Summary
Modification of CG dinucleotides in DNA is part of epigenetic regulation of gene function in vertebrates and is associated with complex human disease. Bisulfite sequencing permits high resolution analysis of cytosine modification in mammalian genomes, however its utility is often limited due to substantial cost. Here, we describe an alternative epigenome profiling approach, named TOP-seq, which is based on covalent tagging of individual unmodified CG sites followed by non-homologous priming of the DNA polymerase action at these sites to directly produce adjoining regions for their sequencing and precise genomic mapping. Pilot TOP-seq analyses of bacterial and human genomes showed a better agreement of TOP-seq with published bisulfite sequencing maps as compared to widely-used MBD-seq and MRE-seq and permitted identification of long-range and gene-level differential methylation among human tissues and neuroblastoma cell types. Altogether, we propose an affordable single CG-resolution technique well-suited for large scale epigenome studies.
Keywords: mTAG labeling, next-generation sequencing, DNA unmethylome profiling, neuroblastoma, differential methylation, DNA polymerase priming
Graphical abstract
Staševskij et al. propose a cost-effective robust approach for high-resolution profiling of mammalian epigenomes, which uses covalent tagging of individual unmodified CpG sites followed by non-homologous priming of the DNA polymerase action at these sites to directly produce adjoining regions for their sequencing and precise genomic mapping.
Introduction
Methylation of cytosine to 5-methylcytosine (5mC) in CG dinucleotides acts as a key epigenetic modification affecting gene regulation and cellular differentiation in higher eukaryotes. Dysregulation of 5mC patterns is associated with various complex human diseases including cancer. The complexity of the human epigenome has further increased with the discovery of other modified forms of cytosine - 5-hydroxymethylcytosine (hmC), 5-formylcytosine and 5-carboxylcytosine - demanding more elaborate analytical approaches. Current techniques for the determination of the modification status of CG sites can be divided into a) bisulfite conversion-based methods, b) restriction endonuclease-based methods, and c) affinity capture-based techniques (Weber et al., 2005; Maunakea et al., 2010; Harris et al., 2010). The gold standard and most widely used method is bisulfite sequencing (BS), which can infer modification information of each cytosine at a single-base resolution. Besides its unique and obvious advantages, BS suffers from experimental artifacts (due to extensive DNA degradation) and obstructed genomic mapping of sequencing reads. Most importantly, whole genome BS sequencing (WGBS) unavoidably generates large amounts of data, and the majority of the reads (50–80%) provide little or no information about CG methylation (Ziller et al., 2013). Although the cost of sequencing and data analysis is on decline, it still remains prohibitively high for large scale case-control studies of epigenomic diseases.
The other two groups of methods provide more affordable inroads into the methylome structure although at significant sacrifice in resolution and/or informativity. Methylated or unmethylated fraction of genomic DNA can be selectively enriched by restriction endonuclease cleavage (MRE-seq) (Maunakea et al., 2010), however, the target specificity of available enzymes confines such analysis to a small subset of the CG sites in the genome. Affinity-based methods (using antibodies or MBD, MeCP2 proteins) utilize the enrichment of modified cytosines by using antibodies or modification-specific binding proteins, which often underrepresent regions of lower modification densities (Weber et al., 2005). Typically, these strategies include end-sequencing of the enriched fragments yielding rather long stretches of genomic DNA corresponding to a detected signal (low resolution, defined by the fragment length). During the past few years, chemo-enzymatic tagging of modification sites has been adapted for in vitro studies of hmC and 5mC residues, permitting incorporation of reactive azide, keto or primary amine groups followed by chemo-selective conjugation of biotin (Song et al., 2011; Zhang et al., 2013). Similar profiling of the unmodified fraction of the genome, so-called DNA unmethylome, has been achieved based on selective covalent derivatization of unmodified CG sites (Kriukiene et al., 2013). However, despite a higher precision and added versatility as compared with the affinity-based techniques, none of the enrichment-based approaches can break the resolution limit of 200–500 bp.
Here we propose an alternative concept in analysis of DNA modification patterns that bridges the existing economy-versus-resolution gap by combining selective covalent tagging and genomic sequencing primed at the epigenetic modification sites. The validity of this concept (proof of principle) is demonstrated by developing a high resolution technique for whole-genome analysis of uCG sites. Pilot studies of model bacterial and human genomic DNA samples using the designed technique (named unmodified CG-specific Tethered-Oligonucleotide-Primed sequencing, uCG-TOP-seq) showed that the technology offers high resolution and the capacity to uncover unique epigenetic features currently approachable only by the gold standard WGBS. Unlike WGBS, it avoids sequencing of the entire genome, thereby providing a good alternative for cost-effective genome-wide profiling of DNA methylation patterns.
Design
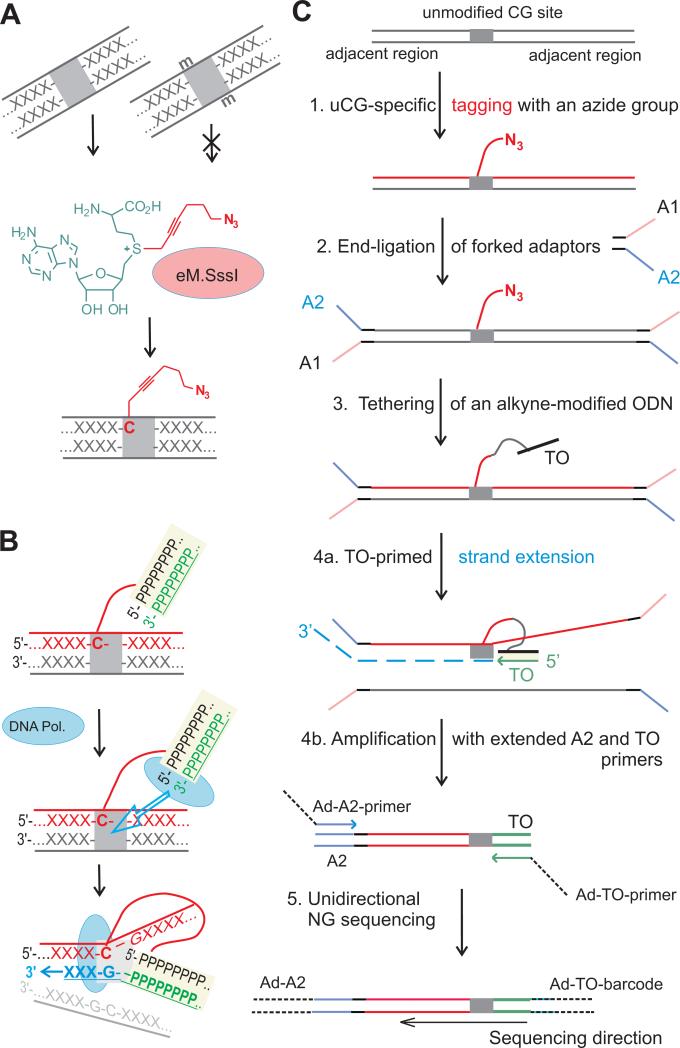
The resolution limits of enrichment-based genome-wide profiling strategies could be potentially increased to a single nucleotide, if the modification status of individual target sites in a fragment could be identified. We, therefore, went on to explore the possibility of whether a chemical tag attached to a DNA epigenetic modification site could be devised to prime the DNA polymerase action. We took advantage of our previously developed mTAG-seq technique (Kriukiene et al., 2013), which uses an engineered version of the M.SssI methyltransferase and a synthetic analog of the AdoMet cofactor to tag the unmodified and hemimethylated CG sites (excluding all modified CG sites) with a reactive azide group (Fig. 1A). We term here the unmodified CG sites as uCGs to distinguish this epigenetic state from genetic CG dinucleotides, which may generally include any epigenetic form of the target cytosine. Analysis of the uCGs, i.e. a smaller fraction of the CG dinucleotides (65–80% of CG sites in the human genome are methylated), may be more sensitive for detecting subtle changes in DNA modification profiles as compared to analysis of methylated CG sites. We chemically tethered an alkyne-bearing DNA oligonucleotide to direct binding of a DNA polymerase at a physical proximity to the target site and thus facilitate the template-dependent polymerase action from the 3’ end of the tethered DNA duplex in the absence of nucleotide complementarity between the primer and the template DNA (Fig. 1B). Initial experiments involving model DNA fragments (Fig. S1A) showed that internal priming can be achieved from a duplex oligonucleotide that is chemically tethered at one of its 5’-terminal nucleotides to a CG site. The detailed mechanism of this non-homologous proximity-driven priming has not yet been investigated, but apparently involves strand invasion and template switching events. Such tethered-oligonucleotide-primed (TOP) template-dependent polymerase action produces nested DNA strands that sequentially include the CG site and its adjacent genomic region. We undertook the development a full analytical procedure for whole-genome TOP-seq analysis of unmodified CG sites as outlined in Fig. 1C. We optimized the TOP-seq reaction conditions in our model DNA system and adapted it for Ion Torrent sequencing (Fig. S1).
Figure 1. Tethered Oligonucleotide-Primed sequencing (TOP-seq) analysis of DNA.
A, Selective tagging of genomic uCG sites with an azide group using an engineered variant of the SssI methyltransferase (eM.SssI) and a synthetic analog of the SAM cofactor (Step 1 in C). B, Tethered Oligonucleotide-primed DNA polymerase activity at an internal covalently tagged CG site (Step 4a). X and P denote generic nucleotides in genomic DNA and the tethered oligonucleotide, respectively. C, TOP-seq procedure for whole-genome mapping of unmodified CG sites using next generation sequencing. For selective amplification of the TOP strands, fragmented genomic DNA is processed to add partially complementary adapters in Step 2. Following TO-primed strand extension (Step 4a), PCR amplification with Ad-A2 and Ad-TO primers containing NGS platform-specific 5’-end adaptor sequences selectively enriches the primed product but not the original DNA strand (Step 4b). Sequencing is initiated at the A adaptor sequence included in the 5’-part of Ad-TO-barcode amplification primer (Step 5). TO, tethered oligodeoxyribonucleotide; A1 and A2, strands of a partially complementary adapter; Ad, extended sections of platform-specific adapters. See also Figure S1.
Results
uCG-TOP-seq analysis of a model bacterial genome
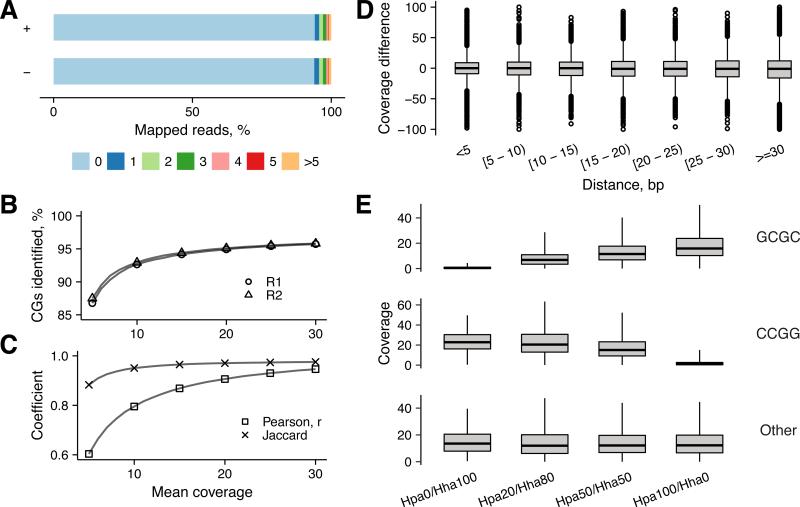
We first examined our developed procedure on a megabase scale genome using a custom Staphylococcus aureus strain that carried no CG-specific MTase. Highly divergent Staphylococcal genomes (~3 Mb) typically contain >70,000 unmodified CG sites; ~1% of these sites can be hemimethylated due to endogenous methylation of GATC sites. To assure high quality read mapping, we de novo assembled the genomic sequence of the custom strain and identified 68,654 CG sites in 321 contigs covering a total of 2,726,458 bp. We applied the TOP-seq procedure on duplicate samples of gDNA followed by next generation sequencing on an Ion Proton sequencer. Read processing and analysis was conducted using our custom pipeline (see STAR Methods). In both replicates, 94% of reads from both strands featured a CG dinucleotide, immediately following the sequence of the priming oligonucleotide (Fig. 2A). For all subsequent data processing routines, we set a uCG read start window to ±3 nt of the target cytosine. With this read start window, 93% and 95% of all CGs were identified with a mean sequencing depth of 10x and 20x, respectively (Fig. 2B). The bacterial data analysis showed a high degree of reproducibility and 95% of common uCG calls between technical replicates (Pearson correlation 0.8 and 0.9 at 10x and 20x mean coverage, respectively; Jaccard 0.95 and 0.97 at 10x and 20x mean coverage, respectively Fig. 2C).
Figure 2. Validation of TOP-seq on a model bacterial genome.
A, Distance distribution of TOP-seq read start positions from a uCG site in the TOP-seq library of S. aureus genome. Top and bottom strands are shown as “+” and “−”. B, Identification of uCGs at different mean CG coverage. C, Pearson correlation and Jaccard coefficient between technical replicates at different mean uCG coverage. D, Difference in coverage between neighbouring CG sites as a function of the distance between them. E, Distribution of uCG coverage at partially methylated CCGG and GCGC sites. Genomic DNA samples containing 0/100, 20/80, 50/50 and 100/0 % methylation of CCGG and GCGC sites, respectively, were prepared and analyzed. Total coverage across technical replicates was summed and divided by total number of reads in a sample and multiplied by 106.
We next performed a direct assessment of how the TOP-seq read counts are affected by the proximity of genomic CG sites. We thus plotted the difference in coverage between closest neighboring CGs as a function of their separation and found that the TOP-seq signal was largely independent of the spatial distribution of the targets sites (Fig 2D). To further explore the quantitative aspects of uCG-TOP-seq we prepared a series of S.aureus DNA samples partially methylated at GCGC sites (3048 occurrences in the genome) or CCGG sites (1222 occurrences) by mixing at defined ratios the genomic DNAs premethylated in vitro with M.HhaI or with M.HpaII MTases, respectively. The TOP-seq libraries for duplicate samples of each partially methylated DNA were produced and sequenced. As expected, an inverse correlation between the read number and the extent of methylation at the GCGC and CCGG sites was observed (Fig. 2E). Altogether, our megabase genome experiments demonstrated a solid reproducibility of the TOP-seq approach even at the single-CG resolution and its high responsiveness to differential methylation levels in model DNA.
TOP-seq analysis of the human genome
Having confirmed the capacity of TOP-seq on a smaller unmethylated genome, we went on to examine its utility to discern the modification profiles of human DNA. We chose two types of human cells: fetal lung fibroblasts IMR90 and the prefrontal brain cortex (referred further on as “Brain”). This selection enabled direct comparison of TOP-seq data with published methylome profiling methods: WGBS; MBD-seq; and MRE-seq (Lister et al., 2009; Ziller et al., 2013; Wen et al., 2014; Bert et al, 2013; Maunakea et al., 2010). Additionally, we were interested to see if TOP-seq is a suitable tool for the identification of subtle tissue-specific differences associated with human disease. Therefore, we performed the TOP-seq analysis of two clonal neuroblastoma (NB) cells, N-type LA1-55n and S-type LA1-5s, both derived from LA-N-1 NB cell line (Ciccarone et al., 1989).
Since uCG-TOP-seq generates reads only from unmethylated CG sites, which constitute a smaller fraction of the human genome, we combined TOP-seq with the medium capacity Ion Proton sequencing, which routinely generates 70–90 M of reads per PI chip. DNA samples were split into technical replicates at two different steps of the procedure (Fig. 1C): at Step 4 generating Brain-1 R1, R2 and IMR90-1 R1, R2 libraries, and at Step 1, generating Brain-2 R1, R2 and IMR90-2 R1 libraries. We obtained on average 87 M raw single-end reads for two technical replicates of a TOP-seq library sequenced on one PI chip. Approximately 18 M reads were obtained for each replicate after mapping and ~96% of those reads started at a CG site immediately following the sequence of the priming oligonucleotide. For basic characteristics see Table S1 and Fig. S2A. On average, 21% of all CGs were identified by at least 1 sequencing read on either strand starting at 0-3 nt distance to a CG dinucleotide. Combining Brain-1 R1/R2 and IMR90-1 R1/R2 technical replicates together (so-called “low coverage” libraries, ~9.4 and ~9 M reads, respectively) increased the fraction of identified CGs to 33.5% in Brain and 31.9% in IMR90. This amounted to 4.1 and 3.9 mean coverage (Table S1) distributed uniformly across all autosomes (Fig. S2B).
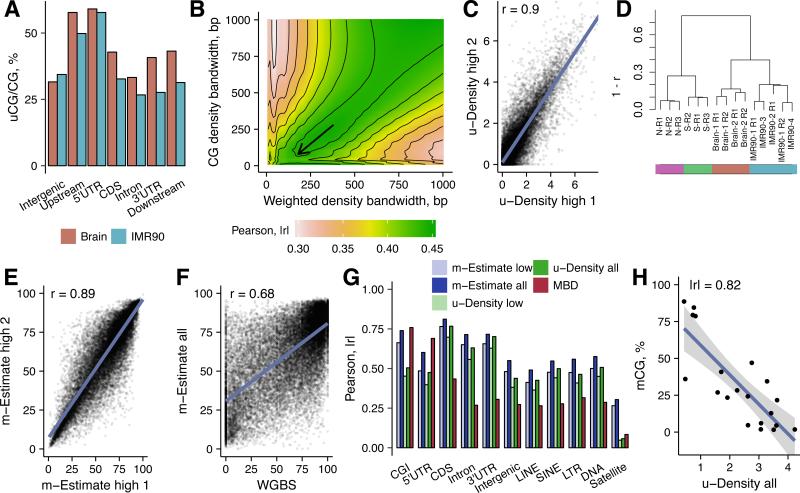
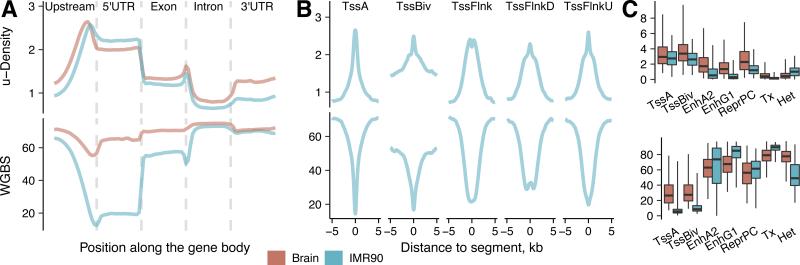
We also subjected the IMR90-1 R1/R2 libraries to deeper sequencing generating ~100 M reads per technical replicate (so-called “high coverage” IMR90-3 and IMR90-4 libraries, Table S1). This increased the mean coverage to ~9x per identified uCG, and led to the identification of 34-35% of total genomic CGs per library (46% of CGs and ~14x coverage in the combined IMR90-3/4 dataset). The number of identified sites monotonically grows with the total number of reads (closely resembling a logarithmic function) (Fig. S2C), suggesting that at increasing sequencing depths, the uCG calling progressively expands from low- to moderate- and even to high-methylation CG sites. ~50% of CGs were identified as uCGs in 5’UTR regions as well as in 2 kb upstream regions from transcription start sites. These were followed by intergenic and coding sequence (CDS) regions (~30% each) for IMR90 and by CDS, downstream sequences and 3’UTRs for Brain (Fig. 3A). TOP-seq signal was detectable in 96% of 26,641 autosomal CGIs. As expected, promoter CGIs were the most enriched in uCGs (50-100% CGs identified in 93-96% of CGIs) indicating their highly unmethylated state (Fig. S2D). The variation of identified uCGs was higher among intragenic and intergenic CGIs attesting their diversity and on average higher methylation levels. These findings showed that TOP-seq data are generally consistent with established genome methylation patterns.
Figure 3. TOP-seq analysis of two human tissues.
A, Percentage of called uCGs of total CGs in a genomic element for the brain cortex and IMR90 cells. CDS - protein coding DNA sequence. Upstream and Downstream regions encompass 2 kb regions from the gene start or end site, respectively. B, Dependence of the WGBS and TOP-seq u-density correlation on the kernel bandwidth parameters. Color scale represent correlation of u-density signal and WGBS. Selected kernel bandwidths: 180 bp for coverage-weighted density and 80 bp for CG density. C, Correlation between technical replicates of the high coverage TOP-seq u-density IMR90 library (IMR90-3 and IMR90-4; ~100 M reads each). D, Hierarchical clustering of TOP-seq u-density profiles of technical replicates of all tissues. E, Correlation between technical replicates of the high coverage TOP-seq m-estimate IMR90 library. F, Scatterplot of the correlation between the TOP-seq m-estimates (‘all’ means combined IMR90 dataset, 16x CG coverage) and WGBS at single CG resolution. G, Correlation between TOP-seq data (u-density and m-estimates), MBD-seq and WGBS across various genomic elements. H, TOP-seq u-density compared to pyrosequencing methylation values in 20 selected genomic regions. Average TOP-seq u-density values (“all” dataset) across the regions were used for comparison. See also Figures S2, S3 and Tables S1, S2.
For a more detailed assessment of the technique, we evaluated the correlations of technical replicates sequenced to different depths. The Pearson correlation did not exceed r=0.53 for the low coverage IMR90 data (r=0.65 for Brain low coverage replicates) and increased to r=0.68 for the higher coverage. To improve the quantification of DNA methylation, we performed a computational adjustment of the TOP-seq coverage data to generate a high-resolution prediction of DNA methylation levels of 26 M autosomal CGs. Using Epanechnikov kernel we computed weighted density estimates (Parzen, 1962) from the coverage signal and divided them by unweighted CG-density to obtain the TOP-seq unmethylome density (u-density) signal. Kernel bandwidth parameters were determined by scanning the TOP-seq u-density correlations at a wide range of kernel windows with the corresponding public IMR90 WGBS signal (Lister et al., 2009) in chromosome 1 (Fig. 3B). This adjustment enhanced Pearson correlation of the low coverage replicates to r=0.8 for IMR90 and r=0.89 for Brain. Correlation of the high coverage IMR90 replicates increased to r=0.9 (Fig. 3C). As expected, the TOP-seq u-density performed equally well across regions of different CG density (Fig. S2E). Finally, hierarchical clustering of samples using TOP-seq densities further confirmed a good agreement among technical replicates and marked differences across tissues and cell types (Fig. 3D).
Comparison of TOP-seq with other epigenome profiling approaches
Cross-platform correlation of the low coverage and high coverage TOP-seq u-density datasets with IMR90 WGBS data was |r|=0.51 and |r|=0.57 respectively. For comparison, we found the correlation of IMR90 WGBS with published MRE-seq and MBD-seq (Bert et al, 2013) to be in the order of r=0.18 and r=0.3, respectively. Weak correlations between WGBS and the enrichment-based methods have previously been noted by us and others (Kriukiene et al., 2013; Zhang et al., 2013), which may in part derive from non-linear relationship between the data produced with different methods (Stevens et al., 2013). In a further adjustment step, we sought to account for possible sequence-specific variations that may influence the TOP-seq signal. We used a small fraction of the WGBS dataset (chromosome 20) to train an exponential decay model containing additional genomic feature-specific covariates which was then used to convert the TOP-seq u-density into so-called CG methylation estimates (m-estimates, methylation values presented in the scale from 0–100). Although the second enhancement step had a minor effect on correlation among the TOP-seq technical replicates (Fig. 3E), it improved the absolute correlation with the IMR90 WGBS (Lister et al., 2009; chromosome 20 excluded) to r=0.63 for low coverage and to r=0.68 for the combined high coverage IMR90 dataset (Table S1; Fig. 3F and Fig. S3A).
Dissection of the whole genome profiles across major genomic features showed a good agreement of the TOP-seq m-estimates and WGBS in CGIs, enhancers, 3’UTRs, CDS, introns and some classes of repeats (Fig. 3G). Importantly, TOP-seq outperformed MBD-seq across the majority of elements spanning a wide range of methylation levels. TOP-seq and WGBS agreed that genic enhancers EnhG1 are hypermethylated relative to active enhancers EnhA1 (Fig. S3B). Correlation of the methods at these enhancer regions was r=0.6 for EnhG1 and r=0.65 for EnhA1 (Fig. S3B). In contrast, correlation of MBD-seq with WGBS at both types of enhancers in our hands was considerably lower (G1 r=0.22, A1 r=0.38). In CGIs, the precision of TOP-seq was comparable with MBD-seq and superior to MRE-seq (r=0.74, r=0.76, r=0.17, respectively) (Fig. S3C). Correlation in repeat elements and in transcribed regions Tx, heterochromatin Het and TssA and TssBiv transcription start sites was lower for all methods concerned (Fig. 3G and Fig. S3D).
Conversion of our experimental u-density data to the m-estimate format was also successful (led to improved correlation with WGBS, r=0.69) using another independently produced IMR90 WGBS map (Ziller et al., 2013) (Fig. S3A to C). However, a similar conversion of the Brain u-density data based on the published brain WGBS map (Wen et al., 2014) did not lead to satisfactory m-estimate maps; that was quite understandable given poor correlation of the u-density and the WGBS dataset (r~0.3), which we tend to attribute to biological diversity of samples obtained from a complex prefrontal brain area in independent experiments. The application of the IMR90-derived model onto the Brain u-density signal did not improve the results either. Altogether, the presented examples suggest that this optional adjustment step is only feasible when a high quality reference WGBS map derived from a related tissue is available. Accordingly, the TOP-seq u-density profiles were used in all further comparative tissue analyses due to lack of a suitable brain WGBS map.
As the ultimate validation of the predictive power of the method, we evaluated how top 10% of unmethylated 1 kb regions in IMR90 cells as well as in Brain identified by TOP-seq, MBD-seq and MRE-seq overlap with top 10% of unmethylated regions derived by WGBS. In IMR90, we observed a very strong association between the TOP-seq u-density and WGBS (Fisher test OR=7; p<2×10−22 and OR=8.1; p<2×10−22 for low and high TOP-seq sequencing depths, respectively). MBD-seq showed a weaker association (OR=3.1; p < 2×10−22) whereas MRE-seq showed significant dissociation (OR=0.2; p < 2×10−22). Differences were not as marked in Brain where TOP-seq scored best (OR=11.1; p < 2×10−22) followed by MRE-seq (OR=10.3; p < 2×10−22). Finally, we used pyrosequencing to examine 20 TOP-seq derived regions representing diverse CG density and methylation levels in CGIs and enhancers (Table S2; methylation levels were chosen according to WGBS data of IMR90 cells). TOP-seq showed a good agreement with the pyrosequencing data (|r|=0.82) (Fig. 3H and Fig. S3E), only slightly behind the gold standard WGBS (r=0.95) (Fig. S3E).
We also compared the TOP-seq u-density profiles with WGBS across different gene-associated elements (Fig. 4A). As expected, the TOP-seq and WGBS profiles of the corresponding tissues showed inverse patterns throughout the analyzed regions. We went further to determine the TOP-seq u-density in and around segments representing a range of chromatin states (Kundaje et al., 2015). Among the active promoter states, active TSS, bivalent/poised TSS promoters and flanking TSS upstream segments showed higher TOP-seq u-density signals indicating their lower methylation levels (Fig. 4B). Subtle methylation differences can further be inferred based on the distribution of the TOP-seq signal in various chromatin segments (Fig. 4C and Fig. S4), which mirrors closely the methylation profiles derived from the WGBS data.
Figure 4. TOP-seq profiles at various gene-associated elements and chromatin states.
A, TOP-seq u-density and WGBS profiles in the brain cortex and IMR90 cells over different gene-associated regions: upstream (2 kb), 5’-untranslated (5’UTR), exons, introns and 3’-untranslated (3’UTR) regions. Smoothed densities were calculated for 20 equally sized bins per region. B, TOP-seq u-density and WGBS (IMR90 data) profiles in 5 kb flanking regions of transcription start sites (TSS) associated chromatin segments: active TSS states (TssA); bivalent/poised TSS states (TssBiv); TSS flanking regions (TssFlnk) and TSS flanking regions divided into upstream and downstream flanking regions (TssFlnkU and TssFlnkD). C, Mean TOP-seq u-density and WGBS methylation values (for the brain cortex and IMR90 data) in various chromatin states: TssA; TssBiv; active enhancers EnhA2 and genic enhancers EnhG1; segments of actively transcribed genes (Tx); heterochromatin, (Het); and repressed polycomb segments (ReprPC). See also Figure S4.
TOP-seq profiling of human neuroblastoma-specific cell types
Neuroblastoma is a malignancy of the developing sympathetic nervous system that is the most common extracranial solid cancer in childhood. A characteristic feature of NB tumors is a varied presence of several distinct cell types (Shimada et al., 1984; Walton et al., 2004). The most abundant are neuroblastic (N-type) cells, which are tumorigenic and have neuronal features. In contrast, non-tumorigenic, (S-type), cells possess marker proteins identifying them as neural crest-derived cells with features of glial/melanocytes/smooth muscle precursor cells. A third cell type, I-type, has been shown to be a stem-like cell type which can differentiate into either N or S cells and is highly tumorigenic. No comprehensive genome-wide methylation data is available for different NB cell types so far. We focused on the N and S type cells as a case-control example to compare their genome-wide u-density maps and to identify differentially methylated regions.
During cancer development and progression, two concurrent epigenetic abnormalities are commonly observed: global hypomethylation and localized hypermethylation of CG islands. We first looked at the relative quantities of modified cytosines in LA1-55n and LA1-5s DNA (referred to as N and S DNA, respectively) using a quantitative HPLC/MS assay. We found that the level of 5mC in N-type DNA (3.57±0.03%) was slightly lower than in S-type DNA (3.89±0.003%) (Fig. S5A). Indeed, both NB tissues were hypomethylated in comparison to Brain and IMR90 (mdC = 5.04±0.02% and 4.5 ±0.01% in Brain and IMR90, respectively).
Whole-genome TOP-seq analysis of N and S DNA comprised three technical replicates each, resulting in ~48 M of mapped reads in total for each cell type (see Table S1). Called uCGs totalled 31.8% and 38.7% of all CGs in the combined S and N datasets, respectively, consistent with a less methylated epigenotype of the N-type cells (Fig. S5A), and showed more pronounced chromosomal variations as compared to Brain and IMR90 (Fig. S2B). The highest TOP-seq signal was found in chromosome 2, and was particularly strongly enriched in a 1.6 Mb region encompassing the MYCN gene (chr2: 15026730-16640120) in both cell types (Fig. S5B). MYCN amplification is a well known aberration in the progenitor LA-N-1 cell line (Spengler et al., 1997), which can give rise to false DMR calls using TOP-seq or other read count-based profiling approaches.
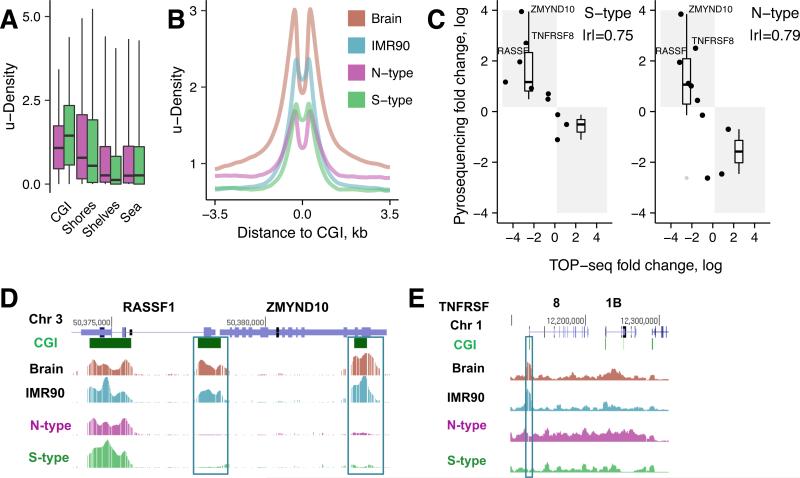
We compared TOP-seq u-density profiles in various genomic elements. CGIs displayed lower TOP-seq u-density, i.e. hypermethylation, in the N cells as compared to the S-type (Fig. 5A). This was further confirmed by TOP-seq u-density profiles around CGIs in all the investigated tissues (Fig. 5B) and agrees well with the methylation studies reporting that the methylation of multiple CGIs is a hallmark of NB with poor prognosis (Abe et al., 2005). When moving into more distant regions, the u-density increases in the N-type cells consistent with their global hypomethylation relative to S-type (Fig. 5B).
Figure 5. TOP-seq analysis of the neuroblastoma N and S cell types.
A, Mean TOP-seq u-density for CGIs, CGI shores (±2 kb around CGIs), CGI shelves (±2 kb around CGI shores) and the rest of the genome (Sea). B, TOP-seq profiles around CG islands. C, Validation of TOP-seq DMRs using pyrosequencing. Fold change of S vs IMR90 and N vs IMR90 were measured (Table S4). For a DMR to validate positive TOP-seq fold change should correspond to negative pyrosequencing fold change and vice versa. Positive fold change in TOP-seq means higher methylation in IMR90, negative fold change means higher methylation in S or N. D, Browser representation of TOP-seq u-density profiles along two NB marker genes: RASSF1 and ZMYND10. Hypermethylated CGIs in NB tissues relative to normal references are boxed. E, Browser view of TOP-seq data along a 86 kb region in Chr1 coding for tumor necrosis factor receptor genes TNFRSF8 and TNFRSF1B. Identified hyperM promoter CGIs (relative to Brain and IMR90) are boxed. See also Figures S5, S6 and Tables S3, S4, S5, S6.
Repeat elements that comprise >40% of the human genome are heavily methylated in somatic tissues (Lister et al., 2009; Sue et at., 2012), but hypomethylation of LINE, Alu, LTR and Satellite repeats has been shown to accompany tumor progression in cancers and is associated with tumor aggressiveness (Rauch et al., 2008). Analysis of the TOP-seq u-density distribution across the most abundant repeat families confirmed WGBS data and revealed distinct TOP-seq u-density in Brain and IMR90 (Fig. S5C), indicating relative hypomethylation of the repeat elements in IMR90 cells as compared to the brain cortex. Importantly, we detected hypomethylation of the most abundant repeat families in NB (and IMR90) cells as compared to Brain, with the highest unmethylation level of Alu repeats in the tumorigenic N cells (Fig. S5C). From this we can conclude that TOP-seq can efficiently reveal subtle methylation differences among hypermethylated genomic elements, such as repetitive sequences.
Differential methylation between NB cell types
We analyzed the TOP-seq data to identify statistically significant methylation differences between the N- and S-type cells relative to the Brain and IMR90 reference across CG islands (CGI-DMR) (Fig. S6A; Table S3). To assess the reliability of the differential TOP-seq u-density values, we performed pyrosequencing validation of the methylation levels of a series of DMRs detected in the N- and S-type NB cells (see Table S4; also Table S2). The selected examples included many subtle and strong DMRs detected between cancerous and IMR90 cells. Pyrosequencing confirmed the methylation status of 10/10 and 8/10 of selected regions (90% total) in the S- and N-samples and showed a good agreement of the corresponding differential methylation values (|r|=0.75 and 0.79) (Fig. 5C). Some uncertainty in the validation of DMRs may have come from potential copy number variations (CNV), which are generally abundant in NB tissues. Altogether, the pyrosequencing experiments confirmed a high predictive power of the method for detection of cell type-specific differential methylation related to human disease.
Given that NB is a neuroendocrine tumor arising from neural crest cells we focused our analysis on promoter and intragenic CGI-DMRs identified between N, S and the Brain reference (Fig. S6A), and assigned them to their host genes (Table S3). As shown above (Fig. 5A and B), N-type cells demonstrated global hypermethylation at CGIs. Accordingly, we identified fewer hypomethylated CGIs in the promoter and intragenic regions of N-type than in S-type cells, while the number of intergenic CGI-DMRs was comparable for both cell types (Fig. S6A).
We performed functional annotation analysis of the genes with the identified CGI-DMRs first focusing on promoter CGI characterization. Gene Ontology (GO) term enrichment analysis for the sets of S/B-hypoM and N/B-hypoM indicated a significant enrichment for components of intracellular organelle lumen and cytoskeleton (Table S5). HyperM promoter CGIs for both N and S cells were significantly enriched in groups of homeobox-domain containing proteins, glycoproteins, signal peptides, and biological processes covering neuron differentiation, development and axonogenesis (Table S5). This is in line with the nature of this developmental tumor, which is associated with the impairment in maturation of the neuronal phenotype. Intriguingly, analysis of the N/B-hyperM CGIs identified hypermethylation of genes involved in neural crest development and migration, which are absent in the S/B-hyperM CGI-DMRs. We then tested whether the detected methylation events matched any group of genes that have been shown to be silenced by aberrant methylation in NB, e.g. CCND1/CCND2, RB1, RASSF1, ZMYND10, HIST1H3C, HOXD3, PCDHB cluster, etc (Caren et al., 2011; Decock et al., 2012; Yanez et al., 2015). Remarkably, hypermethylation of the promoter and/or intragenic CGIs was confirmed in all of the investigated NB marker genes in both NB cell types (genome-browser view of RASSF1 and ZMYND10 are shown in Fig. 5D), as opposed to the IMR90/Brain pair (Table S6). Besides the well-described NB marker genes, we identified promoter CGI hypermethylation in a number of gene groups involved in development, neural functions and in TNF-receptor genes (TNFRSF10B; TNFRSF10D; TNFRSF11A; TNFRSF11B; TNFRSF18, TNFRSF8 etc) (Table S3 and Fig. S6B). These include the transcription factor clusters HoxD and HoxA (methylation of HoxA genes was described in breast cancer (Novak et al., 2006)), the POU class transcription factors involved in neuronal differentiation (POU2F2 was previously described as methylated in NB tumors and cell lines (Caren et al., 2011)), and the protocadherin cluster PCDHG. In NB, epigenetic marks of repression have been described for the PCDHB and PCDHA clusters (Abe et al., 2005), whereas no information is so far available for PCDHG. Beside detected hypermethylation of the HIST1H3C gene in both N- and S-type cells (Table S3 and Table S6), we found promoter CGI methylation in a large cluster of all four types of histone genes (chr 6:cluster 1; 22 genes, Table S3). A half of them were significantly hypermethylated in the N-cells relative to S-type cells, pointing at possible N-cell-specific alterations in DNA packaging and chromatin structure. Among the DMRs selected for pyrosequencing we validated three strong hyperM-CGI DMRs: promoter CGI of NB marker genes RASSF1 and ZMYND10 and one de novo identified promoter CGI residing in the TNFRSF8 gene were hypermethylated in both N- and S-type cells in respect to the normal tissues (Fig. 5C, D and E; Table S4).
It has been reported that tissue- and cell type-specific methylation is present in a small fraction of CGI promoters, whereas a far greater proportion occurs across gene bodies which include potential alternative CGI promoters (Maunakea et al., 2010). Importantly, functional annotation analysis of the intragenic CGI-DMRs of the N and S cells (with respect to Brain and each other) revealed substantial differences between the NB cell types. In contrast to the S/B-hypoM (and S/N-hypoM) comparisons, for N/B-hypoM (and N/S-hypoM) CGI-DMRs, we identified significantly enriched terms related to glycoproteins, extracellular matrix structure, collagens, EGF-like domain proteins, which included many growth factors, developmental and receptor proteins. Comparison of the intragenic N/B-hyperM and S/B-hyperM CGI-DMRs found a strong overlap in GO terms associated with sequence-specific DNA binding proteins, neuron differentiation/development and cell adhesion (Table S5). However, N-specific hypermethylated CGIs with respect to S (N/S-hyperM) fell into large gene clusters involved in neuron differentiation and development, cell-cell signaling, synaptic transmissions and neurological system process, pointing at potential downregulation of these genes as compared to the non-tumorigenic S-type cells (Table S5).
Long-range hypomethylation in neuroblastoma cells
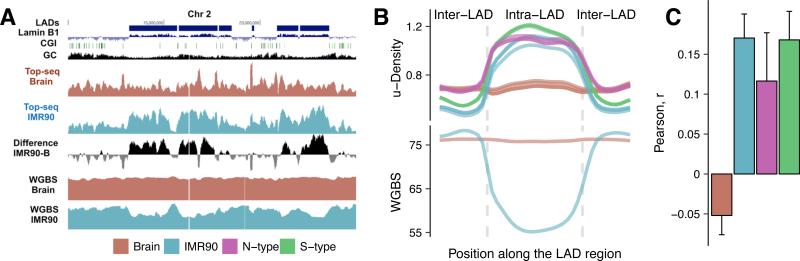
To assess the power of TOP-seq to discern large-scale methylation patterns we investigated long hypomethylated regions (0.1–1 Mb), termed PMDs, or partially methylated domains. PMDs have been detected in IMR90 cells and some cancer lines and tumors (Lister et al., 2009; Berman et al., 2012). A gene silencing role has been suggested for PMDs in IMR90 cells. Moreover, a striking correspondence has been observed between PMDs of IMR90 and nuclear-lamina–associated domains (LADs) (Guelen et al., 2008), which are directly involved in gene repression and usually range from 80 kb to 30 Mb in size (Berman et al., 2012). Besides the presence of PMDs and coincident LADs in the immortalized cell types such as IMR90, PMDs detected in colon tumors also strongly coincided with LAD boundaries (Berman et al., 2012). Since dynamic association with the nuclear lamina has been implicated as a key mechanism in the developmental regulation of long-range gene silencing that can be perturbed in cancer cells, we sought to identify PMDs and investigate their relationship with nuclear LADs in the NB cells. The ability of TOP-seq to detect PMDs was initially tested by analysis of a 15 Mb region containing several LADs (data of TIG3 embryonic fibroblasts, (Guelen et al., 2008)) in IMR90 and Brain DNA (Fig. 6A). The u-density profiles contained scattered peaks originating from hypomethylated CGIs, however, the remaining u-density and differential u-density clearly showed IMR90-specific hypomethylation perfectly matching the LAD boundaries (Fig. 6A). We removed CGIs and their flanking 5 kb regions and calculated mean TOP-seq u-density across a composite LAD in our studied tissues (Fig. 6B). The CGI-devoid analysis showed strong hypomethylation of the LAD regions as compared to inter-LAD regions in the IMR90 cells, while no comparable changes in the TOP-seq u-density were detected in the brain cortex DNA. The observed methylation differences were mirrored by BS-seq data further confirming that LADs (and likely PMDs) are absent in the cells of the adult brain cortex (Fig. 6B). Similar analysis of NB cells revealed the presence of LADs in both of these tissues. This first genome-wide assessment of DNA methylation across LADs and their boundaries in NB-specific cells indicates that large regions of DNA hypomethylation may be characteristic to developing cells of neural crest origin, including the non-tumorigenic S-type cells and tumorigenic neuroblasts.
Figure 6. Lamina-associated domains in somatic and neuroblastoma cell types.
A, Genome browser view of TOP-seq u-density profiles in a 15 Mb region of Chr2. Blue rectangles above the lamin B1 track represent LADs for Tig3 cells. B, Generalized TOP-seq u-density profiles (Top) in LAD and inter-LAD regions for all four tissues compared to WGBS methylation profiles (Bottom) for Brain and IMR90 data. C, Global correlation of the TOP-seq u-density and Lamin-B1 profiles. Error bars show ±SD.
Common features of the chromatin architecture derived by correlation of the TOP-seq u-density (CGI signal removed) with the lamin B1 signal of the TIG3 embryonic fibroblasts were clearly apparent in the N-type, S-type and IMR90 cells but not in the Brain. Strikingly, on average across chromosomes, the correlations of the IMR90 and S-type cells with lamin B1 were higher than those of the N cells (Fig. 6C). Taken together, these observations suggest that an interplay between the DNA methylation and higher-order chromatin organization is a widespread mechanism of epigenetic regulation, which appears to be impaired in the tumorigenic N-type cells.
Discussion
We demonstrate here that certain DNA polymerases can be primed from a covalently tethered oligonucleotide with no required sequence complementarity between the tethered primer and the template DNA, and that such priming can occur with high fidelity with respect to the tagged site (patent no US9347093B2). This general analytical technique generates asymmetric target site-nested amplicons permitting their unidirectional sequencing and precise mapping in a genome. Owing to our previously developed CG-specific chemo-enzymatic labeling of DNA (Kriukiene et al., 2013), its first implementation fell on the unmodified CG sites, however, other known and yet unknown tagging chemistries could be similarly exploited. To this end, our preliminary experiments indicate that the M.SssI-directed tagging of hmCG sites with thiols compounds (Liutkeviciute et al., 2011) is well compatible with the described strategy. The TOP-seq approach is also flexible with respect to selection of downstream sequencing platforms (Ion Torrent, Illumina), and can be configured for whole-genome random fragment profiling, as demonstrated in this work, or for targeted multiplex analysis of selected genomic loci (forked end-adapters to be replaced with a set of locus-specific primers, see Fig. 1C).
We also demonstrate a successful application of the developed TOP-seq technique for high resolution whole genome profiling of unmodified CG sites in human DNA. The gold standard WGBS can directly infer the modification levels of each C nucleotide from experimental data. However, confident estimation of the methylation levels is only achievable at sequencing depths of >10x (Harris et al., 2010) or even 100x (Libertini et al., 2016). While the sequencing costs are on decline and the number of complete high resolution human DNA methylome maps is growing, yet just a few such maps are publicly available, and their need keeps outpacing their production due to prohibitive costs for most laboratories. Many more lower-cost DNA methylomes have been generated across a variety of biological and disease states using sparse sampling (MRE-seq or Infinium arrays) or enrichment-based (MBD-seq or MeDIP-seq) analytical methods. The latter group lack single CG resolution and typically calculate enrichment scores that reflect regional (200–400 bp) DNA methylation levels. In contrast, a TOP-seq u-density (or m-estimate) value for each CG is calculated from a coverage reading of this CG site taking also into account adjacent data points within the kernel window. Thus, each CG in the TOP-seq profile receives an individual experiment-derived value, which can be interpreted separately or combined into a window-resolution profile. Therefore, no other method provides a combination of single CG resolution, genome-wide coverage, and a cost that is affordable for a typical laboratory, particularly when many samples are assayed.
It has been suggested (Stevens et al., 2013) that MBD-seq and MRE-seq libraries of 30–50 M mappable reads approach saturation for the method-targeting CGs, and that such coverage is sufficient for general whole-genome profiling. Similarly, TOP-seq libraries of 30 M of mappable reads and 4x mean CG coverage (Low coverage dataset, Table S1) approach saturation (Fig. S2C), and show both good consistency and agreement with WGBS maps (1.18 billion raw reads and ~28x coverage) (Fig. 3 and Fig. S3). Therefore, low coverage (2–4x) TOP-seq analysis could be applied when the number of samples to be analyzed is important. The number of sequencing reads that gives a qualitatively comparable map by our assay is at least an order of magnitude lower than is required for a WGBS methylome (Harris et al., 2010). Owing to the high informativity of the TOP-seq reads (>90%) and simplicity of data processing, our method bridges this economy-versus-resolution gap.
The presented data demonstrate a higher precision of the TOP-seq technique as compared to the affinity-based profiling approaches, or MRE-seq both on the whole genome scale and across most of the individual genomic elements (Fig. 3F, 3G and Fig. S3). To this end, inferring accurate methylation levels in CG-poor regions is thought to be problematic using MeDIP-seq or MBD-seq (Harris et al., 2010). Owing to robust covalent tagging, TOP-seq can efficiently produce sequencing reads from uCG sites residing in regions of diverse CG or methylation densities (Fig. S2E). Altogether, our results highlight the capacity of the TOP-seq technique to detect both local methylation changes at relatively short genomic elements, such as CGIs, and higher-order chromatin organization in a single assay. The presented results also reinforce the notion that the cytosine modification states at the genomic scale can be revealed by interrogating the unmethylated fraction of CG dinucleotides, which interact with a multitude of regulatory elements of the intricate human epigenome. The presented concepts and tools thus show the potential for tremendously expanding our capabilities in manipulating and harnessing genomic information for a variety of useful applications.
Limitations
In contrast to BS-based methods, TOP-seq cannot directly determine the absolute methylation levels, but can infer u-density or m-estimate profiles from libraries of uCG-derived reads. Akin to other read-count based epigenome profiling approaches, it is sensitive to CNVs, and therefore, de novo discovered DMRs should be verified to fall outside genetic aberrations or validated by an independent method such as clonal bisulfite sequencing. Generally, it is common for cancer-specific and germline-transmitted CNVs to extend over megabase distances (Beroukhim et al, 2010). Therefore, the length of DMRs should be considered when analyzing genetically diverse tissues such as cancers.
STAR METHODS
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Request should be directed and will be fulfilled by Lead Contact S.K. (saulius.klimasauskas@bti.vu.lt.)
METHOD DETAILS
Genomic DNA
DNA from post mortem human brain (the prefrontal brain cortex, Brodmann area 10) of 50-year old healthy men was kindly provided by A. Petronis (CAMH, Toronto). Genomic DNA of human fetal lung fibroblast IMR90 and neuroblastoma specific LA1-55n and LA1-5s cell lines were obtained from the European Collection of Cell Cultures (ECACC, UK). Genomic DNA of Staphylococcus aureus was kindly donated by A. Lubys (Thermo Fisher Scientific Baltics, Vilnius).
Preparation of TOP-seq genomic libraries
Genomic DNA was sonicated on a Bioruptor UCD-200 instrument (Diagenode) in EB buffer (10 mM Tris-HCl (pH 8.5) to yield fragments with a peak size of 150-200 bp.
Step 1
For mTAG labeling, 50-500 ng of gDNA was incubated with eM.SssI (0.5-1 μM) in TNB buffer (10 mM Tris-HCl (pH7.4), 50 mM NaCl, 0.1 mg/ml BSA) supplemented with 200 μM Ado-6-azide cofactor (Kriukienė et al., 2013; Masevicius et al., 2016) for 1 hour at 30 °C followed by Proteinase K treatment (0.2 mg/ml) for 30 min at 55°C and column purification (GeneJet PCR Purification kit, Thermo Scientific (TS)).
Step 2
Azide-tagged DNA was end-filled using a DNA End Repair Kit (TS) according to vendor's recommendations and DNA was purified using the GeneJet Purification Kit. A 3’-dA mononucleotide extension was added to end-repaired DNA by incubating with Klenow exo- polymerase in Klenow Buffer (TS) in the presence of 0.5 mM dATP at 37 °C for 45 min, enzyme inactivated at 75 °C for 15 min followed by purification through GeneJet columns. Partially complementary adapters A1/A2 (4.5 μM) (produced by annealing of partially complementary 32/33 nt oligonucleotides A1/A2, A1 5' PGATTGGAAGAGTGGTTCAGCAGGAATGCTGAG and A2 5' ACACTCTTTCCCTACATGACACTCTTCCAATCT) were ligated by incubating the DNA with 15 u of T4 DNA Ligase (TS) in Ligase buffer at 22 °C overnight in a total volume of 30 μl, followed by thermal inactivation at 65 °C for 10 min and column purification (DNA Clean&Concentrator-5, Zymo Research).
Step 3
DNA eluted in 20 ul of Elution Buffer was supplemented with 10 μM alkyne DNA oligonucleotide (TO, 5’-T(alkyneU)TTATATATTTATTGGAGACTGACTACCAGATGTAACA, Base-click), 3.3 mM of CuBr:TBTA mixture (Sigma) in 60-65% of DMSO, incubated for 2.5 or 6 hours at 45 °C and subsequently diluted to <1% DMSO before purification through a Zymo Clean&Concentrator-25 column.
Step 4
A 50 μl reaction containing 5 ng of TO-DNA, 0.5 μM of a complementary priming strand (EP; 5‘-TGTTACATCTGGTAGTCAGTCTCCAATAAATATAT, with custom LNA modifications (Exiqon)) and 5 u Pfu polymerase (TS) in Pfu buffer with 0.2 mM dNTP was incubated at the following cycling conditions: 1 cycle at 95 °C 1 min, 65 °C 1 min, 72 °C 1 min.
Amplification of a primed DNA library was carried out by adding 5 μl of the TOP reaction mixture to 50 μl of amplification reaction containing 2x Maxima Hot Start PCR Master Mix and barcoded fusion PCR primers A(Ad)-EP-barcode-primer (63 nt) and trP1(Ad)-A2-primer (45 nt) at 0.5 μM each (both primers contained phosphorothioate modifications). The following thermocycler conditions were used for amplification: 94 °C 4 min; 15 cycles at 95 °C 1 min, 60 °C 1 min, 72 °C 1 min. The final libraries were purified through Zymo columns, size-selected for ~250 bp fragments (MagJet NGS Cleanup and Size-selection kit, TS), and were subjected to Ion Proton (TS) sequencing.
Step 5
NG sequencing of TOP-seq genomic libraries using Ion Proton PI chip (TS) and read processing.
Validation of TOP-seq in a model DNA system
To monitor the TO-primed DNA synthesis, 155 bp (1H; 1H-dir 5‘-TGTGTTACTGTGTGGAAAAGACC, 1H-rev 5‘-CCACTCCTTATAGTTTGGCTGA) and 202 bp (2H; 2H-dir 5‘-GCAATGTGTTGTGGAGGAGA and 2H-rev 5‘- CCTACTTGGGTTTGCCCTCT) PCR fragments were made from human BMX gene, each containing a single CG site. The efficiency of mTAG labeling of the model DNA fragments after Hin6I-restriction cleavage (see below) was tested by qPCR with the same primers used to produce the respective PCR fragments. The efficiency of click chemistry reaction was tested by gel-electrophoresis and HPLC–MS analysis of the products (see below). The template-dependent DNA synthesis primed by the priming oligonucleotide and the following amplification of the primed product was monitored on an Agilent Bioanalyzer.
qPCR analysis of eM.SssI-directed alkylation
A mixture of model DNA fragments were mTAG-modified as described in the main TOP-seq protocol. 10 ng of the modified DNA was incubated with the Hin6I restriction endonuclease in Tango Buffer (TS) for 1 hour at 37 °C following thermal inactivation of the enzyme by incubation at 65 °C for 15 min. The amount of intact DNA according to the uncleaved control was calculated for each DNA fragment. qPCR experiments were performed with a Rotor-Gene Q real-time PCR system (Qiagen) using Maxima SybrGreen qPCR Master Mix (TS). 0.3 mM of each primer pair was used in each reaction. The amplification program was set as: 95 °C for 10 min, 40 cycles 95 °C for 15 s, 60 °C for 1 min.
HPLC-MS analysis of oligonucleotide tethering
A model azide-tagged duplex was prepared by incubation of 0.5 μM of pTAACG/pATTCG double-stranded DNA oligonucleotide with eM.SssI MTase (1 μM) and Ado-6-azide cofactor (200 μM) in TEN buffer (Tris-HCl pH 7.4 10 mM, NaCl 50 mM, BSA 0.1 mg/ml) followed by purification with Oligo Clean&Concentrator kit (Zymo Research). Azide-derivatized pTAACG/pATTCG oligonucleotide at 1 μM concentration was combined with IntAlk2 alkyne-modified oligonucleotide (10 μM final concentration) and CuBr:TBTA mix (final concentration 0.33 mM CuBr, 0.67 mM TBTA), using DMSO as a solvent (65-70% of the final reaction mix). The reaction was incubated for 6 h at 45 °C, and then purified using an Oligo Clean&Concentrator kit (Zymo research). Purified DNA was incubated in 40 μl of P1 buffer containing Nuclease P1 (Sigma) 0.5 u for 2 h at 55 °C, and then dephosphorylated by adding 1 μl FastAP phosphatase at 37 °C overnight. Samples were analyzed on an integrated HPLC/ESI-MS system (Agilent 1290 Infinity) equipped with a Supelco Discovery®HS C18 column (7.5 cm × 2.1 mm) by elution with a linear gradient of solvents A (20 mM ammonium formate, pH 3.5) and B (80% aqueous methanol) at a flow of 0.3 ml/min at 30 °C as follows: 0-1 min, 0% B; 1-18 min, 80% B; 18-19 min, 100% B. High-resolution mass spectra of modification products were acquired on an Agilent Q-TOF 6520 mass analyzer (100–2500 m/z range, positive ionization mode).
HPLC-MS/MS analysis of genomic DNA
50 ng of gDNA in two replicates was digested with 0.5 u Nuclease P1 (Sigma) for 2 h at 55 °C in 40 μl of P1 buffer and then dephosphorylated by adding 1 μl FastAP (TS) phosphatase and incubating overnight at 37 °C. Standard d5mC, dhmC, dC and dG nucleosides (Trilink Biotech) were used for external calibration. Varied amounts of standard 5mC and hmC nucleosides were combined with 20 pmol each of dC, dG, dT, dA and samples were analyzed in duplicate. Samples were analyzed on an integrated HPLC/ESI-MS/MS system (Agilent 1290 Infinity/ 6410B triple quadruple) equipped with a Supelco Discovery®HS C18 column (7.5 cm × 2.1 mm, 3 μm) by elution with a linear gradient of solvents A (0.0075% formic acid in water) and B (0.0075% formic acid in acetonitrile) at a flow of 0.3 ml/min at 30 °C as follows: 0-7 min, 0% B; 7-21min, 3% B; 22-25 min, 80% B. Mass spectrometer was operating in the positive ion MRM mode and intensities of nucleoside-specific ion transitions were recorded: d5mC m/z 242.1→126.1; dhmC m/z 258.1→142.1; m/z dG 268.1→152.1. Ionization capillary voltage 2500 V, drying gas temperature 150 °C and flow rate 10 l/min, collision energy 15V.
Pyrosequencing
DNA oligonucleotides for pyrosequencing (Table S2) were designed with PyroMark Assay design 2.0 software (Qiagen) and synthesized by Metabion. Bisulfite conversion of IMR90, N and S type NB DNA was performed with EpiJET Bisulfite Conversion kit (TS). The respective DNA fragments were prepared from the converted DNA using Maxima Master Mix (TS) according to standard PCR conditions. The unmethylated EpiTect Control DNA (human) was acquired from Qiagen. Methylated control DNA was obtained as follows: the unmethylated EpiTec Control DNA was methylated by incubating with M.SssI (TS) and AdoMet 0.3 mM at 30 °C for 10-16 hrs, then incubating with Proteinase K (0.2 mg/ml) at 55 °C for 1 hour and purifying with Genomic DNA purification kit (Zymo). Pyrosequencing was performed on PyroMark Q24 version 2.0 device with PyroMark Gold Q24 kit (Qiagen). Data analysis was done with PyroMark Q24 2.0.6. software.
S.aureus genome assembly
Libraries of S.aureus genomic DNA for Ion PGM sequencing (two replicates) were prepared by using Torrent ClaSeek kit (TS) and amplified with Phusion Hot Start DNA polymerase (TS) according to the vendor's recommendations. The final libraries were size-selected for ~330 bp fragments (MagJet NGS Cleanup and Size-selection kit, TS), evaluated on an Agilent Bioanalyzer, quantified by qPCR and subjected to Ion PGM sequencing (Life Technologies) to give 7,939,365 sequencing reads. Fastq quality trimmer (FASTX toolkit) was used to trim the read ends having quality less than 30. Reads longer than 100 bp were retained. DNA sequence assembler MIRA (v4) with default options was used to construct the assembly (Chevreux et al., 1999). The tool reported 70-80x coverage. Contigs longer than 500 bp were retained yielding 321 contigs spanning 2,726,458 bases.
QUANTIFICATION AND STATISTICAL ANALYSIS
NG sequencing of TOP-seq genomic libraries
TOP-seq samples were sequenced on Ion Proton PI chip (TS). Reads shorter than 80 nt were discarded. Adapter sequences were removed using cutadapt (Martin, 2011) from 5’ end. FASTX (http://hannonlab.cshl.edu/fastx_toolkit/index.html) trimmer was used to trim 33 nt from 3’ ends to remove possible adapter sequences. FASTX quality trimmer was used to trim 3’ ends below phred quality score 20. Finally, 3’ ends were cut to a maximum read length of 70 nt (S.aureus) or 165 nt (human) and reads shorter than 16 nt were discarded. BWA (Li and Durbin, 2009) was used for alignment and only reads with mapping quality score of at least Q30 were retained. TOP-seq libraries of S.aureus were aligned to de novo assembled genome whereas human samples were aligned to hg19 genome. Identical reads (the same original length and starting genomic coordinate) were termed PCR duplicates and only one per group was retained. For the human samples, reads pertaining to sex chromosomes were removed before subsequent computation of u-density.
Post-processing analysis of TOP-seq data
For each mapped read we computed the distance from its start position to the nearest CG dinucleotide. We define CG coverage as the total number of reads, c, on any strand starting within absolute distance, d. We retained only reads with d ≤ 3. For the human samples weighted kernel density estimates of TOP-seq reads were computed using R density function with Epanechnikov kernel over 2^21 points uniformly distributed across each chromosome. Read counts were normalized to sum to 1 within each chromosome and used as weights for the density function. Gaussian kernel smoothing with the same bandwidth implemented in R ksmooth function was used to interpolate respective density values at the exact positions of CG nucleotides. The same approach yet with omission of weights was used to estimate unweighted CG density in the genome. Final TOP-seq unmethylation densities, the u-densities, were obtained by dividing weighted TOP-seq read densities by the unweighted CG density at each CG dinucleotide. Kernel parameters (180 bp for coverage-weighted density and 80 bp for CG density) were selected by evaluating correlation of IMR90 TOP-seq IMR90-1 R1 and WGBS samples at single CG resolution in chromosome 1 (see Fig. 3B) and the same parameters were subsequently used for all samples and all chromosomes.
Methylation estimates
Methylation estimates, m-estimates, were obtained by training an exponential decay model that assumes a linear decrease of WGBS methylation with exponential increase of u-density signal and other genomic feature-specific covariates. The model was computed using R nls procedure and defined as
Covariate values were calculated for each CG using 50 bp windows around each CG. The following covariates were used: GC frequency – percentage of guanine and cytosine bases per window (UCSC gc5BaseBw track); fraction of CG dinucleotides among CN pairs within window; mean sequence mappability value per window (UCSC wgEncodeCrgMapabilityAlign24mer track); percentage of bases per window that belong to SINE, LTR repeats, upstream, UTR 5’ and intergenic regions as separate covariates. Model training was performed using TOP-seq IMR90-1 R1 sample chromosome 20 which was then excluded from subsequent analyses involving m-estimates.
Public sequencing datasets
Brain and IMR90 WGBS datasets were downloaded through the Gene Expression Omnibus (GEO) with accession numbers GSE46710, GSM432687, GSM1204464. We only considered CG methylation and averaged beta values across the strands. GSM1204464 data was filtered for CGs with coverage ≥ 5. Genome wide Brain MRE-seq signal for autosomes was downloaded from the UCSC database (ucsfMreSeqBrainCpG track) in a bigWig format. It was converted to bedGraph format using bigWigToBedGraph conversion tool (Kent et al., 2010). IMR90 MRE-seq dataset was downloaded from GEO (accession number GSM830153). Signals from two strands were summed and genomic coordinates lifted to hg19 using liftOver tool. MBD-seq dataset was downloaded from GEO (accession number GSM947460). MBD windows were lifted to hg19 genomic coordinates and the same value assigned to all CGs that fall into the same window.
Genomic annotations
Repeat element (Repeat Masker annotation, rmsk), CG island (cpgIslandExt), gap location (gap), lamina associated domain (laminB1Lads) and lamin signal (laminB1) tracks were downloaded from the UCSC database (https://genome.ucsc.edu). Reference gene annotation was obtained from the GENCODE encyclopedia of genes (release 19) (https://www.gencodegenes.org). For analysis we used only data from the autosomes.
Chromatin states (expanded 18-state model) were downloaded from the Epigenome Roadmap project (http://egg2.wustl.edu) for the following datasets: cell line IMR90 (E017) and brain dorsolateral prefrontal cortex (E073). Chromatin states were defined according to Kundaje et al. (2015).
CG islands (CGIs) were classified into three classes on the basis of their relation to GENCODE protein coding genes. CGIs overlapping 2 kb region upstream from a transcription start site were classified as promoter CGIs, 9,545 in total. CGIs overlapping gene body of protein coding genes (excluding promoter CGIs) were classified as intragenic, 11,676 in total. All the remaining CGIs were classified as intergenic, 5420 in total.
For each protein coding gene we selected its longest processed transcript and used it as a reference gene. Additionally, we removed genes that were shorter than 1 kb. Upstream regions were defined as 2 kb flanks from the gene start site. When computing generic gene methylation profile each specific upstream, 5’-UTR, exon, intron, 3’-UTR region was divided into 20 equal size bins and individual CG methylation signals were averaged within the bins.
For methylation analysis of specific repeat elements we selected following repeat families: Alu, L1, L2. ERVL-MaLR, ERVL, ERV1, ERVK, ERV repeat families were merged into one family - ERV. Using this approach we calculated 1118248 Alu, 862941 L1, 637751 ERV and 439887 L2 elements. Alu repeats were grouped according to their position to gene body. If Alu element overlapped 2 kb upstream region from the transcription start site it was classified as promoter specific. Alu elements overlapping gene body (but not in promoter specific set) were classified as intragenic. All the remaining Alu's were classified as intergenic. 23835, 572112, 522301 elements were used for promoter, intragenic and intragenic Alu analysis, respectively.
Analysis of TOP-seq and WGBS profiles
For the S.aureus samples overlap of called uCGs was quantified using Jaccard similarity coefficient. For any two sets of uCGs, A and B, Jaccard similarity is defined as .
Pearson correlation coefficient, denoted r, was used as a measure of concordance throughout the paper. In cases where the sign of correlation was not important, we report absolute correlation values, denoted |r|. Overlap between any two sets was measured using two-tailed Fisher's exact test and odds ratio (OR) as well as p values were reported. P value significance threshold was 0.05. For hierarchical clustering Pearson correlation was converted into distance measure and complete linkage was used. Barplots with whiskers denote mean and standard deviation, respectively. In boxplots median was used as the center, the box spans 25th to 75th percentiles, the whiskers indicate 2nd and 98th percentiles.
Analysis of long-range hypomethylated domains
Methylation profiles in lamina associated domains (LADs) and inter-LAD regions were computed as follows. First, from the list of LADs we removed those that overlapped gaps in the genome assembly. LADs were further filtered according to their size retaining only those that are larger than 10% and smaller than 90% quantile of all the LADs. Inter-LAD domains were filtered using the same procedure. Each resulting domain was divided into 10 equally sized bins. Next, we removed CGs that overlapped CGIs or their 5 kb flanking regions. Due to MYCN amplification we also excluded chromosome 2 from analysis. For each CpG we assigned a corresponding bin of LAD or inter-LAD region. The final profile was obtained by averaging the signal in each bin using Gaussian kernel smoother with bandwidth 2. Finally, we evaluated correlation of TOP-seq and lamin B1 signal at each chromosome. Lamin B1 values were interpolated at the positions of used CpGs using Gaussian kernel smoother with 2 kb bandwidth.
Differential methylation of NB cell types
Promoter, intragenic and intergenic CGIs were used to find differentially methylated regions. For each CGI, mean TOP-seq u-density value per sample was computed; if CGI had mean value less than 0.0001 it was not included in analysis. All samples were passed to limma (Ritchie et al., 2015) for multigroup analysis and contrasts interrogated for N vs S, N vs B and S vs B. Regions having FDR adjusted q < 0.01 and absolute fold change above 20% were termed significant. Each CGI was associated with the gene whose promoter or body it overlapped and gene enrichment analysis was performed using DAVID annotation tool (version 6.7) (Huang et al., 2009).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| IMR90, Human Caucasian fetal lung fibroblast, DNA |
the European Collection of Cell Cultures (ECACC), UK https://www.phe-culturecollections.org.uk/ |
Cat# 85020204 |
| LA1-55n, Human neuroblastoma, DNA | the European Collection of Cell Cultures (ECACC), UK https://www.phe-culturecollections.org.uk/ |
Cat# 06041203 |
| LA1-5s, Human Neural Crest-Derived Non- Neuronal Progenitor, DNA |
the European Collection of Cell Cultures (ECACC), UK https://www.phe-culturecollections.org.uk/ |
Cat# 06041204 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CpG Methyltransferase (M.SssI) | Thermo Fisher Scientific | Cat#EM0821 |
| Proteinase K, recombinant, PCR grade | Thermo Fisher Scientific | Cat#EO0491 |
| Pfu DNA polymerase (recombinant) | Thermo Fisher Scientific | Cat#EP0502 |
| Maxima Hot Start PCR Master Mix | Thermo Fisher Scientific | Cat#K1051 |
| Fast DNA End Repair Kit | Thermo Fisher Scientific | Cat#K0771 |
| Klenow Fragment, exo- | Thermo Fisher Scientific | Cat#EP0422 |
| T4 DNA Ligase | Thermo Fisher Scientific | Cat#EL0011 |
| CuBr, 99.999% | Sigma-Aldrich | Cat#254185-10G |
| dNTP Mix (2 mM each) | Thermo Fisher Scientific | Cat#R0241 |
| DNA Clean & Concentrator – 5 kit | Zymo Research | Cat#D4014 |
| DNA Clean & Concentrator – 25 kit | Zymo Research | Cat#D4034 |
| GeneJET PCR Purification Kit | Thermo Fisher Scientific | Cat#K0702 |
| MagJET NGS Cleanup and Size Selection Kit | Thermo Fisher Scientific | Cat#K2821 |
| eM.SssI |
Kriukiene et al., 2013 doi:10.1038/ncomms3190 |
N/A |
| Ado-6-azide cofactor |
Masevicius et al., 2016 doi:10.1002/0471142700.nc0136s64 |
Compound 3a |
| Maxima SYBR Green/ROX qPCR Master Mix (2X) |
Thermo Fisher Scientific | Cat#K0221 |
| Nuclease P1 from Penicillium citrinum | Sigma-Aldrich | Cat#N8630 |
| FastAP Thermosensitive Alkaline Phosphatase | Thermo Fisher Scientific | Cat#EF0654 |
| d5mC, d5hmC | Trilink Biotech | http://www.trilinkbiotech.com/ |
| Agilent High Sensitivity DNA Kit | Agilent | Cat#5067-4626 |
| Qubit® dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat#Q32854 |
| EpiJET Bisulfite Conversion kit | Thermo Fisher Scientific | Cat#K1461 |
| EpiTect Control DNA 1000 (unmethylated) | Qiagen | Cat#59568 |
| Phusion Hot Start II DNA Polymerase | Thermo Fisher Scientific | Cat#F549L |
| Ion PI™ Hi-Q™ OT2 200 Kit | Thermo Fisher Scientific | Cat# A26434 |
| Ion PI™ Hi-Q™ Sequencing 200 | Thermo Fisher Scientific | Cat# A26772 |
| Ion PI™ Chip Kit v3 | Thermo Fisher Scientific | Cat# A26771 |
| PyroMark Gold Q24 kit | Qiagen | Cat#970802 |
| ClaSeek Library Preparation Kit, Ion Torrent compatible |
Thermo Fisher Scientific | Cat# K1351 |
| Sequence-Based Reagents | ||
| TO (tethered oligonucleotide): 5’- (alkyneU)TTATAT ATTTATTGGAGACTGACTACCAGATGTAAC A-3’ |
Base-click | N/A |
| A1 (adapter): 5′ P- GATTGGAAGAGTGGTTCAGCA GGAATGCTGAG-3’ |
Metabion | N/A |
| A2 (adapter) 5′-ACACTCTTTCCCTACATGAC ACTCTTCCAATCT-3’ |
Metabion | N/A |
| EP (Ad-TO-primer): 5’-TGTTACATCTGGTAG TCAGTCTCCAATAAATATAT-3’ |
Exiqon | N/A |
| Ad(A1)-TO-barcode-primer (63 nt): 5’-CCATC TCATCCCTGCGTGTCTCCGACTCAGCTAA GGTAACTGTTACATCTGGTAGTCAGTCTC- 3’ |
Metabion | N/A |
| Ad(A6)-TO-barcode-primer (63 nt) 5’-CCATC TCATCCCTGCGTGTCTCCGACTCAGTAAGG AGAACTGTTACATCTGGTAGTCAGTCTC-3’ |
Metabion | N/A |
| Ad(trP1)-A2-primer (45 nt): 5’-CCTCTCTAT GGGCAGTCGGTGAT CACTCTTTCCCTAC ATGACACT-3’ |
Metabion | N/A |
| pTAACG: 5’- TAATAATAAACGTAATAATAATAAT-3’ |
Metabion | N/A |
| pATTCG: 5’- ATTATTATTATTACGTTTATTATTA-3’ |
Metabion | N/A |
| Software and Algorithms | ||
| BWA | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net |
| R | N/A | https://www.r-project.org |
| limma | Ritchie et al., 2015 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| Cutadapt | Martin, 2011 | http://cutadapt.readthedocs.io/en/stable/index.html |
| FASTX | N/A | http://hannonlab.cshl.edu/fastx_toolkit/index.html |
| MIRA | Chevreux et al., 1999 | https://sourceforge.net/p/mira-assembler/wiki/Home/ |
| bigWigToBedGraph | Kent et al., 2010 | https://genome.ucsc.edu/goldenpath/help/bigWig.html |
| DAVID annotation tool | Huang et al., 2009 | https://david.ncifcrf.gov |
| liftOver | UCSC genome browser store | https://genome-store.ucsc.edu |
| Deposited data | ||
| IMR90 WGBS 1 | Lister et al., 2009 | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM432687 |
| IMR90 WGBS 2 | Ziller et al., 2013 | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1204464 |
| Brain WGBS | Wen et al., 2014 | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE46710 |
| IMR90 MRE-seq | N/A | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM830153 |
| Brain MRE-seq | Maunakea et al., 2010 | https://genome.ucsc.edu |
| IMR90 MBD-seq | Bert et al, 2013 | https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM947460 |
| Human reference genome NCBI build 37, GRCh37 |
Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/human |
| Gene Annotation | Harrow et al., 2012 | https://www.gencodegenes.org |
| Repeat Masker annotation | UCSC database (rmsk) | https://genome.ucsc.edu |
| GC frequency | UCSC database (gc5BaseBw) | https://genome.ucsc.edu |
| Sequence mappability | UCSC database (wgEncodeCrgMapabilityAlign24mer) | https://genome.ucsc.edu |
| CG islands | UCSC database (cpgIslandExt) | https://genome.ucsc.edu |
| Assembly gap locations | UCSC database (gap) | https://genome.ucsc.edu |
| Lamina associated domains | UCSC database (laminB1Lads) Guelen et al., 2008 | https://genome.ucsc.edu |
| Lamin signal | UCSC database (laminB1) Guelen et al., 2008 | https://genome.ucsc.edu |
| Chromatin states | Kundaje et al., 2015 | http://egg2.wustl.edu |
| TOP-seq data | This work | GEO (GSE91023) |
Supplementary Material
Chemo-enzymatic approach to map unmodified CpG sites at single-base resolution
Demonstrates non-homologous proximity-driven priming of DNA polymerases
Offers unidirectional DNA sequencing immediately from covalently tagged CpG sites
Independent of bisulfite conversion and avoids whole-genome sequencing
Acknowledgements
We are grateful to Audronė Rukšėnaitė for assistance with MS analyses and Ion Proton sequencing; Indrė Grigaitytė and Milda Rudytė for technical assistance; Raimonda Kubiliūtė and Sonata Jarmalaitė for help with pyrosequencing. We also thank Artūras Petronis for samples of human brain DNA and Arvydas Lubys for S.aureus DNA. This work was supported by grants from the Research Council of Lithuania (MIP-45/2013 to E.K., MIP-043/2014 to J.G.) and the NIH (HG007200 to S.K.). S.K, Z.S and E.K are inventors on related patents and patent applications.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
S.K. conceived the idea of the technology; E.K. and S.K. directed the development of the protocol; E.K. directed the genome-wide validation studies; J.G. directed bioinformatics analysis; Z.S. performed experiments; P.G. performed bioinformatics analysis; E.K. and S.K. wrote the manuscript with input from J.G. All authors commented on the manuscript.
DATA AND SOFTWARE AVAILABILITY
The sequencing data reported in this paper is available online at GEO (GSE91023). All the software used for analysis is listed in Key Resource Table and is freely available online.
References
- Abe M, Ohira M, Kaneda A, Yagi Y, Yamamoto S, Kitano Y, Takato T, Nakagawara A, Ushijima T. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005;65:828–834. [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina–associated domains. Nat. Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert, et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell. 2013;23:9–22. doi: 10.1016/j.ccr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Müller F, Simmer F, Gu H, Jäge N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carén H, Djos A, Nethander M, Sjöberg RM, Kogner P, Enström C, Nilsson S, Martinsson T. Identification of epigenetically regulated genes that predict patient outcome in neuroblastoma. BMC Cancer. 2011;11:66. doi: 10.1186/1471-2407-11-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevreux B, Wetter T, Suhai S. Genome Sequence Assembly Using Trace Signals and Additional Sequence Information. Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB) 1999;99:45–56. [Google Scholar]
- Ciccarone V, Spengler BA, Meyers MB, Biedler JL, Ross RA. Phenotypic Diversification in Human Neuroblastoma Cells: Expression of Distinct Neural Crest Lineages. Cancer Research. 1989;49:219–225. [PubMed] [Google Scholar]
- Decock A, Ongenaert M, Hoebeeck J, De Preter K, Van Peer G, Van Criekinge W, Ladenstein R, Schulte JH, Noguera R, Stalling RL, et al. Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers. Genome Biology. 2012;13:R95. doi: 10.1186/gb-2012-13-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau E, Martinez F, Orellana C, Canete A, Yañez Y, Oltra S, Noguera R, Hernandez M, Bermúdez JD, Castel V. Hypermethylation of apoptotic genes as independent prognostic factor in neuroblastoma disease. Mol. Carcinog. 2010;50:153–162. doi: 10.1002/mc.20700. [DOI] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat. Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed data sets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriukienė E, Labrie V, Khare T, Urbanavičiūtė G, Lapinaitė A, Koncevičius K, Li D, Wang T, Pai S, Ptak C, et al. DNA unmethylome profiling by covalent capture of CpG sites. Nat. Comm. 2013;4:2190. doi: 10.1038/ncomms3190. [DOI] [PubMed] [Google Scholar]
- Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libertini E, Heath SC, Hamoudi RA, Gut M, Ziller MJ, Herrero J, Czyz A, Ruotti V, Stunnenberg HG, Frontini M, et al. Saturation analysis for whole-genome bisulfite sequencing data. Nat. Biotechnol. 2016 doi: 10.1038/nbt.3524. doi: 10.1038/nbt.3524. [DOI] [PubMed] [Google Scholar]
- Liutkevičiūtė Z, Kriukienė E, Grigaitytė I, Masevičius V, Klimašauskas S. Methyltransferase-directed derivatization of 5-hydroxymethylcytosine in DNA. Angew. Chem. Int. Ed. 2011;50:2090–2093. doi: 10.1002/anie.201007169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. [Google Scholar]
- Masevičius V, Nainytė M, Klimašauskas S. Synthesis of S-Adenosyl-L-Methionine Analogs with Extended Transferable Groups for Methyltransferase-Directed Labeling of DNA and RNA. Curr. Protoc. Nucleic Acid Chem. 2016;64:1.36.1–13. doi: 10.1002/0471142700.nc0136s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P, Jensen T, Oshiro MM, Wozniak RJ, Nouzova M, Watts GS, Klimecki WT, Kim C, Futscher BW. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66:10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- Parzen On Estimation of a Probability Density Function and Mode. The Annals of Mathematical Statistics. 1962;33:1065. [Google Scholar]
- Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc. Natl. Acad. Sci. U. S. A. 2008;105:252–257. doi: 10.1073/pnas.0710735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:E47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Chatten J, Newton WA, Jr., Sachs N, Hamoudi AB, Chiba T, Marsden HB, Misugi K. Histopathologic prognostic factors in neuroblastic tumors: definition of subtypes of ganglioneuroblastoma and an age-linked classification of neuroblastomas. J. Natl. Cancer. Inst. 1984;73:405–416. doi: 10.1093/jnci/73.2.405. [DOI] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler BA, Lazarova DL, Ross RA, Biedler JL. Cell lineage and differentiation state are primary determinants of MYCN gene expression and malignant potential in human neuroblastoma cells. Oncol Res. 1997;9:467–476. [PubMed] [Google Scholar]
- Stevens M, Cheng JB, Li D, Xie M, Hong C, Maire CL, Ligon KL, Hirst M, Marra MA, Costello JF, et al. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 2013;23:1541–1553. doi: 10.1101/gr.152231.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Shao X, Liu H, Liu S, Wu Q, Zhang Y. Genome-wide dynamic changes of DNA methylation of repetitive elements in human embryonic stem cells and fetal fibroblasts. Genomics. 2012;99:10–17. doi: 10.1016/j.ygeno.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Walton JD, Kattan DR, Thomas SK, Spengler BA, Guo HF, Biedler JL, Cheung NK, Ross RA. Characteristics of stem cells from human neuroblastoma cell lines and in tumors. Neoplasia. 2004;6:838–845. doi: 10.1593/neo.04310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wen L, Li X, Yan L, Tan Y, Li R, Zhao Y, Wang Y, Xie J, Zhang Y, Song C, et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome Biol. 2014;15:R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez Y, Grau E, Rodríguez-Cortez VC, Hervás D, Vidal E, Noguera R, Hernández M, Segur V, Cañete A, Conesa A, et al. Two independent epigenetic biomarkers predict survival in neuroblastoma. Clinical Epigenetics. 2015;7:16. doi: 10.1186/s13148-015-0054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Szulwach KE, Hon GC, Song CX, Park B, Yu M, Lu X, Dai Q, Wang X, Street CR, et al. Tet-mediated covalent labelling of 5-methylcytosine for its genome-wide detection and sequencing. Nat Comm. 2013;4:1517. doi: 10.1038/ncomms2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.