template control. At high cell concentrations (1000 cells), target DNA template exceeded the maximum theoretical dynamic range of the SPC digital PCR chip. There is no positive signal in control when no target DNA. (b) Comparison of quantitative results of copy number for β-actin gene from 1000 cells to 1 cell. Bars represent the standard deviation of measurement.
Compared with the results of qPCR, chip-based digital PCR showed a similar quantitative result and error when the sample was 10 and 100 cells, as shown in Fig. 3(b). However, when came to single cell, the digital PCR yielded higher results than qPCR, and the error of digital PCR measurement was much lower than qPCR. The main reason was that the reaction volume of tube-based qPCR was 20 μl while the final reaction volume of digital PCR performed was 5.625 nl (volume of microwells), when β-actin gene expression in a single cell was near the limitation of qPCR precision, the reduced reaction volume of the microchip increased the targeting gene concentration and thus improved the amplification efficiency and brought down errors. Although been considered the gold standard for gene expression analysis, real-time qPCR are thus sometimes inadequate for applications on analysis of single-cell gene expression due to its limited precision (∼20%) and presents difficulties in reliably detecting low-copy-number templates in large volume owing to nonspecific amplification and competitive side reactions. So the SPC digital PCR chip offered the higher quantitative precision and increased detection sensitivity for single cell gene expression measurement.
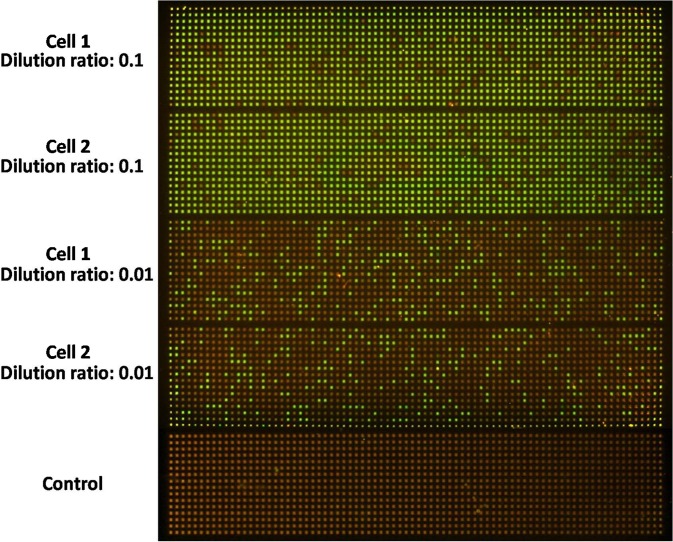
Next, we demonstrated that the SPC digital PCR chip can be accurately applied to gene expression quantification in single cells and evaluated the reproducibility of the SPC chip for single cell gene expression analysis. We applied the SPC digital PCR chip to determine the abundance of 18S rRNA genes in a single A549 cell (Fig. 4). As a housekeeping gene, 18S rRNA gene expression has a high level in single cell and is far beyond the capacity of the SPC digital PCR chip if directly performed digital PCR measurement. We made a tenfold serial dilution of RT product of single A549 cell and used the diluted RT product to perform on-chip digital PCR. The result showed that even diluted 10 times the target template still had a high concentration and when diluted 100 times, the number of positive microwells decreased. By applying the mathematical correction of the Poisson distribution, we found out that there was a linear relationship between the number of positive microwells and sample concentration. Two individual single cell digital PCR results of 18S rRNA on one chip was shown in Fig. 4, both of them have a very close gene copies of 18S rRNA with 1.6%–4.6% of coefficient of variation, and the copy number of 18S rRNA in a single A549 cell was (0.5 ± 0.026) × 106 copies, indicating that the SPC digital PCR chip can offer a consistent and robust result for single cell gene expression measurement.
FIG. 4.
Single cell digital PCR results of 18S rRNA gene expression with a serial dilution ranging from 10 to 100 dilutions on the SPC chip. The results show us that both of them have very closed gene copies of 18S rRNA, indicating the SPC digital PCR chip can offer a consistent and robust result for single cell gene expression measurement.
For single cell gene expression analysis, it is necessary to analyze a couple of genes at the same time. So, the multiplexing capability of the chip was required. One way to achieve better cost efficiency of the SPC chip is to use it in a multiplexing strategy, in other words, that is multiplex digital PCR. To achieve this, first, an expensive detection instrument should be required, which includes different kinds of fluorescence filters to detect multiplex reactions. Second, it is necessary to optimize the primer design, ensuring that different genes can be amplified in one microwell at the same temperature (annealing temperature and amplification efficiency) without primer cross-talk. Third, different kinds of probes labeled with different color fluorophores are used, which will increase the cost extremely. Another way in our strategy is that different panels in the SPC chip can be used to detect different genes at the same time only using the same color probe or SYBR Green, which is much cheaper than the way above mentioned. After single cell lysis and reverse transcription, 20 μl of cDNA for single cell can be split into four panels of each chip, and four different genes of single cell can be detected at the same time. Based on the size of each chip, four chips can be run on one PCR machine block. If four chips were used to detect single cell gene expressions, fourteen genes can be detected at the same time with low-cost, and user-friendly.
Last, we applied the SPC digital chip to detect lung cancer related genes PLAU gene expression of single A549 cell (Fig. 5(a)). PLAU is a gene maker for lung cancer, which has a different expression level in individual cell. In order to demonstrate the single cell heterogeneity, different numbers of cells were analyzed by the SPC chip. The single cell digital PCR results can reflect the PLAU gene expression in a different single cell individually, when we test the multiple single cells, the PLAU gene expression level is (41 ± 18) copies per cell. The multiple cells digital PCR results can give us the average PLAU gene expression level, which is (22 ± 1) copies per cell on average. From the results, some single cell gene expression is close to the population-averaged level, while some single cell gene expression level is three times than the population-averaged result. The difference of measurement results of the PLAU gene expression among the different numbers of cells indicated the single cell heterogeneity. From the corrected digital PCR results (Fig. 5(b)), we observed a large variation among the individual cells. We also noted that the number of cells and the number of target DNA template did not agree with linear relationship. Because PLAU is related to lung cancer, these two findings corresponded with the known cell-to-cell heterogeneity. In order to know the biological noise, the function of individual cell and the interaction of different single cell, single cell gene expression should be detected, respectively. However, the bulk level detection usually gives population-averaged results, obscuring the cell-to-cell heterogeneity. So, the SPC chip can absolutely count the gene expression number, indicating the difference of single cell gene expression.
FIG. 5.
Single cell digital PCR results on the SPC chip. (a) Digital PCR image of lung cancer related genes PLAU gene expression of different numbers of A549 cells from 1000 to 1 cell. (b) Statistical results of digital PCR of PLAU gene expression of different numbers of cells. Bars represent the standard deviation of measurement.
IV. CONCLUSIONS
Single cell gene expression analysis is important to understand the cellular activity and heterogeneity. As an absolute molecule counting method, digital PCR has been widely used in many bioresearch fields and has played a significant role at the cutting-edge technologies as well as single cell analysis. However, there are still many challenges using digital PCR to quantify the single cell gene expression absolutely. Many detection variations existed in terms of different methods including cell lysis, reverse transcription, PCR, and other steps. Besides, RNA is not easy to perform involving degradation, contamination, efficient reverse transcription, and so on. So, the digital PCR device should be very stable and robust for single cell analysis.
In this work, a single cell digital PCR method using the self-priming compartmentalization chip was demonstrated. We use the housekeeping genes (β-actin and 18S rRNA) to assess the ability, precision, and sensitivity of the SPC chip for single cell gene expression analysis. The chip can be used to digitize a sample up to 85% sample digitization. Compared to qPCR, the SPC chip can offer the higher quantification precision, increased detection sensitivity, and reproducibility for single cell gene expression measurement. By detecting lung cancer related genes PLAU gene expression of A549 cells at the single cell level, the single cell heterogeneity was demonstrated. So the SPC chip provides a new tool for single cell analysis. We envision that this chip will enable new studies including bacteria or virus detection, circulating tumour cell (CTC) and cancer stem cell.
V. SUPPLEMENTARY MATERIAL
See supplementary material for the principle and operation procedure of the self-priming compartmentalization chip.
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 31400728 and 31270907), China Postdoctoral Science Foundation (Grant No. 2014M561749), National Key Foundation for Exploring Scientific Instruments (Grant No. 2013YQ470781), and the Autonomous Research Project of State Key Laboratory of Industrial Control Technology, China (Grant No. 1501).
References
- 1. Le Gac S. and van den Berg A., Trends Biotechnol. (2), 55–62 (2010). 10.1016/j.tibtech.2009.10.005 [DOI] [PubMed] [Google Scholar]
- 2. Kalisky T. and Quake S. R., Nat. Methods (4), 311–314 (2011). 10.1038/nmeth0411-311 [DOI] [PubMed] [Google Scholar]
- 3. Sanchez-Freire V., Ebert A. D., Kalisky T., Quake S. R., and Wu J. C., Nat. Protoc. (5), 829–838 (2012). 10.1038/nprot.2012.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Taniguchi K., Kajiyama T., and Kambara H., Nat. Methods (7), 503–U550 (2009). 10.1038/nmeth.1338 [DOI] [PubMed] [Google Scholar]
- 5. Shintaku H., Nishikii H., Marshall L. A., Kotera H., and Santiago J. G., Anal. Chem. (4), 1953–1957 (2014). 10.1021/ac4040218 [DOI] [PubMed] [Google Scholar]
- 6. Eberwine J., Sul J. Y., Bartfai T., and Kim J., Nat. Methods (1), 25–27 (2014). 10.1038/nmeth.2769 [DOI] [PubMed] [Google Scholar]
- 7. Lasken R. S., Curr. Opin. Microbiol. (5), 510–516 (2007). 10.1016/j.mib.2007.08.005 [DOI] [PubMed] [Google Scholar]
- 8. Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B. B., Siddiqui A., Lao K., and Surani M. A., Nat. Methods (5), 377–382 (2009). 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]
- 9. Zong C., Lu S., Chapman A. R., and Xie X. S., Science (6114), 1622–1626 (2012). 10.1126/science.1229164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wheeler A. R., Throndset W. R., Whelan R. J., Leach A. M., Zare R. N., Liao Y. H., Farrell K., Manger I. D., and Daridon A., Anal. Chem. (14), 3581–3586 (2003). 10.1021/ac0340758 [DOI] [PubMed] [Google Scholar]
- 11. Toriello N. M., Douglas E. S., Thaitrong N., Hsiao S. C., Francis M. B., Bertozzi C. R., and Mathies R. A., Proc. Natl. Acad. Sci. U. S. A. (51), 20173–20178 (2008). 10.1073/pnas.0806355106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sims C. E. and Allbritton N. L., Lab Chip (4), 423–440 (2007). 10.1039/b615235j [DOI] [PubMed] [Google Scholar]
- 13. Li Y. Z., Thompson H., Hemphill C., Hong F., Forrester J., Johnson R. H., Zhang W. W., and Meldrum D. R., Anal. Bioanal. Chem. (5), 1853–1859 (2010). 10.1007/s00216-010-3754-0 [DOI] [PubMed] [Google Scholar]
- 14. Zare R. N. and Kim S., Annu. Rev. Biomed. Eng. , 187–201 (2010). 10.1146/annurev-bioeng-070909-105238 [DOI] [PubMed] [Google Scholar]
- 15. Zhong J. F., Chen Y., Marcus J. S., Scherer A., Quake S. R., Taylor C. R., and Weiner L. P., Lab Chip (1), 68–74 (2008). 10.1039/B712116D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marcus J. S., Anderson W. F., and Quake S. R., Anal. Chem. (9), 3084–3089 (2006). 10.1021/ac0519460 [DOI] [PubMed] [Google Scholar]
- 17. White A. K., Van Insberghe M., Petriv O. I., Hamidi M., Sikorski D., Marra M. A., Piret J., Aparicio S., and Hansen C. L., Proc. Natl. Acad. Sci. U. S. A. (34), 13999–14004 (2011). 10.1073/pnas.1019446108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gong Y. A., Ogunniyi A. O., and Love J. C., Lab Chip (18), 2334–2337 (2010). 10.1039/c004847j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lecault V., White A. K., Singhal A., and Hansen C. L., Curr. Opin. Chem. Biol. (3–4), 381–390 (2012). 10.1016/j.cbpa.2012.03.022 [DOI] [PubMed] [Google Scholar]
- 20. Vogelstein B. and Kinzler K. W., Proc. Natl. Acad. Sci. U. S. A. (16), 9236–9241 (1999). 10.1073/pnas.96.16.9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pohl G. and Shih L. M., Expert Rev. Mol. Diagn. (1), 41–47 (2004). 10.1586/14737159.4.1.41 [DOI] [PubMed] [Google Scholar]
- 22. Heyries K. A., Tropini C., Vaninsberghe M., Doolin C., Petriv O. I., Singhal A., Leung K., Hughesman C. B., and Hansen C. L., Nat. Methods (8), 649–651 (2011). 10.1038/nmeth.1640 [DOI] [PubMed] [Google Scholar]
- 23. Baker M., Nat. Methods (6), 541–544 (2012). 10.1038/nmeth.2027 [DOI] [Google Scholar]
- 24. Warren L., Bryder D., Weissman I. L., and Quake S. R., Proc. Natl. Acad. Sci. U. S. A. (47), 17807–17812 (2006). 10.1073/pnas.0608512103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. White A. K., Heyries K. A., Doolin C., Vaninsberghe M., and Hansen C. L., Anal. Chem. (15), 7182–7190 (2013). 10.1021/ac400896j [DOI] [PubMed] [Google Scholar]
- 26. Thompson A. M., Gansen A., Paguirigan A. L., Kreutz J. E., Radich J. P., and Chiu D. T., Anal. Chem. (24), 12308–12314 (2014). 10.1021/ac5035924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu Q. Y., Gao Y. B., Yu B. W., Ren H., Qiu L., Han S. H., Jin W., Jin Q. H., and Mu Y., Lab Chip (22), 4755–4763 (2012). 10.1039/c2lc40774d [DOI] [PubMed] [Google Scholar]
- 28. Zhu Q. Y., Qiu L., Yu B. W., Xu Y. N., Gao Y. B., Pan T. T., Tian Q. C., Song Q., Jin W., Jin Q. H., and Mu Y., Lab Chip (6), 1176–1185 (2014). 10.1039/C3LC51327K [DOI] [PubMed] [Google Scholar]
- 29. Hosokawa K., Omata M., Sato K., and Maeda M., Lab Chip (2), 236–241 (2006). 10.1039/B513424B [DOI] [PubMed] [Google Scholar]
- 30. Dimov I. K., Basabe-Desmonts L., Garcia-Cordero J. L., Ross B. M., Park Y., Ricco A. J., and Lee L. P., Lab Chip (5), 845–850 (2011). 10.1039/C0LC00403K [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the principle and operation procedure of the self-priming compartmentalization chip.