Abstract
Chalcomycin, a 16-membered macrolide antibiotic made by the bacterium Streptomyces bikiniensis, contains a 2,3-trans double bond and the neutral sugar d-chalcose in place of the amino sugar mycaminose found in most other 16-membered macrolides. Degenerate polyketide synthase (PKS)-specific primers were used to amplify DNA fragments from S. bikiniensis with very high identity to a unique ketosynthase domain of the tylosin PKS. The resulting amplimers were used to identify two overlapping cosmids encompassing the chm PKS. Sequencing revealed a contiguous segment of >60 kb carrying 25 putative genes for biosynthesis of the polyketide backbone, the two deoxysugars, and enzymes involved in modification of precursors of chalcomycin or resistance to it. The chm PKS lacks the ketoreductase and dehydratase domains in the seventh module expected to produce the 2,3-double bond in chalcomycin. Expression of PKS in the heterologous host Streptomyces fradiae, from which the tyl genes encoding the PKS had been removed, resulted in production of at least one novel compound, characterized as a 3-keto 16-membered macrolactone in equilibrium with its 3-trans enol tautomer and containing the sugar mycaminose at the C-5 position, in agreement with the structure predicted on the basis of the domain organization of the chm PKS. The production of a 3-keto macrolide from the chm PKS indicates that a discrete set of enzymes is responsible for the introduction of the 2,3-trans double bond in chalcomycin. From comparisons of the open reading frames to sequences in databases, a pathway for the synthesis of nucleoside diphosphate-d-chalcose was proposed.
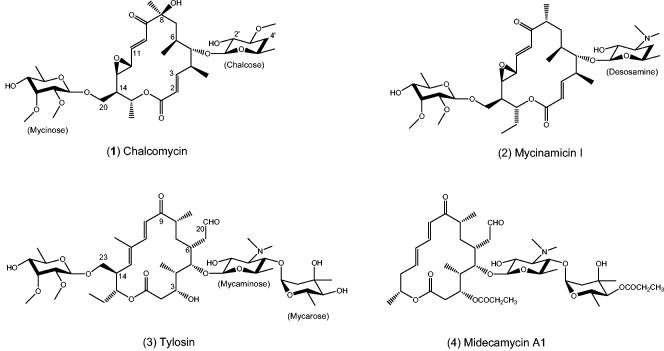
Sixteen-membered macrolide antibiotics have important applications in human and veterinary medicine and are subdivided into three major groups on the basis of the structures of their macrolactone backbones. Chalcomycin (Fig. 1; compound 1), discovered in the late 1950s in a strain of Streptomyces bikiniensis (11), and mycinamicin I (compound 2) represent a group with 2,3-trans double bonds. Three congeners of chalcomycin have been recently identified (2, 13, 21). All contain the same polyketide backbone but differ in the degrees of oxidation or acylation. The other two subgroups are represented by tylosin (compound 3) and midecamycin A1 (compound 4), each of which carries a 3-OH group instead of the 2,3-double bond and which also differ in their C-12 and C-14 substituents. Chalcomycin was determined to have modest antibiotic activity against gram-positive organisms: the MIC at which 50% of 11 susceptible Staphylococcus aureus strains were inhibited was 0.19 μg/ml, with a range of 0.05 to 0.78 μg/ml; the MICs for two susceptible Streptococcus pyogenes strains were 0.19 and 0.78 μg/ml (11). Although its precise mechanism of action was not determined, chalcomycin was found to inhibit protein synthesis and to exhibit cross-resistance with a number of macrolides; hence, chalcomycin was thought to act in the same manner as tylosin (29). In addition, whereas macrolides are not known to inhibit tRNA synthetases, chalcomycin was shown to inhibit the incorporation of [14C]glycine into glycyl-tRNA in S. aureus (18). Chalcomycin also exhibited very potent in vitro activity against a number of Mycoplasma species that were not susceptible to other macrolides, though the basis of this activity was not explored (25). Finally, chalcomycin was also found to inhibit protein synthesis in HeLa cells in culture (14), an activity not commonly associated with 16-membered macrolides. Despite these various activities, however, chalcomycin was not developed as a drug.
FIG. 1.
Structures of selected 16-membered macrolides.
Recent physicochemical and X-ray crystallographic studies (none of which included chalcomycin) have provided detailed information on how macrolides interact with ribosomes and underscore the interesting structural differences between chalcomycin and the commercially important members of the 16-membered macrolide family, tylosin and midecamycin A1. In place of the amino sugar mycaminose in compounds 3 and 4 or desosamine in compound 2, the chalcomycins contain the neutral sugar chalcose substituted at C-5(S) of the macrolactone ring. It is thought that the positive charge of the 3′-substituted amino group together with the 2′-hydroxyl group contribute to the binding of amino sugar-macrolides to domain V of bacterial ribosomes (15, 31, 38). Chalcose lacks the 3′-amino group but has the 2′-hydroxyl group. The chalcomycins also contain a C-6(S) methyl side chain rather than the C-6(R) ethylaldehyde side chain found in compounds 3 and 4 and other clinically useful 16-membered macrolides. Reduction or removal of the aldehyde in compound 3 and other 16-membered macrolides results in significant loss of ribosome binding and potency (26). X-ray analysis has shown a reversible covalent linkage between the aldehyde of compound 3 and the 6-amino group of A-2062 (Escherichia coli numbering) in domain V of the Haloarcula marismortui ribosome (15). In addition, chalcomycin contains an 8(S)-OH group not present in compound 2 or the clinically important 16-membered macrolides, but the contribution of this group to overall potency has not been explored. Finally, the 2,3-trans double bond of chalcomycin probably accounts for the large differences in the conformations of the macrocyclic backbones reported for the crystal and solution structures of chalcomycin and tylosin (44). Whether the differences in the backbones impact binding of the respective macrolides to the ribosome remains to be determined.
An important similarity between the chalcomycins and compound 3 is the presence of the sugar mycinose substituted at C-14(R) of the ring. Chemical footprinting of ribosomes from a number of bacteria and from X-ray data for H. marismortui ribosomes have indicated that the mycinose moiety of compound 3 makes contact with domain II of the ribosome and contributes to enhanced binding of the macrolide (4, 15).
To better understand the biosynthesis of chalcomycin and to determine how its components contribute to antibacterial potency, we isolated the chalcomycin biosynthesis cluster, determined its nucleotide sequence, and expressed some of the genes in heterologous hosts. The nucleotide sequence indicated that the 2,3-trans double bond is introduced by a reductase or dehydratase (DH) separate from the chalcomycin polyketide synthase (PKS), an unprecedented situation. Expression of the chm PKS in a mutant of Streptomyces fradiae from which the tyl genes encoding PKS had been removed confirmed the non-PKS origin of the 2,3-double bond and resulted in the production of compounds containing novel macrolactone-sugar combinations.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Routine DNA manipulations were performed with E. coli DH5α as a cloning host under standard culture conditions (36). S. bikiniensis NRRL2737, the wild-type producer of chalcomycin, was obtained from the ATCC strain collection and grown by a protocol described previously (28). For isolations of total DNA, S. bikiniensis was grown in tryptone soya broth (19). S. fradiae K342-45 was made by conjugal transfer of plasmid pKOS342-45 from E. coli DH5α/pUB307/pKOS342-45 to S. fradiae K159-1 as described previously (34). S. fradiae was maintained on AS-1 agar (8). For shake flask fermentations, seed cultures were grown in tryptone soya broth, and fermentations were performed in R medium [containing, per liter, 15 g of wheat flour, 10 g of corn gluten, 25 g of molasses, 2.5 g of fodder yeast, 1 g of (NH4)2HPO4, 1 g of NaCl, 2 g of CaCO3, and 34 ml of soybean oil] at 30°C for 5 to 7 days. When antibiotic selection of transformants was employed, carbenicillin (100 μg/ml), kanamycin (50 μg/ml), or apramycin (60 μg/ml) was used as required. Micrococcus luteus ATCC 9341 was used as an indicator strain in bioassays of zone of inhibition on medium 11 (Difco).
DNA manipulations.
Total DNA isolation, plasmid DNA preparations, restriction endonuclease digestions, ligations, and other DNA manipulations were done using the standard protocols for E. coli (36) and Streptomyces (19). Digoxigenin labeling of DNA was performed according to protocols supplied by the distributor (Boehringer Mannheim).
DNA sequencing and analysis.
DNA sequencing was performed with cosmids by standard shotgun cloning to obtain at least fourfold coverage. Primer walking was used to close the gaps. The sequence was assembled using the Sequencher software package (Gene Codes) and analyzed with MacVector (Accelerys) and the BLAST server of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST).
Isolation of the chm biosynthesis cluster.
A genomic library of S. bikiniensis NRRL2737 was made by digestion of the total DNA with Sau3AI and ligation of 35- to 47-kb fragments into pSuperKos, a derivative of pSuperCos (Stratagene), which was digested with AfeI and self-ligated to eliminate the neomycin resistance gene. Degenerate primers designed based on conserved regions of ketosynthase (KS) domains for the type I PKS cluster with a codon bias for high-G+C-content organisms are described elsewhere (33). DNA fragments (termed KS amplimers) obtained after PCR amplification were cloned into pLitmus28 (New England Biolabs) cut with EcoRV; 100 were sequenced. Three of the amplimers, Sb3/7-31 (KSQ), Sb1/5-75 (KS3), and Sb1/5-78 (KS7), believed to constitute segments of the chm PKS (Table 1), were used to probe the cosmid library under high-stringency conditions. To isolate both ends of the proposed >35-kb chm PKS cluster, plasmids from the colonies that showed strong hybridization signals were isolated and subjected to Southern blotting with the putative chm KSQ or KS7 amplimers as probes. End sequencing of the plasmids that hybridized to one or both probes revealed two cosmids, pKOS146-185.1 and pKOS146-185.10, that possessed high homology at one end with a segment of the PKS from the tylosin biosynthesis cluster. Specific primer pairs, designed for the putative chm KSQ (Sb3/7-31), KS3 (Sb1/5-75), and KS7 (Sb1/5-78) amplimers, were synthesized and used in PCRs with cosmids pKOS146-185.1 and pKOS146-185.10. pKOS146-185.1 produced correctly sized amplimers with the KSQ- and KS3-specific primers but not with KS7-specific primers, and pKOS146-185.10 produced a correctly sized amplimer with the KS7-specific primers but not with the KSQ- and KS3-specific primers, indicating that pKOS146-185.1 and pKOS146-185.10 contained the 5′ and 3′ regions, respectively, of the chm PKS genes. End sequencing and preliminary mapping suggested that the inserts of the two cosmids overlapped by ca. 4 kb. The complete nucleotide sequences of the inserts in the two cosmids were determined.
TABLE 1.
BLASTP analysis of unique KS amplimers from the genome of S. bikiniensis
| KS amplimer | Best BLASTP match(es)a/organism(s) | % Identity/% Similarity | Accession no. |
|---|---|---|---|
| Sb1/5-60 | MycAIV/Micromonospora griseorubida | 82/91 | BAC57031 |
| Tyl KS6/Streptomyces fradiae | 77/86 | AAB66507 | |
| Sb1/5-62 | MycD/Microcystis aeruginosa | 48/62 | BAB12210 |
| Sb1/5-67 | Put. PKS ORF3/Streptomyces sp. strain GERI155 | 85/89 | AAM81586 |
| Tyl KS1/Streptomyces fradiae | 83/90 | AAB66504 | |
| Sb1/5-68 | Put. PKS ORF1/Streptomyces sp. strain GERI155 | 95/97 | AAM81584 |
| Tyl KS2/Streptomyces fradiae | 80/88 | AAB66504 | |
| Sb1/5-72 | Nid A1/Streptomyces caelestis | 77/86 | AAC46024 |
| Sb1/5-75 | Put. PKS ORF2/Streptomyces sp. strain GERI155 | 94/97 | AAM81585 |
| Ty1 KS3/Streptomyces fradiae | 75/84 | AAB66505 | |
| Sb1/5-76 | Put. PKS/Streptomyces avermitilis MA4680 | 73/85 | NP_822424 |
| Sb1/5-78 | NidA5/Streptomyces caelestis | 74/85 | AAC46028 |
| Ty1 KS7/Streptomyces fradiae | 75/84 | AAB66508 | |
| Sb1/5-80 | Put. PKS ORF3/Streptomyces sp. strain GERI155 | 97/99 | AAM81586 |
| Ty1 KS5/Streptomyces fradiae | 80/88 | ||
| Sb1/5-81 | GdmAI/Streptomyces hygroscopicus | 75/86 | AAO06916 |
| Sb1/5-87 | Put. PKS ORF3/Streptomyces sp. strain GERI155 | 93/95 | AAM81586 |
| Tyl KS4/Streptomyces fradiae | 81/88 | AAB66506 | |
| Sb2/5-A16 | Put. PKS/Nostoc sp. strain PCC7120 | 45/59 | NP_485688 |
| Sb2/5-A17 | NidA1/Streptomyces caelestis | 76/86 | AAC46024 |
| Sb3/7-31 | mycA ORF 1/Micromonospora griseorubida | 72/80 | BAA76543 |
| Ty1 KSO/Streptomyces fradiae | 75/84 | AAB66504 |
Put., putative.
Assembly of the PKS genes on plasmid pKOS342-45.
The chm PKS was pieced together for expression from cosmids pKOS146-185.1 and pKOS146-185.10 as follows. Restriction sites in the chm genes encoding PKS employed are shown in Fig. 2. PCR was used to obtain a 942-bp fragment encompassing the 3′ end of chmGV by use of the following oligonucleotides, with pKOS146-185.10 as the template: 5′-GACACGGCCGGTGAGAGCAGC-3′ and 5′-CTTCTAGATGTCGCGGTGTACGG-3′ (the restriction site is underlined). The PCR product was digested with NcoI and XbaI, and the 309-bp subfragment was ligated into pLitmus29 digested with NcoI plus XbaI to generate pKOS342-33. pKOS342-33 was digested with NcoI and XhoI and ligated with a ca. 2.4-kb fragment from similarly digested pKOS146-185.10 to produce pKOS342-35. This plasmid was digested with BglII and XhoI and ligated with a ca. 6.4-kb BglII- plus XhoI-generated fragment from pKOS146-185.10 to create pKOS342-36. The chmGIII gene was isolated by digesting pKOS146-185.1 with HindIII and PstI to obtain a ca. 5.4-kb fragment, and pKOS146-185.10 was digested with PstI and BglII to obtain a ca. 6.3-kb fragment. These two fragments were ligated into HindIII- plus BglII-digested pLitmus28 to produce pKOS342-38. pKOS342-36 was then cut with BglII and SpeI to produce a ca. 9-kb piece, which was introduced into the corresponding sites of pKOS342-38 to generate pKOS342-39. The first two open reading frames (ORFs) of the chm PKS were isolated from the cosmid pKOS146-185.1 as EcoRI/XhoI and XhoI/BspHI fragments. These two pieces were ligated into pKOS232-165, a pLitmus28 plasmid that had a linker to introduce PacI and NdeI sites at the start of the translation and was cut with EcoRI and NcoI. The plasmid that resulted from this three-piece ligation was designated pKOS232-172. To make the final expression plasmid, pKOS232-172 was cut with NdeI and HindIII to generate a ca. 19-kb fragment, and pKOS342-39 was digested with HindIII and SpeI to produce a ca. 20-kb fragment. These two fragments were ligated into NdeI- plus SpeI-digested pKOS244-20, a pSET152-based vector (5) that was modified to contain the λ cos site and tylGIp, and plasmid pKOS342-45, containing the ca. 40-kb segment of chm encoding PKS, was obtained by use of a λ phage packaging kit (Stratagene).
FIG. 2.
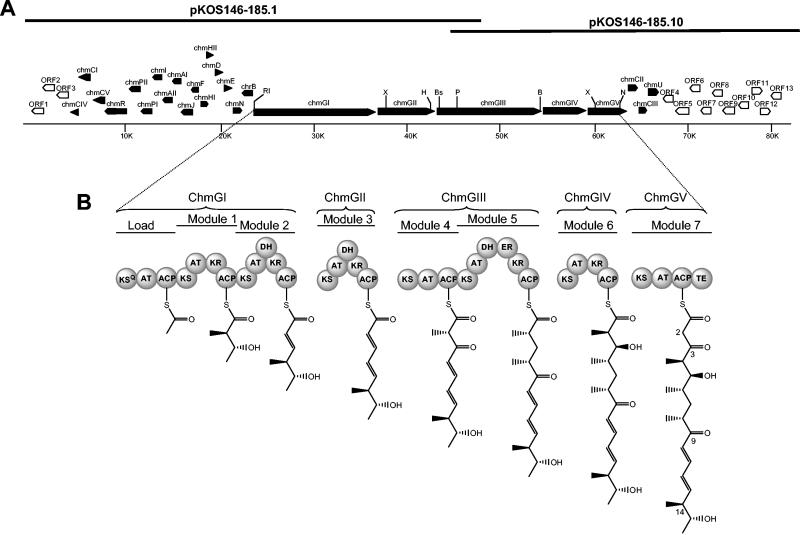
(A) Map of the chm biosynthestic gene cluster on two overlapping cosmids. The chm biosynthesis or resistance genes are shown as closed polygons, and non-chm genes are shown as open polygons. Restriction site abbreviations: B, BglII; Bs, BspHI; H, HindIII; N, NcoI; Nd, NdeI; P, PstI; RI, EcoRI; X, XhoI; Xb, XbaI. (B) Domain organization of the chm PKS and proposed structures of the thioester intermediates at the end of each cycle of elongation. ACP, acyl carrier protein; AT; acyltransferase; ER, enoylreductase; KR, β-ketoreductase; KS, β-ketoacyl ACP synthase; KSQ, KS domain with active site cys replaced by gln; TE, thioesterase.
Detection of macrolides from fermentation broths.
After 7 days of growth at 30°C, the culture broths (10 to 50 ml) were analyzed for 16-membered macrolide production by high-pressure liquid chromatography (HPLC) (Metachem Metasil Basic column; 4.6 by 150 mm; 5-μm-diameter particle) using a linear gradient of 15 to 100% organic phase (56% methanol, 5 mM ammonium acetate) at 1 ml/min over 7 min. The HPLC used simultaneous detection by electrospray mass spectrometry (Turbo Ionspray) and UV absorption at 282 nm.
Isolation and characterization of 5-O-mycaminosylchalcolactone.
Approximately 500 ml of cell-free supernatant of fermentation broth from a culture of S. fradiae K342-45 was adjusted to pH 7.8 with solid NaHCO3. The solution was filtered, and the filtrate was extracted with CHCl3 (500 ml used for each of four extractions). The organic extracts were combined and dried with vigorous stirring over Na2SO4, filtered, and concentrated in vacuo into an amber oil. The crude material was partially purified by flash silica gel chromatography using a gradient of 0 to 35% acetone (plus 2% [vol/vol] triethylolamine [Net3]) in hexane. The crude product (12 mg) was eluted in the 30% fractions. This material was further purified by preparatory HPLC with a 150- by 21.5-mm Polaris C18 column (MetaChem), and elution was monitored at 280 nm. Purification was performed at a flow rate of 10 ml/min, employing a linear 50 to 100% gradient of solvent A (CH3CN-methanol [80/20] buffered with 5 mM NH4 acetate). The compound eluted in 60 to 70% solvent A. The solvent was removed in vacuo, and the residual NH4 acetate was removed by application of the sample to a silica gel plug. The plug was eluted with 30% acetone-2% (vol/vol) NEt3 in hexane, providing purified 3-hydroxy-5-O-mycaminosyl-chalcolactone (5.6 mg) as a white solid. High-resolution mass spectra for C28H45NO8 were calculated to be 523.3140 and observed to be (M + H) 524.3760. Nuclear magnetic resonance (NMR) spectra were collected in CDCl3 with a 400-MHz spectrometer. In CDCl3, the sample was present as a mixture of the C3 enol and keto tautomers (enol/keto ratio of 3:1). NMR results were as follows: for 1H NMR (CDCl3, 400 MHz, data for enol tautomer), δ 0.98 (d, 3H, J = 6.8 Hz), 1.08 (d, 3H, J = 6.8 Hz), 1.15 (d, 3H, J = 6.8 Hz), 1.27 (m, 3H), 1.29 (m, 3H), 1.33 (d, 3H, J = 7.2 Hz), 1.38 (m, 1H), 1.50 (m, 1H), 1.57 (m, 1H), 2.22 (m, 1H), 2.36 (t, 1H, J = 10.0 Hz), 2.49 (s, 6H), 2.51 (m, 1H), 2.54 (m, 1H), 3.09 (t, 1H, J = 9.4 Hz), 3.27 (m, 1H), 3.52 (dd, 1H, J = 7.2, 10.0 Hz), 3.69 (d, 1H, J = 10.4 Hz), 4.28 (d, 1H, J = 7.2 Hz), 4.79 (m, 1H), 4.86 (s, 1H), 5.68 (dd, 1H, J = 15.2, 9.2 Hz), 6.11 (dd, 1H, J = 11.2, 15.2 Hz), 6.23 (d, 1H, J = 14.8 Hz), and 7.10 (dd, 1H, J = 14.8, 11.2 Hz); and for 13C NMR (400 MHz, CDCl3), δ 15.73, 17.16, 17.62, 17.81, 18.06, 18.25, 33.37, 41.67, 43.69, 44.74, 45.17, 70.13, 70.91, 71.05, 72.15, 73.19, 84.74, 89.82, 104.39, 123.39, 132.41, 141.64, 144.16, 171.92, 180.09, and 203.71. High-resolution mass spectra for C28H45NO8 were calculated to be 523.3140 and found to be 524.3760 (M + H).
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited into GenBank under accession no. AY509120.
RESULTS
Identification of chm PKS probes by use of phylogenetic relationships.
Comparisons of previously sequenced 16-membered macrolide PKS clusters suggested that the chm PKS genes are most homologous to the niddamycin or tylosin (tyl) PKS genes. Hence, five degenerate oligonucleotides, designed on conserved regions of KS domains of type I PKS genes from high-G+C-content organisms, were used in three specific combinations in PCR with total DNA from S. bikiniensis as the template. Amplimers of the expected sizes were obtained and sequenced. Fourteen different sequences were identified from 81 amplimers sequenced. Eight amplimers exhibited 71 to 81% identity at the protein level to a corresponding sequence in a KS domain of the tyl PKS (Table 1). Two additional amplimers exhibited very high similarity to segments of the PKS involved in the synthesis of the 16-membered macrolide niddamycin, but these sequences were much less similar to the corresponding tylosin PKS segments. Hence, three of the KS amplimers, which showed the highest degrees of homology to the tylosin KS domains from the loading module or from modules 3 or 7, were used as “perfect probes” (37) to probe a cosmid library of S. bikiniensis DNA, leading ultimately to isolation of the chm biosynthesis cluster on two overlapping cosmids (Fig. 2A).
Analysis of the chm biosynthesis cluster.
Thirty-five ORFs were identified within the ca. 80-kb segment of S. bikiniensis DNA from cosmids pKOS146-185.1 and pKOS146-185.10 (Fig. 2A; Table 2). The chm cluster most likely starts with the gene designated chmCIV, similar to a gene involved in desosamine biosynthesis in the oleandomycin biosynthesis pathway (41), and ends with the gene designated chmU. ORFs 1 to 3 and 4 to 12 appear to have orthologs in the genomes of Streptomyces coelicolor and Streptomyces avermitilis, which are not known to be associated with secondary metabolism, and their proposed functions do not correspond to roles found in the synthesis of macrolides. ORF 13, at the far right end of the sequenced segment, appears to encode a type II thioesterase, homologs of which have been found in many other modular PKS systems and which are thought to play an editing role in the biosynthesis of the polyketide (8, 20). However, the protein encoded by chmI is more similar to the family of type II thioesterases associated with polyketide biosynthesis (57% amino acid identity) than ORF 13 (45% amino acid identity). Therefore, we propose that ChmI probably provides the required editing role in chalcomycin biosynthesis and that ORF 13 likely does not play a role in chalcomycin biosynthesis.
TABLE 2.
ORFs and their proposed functions
| ORF designation | Size of encoded proteina | Proposed function | Closest homolog | % Identity |
|---|---|---|---|---|
| ORF 1 | 343 | Membrane protein | SCO0450 | 56 |
| ORF 2 | 280 | Hypothetical protein | SCO0451 | 70 |
| ORF 3 | 293 | Oxidoreductase | DR1890 | 55 |
| chmCIV | 405 | 3,4-DH; d-chalcose pathway | OleN1 | 66 |
| chmCI | 259 | O-Methyltransferase; d-chalcose pathway | SpnH 250 | 59 |
| chmCV | 485 | d-Chalcose pathway | EryCV | 76 |
| chmR | 836 | β-Glucosidase, extracellular reactivator of chalcomycin | DesR | 58 |
| chmPII | 401 | P450; 12,13-epoxidase | LnmZ | 51 |
| chmPI | 407 | P450; C-8 hydroxylase | OleP | 51 |
| chmI | 282 | Thioesterase (TEII family) | TylO | 57 |
| chmAII | 323 | TDP-glucose 4,6-DH | AveBII | 68 |
| chmAI | 305 | TDP-glucose synthase | DesIII | 69 |
| chmJ | 196 | 3-Epimerase; 6-deoxy-d-allose pathway | TylJ | 64 |
| chmF | 255 | 3-O-Methyltransferase; d-mycinose pathway | TylF | 69 |
| chmHI | 420 | P450; C-20 hydroxylase | TylHI | 60 |
| chmHII | 73 | Ferredoxin | TylHII | 53 |
| chmD | 326 | 4-Ketoreductase; 6-deoxy-d-allose pathway | TylD | 58 |
| chmE | 403 | 2-O-Methyltransferase; d-mycinose pathway | TylE | 71 |
| chmN | 418 | 6-Deoxy-d-allosyltransferase | TylN | 67 |
| chrB | 280 | 23S-rRNA G748-methyltransferase | TlrB | 58 |
| chmGI | 4,441 | PKS, modules 0 to 2 | ||
| chmGII | 1,974 | PKS, module 3 | ||
| chmGIII | 3,788 | PKS, modules 4 and 5 | ||
| chmGIV | 1,612 | PKS, module 6 | ||
| chmGV | 1,350 | PKS, module 7 and thioesterase | ||
| chmCII | 412 | NDP-hexose 3,4-isomerase; d-chalcose pathway | TylMIII | 41 |
| chmCIII | 425 | Chalcosyltransferase | TylMII | 62 |
| chmU | 248 | 3-Ketoreductase | PP2783 | 50 |
| ORF 4 | 382 | Permease | SC0513 | 79 |
| ORF 5 | 487 | Membrane protein | SC2255 | 53 |
| ORF 6 | 231 | d-Alanyl-d-alanine carboxypeptidase | SAV3781 | 49 |
| ORF 7 | 386 | Sensory histidine kinase | SC5304 | 47 |
| ORF 8 | 228 | Response regulator | SAV4704 | 60 |
| ORF 9 | 476 | Permease | SC6214 | 71 |
| ORF 10 | Hypothetical protein | SC5427 | 35 | |
| ORF 11 | 612 | Permease | SC6214 | 52 |
| ORF 12 | 223 | MerR family transcriptional regulator | SCO7698 | 34 |
| ORF 13 | 251 | Thioesterase (TEII family) | RifR | 45 |
Number of amino acids.
Resistance genes.
Twenty-five of the ORFs showed similarities to genes in other macrolide pathways; all but one are designated chm (Table 2). The remaining gene, located immediately upstream of the chm PKS genes and designated chrB, strongly resembles tlrB and myrA in the tylosin and mycinamicin biosynthesis clusters, respectively. TlrB methylates nucleotide G748 (in domain II) of 23S rRNA to prevent tylosin from binding to and inhibiting the host's ribosomes (22). It is very likely, therefore, that ChrB catalyzes methylation of G748 in S. bikiniensis ribosomes, resulting in self-resistance. An erm gene responsible for methylation of A2058 in domain V of the 23S rRNA, a major factor in self-resistance in strains that produce most of the clinically important macrolides (all of which contain either desosamine or mycaminose), was not found in the chm cluster, but whether one is present elsewhere in the genome was not determined. A second putative resistance gene, chmR, strongly resembles oleR in the oleandomycin biosynthesis cluster of Streptomyces antibioticus. OleR is an extracellular β-glucosidase that removes the glucose moiety that is attached to the 2′OH of desosamine by the action of OleI (32). OleI-mediated glucosylation of oleandomycin confers self-resistance; removal of the glucose residue by OleR, after the glucosylated macrolide is excreted from the cell, restores activity to the compound. A counterpart to oleI, however, was not found within the chm cluster.
The chalcomycin PKS.
The chalcomycin PKS is encoded by five genes (chmGI to chmGV) that together contain eight modules, including a loading module (Fig. 2A), and resemble, in both gene and module organization, the PKS genes that encode the macrolactone backbones of other 16-membered macrolides (17). The loading module contains a KSQ domain; biosynthesis of the macrolactone is thus believed to require malonyl CoA as the starter, which is subsequently decarboxylated after attachment to the loading module (6, 43). The scheme representing the progression of the nascent polyketide through the modules of the chm PKS is shown in Fig. 2B. Examination of the sequence revealed that module 7, which determines the composition of carbons 2 and 3 of the macrolactone produced by the chm PKS, does not contain a ketoreductase or a DH domain. How and at what step during the synthesis of the completed molecule the 2,3-double bond in chalcomycin is produced are not immediately apparent from examination of the cluster and are discussed below.
Expression of the chm PKS in S. fradiae results in a 3-ketomacrolactone.
The >40-kb segment of chm encoding the PKS was assembled as a single contiguous DNA fragment from cosmids pKOS146-185.1 and pKOS146-185.10 and then subcloned into a derivative of plasmid pSET152 that placed the PKS genes under the control of the promoter that drives the tyl PKS (tylGp). This plasmid, designated pKOS342-45, was transferred via conjugation from its E. coli cloning host to the streptomycete expression host, S. fradiae K159-1, from which the tylGI to tylGV (PKS) genes had been deleted (34). pSET152 does not carry genes enabling autonomous replication in Streptomyces but carries the attP-int segment of the Streptomyces phage φC31 and integrates, through site-specific recombination, into a unique site (attB) on the chromosome (5). S. fradiae K159-1/pKOS342-45 transconjugants were found to be stable for the maintenance of the aacCIV marker without the continued inclusion of apramycin in the fermentation medium. Ethyl acetate extracts of whole fermentation broths from four transconjugants exhibited antibiotic activity, whereas the extract of the fermentation broth of the parent was not bioactive. One of these strains was chosen for further analysis and designated S. fradiae K342-45.
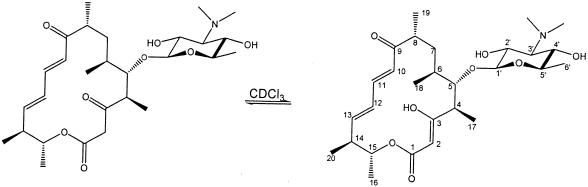
Liquid chromatography-mass spectrometry analysis of extracted whole broths from a small-scale fermentation of K342-45 revealed four prominent compounds not present in the parent, with the following m/z values [(M + H)+]: 525, 669, 843, and 845. Only the compound corresponding to m/z 525 was purified in sufficient amount from a large-scale fermentation to enable a determination of its structure. From the NMR data, it can be seen that this compound, named 5-O-mycaminosylchalcolactone (compound 5), contains a 3-keto group in the macrolactone backbone (present mainly as the 3-enol tautomer in the NMR solvent, CDCl3) as predicted from the domain structure of the chm PKS, along with the sugar mycaminose attached at C-5 (Fig. 3).
FIG.3.
5-O-Mycaminosylchalcolactone.
The finding of the expected macrolactone is consistent with the hypothesis that the genes expressed in S. fradiae correspond to the chalcomycin PKS and that the cluster shown in Fig. 2A is indeed the chalcomycin biosynthesis cluster. Direct evidence that these genes are involved in chalcomycin biosynthesis was obtained from a loss of chalcomycin biosynthesis in S. bikiniensis after insertion of an antibiotic resistance marker into the chm PKS. This insertion was accomplished employing an in vivo cosmid packaging system to produce an S. bikiniensis integration vector and is described elsewhere (P. Revill et al., unpublished results).
Compound 5 was assayed for bioactivity and found to be virtually inactive (MICs, >12.5 μg/ml) against a number of S. aureus and Streptococcus pneumoniae strains. Since the original unpurified extracts of the transconjugants exhibited at least modest antibiotic activity, these results suggested the presence of at least one other more-potent chalcomycin-tylosin hybrid compound. Although structures of additional compounds were not determined, fragmentation patterns of the m/z 669 (M + H) compound are consistent with a structure consisting of the 3-keto macrolactone attached to the disaccharide of tylosin. Similarly, the fragmentation patterns of the m/z 843 (M + H) and m/z 845 (M + H) compounds are consistent with structures comprised of a 16-membered backbone and the three sugars of tylosin. It is conceivable that the m/z 845 (M + H) compounds may explain the antibacterial activity observed in the crude extract.
Deoxysugar genes.
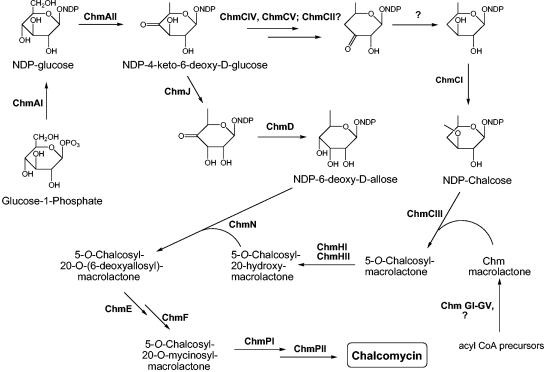
Genes for the synthesis of the two deoxysugar moieties of chalcomycin, d-chalcose and d-mycinose, are found in the cluster. Each is presumed to be derived from the common intermediate nucleoside diphosphate (NDP)-4-keto-6-deoxyglucose, which itself is produced from glucose-1-phosphate through the action of the gene products of chmAI and chmAII (Fig. 4).
FIG. 4.
Proposed pathways of deoxysugar biosynthesis and post-PKS modifications in S. bikiniensis showing roles for the various enzymes encoded by genes in the chm cluster.
Chalcose.
As in the case of desosamine, synthesis of NDP-chalcose involves deoxygenation at C-4, which requires reduction of the radical formed after removal of the oxygen atom. We propose that these reactions are carried out by ChmCIV and ChmCV (Fig. 4), homologs of the desosamine biosynthesis enzymes EryCIV and EryCV, respectively, in the erythromycin pathway and of DesI and DesII, respectively, in the pikromycin pathway (12, 35, 40, 45). It has been proposed that deoxygenation takes place through the formation of a pyridoxamine-5′-phosphate (PMP)-sugar adduct at either C-3 (12, 40) or C-4 (16, 47). Formation of the PMP-sugar adduct at C-3 would require isomerization of the keto group from C-4 to C-3, a role proposed for EryCII in the desosamine pathway in erythromycin biosynthesis (12, 40). ChmCII is highly homologous to EryCII and could catalyze the analogous isomerization in chalcose biosynthesis, if the pathway proceeds through PMP-sugar adduct formation at C-3. If the adduct is formed at C-4, a role for ChmCII is not apparent. The next step after deoxygenation is reduction of the 3-keto group of the pathway intermediate. SpnQ is believed to serve this role in the biosynthesis of the forosamine component of spinosad (23). A homolog of SpnQ was not found within the >80-kb sequenced segment from the chalcomycin producer. The gene chmU, described below, which is presumed on the basis of sequence matching to encode a ketoreductase, lies immediately downstream of chmCIII but is thought to play a role in the introduction of the 2,3-double bond into the macrolactone component of chalcomycin. Hence, we do not presently have a candidate for the required 3-keto reduction step in chalcose synthesis in S. bikiniensis. The penultimate step, 3-O-methylation of NDP-4,6-dideoxy-d-glucose to produce NDP-chalcose, is likely carried out by ChmCI, which is highly similar to SpnH, a probable methyltransferase involved in O methylation of the rhamnose precursor of spinosyn (23, 42). Attachment of chalcose to the aglycone moiety is likely carried out by ChmCIII, a proposed glycosyltransferase.
Lankamycin is a 14-membered macrolide that also contains chalcose. Homologs of ChmCII, ChmCIII, ChmCIV, and ChmCV were found in the lankamycin (lkm) cluster, but their putative roles were not described (24).
Mycinose.
Synthesis of the mycinose precursor NDP-6-deoxy-d-allose from NDP-4-keto-6-deoxyglucose requires two enzymes, a 3-epimerase and a 4-ketoreductase, most likely encoded by chmJ and chmD (Fig. 4); both are highly similar to their counterparts in the tylosin pathway. Attachment of 6-deoxy-d-allose to the macrolactone requires a glycosyltransferase, likely encoded by chmN. Attachment also requires a primary hydroxyl group at C-20 of the macrolactone, hence the need for the P450 enzyme encoded by chmHI and its corresponding ferredoxin encoded by the chmHII gene, which are orthologs of the corresponding proteins, TylHI and TylHII, respectively, that hydroxylate the C-23 position of the tylosin macrolactone. In the biosynthesis of tylosin, attachment of 6-deoxy-d-allose to the macrolactone takes place after attachment of the 5-O-sugar (mycaminose) and cannot take place in mutants that do not carry out this step (for a review, see reference 3). Hence, we propose that attachment of 6-deoxy-d-allose to the chalcose macrolactone also takes place after addition of chalcose to the ring, as shown in Fig. 4. Because of the uncertainty of the step at which the 2,3-double bond is introduced into the ring, we do not present the structure for the intermediates that carry one or both sugars. Conversion of the 6-deoxy-d-allose residue to d-mycinose, after attachment to the macrolactone, requires the action of two O-methyltransferases, likely encoded by chmE and chmF, each ca. 70% identical to their respective counterparts in the tylosin biosynthesis cluster, TylE and TylF, which are also proposed to act after the sugar has been attached to the backbone. As can be seen in Fig. 2A, the seven genes required for the synthesis and attachment of the mycinose unit of chalcomycin are clustered.
Late steps in the pathway.
Two oxidation steps are required to convert the product of ChmF to chalcomycin, i.e., hydroxylation at C-8 and 12,13-epoxidation of the lactone ring, but the order of the reactions is not known. Each of these steps is proposed to be carried out by one of the cytochrome P450 enzymes, ChmPI or ChmPII.
DISCUSSION
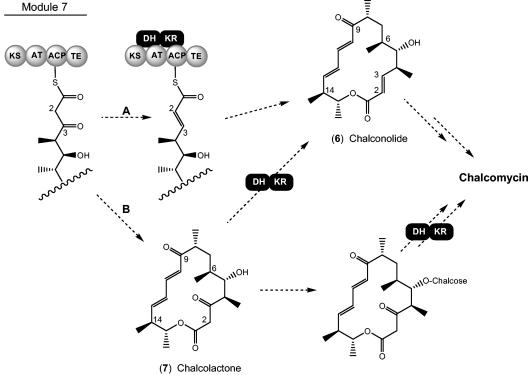
The structure of compound 5 confirms the prediction that the chm PKS does not encode the activities required for introduction of the 2,3-trans double bond and suggests that it is introduced by a 3-ketoreductase and a 2,3-DH that are not components of the chm PKS. This is an unprecedented finding in macrolide biosynthesis. Mycinamicin also contains a 2,3-trans double bond, but the mycinamicin PKS contains the expected DH and β-ketoreductase domains in module 7 (1). As shown in Fig. 5, introduction of the double bond into chalcomycin could take place either during the seventh elongation step of nascent polyketide chain synthesis to release a Δ-2,3-aglycone (chalconolide [compound 6]), or after the chain has been completed and cyclized to produce the 3-keto-aglycone (chalcolactone [compound 7]). Chalconolide would be produced if the required β-ketoreductase and DH enzymes interacted with the nascent polyketide chain while it is attached to the acyl carrier protein domain of module 7 (Fig. 5, scheme A). Interactions between type I PKS systems and discrete enzymes have been reported for acyltransferases (9, 30) but not for ketoreductases or DHs. The gene chmU, which lies ca. 3 kb downstream of the PKS-encoding chm, encodes a protein that appears to belong to the short-chain dehydrogenase/reductase family, which is composed of many ketoreductases, including those that participate in the synthesis of polyketides and fatty acids (as components of type II enzymes) and might fulfill the role of the 3-ketoreductase. A gene encoding a candidate 2,3-DH was not observed in the cluster, however.
FIG. 5.
Possible schemes for formation of the 2,3-trans double bond of the macrolactone produced from the chm PKS. (A) Two-step reduction and dehydration of the full-length polyketide chain prior to release from the PKS enzyme and cyclization; (B) post-PKS reduction and dehydration of the macrolactone showing two different points at which these reactions may occur.
The alternative pathway for introduction of the 2,3-trans double bond in chalcomycin biosynthesis, after the 3-keto acyl chain is released from the PKS and cyclized, is depicted in Fig. 5, scheme B. Because there are no reported examples of post-PKS 3-reduction or 2,3-dehydration in 16- (or 14-) membered macrolides, it is not possible to pinpoint the precise step at which these events would take place along the pathway of synthesis of chalcomycin after the formation of chalcolactone. A number of 16-membered macrolides, including spiramycin, have been identified in which the 9-keto group is reduced to its 9-hydroxy counterpart. The spiramycin PKS does not contain an active β-ketoreductase domain in the fourth module (7), hence reduction is believed to take place through the action of a discrete 9-ketoreductase following release of the 9-keto-macrolactone platenolide from the PKS. The seventh module of the type I PKS of Streptomyces strain HK-803, which produces phoslactomycin B, an antitumor polyketide, contains the required KR domain but lacks the DH domain for 2,3-dehydration of the nascent acyl chain; hence, post-PKS 2,3-dehydration is thought to occur. Again, however, a candidate 2,3-DH was not observed in the phoslactomycin biosynthesis cluster (27). A DH domain required for the biosynthesis of myxalamid is also absent from the corresponding PKS in the producing myxobacterial host (39).
Compound 5 represents the first 3-keto 16-membered macrolide isolated from a fermentation, in this case an unnatural natural product. The 14-membered macrolide pikromycin, produced from Streptomyces venezuelae, contains a 3-keto group which does not exist in the enol form to a significant degree, and a 3-keto derivative of the 14-membered macrolide erythromycin, termed “ketolides,” has been successfully developed as a drug useful against many gram-positive hosts that are resistant to macrolides (for a review, see reference 46). 3-Keto derivatives of tylosin synthesized chemically were also found to adopt the 2,3-trans enol configuration and were shown to have significantly diminished antimicrobial activity compared to their 3-hydroxy counterparts (10). Whether the loss of antibiotic potency is due to the enolization of the 3-keto group remains to be determined.
Acknowledgments
We thank Nina Viswanathan and John Carney for performing liquid chromatograpy-mass spectrometry analyses. DNA sequencing was performed at Kosan by Marigold Manlusoc and at The Institute for Genomic Research by Matthew Lewis and William Nierman. We thank David Hopwood for reviewing the manuscript.
REFERENCES
- 1.Anzai, Y., N. Saito, M. Tanaka, K. Kinoshita, Y. Koyama, and F. Kato. 2003. Organization of the biosynthetic gene cluster for the polyketide macrolide mycinamicin in Micromonospora griseorubida. FEMS Microbiol. Lett. 218:135-141. [DOI] [PubMed] [Google Scholar]
- 2.Asolkar, R. N., R. P. Maskey, E. Helmke, and H. Laatsch. 2002. Chalcomycin B, a new macrolide antibiotic from the marine isolate Streptomyces sp. B7064. J. Antibiot. 55:893-898. [DOI] [PubMed] [Google Scholar]
- 3.Baltz, R. H., and E. T. Seno. 1988. Genetics of Streptomyces fradiae and tylosin biosynthesis. Annu. Rev. Microbiol. 42:547-574. [DOI] [PubMed] [Google Scholar]
- 4.Barianov, P. V., P. V. Sergiev, O. A. Donstova, A. A. Bogdanov, and R. Brimacombe. 1998. The database of ribosomal cross-links. Nucleic Acids Res. 26:187-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Bisang, C., P. F. Long, J. Cortes, J. Westcott, J. Crosby, A. L. Matharu, R. J. Cox, T. J. Simpson, J. Staunton, and P. F. Leadlay. 1999. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 401:502-505. [DOI] [PubMed] [Google Scholar]
- 7.Burgett, S. G., S. A. Kuhstoss, R. N. Rao, M. A. Richardson, and P. R. Rosteck, Jr. Aug. 1999. Platenolide synthase gene. U.S. patent 5,945,320.
- 8.Butler, A. R., N. Bate, and E. Cundliffe. 1999. Impact of thioesterase activity on tylosin biosynthesis in Streptomyces fradiae. Chem. Biol. 6:287-292. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, Y. Q., G. L. Tang, and B. Shen. 2003. Type I polyketide synthase requiring a discrete acyltransferase for polyketide biosynthesis. Proc. Natl. Acad. Sci. USA 100:3149-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creemer, L. C., J. E. Toth, and H. A. Kirst. 2002. Synthesis and in vitro antimicrobial activity of 3-keto 16-membered macrolides derived from tylosin. J. Antibiot. 55:427-436. [DOI] [PubMed] [Google Scholar]
- 11.Frohardt, R. P., R. F. Pitillo, and J. Ehrlich. Nov. 1962. Chalcomycin and its fermentative production. U.S. patent 3,065,137.
- 12.Gaisser, S., G. A. Bohm, J. Cortes, and P. F. Leadlay. 1997. Analysis of seven genes from the eryAI-eryK region of the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol. Gen. Genet. 256:239-251. [DOI] [PubMed] [Google Scholar]
- 13.Goo, Y. M., Y. Y. Lee, and B. T. Kim. 1997. A new 16-membered chalcomycin type macrolide antibiotic, 250-144C. J. Antibiot. 50:85-88. [DOI] [PubMed] [Google Scholar]
- 14.Gupta, R. S., W. Murray, and R. Gupta. 1988. Cross resistance pattern towards anticancer drugs of a human carcinoma multidrug-resistant cell line. Br. J. Cancer 58:441-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen, J. L., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 16.He, X., and H. W. Liu. 2002. Mechanisms of enzymatic C-O bond cleavages in deoxyhexose biosynthesis. Curr. Opin. Chem. Biol. 6:590-597. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda, H., and S. Ômura. 2002. Biosynthesis, regulation, and genetics of macrolide production, p. 286-326. In S. Ômura (ed.), Macrolide antibiotics: chemistry, biology, and practice, 2nd ed. Academic Press, New York, N.Y.
- 18.Jordan, D. C. 1963. Effect of chalcomycin on protein synthesis by Staphylococcus aureus. Can. J. Microbiol. 9:129-132. [Google Scholar]
- 19.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 20.Kim, B. S., T. A. Cropp, B. J. Beck, D. H. Sherman, and K. A. Reynolds. 2002. Biochemical evidence for an editing role of thioesterase II in the biosynthesis of the polyketide pikromycin. J. Biol. Chem. 277:48028-48034. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S. D., I. J. Ryoo, C. J. Kim, W. G. Kim, J. P. Kim, J. Y. Kong, H. Koshino, M. Uramoto, and I. D. Yoo. 1996. GERI-155, a new macrolide antibiotic related to chalcomycin. J. Antibiot. 49:955-957. [DOI] [PubMed] [Google Scholar]
- 22.Liu, M., and S. Douthwaite. 2002. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc. Natl. Acad. Sci. USA 99:14658-14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Madduri, K., C. Waldron, and D. J. Merlo. 2001. Rhamnose biosynthesis pathway supplies precursors for primary and secondary metabolism in Saccharopolyspora spinosa. J. Bacteriol. 183:5632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mochizuki, S., K. Hiratsu, M. Suwa, T. Ishii, F. Sugino, K. Yamada, and H. Kinashi. 2003. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48:1501-1510. [DOI] [PubMed] [Google Scholar]
- 25.Omura, S., Y. Hironaka, A. Nakagawa, I. Umezawa, and T. Hata. 1972. Antimycoplasma activities of macrolide antibiotics. J. Antibiot. 25:105-108. [DOI] [PubMed] [Google Scholar]
- 26.Omura, S., and M. Tishler. 1972. Relationship of structures and microbiological activities of the 16-membered macrolides. J. Med. Chem. 15:1011-1015. [DOI] [PubMed] [Google Scholar]
- 27.Palaniappan, N., B. S. Kim, Y. Sekiyama, H. Osada, and K. A. Reynolds. 2003. Enhancement and selective production of phoslactomycin B, a protein phosphatase IIa inhibitor, through identification and engineering of the corresponding biosynthetic gene cluster. J. Biol. Chem. 278:35552-35557. [DOI] [PubMed] [Google Scholar]
- 28.Parke, Davis and Co. Aug. 1961. A new antibiotic chalcomycin and method for producing the same. United Kingdom patent 875,011.
- 29.Pernodet, J. L., F. Boccard, M. T. Alegre, M. H. Blondelet-Rouault, and M. Guerineau. 1988. Resistance to macrolides, lincosamides and streptogramin type B antibiotics due to a mutation in an rRNA operon of Streptomyces ambofaciens. EMBO J. 7:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piel, J. 2002. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc. Natl. Acad. Sci. USA 99:14002-14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poehlsgaard, J., and S. Douthwaite. 2003. Macrolide antibiotic interaction and resistance on the bacterial ribosome. Curr. Opin. Investig. Drugs. 4:140-148. [PubMed] [Google Scholar]
- 32.Quiros, L. M., I. Aguirrezabalaga, C. Olano, C. Mendez, and J. A. Salas. 1998. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol. Microbiol. 28:1177-1185. [DOI] [PubMed] [Google Scholar]
- 33.Rascher, A., Z. Hu, N. Viswanathan, A. Schirmer, R. Reid, W. C. Nierman, M. Lewis, and C. R. Hutchinson. 2003. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL 3602. FEMS Microbiol. Lett. 218:223-230. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, E., Z. Hu, S. Ou, Y. Volchegursky, C. R. Hutchinson, and R. McDaniel. 2003. Rapid engineering of polyketide overproduction by gene transfer to industrially optimized strains. J. Ind. Microbiol. Biotechnol. 30:480-488. [DOI] [PubMed] [Google Scholar]
- 35.Salah-Bey, K., M. Doumith, J. M. Michel, S. Haydock, J. Cortes, P. F. Leadlay, and M. C. Raynal. 1998. Targeted gene inactivation for the elucidation of deoxysugar biosynthesis in the erythromycin producer Saccharopolyspora erythraea. Mol. Gen. Genet. 257:542-553. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Santi, D. V., M. A. Siani, B. Julien, D. Kupfer, and B. Roe. 2000. An approach for obtaining perfect hybridization probes for unknown polyketide synthase genes: a search for the epothilone gene cluster. Gene 247:97-102. [DOI] [PubMed] [Google Scholar]
- 38.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 39.Silakowski, B., G. Nordsiek, B. Kunze, H. Blocker, and R. Muller. 2001. Novel features in a combined polyketide synthase/non-ribosomal peptide synthetase: the myxalamid biosynthetic gene cluster of the myxobacterium Stigmatella aurantiaca Sga15. Chem. Biol. 8:59-69. [DOI] [PubMed] [Google Scholar]
- 40.Summers, R. G., S. Donadio, M. J. Staver, E. Wendt-Pienkowski, C. R. Hutchinson, and L. Katz. 1997. Characterization of ten genes from the erythromycin biosynthetic cluster of Saccharopolyspora erythraea that are involved in l-mycarose and d-desosamine production. Microbiology 143:3251-3262. [DOI] [PubMed] [Google Scholar]
- 41.Trefzer, A., J. A. Salas, and A. Bechthold. 1999. Genes and enzymes involved in deoxysugar biosynthesis in bacteria. Nat. Prod. Rep. 16:283-299. [DOI] [PubMed] [Google Scholar]
- 42.Waldron, C., P. Matsushima, P. R. Rosteck, Jr., M. C. Broughton, J. Turner, K. Madduri, K. P. Crawford, D. J. Merlo, and R. H. Baltz. 2001. Cloning and analysis of the spinosad biosynthetic gene cluster of Saccharopolyspora spinosa. Chem. Biol. 8:487-499. [DOI] [PubMed] [Google Scholar]
- 43.Witkowski, A., A. K. Joshi, Y. Lindqvist, and S. Smith. 1999. Conversion of a β-ketoacyl synthase to a malonyl decarboxylase by replacement of the active-site cysteine with glutamine. Biochemistry 38:11643-11650. [DOI] [PubMed] [Google Scholar]
- 44.Woo, P. W. K., and J. R. Rubin. 1996. Chalcomycin: single-crystal X-ray crystallographic analysis; biosynthetic and stereochemical correlations with polyoxo macrolide antibiotics. Tetrahedron 52:3857-3872. [Google Scholar]
- 45.Xue, Y., L. Zhao, H. W. Liu, and D. H. Sherman. 1998. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA 95:12111-12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhanel, G. G., M. Walters, A. Noreddin, L. M. Vercaigne, A. Wierzbowski, J. M. Embil, A. S. Gin, S. Douthwaite, and D. J. Hoban. 2002. The ketolides: a critical review. Drugs 62:1771-1804. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, L., S. Borisova, S. M. Yeung, and H. Liu. 2001. Study of C-4 deoxygenation in the biosynthesis of desosamine: evidence implicating a novel mechanism. J. Am. Chem. Soc. 123:7909-7910. [DOI] [PubMed] [Google Scholar]