Abstract
Saccharomyces cerevisiae mating-type switching is initiated by a double-strand break (DSB) at MATa, leaving one cut end perfectly homologous to the HMLα donor, while the second end must be processed to remove a nonhomologous tail before completing repair by gene conversion (GC). When homology at the matched end is ≤150 bp, efficient repair depends on the Recombination Enhancer, which tethers HMLα near the DSB. Thus homology shorter than an apparent “minimum efficient processing segment” can be rescued by tethering the donor near the break. When homology at the second end is ≤150 bp, second-end capture becomes inefficient and repair shifts from GC to break-induced replication (BIR). But when pol32 or pif1 mutants block BIR, GC increases three-fold, indicating that the steps blocked by these mutations are reversible. With short second-end homology, absence of the RecQ helicase Sgs1 promotes synthesis-dependent strand annealing whereas deletion of the FANCM-related Mph1 helicase promotes BIR.
Introduction
DNA double strand breaks (DSBs), if not properly repaired, can result in loss of chromosomal integrity. DSBs arise from exogenous and endogenous sources (Mehta and Haber, 2014) and can be repaired by nonhomologous end-joining (NHEJ) to re-ligate broken ends, or by homologous recombination (HR), which copies an intact template DNA sequence: a sister chromatid, an allelic locus or an ectopically located donor (Haber, 2013; Jasin and Rothstein, 2013).
Two principal mechanisms of HR repair - gene conversion (GC) and break-induced replication (BIR) - are governed by the extent of homology shared between the two DSB ends and a donor template. When a DSB occurs such that both ends share homology with a donor sequence, repair usually occurs by GC, requiring synthesis of a short patch of new DNA to repair the DSB (Figure 1). BIR becomes necessary when only one end shares homology with a donor, e.g. at stalled or broken replication forks or at eroding telomeres (Anand et al., 2013; Davis and Symington, 2004; McEachern and Haber, 2006). Ectopic BIR leads to recombination-dependent DNA replication and creation of a non-reciprocal translocation. When homology to both ends is present in close proximity and in proper orientation, GC strongly out-competes BIR (Jain et al., 2009; Malkova et al., 2005). If the homologies to the DSB ends are separated by an insertion, and the separation of the ends is > 1 kb, gap repair shifts to BIR, initiated from one or both ends. In mitotic cells, GC occurs mainly by synthesis-dependent strand annealing (SDSA) (Gloor et al., 1991), which leads to predominantly non-crossover (NCO) products (Ira et al., 2006). Repair can also occur by formation of a double-Holliday junction (dHJ) (Ira et al., 2003; Szostak et al., 1983). The dHJs can be cleaved by HJ resolvases, and can yield either NCO or crossover (CO) outcomes. As an alternative to dHJ resolution by cleavage, dHJs are ‘dissolved’ by the Sgs1-Top3-Rmi1 (STR) complex to yield only NCOs (Ira et al., 2003; Wu and Hickson, 2003).
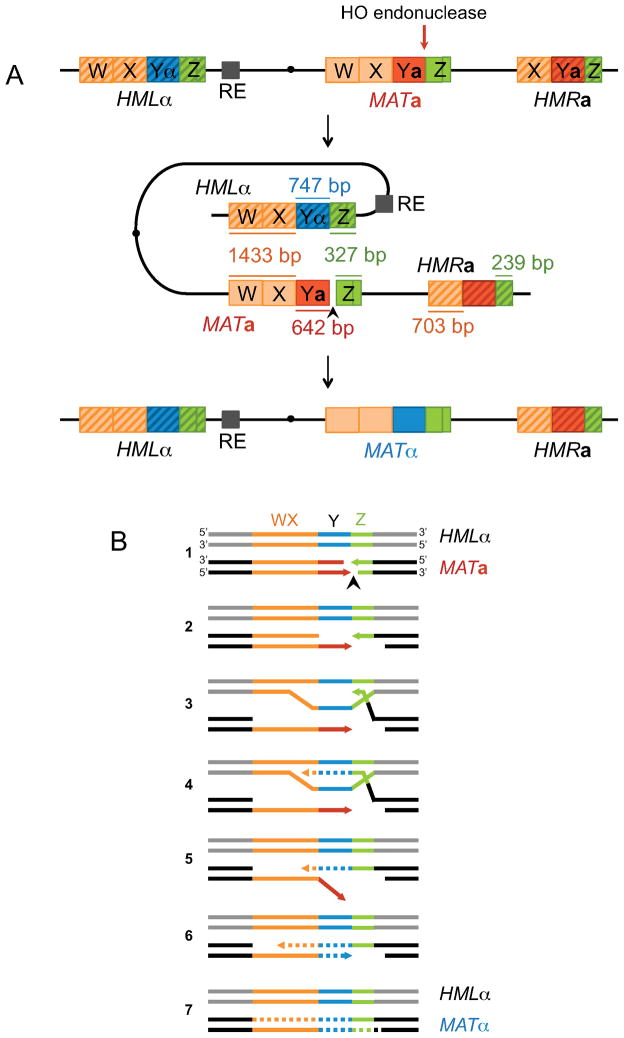
Figure 1. Schematic of S. cerevisiae mating-type loci on chromosome 3 and the SDSA model of MAT switching.
(A) The expressed MAT locus and the silent HML and HMR loci are located on chromosome 3. MAT shares 327 bp of Z1–Z2 homology (invading end, shown in green) and 1433 bp of W-X homology (non-invading end, shown in orange) with HML. The recombination enhancer (RE) is illustrated as a small dark gray box near HML. (B) Individual steps of the SDSA repair mechanism. 1: MATa switching is initiated by an HO endonuclease-induced DSB at the Ya-Z1 junction. 2: 5′–3′ resection creates 3′ ssDNA, which is bound by Rad51 recombinase. 3: The Rad51 nucleoprotein filament searches for homology, allowing strand invasion at the HML donor. 4: New DNA synthesis is primed from the 3′ invading Z end. 5: The newly synthesized strand dissociates from the donor and anneals with the W-X homologous sequences on the left side of the break (non-invading end) after a successful second-end capture. 6: The non-homologous 3′ tail is removed, and new DNA synthesis begins at the second end 3′. 7: Gaps are filled and DSB repair is completed by ligation.
Budding yeast mating-type (MAT) switching has proven to be an excellent model of GC to identify many distinct intermediates of DSB repair (reviewed by Lee and Haber (2015). Haploid mating type is determined by MATa or MATα alleles located on chromosome 3 (Chr3) (Figure 1A). MAT switching from MATa to MATα is initiated by a site-specific DSB at MATa created by HO endonuclease, leading to intrachromosomal repair using HMLα as the donor. HML encodes a complete copy of MATα, but is heterochromatic, transcriptionally silent and refractory to HO cleavage because of the presence of highly positioned nucleosomes (Ercan and Simpson, 2004; Weiss and Simpson, 1998). MAT switching occurs by SDSA gap repair (Ira et al., 2006) and involves replacement of 642 bp of MAT-Ya by 747 bp of Yα sequences, located between W-X and Z1–Z2 shared by MAT and HML (Figure 1B). HMLα shares sufficient homology on both sides of the DSB with MATa to yield efficient (>90%) gene conversion (Figure 2A). A second donor, HMRa, is deleted in our experiments, but serves as the donor to switch MATα to MATa.
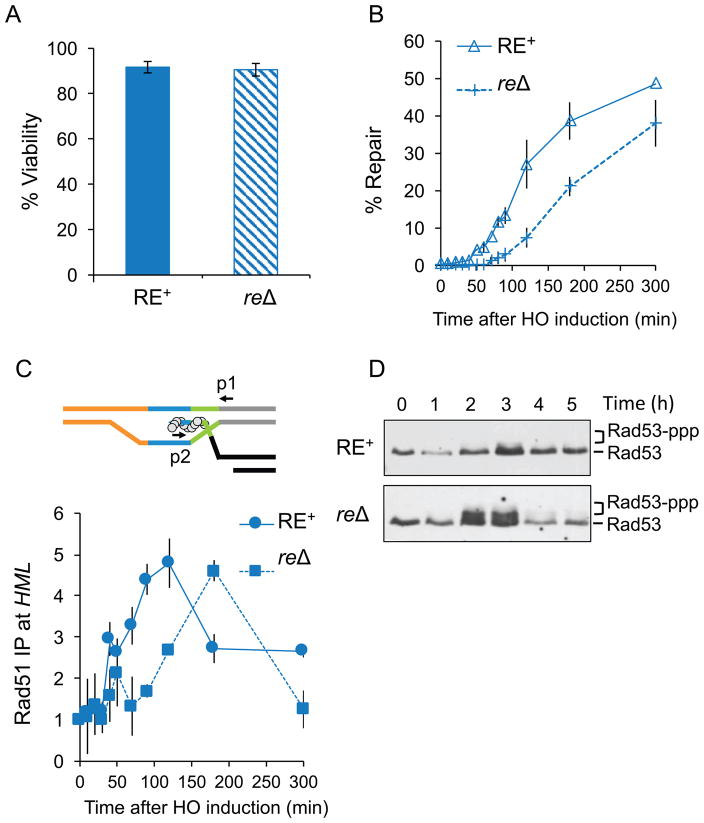
Figure 2. Repair is delayed in the absence of RE due to delay in strand invasion at the donor HML.
(A) Viability of RE+ and reΔ strains, mean ± SD (n ≥ 3). (B) Kinetics of product formation as determined by Southern blots of RE+ (solid line) and reΔ (dotted line). Product from a repaired MATα-inc survivor colony was set to 100%. Data represent mean ± SD (n = 3). (C) Rad51 ChIP signal at the donor HML represent kinetics of strand invasion in RE+ and reΔ. The top panel: synapsis between Rad51 nucleoprotein filament (circles) and HML. Arrows indicate primers p1 and p2 used for qRT-PCR analysis. Bottom panel: quantification of the PCR product following HO induction. IP signal from the ARG5,6 locus was used to normalize for input DNA in ChIP assays. (D) Representative Rad53 Western blots from whole cell extracts of RE+ and reΔ during DSB repair. Phosphorylated Rad53 (Rad53-ppp) migrates slowly compared to unphosphorylated Rad53. See also Figure S1.
MAT switching is highly directional; MATa cells preferentially use HMLα whereas MATα usually recombines with HMRa. Preference is imposed by a small, cis-acting recombination enhancer (RE) sequence located 17 kb proximal to HML (Wu and Haber, 1996). RE binds multiple copies of the Fkh1 protein whose phosphothreonine-binding FHA domains bring RE close to the DSB ends, to tether the left arm of Chr3 physically close to MAT, thus facilitating recombination with HML (Avşaroğlu et al., 2016; Coïc et al., 2006b; Li et al., 2012; Sun et al., 2002). RE is repressed in MATα cells (Wu et al., 1998). Thus in MATα or when RE is deleted, HMR becomes the default donor. RE is “portable” and thus can stimulate the use of an adjacent donor even in inter-chromosomal recombination (Coïc et al., 2006a; Lee et al., 2016; Li et al., 2012; Wu and Haber, 1996).
Synchronous induction of a site-specific DSB at a single locus has made it possible to elucidate the molecular steps and requirements of SDSA in real time (Chen et al., 2011; Connolly et al., 1988; Lee and Haber, 2015; Hicks et al., 2011; Mimitou and Symington, 2008; Sugawara et al., 2003; White and Haber, 1990; Wolner and Peterson, 2005). The DSB ends are processed by 5′-to-3′ resection at a rate of ~1 nt/s, producing 3′-ended ssDNA tails (Figure 1B, step 2). These tails are bound by Rad51 recombinase. The Rad51 nucleoprotein filament then searches for homologous donor sequences elsewhere in the genome and base-pairs to form a displacement loop (D-loop) (Figure 1B, step 3). In MAT switching, strand invasion occurs from the DSB end to the right of the cut, which is fully homologous to the 327 bp Z1–Z2 region in HML (denoted as the invading end). The 3′ end can then be used as a primer to initiate new DNA synthesis from the template (Figure 1B, step 4). Because of the presence of the 650 nt nonhomologous (NH) Ya tail, the opposite DSB end cannot be used to initiate new DNA synthesis until the nonhomologous tail is clipped off. We will refer to this second end as the non-invading end.
Unlike semi-conservative DNA replication, the newly synthesized strand extended from the invading end is displaced from the template and anneals to the homology at the resected, single-stranded W-X-Ya end of the DSB (Ira et al., 2006) (Figure 1B, step 5). This step is known as second-end capture. However, before new DNA synthesis can begin from this end, the nonhomologous single-stranded Ya sequences must be clipped off by a complex consisting of Rad1–Rad10 endonuclease, Msh2–Msh3 mismatch repair proteins and ‘scaffold’ proteins Slx4 and Saw1 (Ivanov and Haber, 1995; Li et al., 2008; Sugawara et al., 1997; Toh et al., 2010). Tail clipping appears to require annealing with the newly copied strand. The trimmed W-X end is then used as a primer to create the second strand of the DSB repair product (Figure 1B, step 6). Ends are ligated, reestablishing unbroken chromatin at MAT (Figure 1B, step 7); MATa is converted to MATα. SDSA and BIR share similar initial repair steps (Figure 1B, steps 1 – 4); however, in the absence of the second-end homology, strand invasion leads to BIR by continued DNA synthesis from the 3′ invading end in a migrating D-loop (Davis and Symington, 2004; Jain et al., 2009; Malkova et al., 2005; Saini et al., 2013). When both DSB ends share homology with a donor, GC out-competes BIR (Malkova et al., 1996).
While RE’s role in directing donor preference has been well demonstrated, we were interested in examining whether RE might influence the kinetics of SDSA repair. Moreover, although the extent of homology required for DSB repair has been assessed using a plasmid substrate (Ira and Haber, 2002), a detailed analysis of how much homology is required to repair chromosomal DSB efficiently has not been examined. We also wished to examine how second-end capture differs from initial strand invasion. Accordingly, we built a series of haploid strains with different lengths of shared homology on each side of the DSB. We find that homology requirements for initial strand invasion and second-end capture are quite different. RE plays a key role in the initial strand invasion step – making short homologies much more efficient – but has no role in later in repair. When homology available to the non-invading end is reduced, second-end capture is inefficient and repair is diverted towards BIR; however blocking BIR improves second end capture. Finally, the RecQ family helicase Sgs1 and the FANCM-related Mph1 helicase direct repair towards opposing HR pathways.
Results
Repair is delayed in the absence of RE
In MATa, RE brings the distal part of the left arm of Chr3 in close proximity to the DSB. To examine in more detail the role of RE in repair, we deleted HMRa so that a DSB can be repaired by GC using HMLα-inc, a single base-pair alteration that, after switching to MATα-inc, prevents HO cutting. The integrated HO gene is regulated by a galactose-inducible promoter (GAL::HO) (Herskowitz and Jensen, 1991; Sandell and Zakian, 1993; Sugawara et al., 2003). To avoid any complications of DSBs re-joined by NHEJ, the NEJ1 gene was deleted (Valencia et al., 2001).
To examine if RE is required to maintain cell viability, we created a 275-bp deletion of the RE region (reΔ). When cells were plated on galactose medium to express HO endonuclease, repair efficiency was ~92% in both the RE+ and reΔ strains (Figure 2A). To assess whether the absence of RE had any effect on the rate of repair, we examined repair kinetics after HO was induced in exponentially growing cultures. Quantitative Southern blot analysis showed product formation in 10% of RE+ cells by 70 min (Hicks et al., 2011) (Figures 2B and S1A). In contrast, in reΔ, 10% repair product appeared only at 120 min, a delay of about 50 min (Figures 2B and S1A), indicating that repair is delayed in the absence of RE.
We used a Rad51 chromatin immunoprecipitation (ChIP) assay to detect Rad51 binding first to the resected ends at MAT and then to HML, indicating strand invasion (Hicks et al., 2011; Sugawara et al., 2003). In both strains, Rad51 recruitment to MAT was detected 20 min after HO induction (Figure S1B); however in the RE+ strain, Rad51 protein was recruited to HML ~30 min prior to its initial detection in reΔ (Figure 2C). Thus, although the absence of RE does not affect survival there is a significant delay in repair at strand invasion.
DSB formation is detected by Mec1ATR and Tel1ATM kinases, which activate the DNA damage checkpoint, causing cell cycle arrest until the break is repaired (Harrison and Haber, 2006). Checkpoint activation and deactivation can be monitored by the hyperphosphorylation of Rad53 kinase (Pellicioli et al., 2001). Wild type (WT) MAT switching is an efficient repair process occurring after modest 5′ to 3′ resection of DSB ends and hence no phosphorylated Rad53 is detected (Figure 2D). However, in absence of RE the delay in repair kinetics (Figure 2B) is accompanied by the appearance of hyperphosphorylated Rad53 about 2 h after HO induction (Figure 2D). As expected from previous studies Rad53 became dephosphorylated as DSBs were repaired (Leroy et al., 2003; Vaze et al., 2002).
Viability and rate of repair decreases as length of shared homology between the invading end and donor is decreased
To examine the role of the invading-end homology length, we created strains with Z homologies ranging from 35 bp to 2216 bp, denoted as δZ strains, while the W-X side retained its WT length of 1433 bp (Figure 3A). When Z homology was reduced below 327 bp, viability decreased slightly in RE+ strains, even when Z was only 50 bp (Figure 3B). However, without RE, viability with short invading homology length was more severely reduced. With Z homology of 35 bp, survival dropped from 56% in the presence of RE and 29% in its absence (Figure 3B). The strong stabilizing effect of RE when Z homology is short suggests that the apparent minimum length of homology needed for strand invasion and repair is dependent on how stably the donor and recipient are tethered.
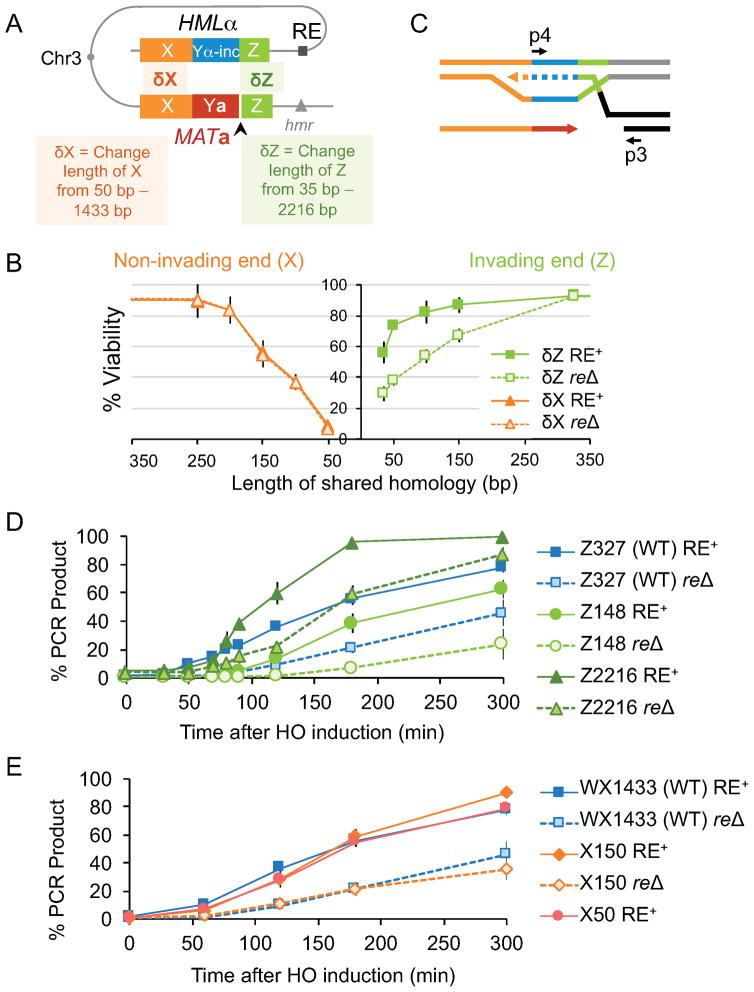
Figure 3. Homology length on two sides of a DSB has different effect on viability and repair kinetics.
(A) Homology length at the invading end (Z; green) or non-invading end (X; orange) was altered at MAT. Arrowhead represents the HO cut site. (B) Viability of δZ (green graphs) and δX (orange graphs) strains with RE (solid line) and without RE (dotted line). Data represent mean ± SD (n ≥ 3). (C) Schematic showing initiation of new DNA synthesis, measured by primer extension. Newly synthesized DNA is shown by dotted lines. Arrows indicate the position of primers used for PCR. (D, E) Kinetics of new DNA synthesis as determined by primer extension. Graphs show qRT-PCR product at intervals following HO induction in various δZ and δX strains. The amount of PCR product obtained from a switched MATα-inc survivor colony = 100%. Data reflect mean ± SD (n = 3).
The initial rate of repair can be assayed by using a PCR-based primer extension assay (White and Haber, 1990), which detects the recombination intermediate created after at least 37 nt of new DNA has been added to the invading 3′ end (Figure 3C). In RE+, 20% of the maximum PCR product was detected 90 min after HO induction (Figure 3D; also Hicks et al., 2011). In reΔ, consistent with the delay in strand invasion, the rate of new DNA synthesis was also slower: 20% PCR product was detected after 150 min (Figure 3D).
When the length of invading-end homology between MAT and HML was reduced to 148 bp, there was a delay in initiation of new DNA synthesis (Figure 3D). There was a further drop in repair kinetics when RE was deleted. When Z homology was increased to 2216 bp in RE+, the rate of initiation of new DNA synthesis was faster than WT (Z327) strain (Figure 3D). Together these data indicate that rate of repair increases with increasing length of invading end homology and becomes still faster in the presence of RE. This correlation between homology length and rate of repair is also reflected in the phosphorylation status of Rad53 kinase (Figure S1C), as when repair is delayed, continuing 5′ to 3′ resection activates the DNA damage checkpoint.
Decreasing the non-invading end homology reduces viability without affecting the rate of repair
We then created strains where the non-invading end (W-X) homology ranged from 50 to 1433 bp (denoted as δX strains). Here, the Z end was maintained at 327 bp (Figure 3A). Viability declined severely as the X homology length was reduced (Figure 3B); the X50 strain repaired only 7% of the level seen with WX1433 (WT). Compared to reducing homology on the Z side, there was a greater impact on viability when homology was shortened at W-X. Unlike at the invading end, deleting RE had no effect on viability (Figure 3B). The lack of impact of RE on the second end is consistent with models where second end synthesis uses the extended first-strand template only after it dissociates from the donor.
The timing of new DNA synthesis from the invading end remained unchanged even when homology at the non-invading end was reduced to X150 or even X50 (Figure 3E). Furthermore, unlike the δZ strains, there was no effect on the rate of new DNA synthesis in the absence of RE (Figure 3E).
The more severe effect of reducing homology on the X side likely reflects the need to clip off the ssDNA Ya tail before new DNA synthesis can be initiated. Tail-clipping may be inefficient when the annealed homologous regions is short, as with single-strand annealing (SSA) (Sugawara et al., 2000). This decrease could also be attributed to possible formation of a second nonhomologous tail formed by copying of HML beyond the short 50 bp X homology. This second nonhomologous tail would also need to be clipped to complete SDSA.
Cells shift from GC to BIR as homology at non-invading end is decreased
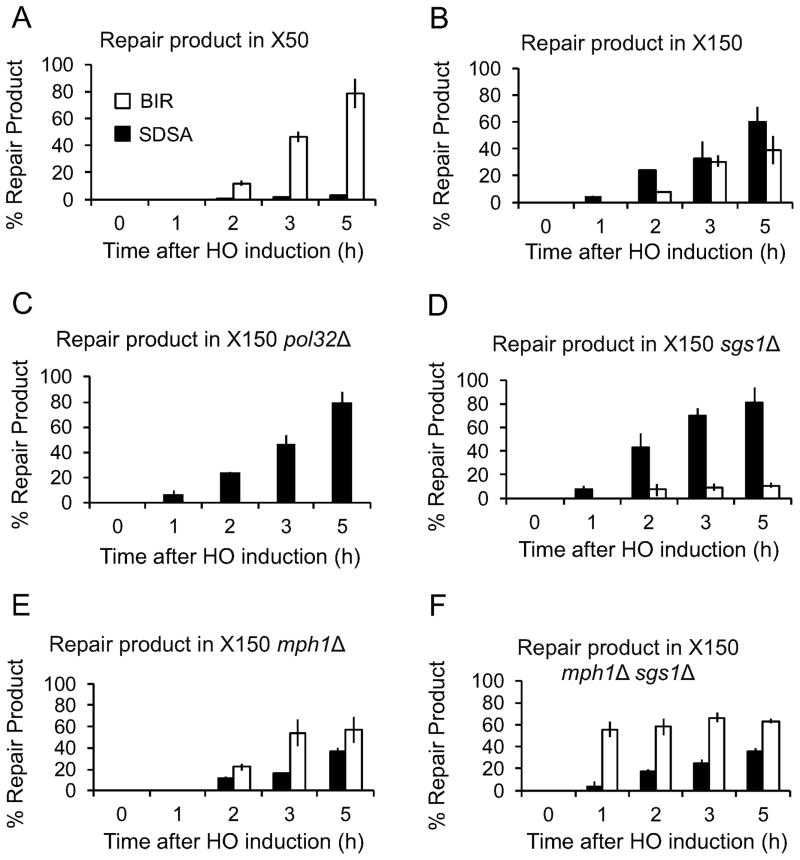
Even when X homology was reduced, strand invasion and initial new DNA synthesis from the invading end were unaltered; nonetheless, a severe drop in viability occurred. We hypothesized that short homology at the second end adversely affected second-end capture. Without such capture, repair might proceed instead through BIR. In this intrachromosomal repair system, BIR leads to an inviable outcome (Figure S2). To monitor repair, we examined Southern blots (Figure 4), which can be compared to normal repair for WX1433 in Figures S1A (RE+) and 2B. In the X150 strain, GC product was seen 1 h after HO induction (Figure 4A, middle panel) while a band corresponding to the expected BIR repair product appeared only at 2 h. To confirm that the new band did indeed result from BIR, we deleted the nonessential Pol32 subunit of DNA polymerase δ, which is required for BIR but not GC (Jain et al., 2009; Lydeard et al., 2007). No BIR product was seen without Pol32 (Figure 4A, right panel). With even less homology length, the X50 strain showed a much higher ratio of BIR product compared to GC (Figure 4A, left panel). The ratios of SDSA v. BIR for X150 and X50 are shown in Figures 4B and S3A.
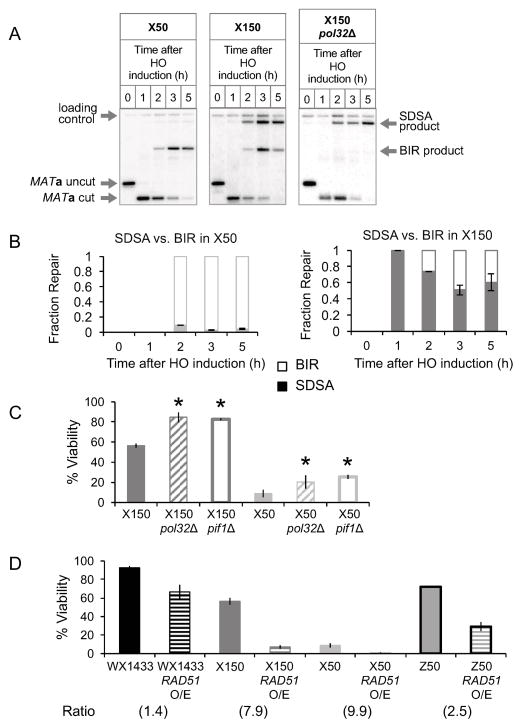
Figure 4. Cells shift from GC to BIR as length of non-invading homology is decreased.
(A) Representative Southern blots showing DSB repair kinetics and products in X50, X150 and X150 pol32Δ strains. DNA was digested with Bsp1286I and probed with a MAT distal fragment. (B) Graphs show quantitative densitometric analysis of repair Southern blots for X50 (left panel) and X150 (right panel). Proportions of SDSA product (solid color graph) and BIR product (unfilled graph) over total product. Data represent mean ± SD (n = 3). (C) Viability of WT (solid color graph), pol32Δ (hatched graph) and pif1Δ (white graph) δX strains. No significant change was observed in WX1433 deletion mutants (Figure S3B). Data represent mean ± SD (n ≥ 3). Asterisks denote significant differences between WT and mutant (P ≤ 0.001). (D) Viability of WT (solid graph) and RAD51 over-expression strains of WX1433, X150, X50 and Z50 (hatched graph). Data represent mean ± SD (n ≥ 3). Ratio of the viability obtained in WT strain : RAD51 overexpression-containing strain is indicated at the bottom of each strain. See also Figure S2, Figure S3, Figure S5 and Figure S6.
Surprisingly, viability of X150 and X50 strains markedly increased without Pol32 (Figure 4C). A similar increase was found by deleting PIF1, a 5′ to 3′ helicase that, like Pol32, is required for BIR but not GC (Chung et al., 2010; Wilson et al., 2013). Viability of the WX1433 control remained high in the absence of these two factors (Figure S3B). Thus, when prohibited from completing BIR, more cells repaired by SDSA. The GC product band intensities in Southern blots correlated with the observed viabilities (Figures 5A, 5B and 5C). Together, these data demonstrate that when the second-end homology length is reduced, cells shift towards BIR repair, as fewer cells manage to capture the second end. Moreover, the blocks in BIR by removing Pol32 or Pif1 must be reversible, allowing an increase in GC. It should be noted that we have measured the ratio of SDSA to BIR after 5 h, when most SDSA repair is complete (Hicks et al. 2011). If cells are harvested at 8 h, the proportion of SDSA noticeably increases (Figure S4) as repaired and viable SDSA cells overgrow the inviable BIR cells.
Figure 5. Sgs1 and Mph1 helicases have opposing roles in repair pathway choice when non-invading homology length is reduced.
Graphs showing quantitative densitometric analysis of repair Southern blots for X50, X150 and mutant X150 strains as denoted under the axes. The amount of product obtained from 5h time point was set to 100%. Data represent mean ± SD (n = 3).
The presence of Rad51 impairs Rad52-mediated annealing of complementary ssDNA, a property particularly important for efficient second-end capture (Shi et al., 2009; Sugiyama and Kantake, 2009; Wu et al., 2008). While overexpression of RAD51 had a modest effect on WX1433 and Z50, it notably reduced the viability of X150 and X50 strains (Figure 4D). This implies that annealing needed for second-end capture is impaired by excess Rad51, similar to Rad51’s effect on SSA (McDonald and Rothstein, 1994).
SGS1 and MPH1 have opposing influence on repair pathway choice when non-invading homology length is reduced
The ability to accomplish successful second-end capture could be mediated by a helicase that can either alter the size of a D-loop or influence heteroduplex rejection. Especially when homology at the non-invading end is short, helicases might play a major role in modulating the balance between GC and BIR.
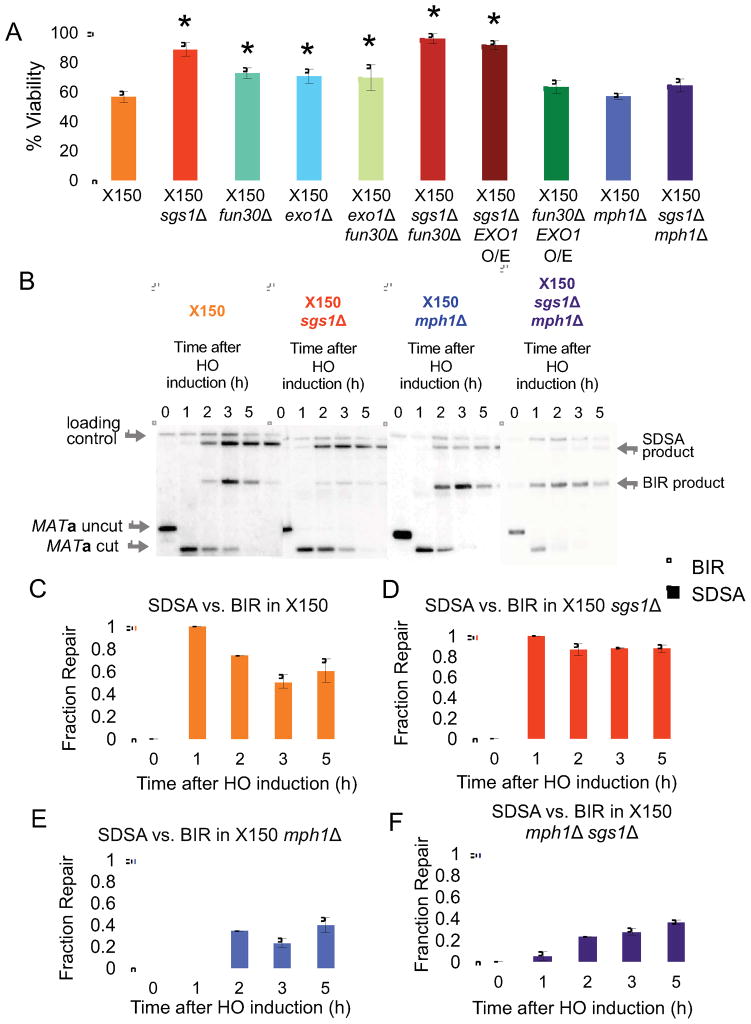
We deleted Sgs1, a 3′-to-5′ helicase related to human BLM, that has been variously implicated in disruption of HR intermediates (Branzei and Foiani, 2007b), resolution of dHJs to yield NCO (Ira et al., 2003; Lo et al., 2006), regression of stalled replication forks (Bachrati and Hickson, 2008; Branzei and Foiani, 2007a), repressing BIR (Jain et al., 2009; Lydeard et al., 2010) and resection of DSB ends (Gravel et al., 2008; Mimitou and Symington, 2008; Zhu et al., 2008). Deletion of SGS1 significantly increased the viability of both the X50 and X150 strain, restoring the viability of the X150 strain to WX1433 levels (Figures 6A and S3B). Physical monitoring of the repair products showed a substantial shift towards repair by GC (Figure 6B; compare Figure 6C with Figure 6D). Quantification of the Southern blots showed strong correlation between the levels of GC repair product and the viability (Figure 5D).
Figure 6. Sgs1 and Mph1 helicases have opposing roles in repair pathway choice when non-invading homology length is reduced.
(A) Viability of WT and mutant X150 strains. Data represent mean ± SD (n ≥ 3). No significant change was observed in the WX1433 deletion mutants (Figure S3B). Asterisks denote significantly different (P ≤ 0.001) viabilities of the WT and marked mutant strains. (B) Representative Southern blots showing DSB repair kinetics and products in X150, X150 sgs1Δ, X150 mph1Δ, and X150 mph1Δ sgs1Δ strains, digested with Bsp1286I and probed with a MAT distal fragment. (C) Quantitative densitometric analysis of repair Southern blots in Figure 6B. Data are represented as ratio of SDSA product (solid color graph) and BIR product (unfilled graph) over total product for a given time-point, which is normalized as 1. Data represent mean ± SD (n = 3). See also Figure S3, Figure S4, Figure S5 and Figure S6.
Slower resection improves ectopic DSB repair (Lee et al., 2016). To assess whether the increase in GC viability was caused by a resection defect in sgs1Δ, we deleted two factors involved in resection: Fun30, a Swi2/Snf2 chromatin remodeler (Chen et al., 2012; Costelloe et al., 2012; Eapen et al., 2012), and exonuclease Exo1 (Mimitou and Symington, 2008; Nimonkar et al., 2011; Zhu et al., 2008). Deletion of FUN30 or EXO1 or a fun30Δ exo1Δ double mutant improved viability, but the increase was not as substantial as observed in sgs1Δ (Figure 6A). Viability remained high when FUN30 was deleted in sgs1Δ. The ratio of SDSA v. BIR remained unaltered in the absence of Fun30 or Exo1 (Figure S5). In agreement with our recent study of interchromosomal ectopic recombination (Lee et al., 2016), overexpression of EXO1 in fun30Δ restored viability back to WT levels; however, overexpressing EXO1 in sgs1Δ had no effect (Figure 6A). Together, these data suggest that the switch in repair pathway in the absence of SGS1 is not principally related to its resection defect.
To test whether the helicase domain of Sgs1 is required to repair by BIR in strains with low non-invading homology length, we complemented the X150 sgs1Δ strain with a plasmid carrying the helicase-dead mutant sgs1-hd (Jain et al., 2009). Although addition of the helicase-dead mutant appeared still to have some ability to complement sgs1Δ compared to an empty vector (Figure S3C), the difference is not statistically significant (p > 0.0125, with Bonferroni correction). We conclude that the helicase domain of Sgs1 is important to increase repair by BIR, although it is possible that some of the repair functions of Sgs1 are helicase-independent (Lo et al., 2006).
We also examined the effect of deletion of another ATP-dependent 3′ to 5′ helicase, Mph1, an ortholog of human Fanconi anemia protein FANCM. Mph1 dissociates Rad51-dependent D-loops and channels recombination intermediates into the NCO pathway (Jain et al., 2016; Luke-Glaser and Luke, 2012; Prakash et al., 2009). As shown in Figures 6B and 6E, the intensity of the BIR band was significantly higher in X150 mph1Δ as compared to X150. Unlike pol32Δ and pif1Δ, where a decrease in BIR product was reflected in an increase in viability, despite an increase in BIR, mph1Δ did not show the expected drop in viability (Fig. 6A). We have no clear explanation for this discrepancy. It is possible that some apparent Mph1 events are reversed, even after completing enough DNA synthesis to create the restriction fragment assayed in Figure 5; complete BIR to the end of the chromosome would require another ~10 kb of synthesis. Nevertheless, it is clear that deleting MPH1 channels repair away from SDSA and into a BIR pathway (see Discussion).
Since Sgs1 and Mph1 appeared to play opposing roles, we made a double deletion in X150. Recently we have shown that sgs1Δ mph1Δ double mutants increased the efficiency of BIR and also eliminated the delay in product appearance that is normally seen for BIR (Jain et al., 2016). Here too, simultaneous deletion of both MPH1 and SGS1 increased BIR and also accelerated the appearance of this product (Figures 5F, 6B, and 6F), but again there was no concomitant reduction in viability.
Loss of Sgs1 or Mph1 has been reported to channel DSB repair events into the dHJ pathway with an increase in CO-associated GC repair (Ira et al., 2003; Mitchel et al., 2013; Prakash et al., 2009). However, in this intrachromosomal recombination system, few of the repair events proved to be reciprocal COs (Figure S6).
Discussion
We have analyzed homology requirements for GC repair in strains with varying homology lengths in budding yeast MAT switching, an intrachromosomal DSB repair event in which one of the two DSB ends almost always initiates recombination. Even in the absence of the assistance of the RE, recombination is ~30% efficient with only 35 bp of homology between the DSB end and the HML donor, but when the RE becomes tethered in the vicinity of the DSB, successful repair almost doubles.
RE improves the efficiency of short homology at the invading end, but also accelerates the rate of repair (Figure 3). RE acts as a tether to enhance the proximity between the donor and the DSB (Figure 7A, step 3), apparently providing more opportunities for Rad51 to identify a small region of homology to achieve strand invasion. Shen and Huang (1986) first articulated the notion of a minimum efficient processing segment. Our results suggest that the definition of this length can be altered by increasing the tethering between the DSB and a donor. The fact that tethering a donor sequence close to the site of a DSB can improve both the efficiency and the kinetics of repair suggests that adopting such an approach may be important in gene editing. Indeed, Ruff et al. (2014) have shown that aptamer-guided tethering of a ssDNA sequence improves gene targeting in yeast and mammals. As expected, although RE affects repair of a short Z sequence, reducing X homology did not affect the kinetics of strand invasion or primer extension of the Z end.
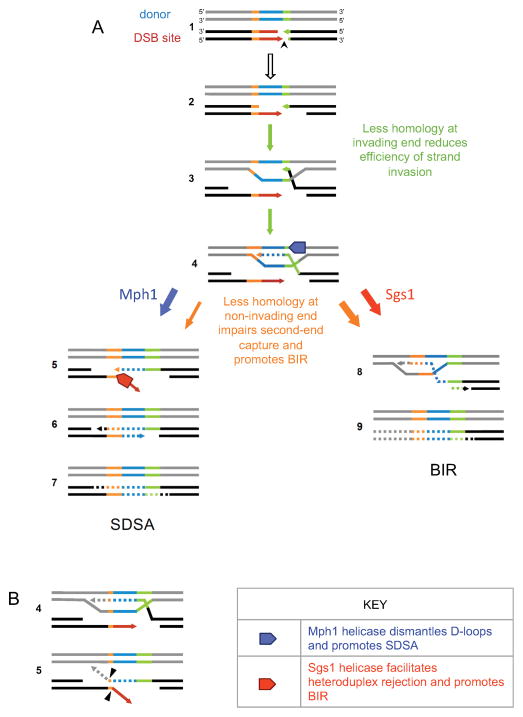
Figure 7. Proposed model for effect of reduced homology length on DSB repair pathway choice.
(A) Intermediate steps of repair of a DSB (black arrowhead) are illustrated. Less homology at the invading end (green) reduces the efficiency of strand invasion and kinetics of new DNA synthesis (steps 3 and 4, green arrows). Changes in non-invading homology (orange) has no effect on these steps. Short homology at the non-invading end affects second-end capture (step 5, orange arrows), reducing SDSA and diverting it towards BIR (steps 8 and 9). With short second-end homology, Sgs1 promotes heteroduplex rejection and channels repair towards BIR (red arrow) while Mph1 helicase disassembles the D-loop required for BIR and consequently promotes SDSA (blue arrow). (B) After Rad51 forms a stable strand invasion intermediate, repair DNA synthesis is initiated and likely proceeds beyond the shared non-invading homology in the X50 strain (step 4). This could lead to formation of a second nonhomologous tail (dotted gray line) after second-end capture, which would need to be removed along with the Ya tail, before this first 3′ end can be used as a primer to initiate gap filling synthesis. Arrowheads show clipping by the Rad1–Rad10 endonucleases.
When homology at the non-invading end is reduced below 200 bp, second-end capture is inefficient and increasingly results in repair by BIR; with only 50 bp, 7% of the cells repair by SDSA (Figure 4). This is much less than the 38% observed for the Z end in the absence of RE. We attribute this difference to the need not only to pair the two strands (which should be easier than Rad51-mediated strand invasion per se) but to clip off the nonhomologous Ya tail. This tail removal may become more difficult when the length of the annealed region is reduced, as we have inferred in studies of SSA, where several additional proteins are required when homologies are short (Sugawara et al., 2000). Moreover the process may be more complex when the length of Z is very short, because when the primer-extended first strand anneals it may produce a second nonhomologous tail (Figure 7B). Indeed, (McCulloch et al., 2003) showed that synthesis can proceed beyond HML W region in WT cells. However, second end capture most likely occurs much sooner than when the entire 1433 bp W-X regions is copied, given that mutated sites in HML X are transferred to MAT much more often than those in HML W (McGill et al., 1989).
Although GC dominates repair when the length of homology on either side of the DSB is >300 bp, the fact that GC and BIR are in competition becomes increasingly evident as the X homology decreases, so that with X50 BIR occurs ten times as often as GC. Unexpectedly, mutations that impair BIR (pol32Δ, pif1Δ) do not simply arrest BIR, they allow increased GC and a 3-fold increased viability. Thus, when an initial strand invasion has been accomplished it is possible to re-direct intermediates back to GC when key requirements for BIR have been eliminated.
With short X homologies, deletion of SGS1 increased SDSA while reducing BIR, whereas deletion of MPH1 actively promoted BIR. These outcomes inform a hierarchy of effects of these mutants on repair. When BIR is the only possible outcome, deleting SGS1 or MPH1 each increase BIR whereas overexpressing either helicase depresses BIR (Luke-Glaser and Luke, 2012; Lydeard et al., 2010; Prakash et al., 2009); but here mph1Δ leads to an increase in BIR over GC. When GC is the predominant outcome, deleting MPH1 led to an increase in associated crossovers (COs) (Mitchel et al., 2013; Prakash et al., 2009), which was interpreted to mean that there was a shift from SDSA to dHJ intermediates that could then be resolved by HJ resolvases to produce more COs. But here we also fail to see an increase in COs (Figure S6). We postulate that the very short X homology (50 – 150 bp) might be incapable of forming/sustaining a stable double Holliday junction required for crossing-over. We conclude that the predominant effect of deleting MPH1 is to shift intermediates away from SDSA, either toward dHJ intermediates or – as in this case - to a higher level of BIR. If the homology at the second end is very short, the absence of Sgs1 could lead to formation of a more stable heteroduplex structure by second-end capture, much as sgs1Δ improves SSA when these annealing strands have 3% mismatches (Sugawara et al., 2004). We note that despite the obvious increase in the proportion of BIR events in mph1Δ, there was not a concomitant decrease in viability. When cells are grown in liquid culture we see clear evidence that cells that repaired by SDSA outgrow the inviable BIR recombinants. When cells are plated directly on galactose-containing plates, as we do to measure viability, this competition should not happen, but it is possible that mph1Δ cells only repair after some cells have divided, increasing the proportion of colonies that will harbor any SDSA event.
Recently we reported that the mph1Δ sgs1Δ double mutant exhibited a synergistic increase in the efficiency and kinetics of BIR (Jain et al., 2016). The results here are in agreement with this finding; Southern blots showed a more rapid appearance of the BIR product (Figures 5 and 6); however, the level of BIR was not greater than that seen for mph1Δ alone. The viability of the double mutant was similar to mph1Δ, indicating that the ability to repair by GC was similar to mph1Δ (Figure 6A). Thus, at short non-invading homology lengths, Mph1’s role in dismantling D-loops (Figure 7A, step 4) and blocking BIR appears to take precedence over Sgs1’s effect on heteroduplex rejection (Figure 7A, step 5) (Mitchel et al., 2013).
We showed previously that the overall sequence of events at MAT is the same as seen when an HO-induced DSB is made in other contexts (Ira and Haber, 2002; Jain et al., 2009; Jain et al., 2016; Kim and Haber, 2009; Lee et al., 2016); hence, we believe that the effect of limited homology at the non-invading end will influence competition between GC and BIR in all DSB-induced HR events.
To ensure GC repair, both ends of the DSB need to communicate with the homologies at the donor before new DNA synthesis can begin (Jain et al., 2009; Jain et al., 2016). This recombination execution checkpoint (REC) confers a kinetic delay in primer extension of several hours in BIR repair (Jain et al., 2009; Lydeard et al., 2007; Malkova et al., 2005) compared with GC. Nevertheless, there is evidence to suggest that both ends of the DSB might still be involved in preliminary communication. Rad51 is recruited to both the HML Z and HML X regions 30–45 min after HO induction, even in the presence of the NH-Ya tail (Hicks et al., 2011). Thus the second end was able to interact with the donor despite being unable to effectively instigate new DNA synthesis because of the hindrance caused by the NH-tail. We propose that the second end might form transient side-by-side paranemic base pairings (Christiansen and Griffith, 1986), which might be sufficient for the REC to direct repair towards GC instead of BIR. It is interesting to note that the budding yeast MAT locus has evolved to share 4 times more homology at the non-invading end than at the invading end. This might be a way to prevent BIR, which could lead to deleterious genome rearrangements and translocations and in this case an acentric chromosome. This large homology might also safeguard against Sgs1’s role in promoting heteroduplex rejection and hence ensure the absence of BIR repair during MAT switching.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author, Dr. James E. Haber (haber@brandeis.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains were freshly thawed from frozen stocks and grown at 30 °C using standard practices.
METHOD DETAILS
Strains and Plasmids
All strains except when noted are isogenic to YAM033 (hoΔade3::GAL-HO HMLα-inc MATa hmr::ADE1 bar1Δ::ADE3 nej1Δ::KANMX ade1 leu2,3-112 trp1::hisG ura3-52 thr4 lys5) a meiotic segregant of a diploid created by mating YJK139 (Kim and Haber, 2009) and MAV015 (Valencia et al., 2001). The Z2216 strain was a segregant of a diploid from mating YAM033 and ECY490 (Coïc et al., 2011). HMR was deleted in ECY490 using a LEU2 marker. All other δZ strains were constructed by insertion of Candida glabrata TRP1 from plasmid pYO2242 (Sekiya-Kawasaki et al., 2002) at different distances distal to MAT to create different Z length homologies (primers listed in Table S2). δX strains were constructed by complete deletion of MAT-W and partial deletion of MAT-X regions by introducing the C. glabrata TRP1 from pYO2242 (Sekiya-Kawasaki et al., 2002) at different distances proximal to MAT (primers listed in Table S2). reΔ, sgs1Δ, pol32Δ, pif1Δ, exo1Δ, fun30Δ, mph1Δ and all double mutants were made by the standard PCR-based gene disruption method. X150 sgs1Δ + sgs1-hd strain was built by introducing plasmid pSJ23 (Jain et al., 2009) into X150 sgs1Δ. To overexpress EXO1, yeast strains were transformed with ClaI digested plasmid pJH1944 (a gift from Achille Pellicioli) that integrates a galactose-inducible full copy of the gene at the LEU2 locus. To overexpress RAD51, yeast strains were transformed with plasmid pDBL(RAD51) (Milne et al., 1995) expressing RAD51 under the ADH1 promoter. A list of all strains used in this study is in Table S1.
HO induction time courses
Yeast cells were grown in YEP-lactate to 5 X 106 cells/ml. HO endonuclease was induced by adding 2% galactose. Samples were collected for DNA/protein analysis just prior to and at different time points following addition of galactose, as described before (Holmes and Haber, 1999).
Viability measurements
Yeast cells were grown in YEP-lactate to a density of 5 × 106 cells/ml. Equal number of cells were plated on YEP containing 2% galactose (YEPGal) and YEP with 2% dextrose (YEPD). Viability was determined from the ratio of colony-forming units (CFUs) on YEPGal to CFUs on YEPD.
Southern blot Southern blot
Genomic DNA (gDNA) was digested using Bsp1286I following NEB reaction conditions. Digested DNA was precipitated using the MasterPure Yeast DNA Purification Kit protocol, separated using 1% agarose gel electrophoresis for 2 hours at 100V and transferred to GeneScreen Plus nylon membranes by vacuum blotting using standard procedures. Southern blots were probed with a 32P-labeled MAT distal fragment. All quantitative densitometric analysis was done using ImageQuant TL 5.2 software. To control for differences in gDNA loaded into each lane, each fragment of interest was normalized to a loading control fragment: ACT1 fragment was used for RE+ and reΔ; and MLH1 gene fragment (restriction digest fragment) for the rest of the blots. DNA from RE+ and reΔ was digested with AseI and DNA from all δX strains with Bsp1286I. For Figure S6, Southern blots were stripped using 2% SDS and re-probed with a MAT proximal probe. All primers used to synthesize the probes are listed in Table S2.
ChIP assay
Rad51 ChIP samples were prepared with lysis buffer containing 50mM HEPES pH7.5, 0.5M EDTA, 5M NaCl, 10% v/v Triton X-100, 0.1% NaDOC, 1mg/ml Bacitracin, 1mM Benzamidine and 1mM PMSF. Samples were incubated with antibody for 1 hour at 4°C and precipitated using Protein G agarose beads for anti-rfa1 and Protein A agarose beads for anti-rad 51. All ChIP data were analyzed by quantitative real-time (qRT) PCR using Qiagen Rotor Gene Q. All ChIP primer sequences are listed in Table S2. The IP signal from the donor locus was normalized to the IP signal from the ARG5,6 locus. ChIP data are presented as a ratio of ratios (HML IP/ARG5,6 IP) (Figure 2C) and (MAT IP/ARG5,6 IP) (Figure S1B).
Primer extension and non-homologous tail clipping assay
Primer extension assay and non-homologous tail clipping assay was performed. They were analyzed by qRT-PCR using Qiagen Rotor-Gene Q and normalized to ARG5,6. For Z2216 RE+ and Z2216 reΔ strain time courses, PCR reactions were run on 1% agarose gels and the repair product was quantified using Bio-Rad Quantity One software, normalized to PCR signal from the ADE2 locus. Primers used are described in Table S2. Data are presented as a percentage of repaired MATα colony.
Western blot analysis
Sample prep was carried out using using 5% trichloroacetic acid (TCA) with glass beads and resuspended in 200 ul of 6X Laemmli Buffer and 100ul of Tris pH8, boiled for 5 minutes at 100°C and spun down for 1 minute at 13000 rpm. Samples were resolved in 6% SDS-PAGE gel using the Biorad Mini PROTEAN Tetra System for 20 minutes at 60V to stack samples then shifted to 100V. The resolved gel was transferred to PVDF membranes using the Biorad Mini Trans-Blot Cell Tetra System run at 100V for 1 hour and 15 minutes at 4°C. Blocking was performed using Li-Cor blocking buffer/5% Milk TBS Tween and blotted with anti-Rad53 antibody purchased from Abcam (ab166859). Analysis was performed using Bio-rad Quantity One Software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Southern blot analysis
Quantitative densitometric analysis of the Southern blots was carried out using ImageQuant software as previously described (Hicks et al., 2011). Fragments of interest were normalized to a loading control fragment: ACT1 fragment was used for RE+ and reΔ; and MLH1 gene fragment for other blots.
KEY RESOURCES TABLE
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Rad53 antibody | Abcam | ab166859 |
| Chemicals, Peptides, & Recombinant Proteins | ||
| Taq DNA Polymerase | Promega | M300F |
| Fail Safe 2X PCR Mix E - Buffer E | Epicenter | FSP995E |
| AseI | New England Biolabs | R0526M |
| NEB Buffer 3.1 | New England Biolabs | B7203S |
| Bsp1286I | New England Biolabs | R0120L |
| ClaI | New England Biolabs | R0197S |
| 10X Cut Smart Buffer | New England Biolabs | B7204S |
| Easy Tides Deoxyadenosine 5′ triphosphate (32P) | Perkin Elmer | BLU512H250HC |
| GeneScreen Plus Nylon Hybridization Transfer Membrane | Perkin Elmer | NEF988001PK |
| Agarose Low EEO (EP) Electrophoresis Grade | US Biological | A1016 |
| Critical Commercial Assays | ||
| AccuPrep Plasmid Mini Extraction Kit | Bioneer | K3030 |
| Gene Jet PCR Purification Kit | Thermo Scientific | K0702 |
| MasterPure Yeast DNA Purification Kit | Epicentre | MPY80200 |
| DECA primer II - Random Primed DNA Labeling Kit | Ambion | AM1456 |
| Experimental Models: Organisms/Strains | ||
| See Table S1 for a list of yeast strains used in this study | ||
| Sequence Based Reagents | ||
| See Table S2 for a list of all oligonucleotides used in this study | IDT (Integrated DNA Technologies) | |
| Software and Algorithms | ||
| Bio-Rad Quantity One Software Version 4.5.2 | Bio-Rad | www.bio-rad.com |
| Image Quant TL Version 5.2 | GE Healthcare | www.gelifesciences.com |
| Lasergene SeqBuilder | DNAStar | www.dnastar.com/t-seqbuilder.aspx |
| Rotor Gene Q software Version 2.2.3 | Qiagen | www.qiagen.com |
| Equipment | ||
| Bio-Rad Mini PROTEAN Tetra System | Bio-Rad | #165-8004 |
| Bio-Rad Mini Trans-Blot Cell | Bi0-Rad | #1703930 |
Supplementary Material
Acknowledgments
We are grateful to Akira Shinohara for anti-Rad51 antibody, Yoshikazu Ohya for the Candida glabrata plasmids and Achille Pellicioli for the EXO1 plasmid. We thank members of Haber laboratory for helpful comments and suggestions. This work was supported by NIH grants GM20056 and GM76020.
Footnotes
Author Contributions
Conceptualization and Design of Experiments: A.M. and J.E.H.; Formal Analysis: A.M.; Writing, review and editing: A.M. and J.E.H.; Experimentation: A.M. and A.B.; Funding Acquisition: J.E.H.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anand RP, Lovett ST, Haber JE. Break-induced DNA replication. Cold Spring Harbor perspectives in biology. 2013;5:a010397. doi: 10.1101/cshperspect.a010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avşaroğlu B, Bronk G, Li K, Haber JE, Kondev J. Chromosome refolding model of mating-type switching in yeast. Proc Natl Acad Sci USA. 2016 doi: 10.1073/pnas.1607103113. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA repair. 2007a;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev. 2007b;21:3019–3026. doi: 10.1101/gad.1624707. [DOI] [PubMed] [Google Scholar]
- Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Niu H, Chung WH, Zhu Z, Papusha A, Shim EY, Lee SE, Sung P, Ira G. Cell cycle regulation of DNA double-strand break end resection by Cdk1-dependent Dna2 phosphorylation. Nature structural & molecular biology. 2011;18:1015–1019. doi: 10.1038/nsmb.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G, Griffith J. Visualization of the paranemic joining of homologous DNA molecules catalyzed by the RecA protein of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:2066–2070. doi: 10.1073/pnas.83.7.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Zhu Z, Papusha A, Malkova A, Ira G. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS genetics. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coïc E, Martin J, Ryu T, Tay SY, Kondev J, Haber JE. Dynamics of homology searching during gene conversion in Saccharomyces cerevisiae revealed by donor competition. Genetics. 2011;189:1225–1233. doi: 10.1534/genetics.111.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coïc E, Richard GF, Haber JE. Saccharomyces cerevisiae donor preference during mating-type switching is dependent on chromosome architecture and organization. Genetics. 2006a;173:1197–1206. doi: 10.1534/genetics.106.055392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coïc E, Sun K, Wu C, Haber JE. Cell cycle-dependent regulation of Saccharomyces cerevisiae donor preference during mating-type switching by SBF (Swi4/Swi6) and Fkh1. Mol Cell Biol. 2006b;26:5470–5480. doi: 10.1128/MCB.02443-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly B, White CI, Haber JE. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2342–2349. doi: 10.1128/mcb.8.6.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol Cell Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol Cell Biol. 2012;32:4727–4740. doi: 10.1128/MCB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan S, Simpson RT. Global chromatin structure of 45,000 base pairs of chromosome III in a- and alpha-cell yeast and during mating-type switching. Mol Cell Biol. 2004;24:10026–10035. doi: 10.1128/MCB.24.22.10026-10035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor GB, Nassif NA, Johnson-Schlitz DM, Preston CR, Engels WR. Targeted gene replacement in Drosophila via P element-induced gap repair. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Genome Stability: DNA Repair and Recombination. Garland Science; 2013. [Google Scholar]
- Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- Herskowitz I, Jensen RE. Putting the HO gene to work: practical uses for mating-type switching. Methods in enzymology. 1991;194:132–146. doi: 10.1016/0076-6879(91)94011-z. [DOI] [PubMed] [Google Scholar]
- Hicks WM, Yamaguchi M, Haber JE. Real-time analysis of double-strand DNA break repair by homologous recombination. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3108–3115. doi: 10.1073/pnas.1019660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol Cell Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Satory D, Haber JE. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol Cell Biol. 2006;26:9424–9429. doi: 10.1128/MCB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Haber JE. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23:291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Sugawara N, Mehta A, Ryu T, Haber JE. Sgs1 and Mph1 Helicases Enforce the Recombination Execution Checkpoint During DNA Double-Strand Break Repair in Saccharomyces cerevisiae. Genetics. 2016;203:667–675. doi: 10.1534/genetics.115.184317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harbor perspectives in biology. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1151–1156. doi: 10.1073/pnas.0812578106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Haber JE. Mating-type Gene Switching in Saccharomyces cerevisiae. Microbiology spectrum. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0013-2014. MDNA3-0013-2014. [DOI] [PubMed] [Google Scholar]
- Lee CS, Wang RW, Chang HH, Capurso D, Segal MR, Haber JE. Chromosome position determines the success of double-strand break repair. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E146–154. doi: 10.1073/pnas.1523660113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy C, Lee SE, Vaze MB, Ochsenbein F, Guerois R, Haber JE, Marsolier-Kergoat MC. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Molecular cell. 2003;11:827–835. doi: 10.1016/s1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Li F, Dong J, Pan X, Oum JH, Boeke JD, Lee SE. Microarray-based genetic screen defines SAW1, a gene required for Rad1/Rad10-dependent processing of recombination intermediates. Molecular cell. 2008;30:325–335. doi: 10.1016/j.molcel.2008.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Coïc E, Lee K, Lee CS, Kim JA, Wu Q, Haber JE. Regulation of budding yeast mating-type switching donor preference by the FHA domain of Fkh1. PLoS genetics. 2012;8:e1002630. doi: 10.1371/journal.pgen.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YC, Paffett KS, Amit O, Clikeman JA, Sterk R, Brenneman MA, Nickoloff JA. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol Cell Biol. 2006;26:4086–4094. doi: 10.1128/MCB.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B. The Mph1 helicase can promote telomere uncapping and premature senescence in budding yeast. PloS one. 2012;7:e42028. doi: 10.1371/journal.pone.0042028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE. Sgs1 and exo1 redundantly inhibit break-induced replication and de novo telomere addition at broken chromosome ends. PLoS genetics. 2010;6:e1000973. doi: 10.1371/journal.pgen.1000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Cell Biol. 2005;25:933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch RD, Read LR, Baker MD. Strand invasion and DNA synthesis from the two 3′ ends of a double-strand break in Mammalian cells. Genetics. 2003;163:1439–1447. doi: 10.1093/genetics/163.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JP, Rothstein R. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics. 1994;137:393–405. doi: 10.1093/genetics/137.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- McGill C, Shafer B, Strathern J. Coconversion of flanking sequences with homothallic switching. Cell. 1989;57:459–467. doi: 10.1016/0092-8674(89)90921-5. [DOI] [PubMed] [Google Scholar]
- Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harbor perspectives in biology. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne GT, Ho T, Weaver DT. Modulation of Saccharomyces cerevisiae DNA double-strand break repair by SRS2 and RAD51. Genetics. 1995;139:1189–1199. doi: 10.1093/genetics/139.3.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel K, Lehner K, Jinks-Robertson S. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS genetics. 2013;9:e1003340. doi: 10.1371/journal.pgen.1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Molecular cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff P, Koh KD, Keskin H, Pai RB, Storici F. Aptamer-guided gene targeting in yeast and human cells. Nucleic acids research. 2014;42:e61. doi: 10.1093/nar/gku101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Sekiya-Kawasaki M, Abe M, Saka A, Watanabe D, Kono K, Minemura-Asakawa M, Shen P, Huang HV. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi I, Hallwyl SC, Seong C, Mortensen U, Rothstein R, Sung P. Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. The Journal of biological chemistry. 2009;284:33275–33284. doi: 10.1074/jbc.M109.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9315–9320. doi: 10.1073/pnas.0305749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Paques F, Colaiacovo M, Haber JE. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Molecular cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kantake N. Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates. Journal of molecular biology. 2009;390:45–55. doi: 10.1016/j.jmb.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Sun K, Coïc E, Zhou Z, Durrens P, Haber JE. Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer. Genes Dev. 2002;16:2085–2096. doi: 10.1101/gad.994902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Toh GW, Sugawara N, Dong J, Toth R, Lee SE, Haber JE, Rouse J. Mec1/Tel1-dependent phosphorylation of Slx4 stimulates Rad1–Rad10-dependent cleavage of non-homologous DNA tails. DNA repair. 2010;9:718–726. doi: 10.1016/j.dnarep.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia M, Bentele M, Vaze MB, Herrmann G, Kraus E, Lee SE, Schar P, Haber JE. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature. 2001;414:666–669. doi: 10.1038/414666a. [DOI] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Molecular cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLalpha. Mol Cell Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CI, Haber JE. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. The EMBO journal. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung WH, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, et al. Pif1 helicase and Poldelta promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolner B, Peterson CL. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. The Journal of biological chemistry. 2005;280:10855–10860. doi: 10.1074/jbc.M414388200. [DOI] [PubMed] [Google Scholar]
- Wu C, Weiss K, Yang C, Harris MA, Tye BK, Newlon CS, Simpson RT, Haber JE. Mcm1 regulates donor preference controlled by the recombination enhancer in Saccharomyces mating-type switching. Genes Dev. 1998;12:1726–1737. doi: 10.1101/gad.12.11.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wu X, Haber JE. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls Rad52-mediated DNA annealing. The Journal of biological chemistry. 2008;283:14883–14892. doi: 10.1074/jbc.M801097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.