Abstract
Recently, the development of antibiotic resistance emerged as a significant clinical problem in the eradication of Helicobacter pylori. We investigated the MICs of antibiotics for 135 H. pylori isolates from adults in Seoul, South Korea, over the past 16 years. The MICs of amoxicillin, clarithromycin, metronidazole, tetracycline, azithromycin, and ciprofloxacin increased from 1987 to 2003. Rates of primary resistance to clarithromycin increased from 2.8% in 1994 to 13.8% in 2003. The A2144G mutation was frequently observed in the 23S rRNA gene in clarithromycin-resistant isolates. The increase in resistance to clarithromycin seems to result in a decrease in eradication efficacy for H. pylori. These results suggest that the MICs of several antibiotics for H. pylori have increased over the past 16 years in Seoul.
Helicobacter pylori infection is recognized as a causal factor of chronic gastritis, peptic ulcer, and gastric cancer (23). Thus, H. pylori should be eradicated in patients with peptic ulceration (23), as eradication not only accelerates ulcer healing (7) but also prevents long-term ulcer relapse (6). Treatment regimens containing a proton pump inhibitor (PPI) and a combination of two or more antibiotics, such as amoxicillin, clarithromycin, metronidazole, and tetracycline, are considered to be most efficacious (17), but drug resistance is a growing problem (1, 8, 15, 16).
The frequencies of resistance to antibiotics have varied widely according to geographical regions and subgroups within study populations (1, 4). For example, metronidazole resistance is found in 10 to 50% of strains from H. pylori-positive adults in the developed world (1, 4, 5), whereas virtually all strains from developing countries are resistant (19). The clarithromycin resistance rate, which is relatively low and ranges from 2 to 15% (1, 3, 5, 12, 21), has also been increasing. This background information makes it clear that there is a need to explore the status of and changes in H. pylori resistance to antibiotics in recent years. Moreover, treatment for H. pylori infection is usually started on an empirical basis, and if an infecting strain is resistant, its successful eradication is hampered. Nevertheless, little information is available regarding the prevalence of primary antibiotic resistance of H. pylori and the mechanism of clarithromycin resistance in South Korea. The aim of this study was to assess the MICs of several antibiotics for H. pylori isolates from patients in Seoul, South Korea, over the past 16 years. The mutations in clarithromycin-resistant H. pylori strains, which could explain the organism's resistance mechanism, especially were investigated in these clinical isolates.
MATERIALS AND METHODS
Patients and H. pylori strains.
One hundred thirty-five strains of H. pylori were isolated from antral gastric mucosal biopsy specimens obtained from adults in Seoul in 1987 (34 strains from Hanyang University Hospital; mean age of patient ± standard deviation, 55.9 ± 17.4 years; male/female ratio, 18:16), in 1994 (36 strains from Seoul National University Hospital; patient age, 50.3 ± 11.2 years; male/female ratio, 17:19), and in 2003 (65 strains from Seoul National University Hospital; patient age, 58.1 ± 10.2 years; male/female ratio, 37:28). The diagnoses in 1987 included gastritis (n = 10 patients), gastric ulcer (n = 8), duodenal ulcer (n = 13), and gastric cancer (n = 3). The diagnoses in 1994 included gastritis (n = 12), gastric ulcer (n = 10), duodenal ulcer (n = 9), and gastric cancer (n = 5). The diagnoses in 2003 included gastritis (n = 4), gastric ulcer (n = 13), duodenal ulcer (n = 47), and gastric cancer (n = 1). No patients had taken antibiotics, PPIs, or nonsteroidal anti-inflammatory drugs during the preceding 3 months. The H. pylori strains were cultured under microaerophilic conditions (5% O2, 10% CO2, and 85% N2) as previously described (13). We obtained one isolate per patient, and all tested strains were the first isolates. All stock cultures were maintained at −70°C in brucella broth supplemented with 15% glycerol. These preparations were thawed and subcultured for experiments.
Antibiotic MIC testing.
The determination of MICs of amoxicillin (Sigma Chemical Co., St. Louis, Mo.), clarithromycin (Abbott Laboratories, Abbott Park, Ill.), metronidazole (Sigma), tetracycline (Sigma), azithromycin (Groton Laboratories, Pfizer Inc., Groton, Conn.), and ciprofloxacin (Sigma) for the H. pylori isolates were examined by use of the serial twofold agar dilution method as described previously (22). Briefly, the bacteria were subcultured on Mueller-Hinton agar supplemented with 5% defibrinated sheep blood for 48 h. A bacterial suspension adjusted to 107 CFU was inoculated directly onto each antibiotic-containing agar dilution plate. After incubation of the plate for 72 h, the MIC of each antibiotic was determined. Quality control was performed with H. pylori ATCC 43504. The MIC breakpoint of clarithromycin was set at >1.0 μg/ml (22).
Restriction fragment length polymorphism analysis and DNA sequencing.
The extraction of H. pylori genomic DNA was performed as reported previously (13). To detect the mutation in the 23S rRNA gene that resulted in clarithromycin resistance, we used oligonucleotide primers (sense, 5′-CGT AAC TAT AAC GGT CCT AAG-3′; antisense, 5′-TTA GCT AAC AGA AAC ATC AAG-3′) and a thermal cycler (GeneAmp PCR system 9600; Perkin-Elmer Cetus, Norwalk, Conn.). The PCR profile consisted of 35 cycles of 1 min of denaturation at 94°C, 1 min of annealing at 57°C, and 1 min of extension at 72°C. Amplicons (291 bp each) of the 23S rRNA gene were either digested with BsaI (New England BioLabs, Beverly, Mass.) for 18 h at 50°C to detect the adenine-to-guanine mutation at position 2144 or digested with BbsI (New England BioLabs) for 18 h at 37°C to detect the A2143G mutation (27). Digested fragments were separated on a 1.5% agarose gel and viewed on a UV transilluminator. Sequencing was performed with the two strands of the nonrestricted amplicons by use of an ABI PRISM 377 DNA sequencer (Applied Biosystems, Foster City, Calif.).
RESULTS AND DISCUSSION
Distribution of antibiotic MICs and prevalence of clarithromycin resistance.
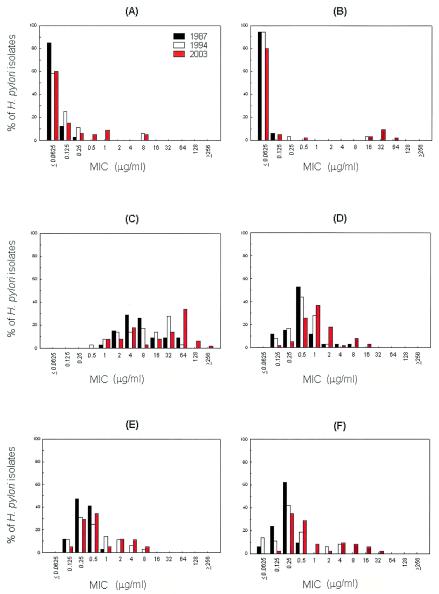
Several studies have found no amoxicillin-resistant strains among primary isolates in Japan (11), Korea (12), or Bulgaria (2), and many studies have shown that all strains examined are susceptible to amoxicillin (1, 10). The official breakpoint for amoxicillin resistance has not been designated for H. pylori isolates by the NCCLS. Therefore, we cannot assign any provisional breakpoint to amoxicillin. However, H. pylori strains for which the amoxicillin MICs were ≥0.5 μg/ml were found in 1994 and 2003; the MICs for 2 strains in 1994 (5.6%) and for 12 strains in 2003 (18.5%) were ≥0.5 μg/ml. Furthermore, the MICs for two strains in 1994 and for three strains in 2003 were also 8 μg/ml (Fig. 1A).
FIG. 1.
Distributions of antibiotic MICs for H. pylori over a 16-year period. Thirty-four strains were examined in 1987, 36 strains were examined in 1994, and 65 strains were examined in 2003. The color key in panel A applies to all panels. MICs were determined by use of the agar dilution method. MICs of amoxicillin (A), clarithromycin (B), metronidazole (C), tetracycline (D), azithromycin (E), and ciprofloxacin (F) are shown.
The patterns of clarithromycin MICs in 1987, 1994, and 2003 were similar (range, 0.0625 to 0.0.5 μg/ml), but MIC ranges broadened over the study period, from 0.0625 to 0.25 μg/ml in 1994 to 0.0625 to 64 μg/ml in 2003 (Fig. 1B), meaning that the prevalence of clarithromycin resistance had increased from 0% (0 of 34 isolates were resistant) to 2.8% (1 of 36 isolates were resistant) in 1994 to 13.8% (9 of 65 isolates were resistant) in 2003 (the resistance breakpoint for clarithromycin was defined as an MIC of 1.0 μg/ml). Interestingly, the clarithromycin MICs showed a bimodal distribution in 2003 (Fig. 1B), when the MICs for clarithromycin-resistant strains ranged between 16 and 64 μg/ml. With respect to clarithromycin, the resistance of H. pylori in vitro was found to be equal to its resistance in vivo (5). Moreover, clarithromycin administration may result in selection for a resistant strain, and this acquired resistance to clarithromycin is stable (19), which could explain the bimodal distribution of clarithromycin MICs found in 2003.
The distribution of metronidazole MICs demonstrated a continuous spectrum ranging between 1 and ≥256 μg/ml and showed a definite shift to high concentrations in 2003 (Fig. 1C). The prevalence of H. pylori isolates for which the metronidazole MICs were ≥8 μg/ml increased from 52.9% in 1987 to 66.2% in 2003. This phenomenon has also been reported in other countries (25). Metronidazole has been widely prescribed for infections such as parasitic or female genital infections in Korea, and the growing use or abuse of this inexpensive drug may have contributed to these increased MICs, also explaining the finding that H. pylori strains for which metronidazole MICs are ≥8 μg/ml are more common in women (Table 1).
TABLE 1.
Distribution of H. pylori isolates for which metronidazole MICs were ≥8 μg/ml
| Yr | No. (%) of infected patientsa
|
||
|---|---|---|---|
| Male | Female | Total | |
| 1987 | 8/18 (44) | 10/16 (63) | 18/34 (53) |
| 1994 | 9/17 (53) | 13/19 (68) | 22/36 (61) |
| 2003 | 22/37 (59) | 21/28 (75) | 43/65 (66) |
| Total | 39/72 (54) | 44/63 (70) | 83/135 (61) |
Number of male or female patients infected with H. pylori isolates for which metronidazole MICs were ≥8 μg/ml/total number of male and/or female patients.
A report showed that primary resistance to tetracycline is rare in H. pylori isolates (20). However, no official breakpoint for tetracycline resistance has been designated for H. pylori isolates. In our study, H. pylori strains for which tetracycline MICs were ≥4 μg/ml were found in 1987 (5.9%) and 2003 (12.3%). Interestingly, H. pylori strains for which tetracycline MICs were ≥4 μg/ml were not found in 1994 (0%); the range of tetracycline MICs in 1994 was 0.125 to 1.0 μg/ml (Fig. 1D). This temporary reduction in MICs might have resulted from reduced drug consumption, as the production of tetracycline in 1995 was ∼12% lower than it was in 1990 (14).
Azithromycin and ciprofloxacin have been proposed as components of a triple PPI-based regimen (9, 27). The MIC patterns of azithromycin and of ciprofloxacin in 1987 were similar, but the MIC patterns changed to a bimodal distribution in 2003, as shown in Fig. 1E and F. Although the official breakpoints for azithromycin and ciprofloxacin resistance have not been designated for H. pylori isolates, the number of H. pylori strains for which MICs were ≥1.0-μg/ml increased from 1994 to 2003. Thus, the prevalence of H. pylori isolates for which azithromycin MICs were ≥1 μg/ml increased rapidly from 5.9% in 1987 to 33.3% in 1994 (Fig. 1E). In addition, the prevalence of H. pylori isolates for which ciprofloxacin MICs were ≥1 μg/ml increased more gradually, from 0% in 1987 to 33.3% in 2003 (Fig. 1F). However, we did not find any significant correlation between MICs of antibiotics and diagnosis.
These results demonstrate that, from 1994 to 2003, the MICs of amoxicillin, clarithromycin, metronidazole, tetracycline, azithromycin, and ciprofloxacin have been increasing for H. pylori strains isolated from the Korean population in Seoul. The results also indicate the need for the continuous surveillance of antibiotic susceptibilities. Moreover, they indicate that susceptibility testing should be conducted before treatment to maximize therapeutic efficacy.
Gene mutations resulting in clarithromycin resistance.
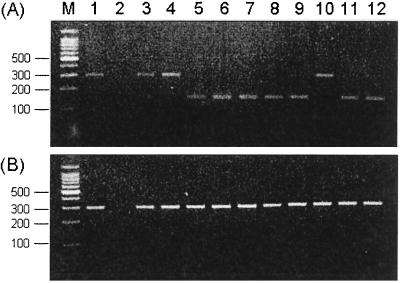
Clarithromycin-resistant strains frequently carry mutations in the 23S rRNA gene (17). Versalovic et al. (26) showed that A-to-G point mutations at positions 2143 and 2144 within domain V of the 23S rRNA gene are a cause of clarithromycin resistance. An A2143C mutation was also reported in the same gene, although it is rare (24). These mutations reduced the affinity between clarithromycin and the 23S ribosomal component, resulting in impaired activity against H. pylori (18). In the present study, the A2144G mutation in the 23S rRNA gene was detected in 7 of 10 clarithromycin-resistant strains (70%) by BsaI digestion (Fig. 2A). In contrast, this mutation was not detected in strains for which clarithromycin MICs were ≤0.5 μg/ml, and none of the PCR products of these strains were digested with BbsI (Fig. 2B). Sequencing of the remaining three clarithromycin-resistant strains (30%) revealed a T-to-C mutation at position 2183 (T2183C). However, we did not find any correlation between the mutation type and the clarithromycin MICs.
FIG. 2.
Restriction endonuclease analysis of 23S rRNA amplicons. (A) Digestion with BsaI; (B) digestion with BbsI. The A2144G mutation was found in lanes 5 to 9 and in lanes 11 and 12 but not in lanes 3, 4, and 10. Note that the A2143G mutation detected by digestion with BbsI was not detected in any of the strains studied. Lanes 3, 4, and 10 revealed the T2183C mutation, as assessed by DNA sequencing. Lane M, DNA size markers (indicated to the left of the gels in base pairs); lane 1, H. pylori ATCC 43504; lane 2, Escherichia coli DNA; lanes 3 to 12, clarithromycin-resistant H. pylori strains. Clarithromycin MICs are 16 (lane 3), 32 (lane 4), 32 (lane 5), 32 (lane 6), 32 (lane 7), 32 (lane 8), 16 (lane 9), 16 (lane 10), and 64 (lane 11) μg/ml.
Clinical outcome.
In the present study, 43 of 65 patients in 2003 were followed up to determine H. pylori eradication after PPI triple therapy, consisting of amoxicillin (1,000 mg twice daily) and clarithromycin (500 mg twice daily) for 1 week. It was found that the therapy successfully eradicated H. pylori in 32 of the 43 patients (74%); the rates of eradication were 97% for the clarithromycin-susceptible strains (for which clarithromycin MICs were ≤1.0 μg/ml and for which amoxicillin MICs were <0.5 μg/ml) and 0% for the clarithromycin-resistant strains (for which clarithromycin MICs were >1.0 μg/ml) (Table 2). These results suggest that the decrease in eradication efficacy for H. pylori might originate from the increase in resistance to clarithromycin over 16 years. Further surveillance regarding the effect of antibiotic resistance on the eradication rate is necessary to establish the appropriate treatment for H. pylori infection.
TABLE 2.
Effect of clarithromycin resistance on eradication rate of H. pylori after PPI triple therapya
| Amoxicillin MIC (μg/ml) | Susceptibility to clarithromycin | No. of patients | Eradication rate of H. pylori (%)
|
|
|---|---|---|---|---|
| Success | Failure | |||
| <0.5 | Susceptible | 31 | 97 | 3 |
| ≥0.5 | Susceptible | 5 | 40 | 60 |
| <0.5 | Resistant | 5 | 0 | 100 |
| ≥0.5 | Resistant | 2 | 0 | 100 |
Resistance breakpoint for clarithromycin was defined as 1.0 μg/ml.
In conclusion, these results suggest that the MICs for H. pylori of several antibiotics have increased over the past 16 years in Seoul.
Acknowledgments
We thank Mi-Soon Kim, Soo Jin Cho, Joo Hyoung Lee, Youn-Sung Son, and Shin-Jai Kang for their excellent technical help.
We also thank Abbott Laboratories for gifts of clarithromycin and Groton Laboratories for gifts of azithromycin.
REFERENCES
- 1.Adamek, R. J., S. Suerbaum, B. Pfaffenbach, and W. Opferkuch. 1998. Primary and acquired Helicobacter pylori resistance to clarithromycin, metronidazole, and amoxicillin—influence on treatment outcome. Am. J. Gastroenterol. 93:386-389. [DOI] [PubMed] [Google Scholar]
- 2.Boyanova, L., R. Koumanova, G. Gergova, M. Popova, I. Mitov, Y. Kovacheva, S. Derejian, N. Katsarov, R. Nikolov, and Z. Krastev. 2002. Prevalence of resistant Helicobacter pylori isolates in Bulgarian children. J. Med. Microbiol. 51:786-790. [DOI] [PubMed] [Google Scholar]
- 3.Cabrita, J., M. Oleastro, R. Matos, A. Manhente, J. Cabral, R. Barros, A. I. Lopes, P. Ramalho, B. C. Neves, and A. S. Guerreiro. 2000. Features and trends in Helicobacter pylori antibiotic resistance in Lisbon area, Portugal (1990-1999). J. Antimicrob. Chemother. 46:1029-1031. [DOI] [PubMed] [Google Scholar]
- 4.Glupczynski, Y., F. Megraud, M. Lopez-Brea, and L. P. Andersen. 2001. European multicentre survey of in vitro antimicrobial resistance in Helicobacter pylori. Eur. J. Clin. Microbiol. Infect. Dis. 20:820-823. [DOI] [PubMed] [Google Scholar]
- 5.Graham, D. Y. 1998. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology 115:1272-1277. [DOI] [PubMed] [Google Scholar]
- 6.Graham, D. Y., G. M. Lew, P. D. Klein, D. G. Evans, D. J. Evans, Jr., Z. A. Saeed, and H. M. Malaty. 1992. Effect of treatment of Helicobacter pylori infection on the long-term recurrence of gastric or duodenal ulcer: a randomized, controlled study. Ann. Intern. Med. 116:705-708. [DOI] [PubMed] [Google Scholar]
- 7.Hentschel, E., G. Brandstatter, B. Dragosics, A. M. Hirschl, H. Nemec, K. Schutze, M. Taufer, and H. Wurzer. 1993. Effect of ranitidine and amoxicillin plus metronidazole on the eradication of Helicobacter pylori and the recurrence of duodenal ulcer. N. Engl. J. Med. 328:308-312. [DOI] [PubMed] [Google Scholar]
- 8.Huang, J.-Q., and R. H. Hunt. 1999. Treatment after failure: the problem of “non-responders.” Gut 45(Suppl. 1):I40-I44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivashkin, V. T., T. L. Lapina, O. Y. Bondarenko, O. A. Sklanskaya, P. Y. Grigoriev, Y. V. Vasiliev, E. P. Yakovenko, P. V. Gulyaev, and V. I. Fedchenko. 2002. Azithromycin in a triple therapy for H. pylori eradication in active duodenal ulcer. World J. Gastroenterol. 8:879-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalach, N., M. Bergeret, P. H. Benhamou, C. Dupont, and J. Raymond. 2001. High levels of resistance to metronidazole and clarithromycin in Helicobacter pylori strains in children. J. Clin. Microbiol. 39:394-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato, S., S. Fujimura, H. Udagawa, T. Shimizu, S. Maisawa, K. Ozawa, and K. Iinuma. 2002. Antibiotic resistance of Helicobacter pylori strains in Japanese children. J. Clin. Microbiol. 40:649-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, J. J., R. Reddy, M. Lee, J. G. Kim, F. A. El-Zaatari, M. S. Osato, D. Y. Graham, and D. H. Kwon. 2001. Analysis of metronidazole, clarithromycin and tetracycline resistance of Helicobacter pylori isolates from Korea. J. Antimicrob. Chemother. 47:459-461. [DOI] [PubMed] [Google Scholar]
- 13.Kim, J. M., J. S. Kim, H. C. Jung, I. S. Song, and C. Y. Kim. 2000. Virulence factors of Helicobacter pylori in Korean isolates do not influence proinflammatory cytokine gene expression and apoptosis in human gastric epithelial cells, nor do these factors influence the clinical outcome. J. Gastroenterol. 35:898-906. [DOI] [PubMed] [Google Scholar]
- 14.Kim, S. I., J. M. Park, S. H. Wie, Y. R. Kim, and M. W. Kang. 2000. The trend of antibiotics usage in Korea during 1981-1998. Korean J. Infect. Dis. 32:439-448. [Google Scholar]
- 15.Korean College of Helicobacter Research and Practice. 2001. Helicobacter pylori: basic and clinical practice. Gunja Publishing Co., Seoul, South Korea.
- 16.Lind, T., F. Megraud, P. Unge, E. Bayerdorffer, C. O'Morain, R. S. Spiller, S. Veldhuyzen Van Zanten, K. D. Bardhan, M. Hellblom, M. Wrangstadh, L. Zeijlon, and C. Cederberg. 1999. The MACH2 study: role of omeprazole in eradication of Helicobacter pylori with 1-week triple therapies. Gastroenterology 116:248-253. [DOI] [PubMed] [Google Scholar]
- 17.Maeda, S., H. Yoshida, K. Ogura, F. Kanai, Y. Shiratori, and M. Omata. 1998. Helicobacter pylori specific nested PCR assay for the detection of 23S rRNA mutation associated with clarithromycin resistance. Gut 43:317-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuoka, M., Y. Yoshida, K. Hayakawa, S. Fukuchi, and K. Sugano. 1999. Simultaneous colonisation of Helicobacter pylori with and without mutations in the 23S rRNA gene in patients with no history of clarithromycin exposure. Gut 45:503-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Megraud, F. 1998. Epidemiology and mechanism of antibiotic resistance in Helicobacter pylori. Gastroenterology 115:1278-1282. [DOI] [PubMed] [Google Scholar]
- 20.Mégraud, F., N. Lehn, T. Lind, E. Bayerdörffer, C. O'Morain, R. Spiller, P. Unge, S. Veldhuyzen van Zanten, M. Wrangstadh, and C. F. Burman. 1999. Antimicrobial susceptibility testing of Helicobacter pylori in a large multicenter trial: the MACH 2 study. Antimicrob. Agents Chemother. 43:2747-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mendonca, S., C. Ecclissato, M. S. Sartori, A. P. Godoy, R. A. Guerzoni, M. Degger, and J. Pedrazzoli, Jr. 2000. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxicillin, tetracycline, and furazolidone in Brazil. Helicobacter 5:79-83. [DOI] [PubMed] [Google Scholar]
- 22.NCCLS. 2002. Acceptable limits for quality control strains used to monitor accuracy of minimal inhibitory concentrations (MICs) (μg/ml) of fastidious organisms, p. 117-121. In Performance standards for antimicrobial susceptibility testing, 12th informational supplement M100-S12, vol. 22, no. 1. NCCLS, Wayne, Pa. [Google Scholar]
- 23.NIH Consensus Development Panel on Helicobacter pylori in Peptic Ulcer Disease. 1994. NIH Consensus Conference. Helicobacter pylori in peptic ulcer disease. JAMA 272:65-69. [PubMed] [Google Scholar]
- 24.Stone, G. G., D. Shortridge, J. Versalovic, J. Beyer, R. K. Flamm, D. Y. Graham, A. T. Ghoneim, and S. K. Tanaka. 1997. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob. Agents Chemother. 41:712-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Wouden, E. J., A. A. van Zwet, G. D. Vosmaer, J. A. Oom, A. de Jong, and J. H. Kleibeuker. 1997. Rapid increase in the prevalence of metronidazole-resistant Helicobacter pylori in the Netherlands. Emerg. Infect. Dis. 3:385-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Versalovic, J., M. S. Osato, K. Spakovsky, M. P. Dore, R. Reddy, G. G. Stone, D. Shortridge, R. K. Flamm, S. K. Tanaka, and D. Y. Graham. 1997. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J. Antimicrob. Chemother. 40:283-286. [DOI] [PubMed] [Google Scholar]
- 27.Xia, H. H., B. C. Yu Wong, N. J. Talley, and S. K. Lam. 2002. Alternative and rescue treatment regimens for Helicobacter pylori eradication. Expert Opin. Pharmacother. 3:1301-1311. [DOI] [PubMed] [Google Scholar]