Abstract
This paper reports on the use of a digital microfluidic platform to perform multiplex automated genetic engineering (MAGE) cycles on droplets containing Escherichia coli cells. Bioactivated magnetic beads were employed for cell binding, washing, and media exchange in the preparation of electrocompetent cells in the electrowetting-on-dieletric (EWoD) platform. On-cartridge electroporation was used to deliver oligonucleotides into the cells. In addition to the optimization of a magnetic bead-based benchtop protocol for generating and transforming electrocompetent E. coli cells, we report on the implementation of this protocol in a fully automated digital microfluidic platform. Bead-based media exchange and electroporation pulse conditions were optimized on benchtop for transformation frequency to provide initial parameters for microfluidic device trials. Benchtop experiments comparing electrotransformation of free and bead-bound cells are presented. Our results suggest that dielectric shielding intrinsic to bead-bound cells significantly reduces electroporation field exposure efficiency. However, high transformation frequency can be maintained in the presence of magnetic beads through the application of more intense electroporation pulses. As a proof of concept, MAGE cycles were successfully performed on a commercial EWoD cartridge using variations of the optimal magnetic bead-based preparation procedure and pulse conditions determined by the benchtop results. Transformation frequencies up to 22% were achieved on benchtop; this frequency was matched within 1% (21%) by MAGE cycles on the microfluidic device. However, typical frequencies on the device remain lower, averaging 9% with a standard deviation of 9%. The presented results demonstrate the potential of digital microfluidics to perform complex and automated genetic engineering protocols.
I. INTRODUCTION
Large-scale genome editing is currently a time consuming and labour-intensive process executed manually, in most cases, at the benchtop by a laboratory technician. Until now, the integration of convenient fluid handling and gene transfer technologies has been a major barrier to full automation of the genome engineering process, required important investment, and a large floor space. The marriage of digital microfluidics and electroporation hardware offers a scalable device architecture that overcomes the technological barriers to process automation. The use of digital microfluidics holds the promise of improving common laboratory protocols, by reducing reagent volumes, increasing fluid handling precision, enabling programmable sample manipulation, and allowing for simple sharing of software protocols for genome engineering between laboratories.1,2 This paper describes the development of a protocol for multiplex automated genome engineering (MAGE) of Escherichia coli, which can be performed by a droplet-based microfluidic platform.
Wang et al. introduced the MAGE method for genome manipulation, allowing rapid, genome-scale modification of a microbial population. MAGE was used to improve by over five-fold the production of lycopene in an E. coli strain, within three days.3 The MAGE protocol developed by Wang et al. serves as the foundation for the digital microfluidic protocol described in this present communication. A MAGE cycle consists of the following steps:3
-
1.
An E. coli strain (with the ΔmutS λ-Red+ genotype, such as EcNR2) is grown to mid-log phase.
-
2.
The cell population is brought to 42 °C for 15 min to induce the production of the λ-Red recombination proteins Exo, Betaβ, and Gam. The protein Betaβ will bind to ssDNA and mediates annealing of ssDNA to complementary strands during DNA replication.4–6
-
3.
Next, cells are cooled to 4 °C to prevent loss of cell viability.
-
4.
Cells are washed in a non-ionic medium to make them electrocompetent.
-
5.
ssDNA, in the form of 90 nucleotides long oligodeoxynucleotides (ODNs) are introduced and mixed into the cell sample.
-
6.
Cells are electroporated with a high electric field strength pulse (18 kV/cm, with RC time constant of approximately 5 ms).
-
7.
The cells are then added to a growth medium to recover and grow.
To automate the process, Wang et al. built a bench-scale robotic system to automate the fluid handling, cell growth, and electroporation steps required for the MAGE process. The MAGE process has been further developed and combined with other techniques to provide single to megabase genome reengineering, simultaneous modification of many separate sites on the genome, and expanded biological function.7–9 While the result demonstrated a vast improvement in the time required for large-scale genome editing, the solution comprised a cumbersome robotic assembly of conventional instruments that was prone to error and difficult to operate. The recent addition of electroporation capability to the digital microfluidic toolset represents a step toward optimization of the MAGE process that aims to further streamline large-scale genome engineering.10 Our work focuses on adapting benchtop media exchange and electroporation methods to the digital microfluidics format for implementing MAGE. The iterative nature of the MAGE cyclic process essentially dictates the complexity of the modifications that can rapidly be performed. The optimization of the lycopene pathway involved the combination of 24 oligo and 35 MAGE cycles, before screening a variant with 5-fold improvement over the previous strain. An automated, affordable, and scalable platform would facilitate the design and execution of complex protocols requiring multiple rounds of induction, transformation, and selection. It would also expedite testing multiple protocols in parallel, which is an integral part of any genome engineering and optimization project. With the decreasing cost of oligo synthesis, strategies involving large libraries of DNA constructs to be combinatorial incorporated into the genome will be more easily assayed in an automated and low cost platform. Beyond MAGE, once completed and optimized, our electrowetting-on-dielectric (EWoD) platform would expedite complex and large scale projects, such as whole genome recoding,11 which involve multiple steps of large scale assemblies, genome integrations, and optimizations, which are best done in parallel and then combined in an iterative process. The device and methods presented in this paper should be suitable for a variety of non-MAGE electroporation applications as well, including CRISPR/Cas9 based genome editing.
The large variety of programmable fluid handling operations that can be performed on electrowetting on dielectric (EWoD) digital microfluidic platforms make these systems attractive for a wide array of lab-on-a-chip (LoC) applications.12 Additionally, digital microfluidic devices, which use oil to surround sample droplets, are compatible with biological samples and boast minimal contamination of the electrowetting surface.13,14 This attribute enables reliable fluid handling, preparation, and analysis of biological samples. Further, digital microfluidic devices have been used for polymerase chain reactions (PCR),15,16 DNA sequencing,17 assays for clinical diagnosis18 (cell assays,19 enzyme assays,20–22 and immunoassays23–25), SPRi detection of DNA hybridization,26 micro RNA analysis,27 cell studies,28 protein analysis,29 and chemical separation.15–30
Our co-authors first presented on the use of a commercial platform (Advanced Liquid Logic, Inc.) to perform successful electrotransformation of E. coli on an EWoD digital microfluidic platform10 and characterized the impact of integrated electroporation devices to fluid transport in the EWoD format.10,31 Sandahl et al. also demonstrated that cell samples remain viable on the EWoD cartridge through 90 cycles (27 days) of dilution and re-growth (mimicking MAGE cycles) on the device. These results revealed the promise of the EWoD platform for continuous large-scale genome editing of a cell population. The present work offers an optimized protocol on the same platform with potential for high transformation frequencies, using an inexpensive, commercially available bioactivated bead system.
While digital microfluidic devices have been used to deliver electroporation pulses to cells,10,32 the goal of the present work is to uncover optimal conditions for automating sample preparation and pulse delivery. Experiments were performed to find suitable lectin coatings and bead blocking agents required for employing lectin-activated magnetic beads in complex cell handling procedures on the digital microfluidic platform. Use of magnetic beads enabled cell isolation from growth medium, and ultimately facilitated media exchange, a critical step for generating electrocompetent cells. Benchtop experiments were employed to optimize the electric field strength of pulses for transformation frequency, explore the effects of the temperature of samples during the electroporation pulse, and to develop a cell preparation procedure that can be automated on the digital microfluidic platform. The digital microfluidic device used in this study employs an array of electrodes which, when actuated, transport droplets of precise fluid volumes (multiples of 350 nl) through electrowetting. Finally, using the results of the benchtop optimization and magnetic bead experiments, trial MAGE cycles were performed on the digital microfluidic platform.
II. MATERIALS AND METHODS
A. Bead coating and binding
Conventional benchtop preparation of electrocompetent cells for high frequency electrotransformation involves suspension of cells in de-ionized (DI) water often with glycerol or sucrose.33 Washes and media exchange are performed by centrifugation of cell samples in growth medium, followed by removal of the supernatant and suspension of the cell pellet in a non-ionic medium. However, separation by centrifugation is not a technique that scales into the digital microfluidic format. Thus, our investigation aims at improving magnetic bead-based capture and retention of E. coli cells, which are imperative for on-cartridge MAGE operation specifically and LoC-based protocol development in synthetic biology in general. Bioactivated magnetic beads have been used to separate, wash, and concentrate biological materials where centrifugation is impossible, such as on a microfluidic platform.10,24 On our platform, magnetic beads and an externally actuated magnet were used to pellet cells within droplets for washing and media exchange, which is critical to preventing arc discharge during electroporation pulse delivery and therefore key to high-efficiency electro-transformation.
Various lectins (Vector Lab's lectin screening kits) were tested for promotion of binding between magnetic beads (i.e., Thermo Fisher Scientific's Dynabeads® MyOneTM Streptavidin C1 superparamagnetic beads of 1 μm diameter) and E. coli. Beads were first washed two times in phosphate buffered saline (PBS). Washes included 200 μl of the stock concentration of beads (10 mg/ml) being pelleted by a permanent magnet. Once out of suspension, the supernatant was removed and beads were suspended into 400 μl of PBS. After two washes in 400 μl of PBS, beads were suspended into 750 μl of PBS. A volume of 25 μl of the lectin of interest (at a stock concentration of 2 mg/ml, from Vector Labs) was added to the 750 μl bead suspension and slowly rotated for 30 min. The beads were then washed three times with 500 μl of the defined growth medium of choice (e.g., MOPS EZ supplemented with biotin, Teknova). Finally, the beads were suspended into 200 μl of the binding medium and stored at 4 °C. This procedure efficiently coupled the bead to the lectin of choice.
To test cell capture by the beads, EcNR2 cultures were grown to mid-log phase (OD600 = 4.5–6.0), and 1 ml samples were subsequently centrifuged to pellet cells. The supernatant was removed and replaced with an equal volume of the defined growth medium of choice, such as MOPS EZ supplemented with biotin (Teknova), which do not contain cell-wall sugars from the “yeast extract” present in complex growth media such as Lysogeny Broth (LB), which are likely to interact with the lectins. While LB is more commonly used as a growth medium for electroporation experiments, it was found to inhibit binding between lectin beads and E. coli (data not shown). Once in the binding solution, 100 μl of coated beads were added to the cell suspension, providing a ratio of approximately one bead per cell. The mixture was put on a rotator to bind for 15 min. After binding, the efficiency of cell capture was determined, as described below.
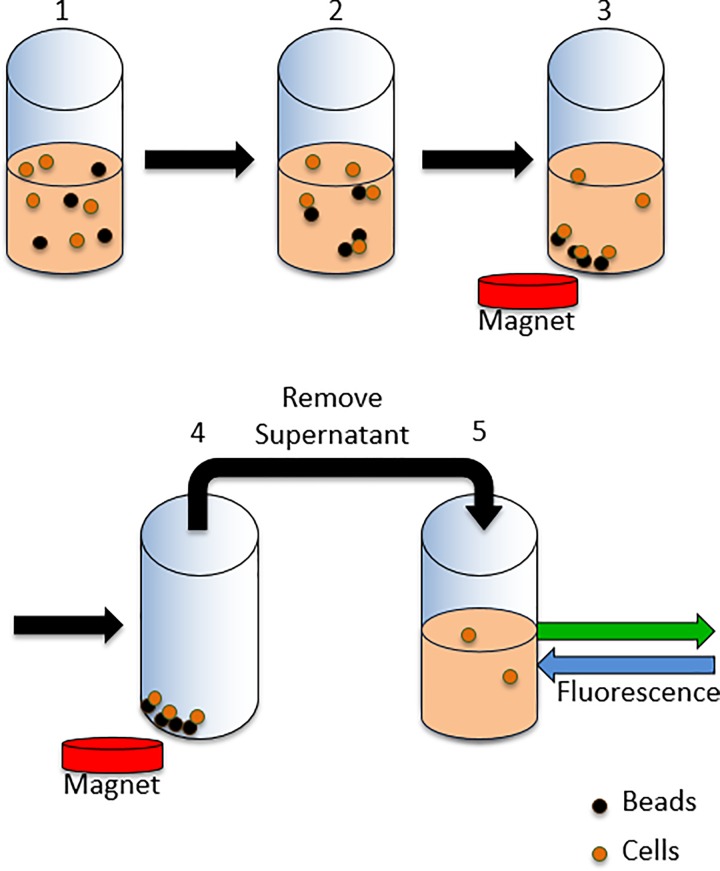
To facilitate detection, we had transformed EcNR2 with a plasmid producing the super-folder green fluorescent protein (GFP),34 allowing fluorescence measurements to determine relative cell numbers in samples. Figure 1 illustrates the steps of the assay to determine the binding efficiency of each bead coating: (1) binding of cells to beads, (2) application of permanent magnet to pull bead-bound cells out of suspension, and (3) the removal and fluorescence measurement of the supernatant. By comparing the fluorescence of the supernatant (FS) to the fluorescence of the initial mid-log phase growth to which the beads were added (FM), the percentage of cells bound to beads was determined using the following equation:
| (1) |
Cell retention of the beads after washing was determined by plating both the mid-log phase cell growth and a sample that was prepared for pulsing. Comparing colony counts of these two samples using Equation (2) yields the proportion of original cells retained by the beads following the entire cell preparation protocol
| (2) |
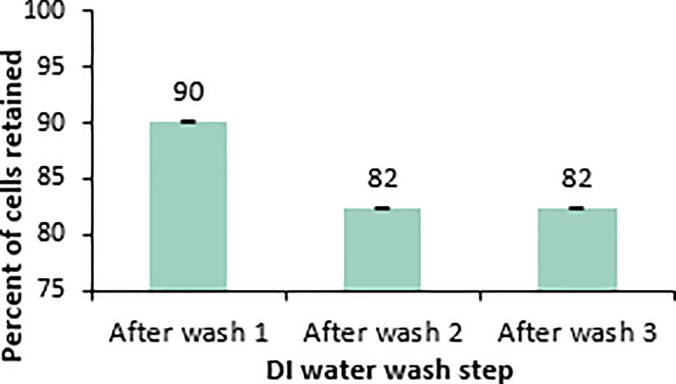
where CFUP and CFUM represent the colony forming units of the post preparation sample and the mid-log phase growth, respectively. Fluorescence was not used in this case, since washing of cells in DI water was found to reduce the fluorescent signal of the cells. The fluorescent signal can be recovered through the addition of small amounts of growth medium (data not shown), so there is no indication of loss of GFP, but rather just loss of the signal. Of all the tested lectins and antibodies, Wheat Germ Agglutinin (WGA) lectin was found to be the most effective at mediating binding between the magnetic beads and E. coli. WGA beads were able to capture over 95% of the cell population and retain around 15% following preparation for electrocompetence. For this reason, WGA beads were used during all experiments involving bead bound cells presented in this paper. While typical cell washing involves resuspension of cells after each wash step, it was found that cell retention of the beads was higher when bound cells remained in a pellet throughout the washing. Figure 2 shows the percentage of the mid-log phase cells that are retained following each of three gentle DI water washes. This method produced adequately electrocompetent cells. While 82% of cells are retained following three gentle DI water washes, only around 15% of the cells are retained when the sample goes through the blocking step and heat shock required for a MAGE cycle. The loss of cells from beads, before the electroporation pulse step, will limit the number of variants generated when many genome edits are attempted per MAGE cycle. However, for applications involving one or two genome edits per cycle, the transformation frequency is the important parameter. In these cases, 15% retention is sufficient, and can be compensated by longer cell growth doubling times between MAGE cycles.
FIG. 1.
Illustration of assay for binding efficiency. Steps include (1) binding of cells to beads, (2) introducing a magnet to bring bound cells out of suspension, and (3) removal and recording fluorescence of supernatant.
FIG. 2.
Percentages of the original cell population retained following each DI water wash where bound cells remain pelleted during the media exchange.
In summary, the capability to manipulate magnetic beads and a robust method for cell attachment are essential components of cellular engineering on a microfluidic platform, enabling pelleting of cells and washing and media exchange analogous to centrifugation on the benchtop.
B. Bead dispersion
During preliminary incubation trials, clumps formed between beads and cells that were magnetically pulled out of suspension on the microfluidic platform. Bead clumping prevented re-suspension of bead-bound cells following magnetic pelleting, and shear forces of fluid flow induced by droplet transport alone were found to be insufficient for dispersion of bead pellets. Moreover, dielectric shielding resulting from close proximity of cells and beads to each other in these bead aggregates reduces the local electric field experienced by individual cells when pulsed. Since dielectric shielding of the electroporation field negatively affects transformation frequency, various proteins and sugars were tested for their ability to prevent bead clumping. It was hypothesized that blocking the bead binding sites with proteins or sugars may prevent bead-cell aggregation.
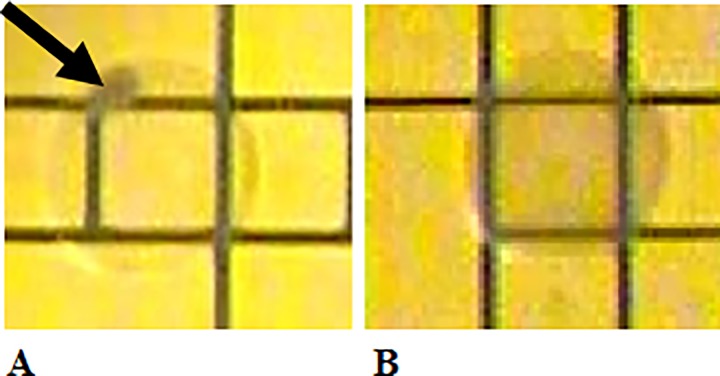
This hypothesis was tested by executing the bead binding protocol on the microfluidic platform with volumes scaled to match that of the microfluidic device. Before the bead-bound cells were pulled out of suspension, various concentrations of blocking agents were mixed with the bound cell samples and incubated for 1 or 3 min. Once mixed, the external permanent magnet was applied to pull the bound cells out of suspension. EWoD electrodes adjacent to the bead pellet were activated to pull the supernatant away from the bound cell pellet. A droplet of DI water with Tween 20 (0.05%) was transported over the bound-cell pellet and the magnet was removed. As the external magnet receded, the bound cells dispersed into the droplet of DI water containing Tween 20. The resulting droplet was then transported back and forth to re-suspend the bound cells. Successful bead blocking was determined visually by the absence of bead aggregates after 5 min of mixing. Visual indications of aggregation and dispersion are shown in Figures 3(a) and 3(b), respectively.
FIG. 3.
Image showing indications of (a) bead aggregation and (b) uniform bead dispersion.
Table I lists blocking agents tested and indicates whether bead aggregates were observed after either 1 or 3 min of mixing in the bound cell suspension. Each blocking agent was tested and observed four times (in a different lane of the microfluidic cartridge for each trial) for indications of bead aggregation. Blocking agents containing casein all successfully promoted dispersion of bound cells within 5 min of re-suspension and mixing. Fat-free powdered milk required the least amount of time to fully disperse bound cells. A concentration of 2.5 mg/ml fat free powdered milk was the lowest tested concentration able to fully disperse the bound cells. There was no observed difference in the rate of dispersion between the 2.5 mg/ml and higher concentrations of fat-free powdered milk. Accordingly, the fat-free powdered milk at 2.5 mg/ml was chosen for use in the pulse optimization experiments and microfluidic trials.
TABLE I.
Bead aggregation observations with tested blocking solutions. (NA: no visible aggregate and A: visible aggregate).
| Blocking agent | Concentration | Observation after 5 min of droplet mixing |
|---|---|---|
| Fat free powdered milk | 10 mg/ml | NA |
| 5 mg/ml | NA | |
| 2.5 mg/ml | NA | |
| 1.3 mg/ml | A | |
| Casein Hammarsten bovine | 2.5 mg/ml | NA |
| Casein | 2.5 mg/ml | NA |
| 10× casein blocking buffer | 2.5 mg/ml | NA |
| Bovine Serum Albumin (BSA) | 0.1 mg/ml | A |
| D mannose | 0.9 mg/ml | A |
| NAG | 2.2 mg/ml | A |
| 1.1 mg/ml | A | |
| Tween 20 | 1 mg/ml (0.10%) | A |
| 0.8 mg/ml (0.08%) | A | |
| D mannose + casein | 0.9 mg/ml mannose, 2.5 mg/ml casein | NA |
| D mannose + BSA | 0.9 mg/ml mannose, 0.1 mg/ml BSA | A |
C. Electrotransformation optimization
Along with automated preparation of electrocompetent cells, conditions affecting electrotransformation were optimized to maximize transformation frequency. Since MAGE is designed for multiple transformation cycles, high transformation frequency at each cycle is required for successful propagation of transformed fractions of the original cell population.
Since its inception, electrotransformation has developed into both an extensively studied and widely used method for genome editing. Cells subjected to high strength electric fields experience increased permeability35 through dielectric breakdown of the cell membrane.36 Pores form in the membrane at sites of dielectric breakdown, a phenomenon called electroporation. Subsequently, the genetic material enters the cell through the transient electropores.37 Adding to the popularity of the method, electroporation is a versatile technique for gene delivery as it is adaptable to many types of cells and genetic vectors.38
Dower et al. established the standard protocol for E. coli electrotransformation, upon which many E. coli electroporation studies have been based. The conventional procedures for E. coli electroporation include the following steps:33
-
1.
Cells are grown to mid-log phase in LB.
-
2.
Cells are washed in a non-ionic solution (commonly DI water, with or without glycerol).
-
3.
Cells are suspended in a non-ionic solution at ten times the concentration of the mid-log phase growth.
-
4.
Genetic material is mixed into the cell suspension.
-
5.
The cell suspension is placed between parallel electrodes in an electroporation cuvette and pulsed with a short, high strength electric field.
-
6.
Cells are then added to a larger volume of LB and incubated for recovery.
Numerous studies were dedicated to determining factors that affect transformation efficiency and frequency of E. coli electrotransformation. Transformation efficiency is a measure of the total number of successfully transformed colony forming units produced from the transformation procedure. Frequency, however, is the proportion of the surviving cells which have been successfully transformed.33,39–42 For MAGE, where a cell population goes through several transformations without selection, it is important to have high transformation frequency to minimize the rate of exponential decline in the proportion of the cell population which has been successfully transformed. Dower et al. showed that efficiency and frequency have a strong dependence upon electric field strength and pulse duration. In their study, cells were grown and recovered in LB, washed in a non-ionic medium, then pulsed while in the presence of plasmid DNA.33 With an exponentially decaying pulse of amplitude 12.5 kV/cm and RC time constant of 5 ms, the protocol yielded an efficiency of up to 1010 cfu/μg DNA and a frequency as high as 80%.33 Other factors shown to influence transformation efficiency and frequency included: DNA concentration, incubation time of cells with DNA, number of pulses, number of washes, current density, salt content, growth temperature, and time taken to add the recovery medium after pulse.33,39–42
Despite identification of several variables affecting transformation, the efficiency reached by Dower et al. remains to be exceeded. Prior to trials on the digital microfluidic platform, experiments for optimization of cell preparation steps and pulse conditions were carried out on benchtop with bead-bound cells and free cells. Electroporation experiments were performed on an EcNR2 bla-E. coli using an oligonucleotide from Integrated DNA Technologies to restore the bla gene, which encodes a beta-lactamase and imparts carbenicillin resistance to E. coli. Electric pulses were applied using a Bio-Rad MicroPulserTM Electroporator on 1 mm gap aluminum cuvettes (USA Scientific). The cell preparation protocol was designed such that electroporation could be performed using available functions on the digital microfluidic platform. The cell preparation and electroporation steps used in the electrotransformation optimization are summarized in Figure 4. For electroporation experiments, 0.05% Tween 20 was present in all solutions, as it facilitates electrowetting on the microfluidic platform. The presence of 0.05% Tween 20 does not affect E. coli growth.10 Variables studied for optimization of transformation frequency included: electric field strength of the pulse, type of growth/recovery medium, and temperature during the pulse. Transformation frequency was determined by plating the pulsed and recovered cell samples on both LB agar and LB with 100 μg/ml Carbenicillin plates. Transformation frequency was determined using the following equation:
| (3) |
where CFULB+Carb and CFULB are the colony forming units on the LB with carbenicillin and LB plates, respectively.
FIG. 4.
Protocol used for benchtop electroporation optimization experiments. All solutions contain 0.05% Tween 20 throughout the procedure.
Since the optimal electric field strength may change as variables in the electroporation protocol are altered, a range of electric field strengths were employed for each set of conditions tested. Varying the electric field allowed for the determination of optimal transformation frequencies for each set of test conditions, which was important for observing any effect that the magnetic beads impart to the electrotransformation process.
While bead-bound cells were prepared using the protocol outlined in Figure 4, free cell samples were prepared with slightly altered volumes in order to obtain large enough cell pellets during the wash steps. Free cells were prepared for electroporation on benchtop to compare the digital microfluidic protocol with conventional cuvette-based protocols. Cells were grown to mid-log phase. Eppendorf tubes, each containing 600 μl of the cell suspension, were centrifuged to pellet cells. The supernatant was removed and the cells were suspended into 600 μl of LB. A 15-min heat shock was conducted by submerging the Eppendorf tubes in a shaking heat bath held at 42 °C. Following the heat shock step, each sample was washed three times with DI water using a centrifuge. After washing, each sample was suspended into 120 μl of DI water. The 120 μl sample was then divided into four tubes of 30 μl each. Oligo (2 μl of 5 mM) was added to each sample and mixed for 30 s. After mixing, each sample was pulsed (electric field strengths ranged from 18 to 26 kV/cm) and immediately mixed with 1 ml of growth medium for 2.5 h of recovery growth at 30 °C. By comparing free cell electroporation to the bead bound cell protocol, any detrimental effects of the binding of cells to beads, blocking of bound cells, or washing with magnet pelleting could be determined.
Bound cells grown and recovered in defined growth medium were compared to free cells for transformation frequency. The protocol from Figure 4 was followed to produce bound electrocompetent cells. LB was also tested as a growth and recovery medium for bound cells, since it is a standard medium used in electroporation experiments. Bound cells were pulsed both at room temperature (∼20 °C) and 4 °C, over a range of voltages to determine the effects of temperature on transformation frequency. Due to the difficulty of controlling the temperature of the electroporation electrode (which was not placed on a controlled temperature region of the cartridge), room temperature electroporation was tested as a more consistent alternative to externally controlling the temperature.
D. Microfluidic platform
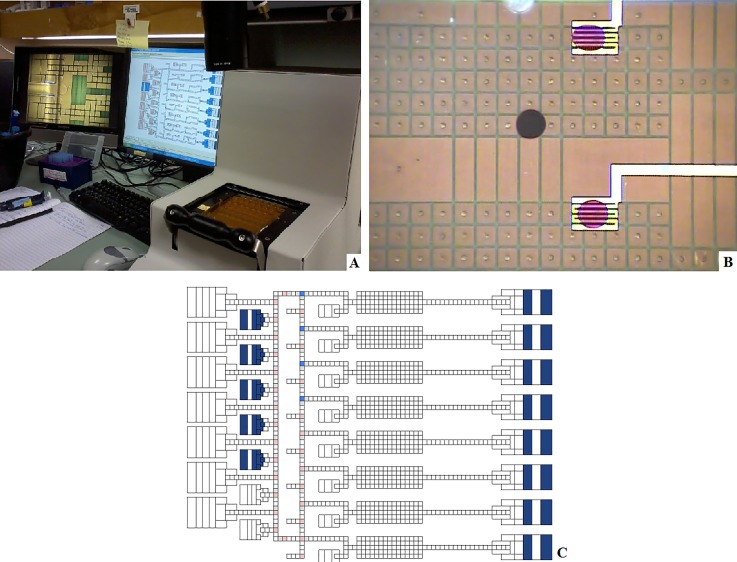
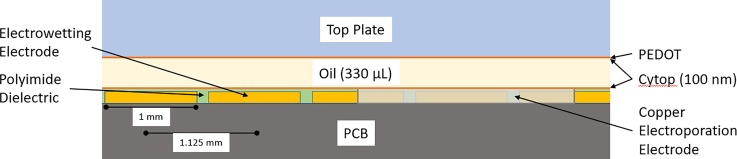
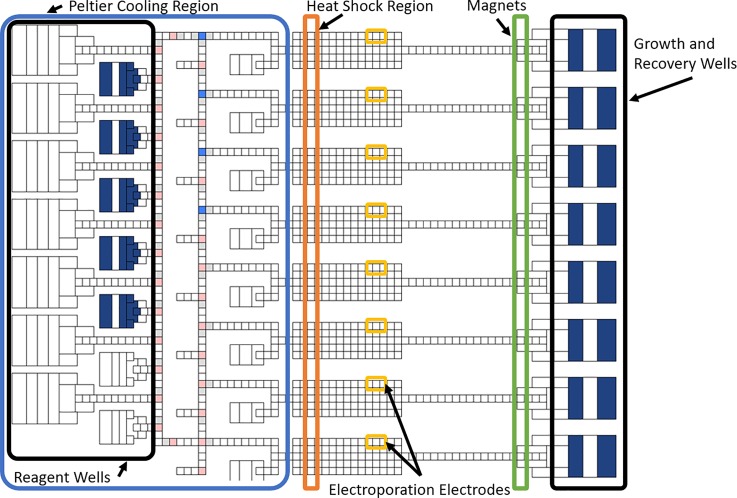
As shown in Figure 5(a), the digital microfluidic system consists of an EWoD cartridge, a controller, and a computer. The cartridge consists of a fluid chamber enclosed by an electrically grounded top plate with wells for fluid input and a bottom plate consisting of a pattern of electrodes positioned beneath a thin polyimide dielectric layer. The top plate contains a layer of Poly(3,4-ethylenedioxythiophene) (PEDOT), used as a ground electrode. Both top and bottom plates are coated with a thin hydrophobic surface coating (approximately 100 nm of Cytop). The chamber between plates is filled with a non-ionic silicone oil to promote electrowetting, prevent contamination, and prevent evaporation of aqueous phase droplets. The electrodes on the bottom plate are part of a printed circuit board (PCB) that connects the electrodes to external pins allowing actuation from the EWoD controller. This rigid PCB layer has an additional commercial flexible PCB layer containing a standard thin film polyimide laminate with bare copper electroporation electrodes attached to the bottom rigid PCB. The layers are displayed in the cross-section diagram of Figure 6, with thicknesses of and materials used for certain layers. Each cartridge has serpentine-shaped electroporation electrodes that extend outside of the fluid chamber and connect to a pulse generator for the delivery of electroporation pulses. The electroporation electrodes are shown amongst a grid of EWoD electrodes in Figure 5(b). The serpentine shape was chosen to provide a large area of direct contact between the electroporation electrode and droplet while minimizing electromagnetic shielding of the underlying electrowetting electrode (the serpentine electrode covers approximately 35% of the electrowetting electrodes in which the serpentine electrode is embedded. These serpentine electrodes span a 2.5 mm × 1.75 mm area, partially covering several of the electrowetting electrodes. Slower actuation frequencies are used to pull droplets onto and off of the serpentine electrodes, as less of the surface can be used for electrowetting. The controller can be programmed to perform actuation of cartridge electrodes through an applied AC voltage, movement of a permanent magnet towards or away from the cartridge, heating through heater bars below the cartridge, and Peltier cooling from below the cartridge. These features allow for sample control through electrowetting, magnetic bead immobilization, heating of three independent regions of the fluid chamber, and cooling of a large area of the fluid chamber. Fluid transport performed by the controller is programmed on a connected computer, showing simulated droplet movement as seen in Figure 5(c). The design of the electrodes in Figure 5(c) was originally for sample prep for sequencing applications. The flexible software allows for repurposing of the cartridge to prototype other applications, such as the MAGE application developed in the present work. Figure 7 shows the electrode design from the simulation software, with regions labelled per the purpose for the MAGE application. On the right side of Figure 7, there are 50 μl wells used for growth and recovery of cells. These wells each have an opening in the top plate in which the aqueous phase liquids can be pipetted into or out of the cartridge. The left side above the Peltier cooler are inlet wells for reagents such as washing solutions, and blocking agents. In the center region, there are zones for heating, magnetic pelleting, and electroporation.
FIG. 5.
(a) A cartridge installed in the controller, with connected computer and camera imaging monitor. (b) Dyed droplets sitting on top of serpentine electroporation electrodes. (c) Image of computer simulation of droplets moving on the cartridge.
FIG. 6.
Cross-section diagram displaying the layers making up the EWoD cartridge.
FIG. 7.
Diagram of electrode layout of the electrowetting cartridge, outlining regions per functionality for MAGE applications.
III. RESULTS AND DISCUSSION
Since optimized pulse parameters depend heavily on the cell preparation procedures, a preparation protocol must be determined before pulse optimization experiments are conducted. First, WGA was selected as a bead coating. This choice was based on preliminary experiments in which WGA captured the greatest portion of mid-log phase E. coli. Then, dry powdered milk at a concentration of 2.5 mg/ml was selected as a blocking agent to prevent in-droplet bead clumping. With a completed preparation protocol, the cell retention over the entire preparation was determined. Free cells were electrotransformed using conventional, benchtop electroporation methods and compared to electrotransformation of bound cells to determine how bead-binding may affect electrotransformation. Next, a defined media was selected as the growth medium, since LB inhibited cell binding. Subsequently, selection of the pulse temperature was made. With the optimized benchtop protocol as a foundation, trials were performed on the digital microfluidic device (Figure 5).
A. Benchtop electroporation of free and bound cells grown and recovered in MOPS EZ
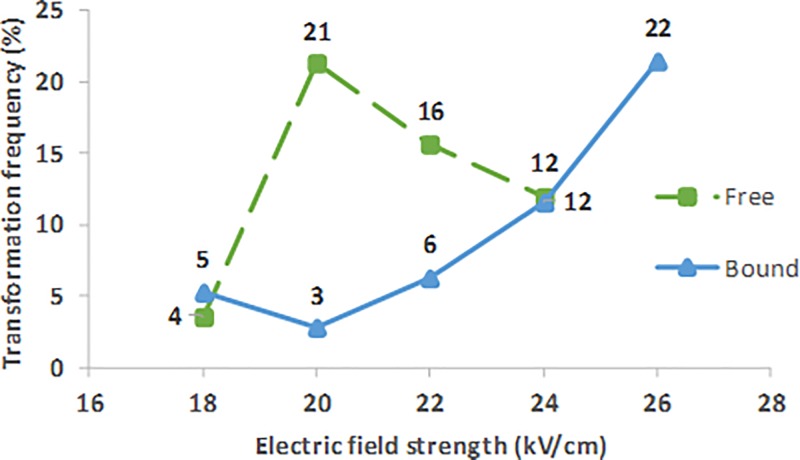
Figure 8 presents typical transformation frequencies of free cells (green) using conventional benchtop electroporation methods (centrifugation for washing). Cells were grown and recovered in growth medium. Samples were held at 4 °C during the pulse. RC time constants for the free cell samples ranged from 5.9 to 6.0 ms.
FIG. 8.
Benchtop transformation frequency over a range of electric field strengths for free cells (green) and bound cells (blue) grown and recovered in MOPS EZ with biotin.
Free cells reached transformation frequencies of up to 21% with electric field strength pulses of 20 kV/cm. The average cell survival at 20 kV/cm was found to be 5.0%. Free cells were compared to bound cells, using the electroporation protocol outlined in Figure 4, with cells at 4 °C during the electroporation pulse. Figure 8 also presents the typical transformation frequency results for growth and recovery in MOPS EZ with biotin (blue), over a range of electric field strengths used to pulse the cells. Samples that comprise the bound cells of Figure 8 had RC time constants ranging from 5.7 to 5.9 ms.
Bound cells grown and recovered in this media showed increasing transformation frequencies as electric field strength increased up to 26 kV/cm. Above 26 kV/cm electric field strengths, data are not available due to arcing (discharge across the top of the sample). At 26 kV/cm, up to 22% transformation frequency is reached, marking a greater than sevenfold improvement over the transformation frequency achieved using the more common LB growth medium with cells attached to beads (data not shown). The cell survival at 26 kV/cm is 1.3% for the bound cell sample, and less than half of the survival of free cells pulsed at 20 kV/cm (the optimal electric field strength for free cells).
To reach peak transformation frequency, bead bound cells grown and recovered in MOPS EZ with biotin require higher electric field strengths than free cells when pulsed at 4 °C. To gain insight into the impact of bead presence on transformation frequency, an electrostatic simulation of induced transmembrane voltage in the presence and absence of magnetic beads was developed with Comsol Multiphysics modelling software. Relevant electric parameters of the electroporation solution, cell membrane, cytosol, and magnetic bead are summarized in Table II.
TABLE II.
Electrical parameters used in the simulation of bead-based dielectric shielding of E. coli during electroporation.
| Conductivity (S/cm) | Dielectric constant | |
|---|---|---|
| Electroporation solution | 4 × 10−5 | 75 |
| Cytosol | 3 × 10−3 | 80 |
| Cell membrane | 1 × 10−8 | 5.0 |
| Bead | 1 × 10−16 | 2.6 |
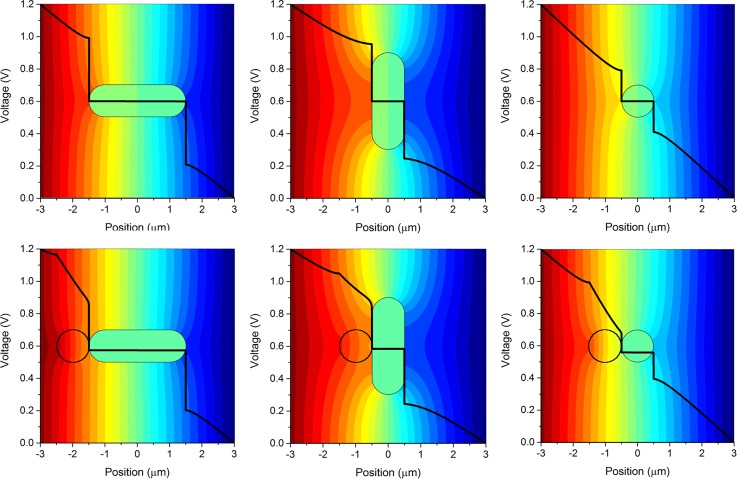
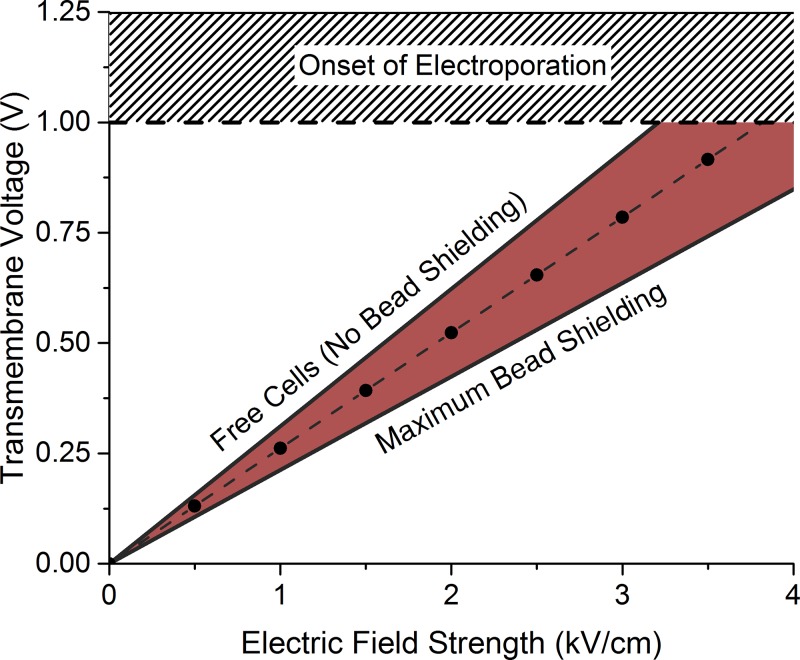
Figure 9 shows three orientations of the potential field around a rod-shaped cell in a uniform electric field in the presence (bottom row) and absence (top row) of a 1 μm dielectric bead. The black line in Figure 9 represents the voltage computed from left to right, through the center of the cell. Figure 10 shows plots of the transmembrane voltage (averaged over the three cell orientations) as electric field strength is increased, for the free cells and cells shielded by beads. The average induced transmembrane voltages of the free and bound cells are plotted in Figure 10. The steepest slope in Figure 10 corresponds to free cells while the shaded region corresponds to cases in which bound cell preparations may participate in dielectric shielding. As expected, the electrostatic model predicts that membranes of free, unbound cells reach the onset of electroporation at lower applied electric fields than those of cells bound to the beads.
FIG. 9.
Transmembrane voltage across the cell using three orthogonal cell orientations, each shown with (bottom) and without (top) the presence of a bead. The black line represents the voltage drop calculated through the sample from left to right, through the center of the cell (and bead if present).
FIG. 10.
Transmembrane voltages at the cell membrane for free cells, and cells in the presence of beads.
The model was used to compute the orientation-specific induced transmembrane voltage of free cells and cells bound to beads. A 1 V transmembrane voltage was used as an approximation of the voltage required for inducing formation of electropores in the cell membrane.43–47 The simulation estimates that a 23% increase in electric field strength is required to induce the same transmembrane voltage for bead-bound cells compared to free cells. Extrapolating this result, the model predicts a peak electric field strength of 24.6 kV/cm based on the 20 kV/cm peak of free cells. Thus, the electrostatic model of bead shielding explains the shift in optimum transformation field strength observed for electrotransformation in the presence and absence of the magnetic beads, as shown in Figure 8. This suggests that dielectric shielding by the beads is likely a major contributor to the increase in electric field strength required for bound cells. Although there is an increased electric field strength required for maximum transformation frequency, these results demonstrate that the maximum transformation frequency was not significantly reduced by (1) binding cells to beads, (2) blocking bound cells with fat-free powdered milk, nor (3) by washing with a magnet rather than a centrifuge. These findings suggest that the bench-scale MAGE protocol will translate well to the digital microfluidic platform and has the potential to yield high frequency transformation.
B. Benchtop electroporation of cells pulsed at room temperature
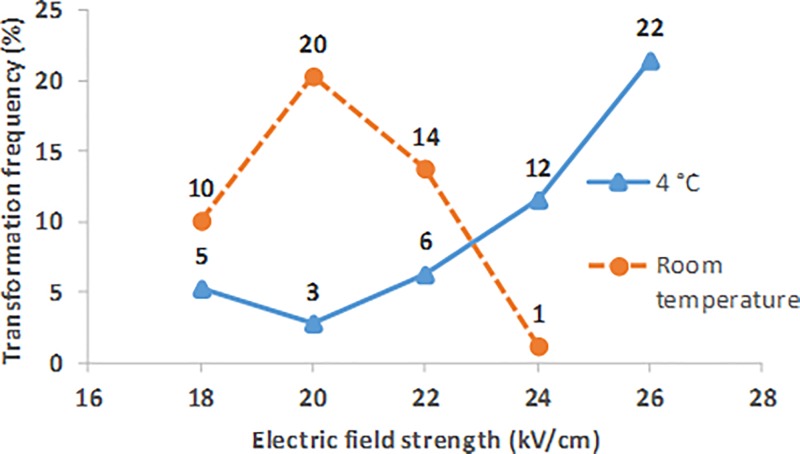
Figure 11 presents typical transformation frequencies over a range of electric field strengths, for bound cells grown and recovered in growth medium, where the sample was at room temperature during the electroporation pulse (orange). Transformation frequencies of the cell samples pulsed at room temperature were more sensitive to factors affecting the RC time constant of the pulse than for samples pulsed at 4 °C. All of the room temperature data from Figure 11 were recorded from the samples pulsed at the most commonly occurring time constants for this protocol (5.8 and 5.9 ms).
FIG. 11.
Benchtop transformation frequency over a range of electric field strengths for bound cells grown and recovered in MOPS EZ, where samples were pulsed at 4 °C (blue) and room temperature (orange).
Transformation frequencies of up to 20% were reached at room temperature; however, a lower electric field strength was required for reaching the peak frequency, compared to pulses at 4 °C. An electric field strength pulse of 20 kV/cm yielded the highest transformation frequency. The room temperature pulses yielded lower survival than those performed at 4 °C, with a survival of 0.2% at 20 kV/cm.
Conventionally, samples are kept cold (4 °C) during the pulse to increase cell stability. At higher temperatures, loss of stability may result in a lower electric field strength requirement for high frequency electroporation. Lower electric field strengths are beneficial for reducing the risk of damaging the surface coatings in a microfluidic device and potentially enable an integrated power supply to perform both electroporation and electrowetting on the same platform.
C. Microfluidic trials
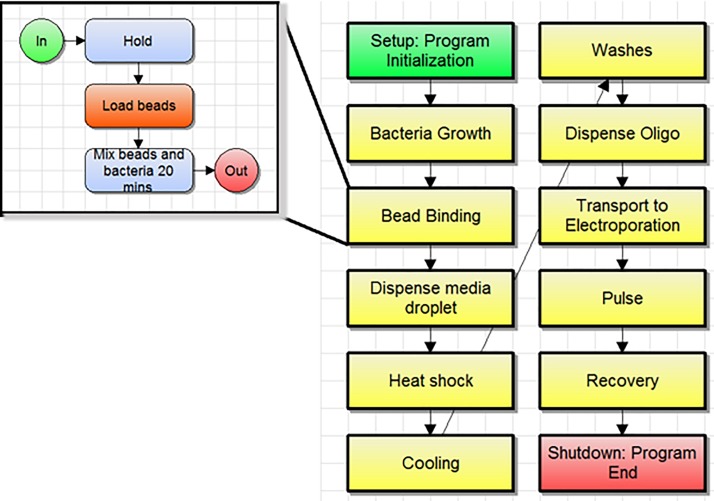
Trials of an automated MAGE cycle on the digital microfluidic platform were performed using the protocol developed on the benchtop as a foundation. On-cartridge preparation of EcNR2 electrocompetent cells was successfully performed with only minor adjustments from the protocol outlined in Figure 4. Figure 12 shows an image of the flow-chart style software script used to organize programmed sample processing tasks. Each block contains a series of sub-routines sent to the controller to be performed on the cartridge. Videos of some of these sub-routines were included in the supplementary material. Supplementary material Figure 1 shows collection of bead bound cells onto the magnet, followed by removal of the supernatant, then pickup by a droplet. Supplementary material Figure 2 shows the re-blocking step for several lanes, followed by washing and resuspension. Differences in bead dispersion homogeneity can be seen between different lanes.
FIG. 12.
Software script showing MAGE cycle workflow and subroutine for bead binding.
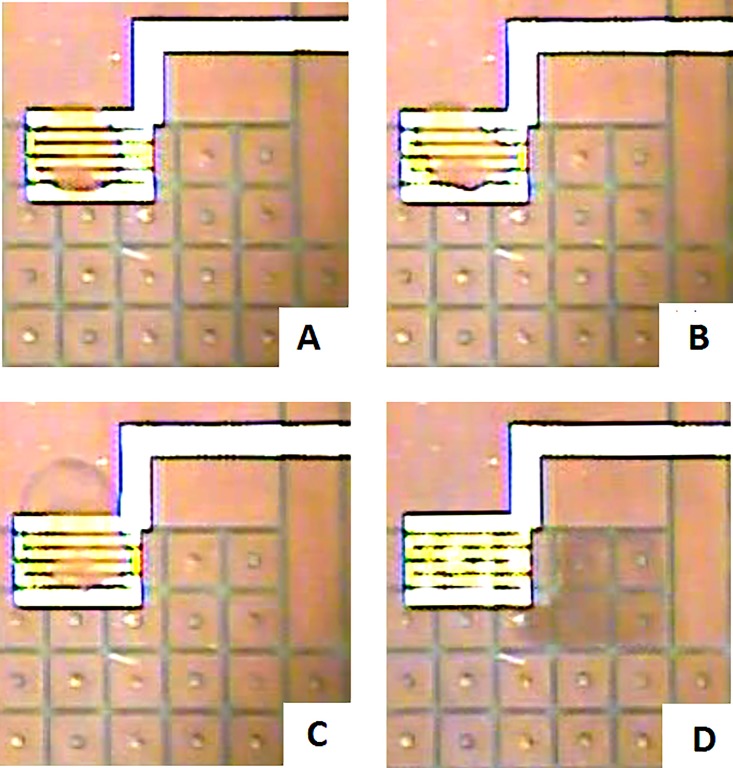
Adjusting the protocol to add a second 1-min bead-blocking step with 2.5 mg/ml fat-free powdered milk after the heat shock was required for bound cells to uniformly disperse after final washes with DI water. Due to the longer time required for droplet transport from the electroporation electrode to the recovery reservoir, a 4× (1.4 μl) droplet of recovery media (four times the volume of the sample droplet) is positioned near the electroporation electrode. After the pulse, this recovery droplet merges with the sample droplet to reduce the time between the electroporation pulse and recovery of the cells. Figure 13(a) shows one of the sample droplets resting on top of the electroporation electrode before pulsing. In Figure 13(b), the droplet had undergone two alternating polarity electric field pulses. The 4× droplet of the recovery medium had merged with the sample droplet in Figure 13(c). Finally, Figure 13(d) shows the 6× droplet being transported to the right, away from the electroporation electrode. Figure 13 shows slight aggregation of beads and cells that is sometimes still present, but not visible unless the droplet sits on top of the serpentine electrode. Although the aggregate appears to be thin, flexible, and not involving all of the beads in the droplet, it indicates that some further optimization of cell dispersion is needed. Antibiotic selection can be performed on the cartridge in various ways depending on the application. A droplet containing the antibiotic can be merged with the sample droplet and mixed on the cartridge for recovery and selection, or the sample droplet can be transported into a larger volume well containing a larger volume (up to 50 μl) of media for recovery and selection.
FIG. 13.
(a) A 2× sized droplet containing E. coli, sitting on top of the electroporation electrode before a double pulse. (b) The 2× droplet sitting on top of the electroporation electrode, immediately after a double pulse. (c) A 6× droplet formed from the 4× recovery droplet merged with the 2× sample droplet. (d) The 6× recovered droplet being transported off the electroporation electrode.
Samples were tested with variations on the foundational parameters developed in the benchtop experiments. Variations in electric field strength, number of pulses, and oligo concentration were tested to account for differences between the microfluidic and benchtop systems. Using these parameters, transformation frequencies averaged 9% with a standard deviation of 9% rounded to the nearest percent, with cell survival averaging 1% with a standard deviation of 2%. On-cartridge trials that yielded transformants resistant to the antibiotic Carbenicillin at an average frequency of 9% were grown and recovered in defined media, went through a single wash of DI water with oligo (bla-restore oligo concentration of 9 mM), and were pulsed at room temperature with two, alternating polarity, 24 kV/cm electric field strength pulses. Frequencies as high as 21% were achieved on the device, using this altered version of the Figure 4 protocol. The cell survival of these high frequency samples was abnormally low (less than 1%). While low cell survival can be overcome with longer growth between MAGE cycles, this low of cell survival is an indication that there may be some unknown or not well understood differences between benchtop and microfluidic MAGE cycles and electroporation. Transformation frequencies of 9% are sufficient when selecting for transformed cells, however higher transformation frequencies are desirable for allowing multiple cycles without the need for selection. Although these results show that automated high frequency transformation is possible on a digital microfluidic device, they also indicate high variability, as a result of an unknown variation between experiments. Because the serpentine electroporation electrodes were not on the cooled Peltier region of the cartridge, there was an uncontrolled temperature variable in our experiments. Efforts to externally cool the surface of the cartridge using cool packs or ice water did not give consistently reproducible results.
IV. CONCLUSIONS
A protocol for automated genetic transformation of E. coli on a digital microfluidic device is presented in this paper. WGA-lectin coated magnetic beads allowed for successful cell immobilization, washing, and media exchange on the microfluidic platform. Our EWoD compatible media exchange protocol, where bound cells remain pelleted during washing, boasts over 95% cell capture and around 15% retention over three media exchanges and was shown to adequately produce electrocompetent cells in conventional and digital microfluidic formats.
Proof of concept trials demonstrated successful preparation of electrocompetent E. coli and electrotransformation automation for a MAGE cycle on a droplet based, EWoD, digital microfluidic platform. Transformation frequencies averaging 9% with some samples yielding as high as 21% were achieved with little on-cartridge optimization. The on-cartridge protocol opens the possibility for multiple transformation cycles to be performed upon a cell population using a commercial digital microfluidics platform. Future trials using a higher total number of cells within the droplet are required to yield more genetic variants and explore the potential of automating MAGE on a digital microfluidic platform. Benchtop experiments using the developed protocol achieved up to 22% transformation frequency, with cells grown and recovered in defined media, and samples kept at room temperature when the electroporation pulse occurs. These results show potential for achieving higher transformation frequency through further optimization of electroporation pulse parameters used on the digital microfluidic platform.
V. SUPPLEMENTARY MATERIAL
See supplementary material for Figure 1 a video showing the collection of bound cells onto a magnet. Cells were grown in the 50 μl chamber on the right side of the screen, to which beads were added to incubate with the cells for binding. Following the collection of beads onto the magnet, the supernatant is removed through electrowetting. Finally, a droplet is moved onto the space above the magnet, the magnet is lowered, and the droplet can move the bound cells away and re-suspend them. Figure 2 shows each lane during the re-blocking step. The milk blocking agent and bound cells are transported for mixing. The magnet is raised to pull the bound cells out of suspension and the supernatant is removed through electrowetting. DI-water wash droplets move across the pelleted cells to promote electrocompetence. Finally, a droplet containing oligo moves above the pellet, the magnet is lowered, and the droplet transports the cells away while re-suspending them. In Figure 2, some lanes suspend the cells homogenously, while others do not, illustrating the need for further optimization of the bead blocking procedure.
ACKNOWLEDGMENTS
This work was supported by Grant No. HR0011-12-C-0057 from the DARPA Living Foundries Program. The authors also thank J. Harrington, M. Royal, and A. Sudarsan for their foundational work on the development of an electrowetting microfluidic cartridge for MAGE.
References
- 1. Gach P. C., Shih S. C. C., Sustarich J., Keasling J. D., Hillson N. J., Adams P. D., and Singh A. K., ACS Synth. Biol. , 426–433 (2016). 10.1021/acssynbio.6b00011 [DOI] [PubMed] [Google Scholar]
- 2. Nielsen A. A. K., Der B. S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E. A., Ross D., Densmore D., and Voigt C. A., Science , aac7341 (2016). 10.1126/science.aac7341 [DOI] [PubMed] [Google Scholar]
- 3. Wang H. H., Isaacs F. J., Carr P. A., Sun Z. Z., Xu G., Forest C. R., and Church G. M., Nat. Lett. , 894–898 (2009). 10.1038/nature08187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yu D., Ellis H. M., Lee E. C., Jenkins N. A., Copeland N. G., and Court D. L., Proc. Natl. Acad. Sci. , 5978–5983 (2000). 10.1073/pnas.100127597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ellis H. M., Yu D., DiTizio T., and Court D. L., Proc. Natl. Acad. Sci. , 6742–6746 (2001). 10.1073/pnas.121164898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Costantino N. and Court D. L., Proc. Natl. Acad. Sci. , 15748–15753 (2003). 10.1073/pnas.2434959100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Isaacs F. J., Carr P. A., Wang H. H., Lajoie M. J., Sterling B., Kraal L., Tolonen A. C., Gianoulis T. A., Goodman D. B., Reppas N. B., Emig C. J., Bang D., Hwang S. J., Jewett M. C., Jacobson J. M., and Church G. M., Science , 348–353 (2011). 10.1126/science.1205822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lajoie M. J., Rovner A. J., Goodman D. B., Aerni H.-R., Haimovich A. D., Kuznetsov G., Mercer J. A., Wang H. H., Carr P. A., Mosberg J. A., Rohland N., Schultz P. G., Jacobson J. M., Rinehart J., Church G. M., and Isaacs F. J., Science , 357–360 (2013). 10.1126/science.1241459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carr P. A., Wang H. H., Sterling B., Isaacs F. J., Lajoie M. J., Xu G., Church G. M., and Jacobson J. M., Nucleic Acids Res. , e132 (2012). 10.1093/nar/gks455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sandahl M., Punnamaraju S., Madison A., Harrington J., Royal M., Fair R., Eckhardt A., Sudarsan A., and Pollack M., in 17th International Conference on Miniaturized Systems for Chemistry and Life Sciences (2013), Vol. 17, pp. 1260–1263. [Google Scholar]
- 11. Ostrov N., Landon M., Guell M., Kuznetsov G., Teramoto J., Cervantes N., Zhou M., Singh K., Napolitano M. G., Moosburner M., Shrock E., Pruitt B. W., Conway N., Goodman D. B., Gardner C. L., Tyree G., Gonzales A., Wanner B. L., Norville J. E., Lajoie M. J., and Church G. M., Science , 819–822 (2016). 10.1126/science.aaf3639 [DOI] [PubMed] [Google Scholar]
- 12. Fair R. B., Khlystov A., Tailor T. D., Ivanov V., Evans R. D., Srinivasan V., Pamula V. K., Pollack M. G., Griffin P. B., and Zhou J., IEEE Des. Test Comput. , 10–24 (2007). 10.1109/MDT.2007.8 [DOI] [Google Scholar]
- 13. Fair R. B., Srinivasan V., Ren H., Paik P., Pamula V. K., and Pollack M. G., IEEE , 779–782 (2003). 10.1109/IEDM.2003.1269396 [DOI] [Google Scholar]
- 14. Srinivasan V., Pamula V. K., and Fair R. B., Lab Chip , 310–315 (2004). 10.1039/b403341h [DOI] [PubMed] [Google Scholar]
- 15. Schell W. A., Benton J. L., Smith P. B., Poore M., Rouse J. L., Boles D. J., Johnson M. D., Alexander B. D., Pamula V. K., Eckhardt A. E., Pollack M. G., D. K. Benjamin, Jr. , Perfect J. R., and Mitchell T. G., Eur. J. Clin. Microbiol. Infect. Dis. , 2237–2245 (2012). 10.1007/s10096-012-1561-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hua Z., Rouse J. L., Eckhardt A. E., Srinivasan V., Pamula V. K., Schell W. A., Benton J. L., Mitchell T. G., and Pollack M. G., Anal. Chem. , 2310–2316 (2010). 10.1021/ac902510u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boles D. J., Benton J. L., Siew G. J., Levy M. H., Thwar P. K., Sandahl M. A., Rouse J. L., Perkins L. C., Sudarsan A. P., Jalili R., Pamula V. K., Srinivasan V., Fair R. B., Griffin P. B., Eckhardt A. E., and Pollack M. G., Anal. Chem. , 8439–8447 (2011). 10.1021/ac201416j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pollack M. G., Pamula V. K., Srinivasan V., and Eckhardt A. E., Expert Rev. Mol. Diagn. , 393–407 (2011). 10.1586/erm.11.22 [DOI] [PubMed] [Google Scholar]
- 19. Barbulovic-Nad I., Yang H., Park P. S., and Wheeler A. R., Lab Chip , 519–526 (2008). 10.1039/b717759c [DOI] [PubMed] [Google Scholar]
- 20. Miller E. M. and Wheeler A. R., Anal. Chem. , 1614–1619 (2008). 10.1021/ac702269d [DOI] [PubMed] [Google Scholar]
- 21. Sista R. S., Eckhardt A. E., Wang T., Graham C., Rouse J. L., Norton S. M., Srinivasan V., Pollack M. G., Tolun A. A., Bali D., Millington D. S., and Pamula V. K., Clin. Chem. , 1444–1451 (2011). 10.1373/clinchem.2011.163139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Srinivasan V., Pamula V. K., and Fair R. B., Anal. Chim. Acta , 145–150 (2004). 10.1016/j.aca.2003.12.030 [DOI] [Google Scholar]
- 23. Sista R. S., Eckhardt A. E., Srinivasan V., Pollack M. G., Palanki S., and Pamula V. K., Lab Chip , 2188–2196 (2008). 10.1039/b807855f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ng A. H. C., Choi K., Luoma R. P., Robinson J. M., and Wheeler A. R., Anal. Chem. , 8805–8812 (2012). 10.1021/ac3020627 [DOI] [PubMed] [Google Scholar]
- 25. Sista R., Hua Z., Thwar P., Sudarsan A., Srinivasan V., Eckhardt A., Pollack M., and Pamula V., Lab Chip , 2091–2104 (2008). 10.1039/b814922d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malic L., Veres T., and Tabrizian M., Biosens. Bioelectron. , 2218–2224 (2009). 10.1016/j.bios.2008.11.031 [DOI] [PubMed] [Google Scholar]
- 27. Shamsi M. H., Choi K., Ng A. H. C., Chamberlain M. D., and Wheeler A. R., Biosens. Bioelectron. , 845–852 (2016). 10.1016/j.bios.2015.10.036 [DOI] [PubMed] [Google Scholar]
- 28. Shah G., Ohta A., Chiou E., Wu M., and Kim C. J., Lab Chip , 1732–1739 (2009). 10.1039/b821508a [DOI] [PubMed] [Google Scholar]
- 29. Moon H., Wheeler A. R., Garrell R. L., Loo J. A., and Kim C.-J., Lab Chip , 1213–1219 (2006). 10.1039/b601954d [DOI] [PubMed] [Google Scholar]
- 30. Abdelgawad M., Watson M. W. L., and Wheeler A. R., Lab Chip , 1046–1051 (2009). 10.1039/b820682a [DOI] [PubMed] [Google Scholar]
- 31. Madison A. C., Royal M. W., and Fair R. B., J. Microelectromech. Syst. , 593–605 (2016). 10.1109/JMEMS.2016.2561699 [DOI] [Google Scholar]
- 32. Wang S. and Lee L. J., Biomicrofluidics , 1–14 (2013). 10.1063/1.4774071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dower W. J., Miller J. F., and Ragsdale C. W., Nucleic Acids Res. (13), 6127–6146 (1988). 10.1093/nar/16.13.6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pedelacq J. D., Cabantous S., Tran T., Terwilliqer T. C., and Waldo G. S., Nat. Biotechnol. , 79–88 (2006). 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- 35. Neumann E. and Rosenheck K., J. Membr. Biol. , 279–290 (1972). 10.1007/BF01867861 [DOI] [PubMed] [Google Scholar]
- 36. Zimmermann U., Pilwat G., and Riemann F., Biophys. J. , 881–899 (1974). 10.1016/S0006-3495(74)85956-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Neumann E., Schaefer-Ridder M., Wang Y., and Hofschneider P. H., EMBO J. , 841–845 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lurquin P. F., Mol. Biotechnol. , 5–35 (1997). 10.1007/BF02821542 [DOI] [PubMed] [Google Scholar]
- 39. Brambach B. C., Michels A., Franzke J., and Kettler R., Prog. Biophys. Mol. Biol. , 46–54 (2013). 10.1016/j.pbiomolbio.2012.12.006 [DOI] [PubMed] [Google Scholar]
- 40. Xie T. D., Sun L., and Tsong T. Y., Biophys. J. , 13–19 (1990). 10.1016/S0006-3495(90)82349-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xie T. D. and Tsong T. Y., Biophys. J. , 897–903 (1990). 10.1016/S0006-3495(90)82434-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chuang S. E., Chen A. L., and Chao C. C., Nucleic Acids Res. , 1641 (1995). 10.1093/nar/23.9.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barnett A. and Weaver J. C., Bioelectrochem. Bioenerg. , 163–182 (1991). 10.1016/0302-4598(91)87001-W [DOI] [Google Scholar]
- 44. Neu J. C. and Krassowska W., Phys. Rev. E , 3471–3482 (1999). 10.1103/PhysRevE.59.3471 [DOI] [Google Scholar]
- 45. DeBruin K. A. and Krassowska W., Biophys. J. , 1213–1224 (1999). 10.1016/S0006-3495(99)76973-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith K. C., Neu J. C., and Krassowska W., Biophys. J. , 2813–2826 (2004). 10.1016/S0006-3495(04)74334-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Krassowska W. and Filev P. D., Biophys. J. , 404–417 (2007). 10.1529/biophysj.106.094235 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for Figure 1 a video showing the collection of bound cells onto a magnet. Cells were grown in the 50 μl chamber on the right side of the screen, to which beads were added to incubate with the cells for binding. Following the collection of beads onto the magnet, the supernatant is removed through electrowetting. Finally, a droplet is moved onto the space above the magnet, the magnet is lowered, and the droplet can move the bound cells away and re-suspend them. Figure 2 shows each lane during the re-blocking step. The milk blocking agent and bound cells are transported for mixing. The magnet is raised to pull the bound cells out of suspension and the supernatant is removed through electrowetting. DI-water wash droplets move across the pelleted cells to promote electrocompetence. Finally, a droplet containing oligo moves above the pellet, the magnet is lowered, and the droplet transports the cells away while re-suspending them. In Figure 2, some lanes suspend the cells homogenously, while others do not, illustrating the need for further optimization of the bead blocking procedure.