Abstract
As part of the SENTRY Antimicrobial Surveillance Program in 2002, five multidrug-resistant Pseudomonas aeruginosa clinical isolates were detected with metallo-β-lactamase (MβL) activity. The isolates were recovered from different patients in a medical center located in Dusseldorf, Germany. The resistant determinant was isolated amplifying the region between the integrase and the aacA4 gene cassette. Sequencing revealed a novel MβL gene, designated blaGIM-1. Additional analysis showed that GIM-1, comprising 250 amino acids and with a pI value of 5.4, differs in its primary sequence from that described for IMP, VIM, and SPM-1 enzymes by 39 to 43%, 28 to 31%, and 28%, respectively. The enzyme possesses unique amino acids within the major consensus sequence (HXHXD) of the MβL family. Kinetics analysis revealed that GIM-1 has no clear preference for any substrate and did not hydrolyze azlocillin, aztreonam, and the serine-β-lactamase inhibitors. blaGIM-1 was found on a 22-kb nontransferable plasmid. The new MβL gene was embedded in the first position of a 6-kb class 1 integron, In77, with distinct features, including an aacA4 cassette downstream of the MβL gene that appeared to be truncated with blaGIM-1. The aacA4 was followed by an aadA1 gene cassette that was interrupted by a copy of the IS1394. This integron also carried an oxacillinase gene, blaOXA-2, before the 3′-CS region. GIM-1 appears to be a unique MβL, which is located in a distinct integron structure, and represents the fourth subclass of mobile MβL enzymes to be characterized.
Pseudomonas aeruginosa is a leading cause of nosocomial infections, giving rise to a wide range of life-threatening conditions. Its intrinsic resistance to many antimicrobial agents and its ability to develop multidrug resistance imposes a serious therapeutic problem (8). Carbapenems are very useful antimicrobial agents for the treatment of infections caused by P. aeruginosa; however, increasing use of these compounds has resulted in the development of carbapenem-resistant P. aeruginosa. Mechanisms of resistance to carbapenems in P. aeruginosa are associated with reduced uptake of the agent resulting from the loss or reduced expression of the OprD porin, combined with derepression of the chromosomal ampC β-lactamase gene (23); overexpression of an efflux pump system (22, 37); and production of a metallo-β-lactamase (MβL) (16).
The first mobile MβL (IMP-1) was found in P. aeruginosa in Japan in the early 1990s (33). For many years, the occurrence of IMP-1-producing bacteria was confined to Japan. More recently, however, IMP-1 and IMP-variant enzymes have been reported from many other countries and across four continents (6, 9, 12, 13, 24, 27). Since the emergence of IMP-1, two other subclasses of clinically relevant MβLs have been described: the VIM series (15) and SPM-1 (29). VIM variants are now found throughout the world as well (2, 13, 17, 20, 21, 30, 32, 34, 35), whereas SPM-1 thus far seems to be restricted to Brazil.
We characterize here a novel subclass of Ambler class B enzyme, GIM-1, which is encoded by a gene cassette located in the first position of a class 1 integron. The carbapenem-resistant P. aeruginosa clinical isolates that produce GIM-1 were recovered in Germany and were detected by The SENTRY Antimicrobial Surveillance Program in 2002.
MATERIALS AND METHODS
Bacterial strains and susceptibility testing.
In 2002, five clinical isolates of P. aeruginosa (73-5671, 73-12198, 73-15553, 73-15574, and 73-15480) were collected in a medical site located in Dusseldorf, Germany, and submitted to the SENTRY Program monitored in North Liberty, Iowa (JMI Laboratories). The clinical isolates were susceptibility tested by using the reference broth microdilution method as described by the National Committee for Clinical Laboratory Standards (19). Rifampin-resistant (Rifr) derivatives of Escherichia coli K-12 and P. aeruginosa PA01 (28) were used as recipients in conjugation experiments. E. coli DH5α and Rifr PA01 were used for transformation.
Phenotypic detection of MβL.
MβL Etest strips (AB Biodisk, Solna, Sweden) were used to screen for class B β-lactamase production according to the manufacturer's instructions. In addition, carbapenemase activities of cell sonicates from overnight broth cultures were determined by spectrophotometric assays. These were carried out with 150 μM imipenem and meropenem as substrates at 299 nm. Assays were performed with or without EDTA (25 mM) to ascertain whether activity is inhibited by chelating agents.
DNA analysis methodology.
Molecular screening for blaVIM, blaIMP, and blaSPM-1 was performed by using PCR, as previously described (29), with primers targeting conserved regions of the MβL genes (Table 1). Strains known to harbor MβL genes were used as positive controls for the reactions. Since several β-lactamases are encoded on gene cassettes, embedded in class 1 integrons, primers targeting the 5′ conserved sequence and the aacA4 gene cassette were used in additional PCR screening reactions. The integron structure was revealed with a walking sequencing strategy, with the primers described in Table 1. Plasmid extraction from P. aeruginosa 73-5671 was undertaken with a QIAprep Spin Miniprep kit (Qiagen, West Sussex, United Kingdom). Restriction endonuclease analysis of the plasmid was carried out with seven different restriction enzymes, namely, BamHI, EcoRI, HindIII, HincII, SpeI, SmaI, and XbaI (Invitrogen, Carlsbad, Calif.), singly and in pairs, to determine the size of the plasmid. Southern blot analysis was performed on agarose gels by standard methods, and hybridization was undertaken with a blaGIM-1 probe labeled with 32P by using a random primer technique (25). Transformation was carried out by using electroporation, and selection for transformants was performed on nutrient agar plates containing ceftazidime at 4 μg/ml. Conjugation experiments were carried out in liquid medium, and selection was performed on nutrient agar with ceftazidime (4 μg/ml) and rifampin (200 μg/ml). Primers intI1F and aacA4FR (Table 1) were used to amplify the integron structure harboring the resistant determinant, and the purified amplicons were cloned into PCRScriptCam SK+ (Stratagene Cloning Systems, La Jolla, Calif.). DHα was used as a host for the recombinant plasmid.
TABLE 1.
Oligonucleotides used as primers for PCR amplification and sequencing
| Primer | Target | Sequence (5′-3′) | Accession no. |
|---|---|---|---|
| IMP1F | blaIMP-like | TGAGCAAGTTATCTGTATTC | AJ223604 |
| IMP1R | blaMP-1 59-be | GCTGCAACGACTTGTTAG | AJ223604 |
| ATT1F | attI1 | TTATGGAGCAGCAACGATGT | AJ515707 |
| VIMR | blaVIM-like | CGAATGCGGCAGCACCAGG | AJ515707 |
| SPM1F | blaSPM-like | CCTACAATCTAACGGCGACC | AJ492820 |
| SPM1R | blaSPM-like | TCGCCGTGTCCAGGTATAAC | AJ492820 |
| intl1F | Integrase | GCCGTAGAAGAACAGCAAGG | AJ515707 |
| intl2F | Integrase | TCAATCTCCGCGAGAAGTGC | AJ515707 |
| ATT1R | attI1 | GCCTGTTCGGTTCGTAAGCT | AJ515707 |
| GIMF | blaGIM-1 | AGAACCTTGACCGAACGCAG | This study |
| GIMR | blaGIM-1 | ACTCATGACTCCTCACGAGG | This study |
| GIMFF | blaGIM-1 flanking region | CTACGTGACCAACAGCAACG | This study |
| aacA4F | aacA4 | TGCGATGCTCTATGAGTGGC | AJ515707 |
| aacA4R | aacA4 | ATGTACACGGCTGGACCATC | AJ515707 |
| aacA4FF | aacA4 flanking region | AACTTGCGAGCGATCCGATG | AJ515707 |
| aacA4FR | aacA4 flanking region | AGCCACTCATAGAGCATCGC | AJ515707 |
| aadA1F | aadA1 | CGCCGAAGTATCGACTCAAC | AJ584652 |
| IS1394R | IS1394 | ACAGAGGTAGTGGCGTTGC | U37284 |
| ISF | IS1394 | CGGTCTTCTGGGTGATTTCC | U37284 |
| ISFR | IS1394 | CGCGCTTAGCTGGATAACG | U37284 |
| aadA1R | aadA1 | GACTACCTTGGTGATCTCGC | AJ584652 |
| aadA1FF | aadA1 flanking region | GAGATCACCAAGGTAGTCGG | AJ584652 |
| oxa2F | blaOXA-2 | TTCAAGCCAAAGGCACGATAG | AF300985 |
| oxa2R | blaOXA-2 | TCCGAGTTGACTGCCGGGTTG | AF300985 |
| oxa2FF | blaOXA-2 flanking region | AAGCGTTACCGCCCAACC | AF300985 |
| oxa2FR | blaOXA-2 flanking region | ATGCGCGAAAGTGGCAAGAG | AF300985 |
| QACR | qacEΔ1 | CGGATGTTGCGATTACTTCG | AJ515707 |
| SUL2R | sul1 | GGCTCTCATCGAAGAAGGAG | AJ515707 |
| Sul1R | sul1 | GGCTCTCATCGAAGAAGGAG | AJ515707 |
DNA sequencing and computer analysis.
The PCR products were sequenced on both strands by using a Perkin-Elmer systems 377 DNA sequencer. The nucleotide sequences, deduced amino acid sequences, and phylogenetic relationships were analyzed by using the Lasergene software package (DNAStar, Madison, Wis.). Obtained sequences were compared to sequences available on the internet (http://www.ebi.ac.uk/fasta33/).
Analytical IEF.
P. aeruginosa extracts showing β-lactamase activity were obtained by cell lyses with BugBuster (Novagen, Nottingham, United Kingdom). Isoelectric focusing (IEF) was performed with a pH 3 to 10 pre-cast vertical IEF gel with the Novex system (Invitrogen). The conditions applied were according to the manufacturer's instructions. The focused β-lactamases were detected by overlaying the gel with nitrocefin solution (Microbiology Systems, Cockeysville, Md.). Isoelectric points were estimated by using linear regression with Graf Prism software (GraphPad Software, Inc., San Diego, Calif.) and by comparison to reference proteins provided by a pI 4.5 to 9.5 Standard IEF Marker (Bio-Rad, Richmond, Calif.) and the TEM-1 β-lactamase (31).
Pulsed-field gel electrophoresis (PFGE).
Genomic DNA was prepared in agarose blocks and digested in situ with the restriction enzyme SpeI (Invitrogen). DNA fragments were separated by electrophoresis in the CHEF-DR III apparatus (Bio-Rad). The band pattern was interpreted according to the recommendation of Tenover et al. (26).
β-Lactamase purification.
Cultures of P. aeruginosa 73-5671 were grown overnight at 37°C in 4 liters of Terrific Broth (12% tryptone, 20% yeast extract, 0.4% glycerol, 0.17 M monopotassium phosphate, 0.72 M dipotassium phosphate). A periplasmic protein preparation was obtained as previously described (1). This protein solution was treated with 60% saturated ammonium sulfate solution to precipitate most of the proteins, which were removed by centrifugation. The clarified supernatant was loaded onto a Q-Sepharose column (Amershan Pharmacia Biotech, Uppsala, Sweden) preequilibrated with buffer A (50 mM cacodylate, 100 μM zinc chloride, 1 mM β-mercaptoethanol, 3 mM sodium azide [pH 6.8]). The proteins were eluted with a linear gradient of 300 mM to 800 mM sodium chloride. Fractions showing the highest degree of β-lactamase activity against nitrocefin were pooled, and proteins in the mix were subjected to a Superdex-200 (Amersham Pharmacia Biotech) gel filtration column at 0.3 ml/min. During the purification procedure, the β-lactamase activity was tracked by nitrocefin hydrolysis.
The protein recovered in the flowthrough was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis, and analytical IEF was applied to confirm the purity of the preparation.
Kinetic measurements.
Purified β-lactamase was used to determine the kinetic parameters kcat and Km. Reactions were performed at 25°C in 2 ml of assay buffer (50 mM cacodylate, 100 mM sodium chloride, 100 μM zinc chloride [pH 7.0]). The rate of hydrolysis of each β-lactam was calculated with triplicate reactions for at least five different concentrations of substrate based on the extinction coefficients for each substrate. The assays were performed in a Lambda 35 spectrophotometer (Perkin-Elmer, Cambridge, United Kingdom) by observing the changes in absorption resulting from the opening the β-lactam ring at the specific wavelengths for each of the 20 antimicrobial agents evaluated, as previously described (18).
Nucleotide sequence accession number.
The nucleotide sequence data reported in the present study have been assigned EMBL/GenBank nucleotide accession number AJ620678.
RESULTS
Properties of the P. aeruginosa clinical isolates.
The five carbapenem-resistant P. aeruginosa isolates were recovered from different patients in the same hospital ward in Dusseldorf, Germany, between February and July 2002. The index strain, 73-5671, was recovered from a 37-year-old male intensive care unit patient with nosocomial pneumonia in February 2002. All five isolates were submitted to the SENTRY Program as respiratory tract specimens. The susceptibility patterns of the five isolates were found to be nearly identical. The isolates were highly resistant to 19 of the 20 antimicrobial agents tested (including all β-lactams, aminoglycosides, and quinolones), as shown in Table 2. The isolates showed particular resistance to imipenem (MIC, >8 μg/ml), meropenem (MIC, >8 μg/ml), ceftazidime (MIC, >16 μg/ml), cefepime (MIC, ≥16 μg/ml), and piperacillin-tazobactam (MIC, >128 μg/ml). Of the antimicrobial agents tested, the five isolates were only susceptible to polymyxin B.
TABLE 2.
Antimicrobial susceptibility patterns of the five GIM-1 producer isolates from Dusseldorf, Gemany, submitted to the SENTRY Antimicrobial Surveillance Program in 2002
| Antimicrobial agents | MIC (μg/ml) for strain:
|
||||||
|---|---|---|---|---|---|---|---|
| P. aeruginosa 73-5671 | P. aeruginosa 73-12198 | P. aeruginosa 73-15553 | P. aeruginosa 73-15574 | P. aeruginosa 73-15480 | E. coli DH5α(pGIM-1) | E. coli DH5α(pSCRIPT) | |
| Piperacillin | >128 | >128 | >128 | >128 | >128 | 16 | 0.5 |
| Piperacillin-tazobactam | >64 | >64 | >64 | >64 | >64 | - | - |
| Ticarcillin | >128 | >128 | >128 | >128 | >128 | - | - |
| Ticarcillin-clavulanic acid | >128 | >128 | >128 | >128 | >128 | - | - |
| Ceftriaxone | >32 | >32 | >32 | >32 | >32 | 128 | 0.125 |
| Ceftazidime | >16 | >16 | >16 | >16 | >16 | 256 | 0.125 |
| Cefepime | 16 | 16 | >16 | 16 | 16 | 4 | 0.06 |
| Imipenem | >8 | >8 | >8 | >8 | >8 | 0.5 | 0.06 |
| Meropenem | >8 | >8 | >8 | >8 | >8 | 0.125 | 0.06 |
| Aztreonam | 8 | 8 | 16 | 16 | 16 | 0.125 | 0.06 |
| Ciprofloxacin | >4 | >4 | >4 | >4 | >4 | - | - |
| Levofloxacin | >4 | >4 | >4 | >4 | >4 | - | - |
| Gatifloxacin | >4 | >4 | >4 | >4 | >4 | - | - |
| Amikacin | 4 | 4 | 16 | 16 | 16 | - | - |
| Gentamicin | >8 | >8 | >8 | >8 | >8 | - | - |
| Tobramycin | 16 | 16 | >16 | 16 | >16 | - | - |
| Netilmicin | >32 | >32 | >32 | >32 | >32 | - | - |
| Tetracycline | >8 | >8 | >8 | >8 | >8 | - | - |
| Trimethoprim-sulfamethoxazole | >2 | >2 | >2 | >2 | >2 | - | - |
| Polymyxin B | 2 | 2 | 4 | 1 | 1 | - | - |
-, Not tested.
By PFGE analysis, the five P. aeruginosa isolates were indistinguishable (clonal). These strains were compared to six carbapenem-susceptible isolates recovered at the same time from the same medical site in Germany. All susceptible isolates were, by PFGE analysis, significantly different from the carbapenem-resistant isolates and also distinct from each other (data not shown).
MβL screening and PCR experiments.
Initial MβL screening experiments showed that each of the five isolates produce an MβL. Analysis with MβL Etest (ABBIODISK) strips showed decreases in the MICs of imipenem in the presence of EDTA (from >256 μg/ml for imipenem to 2 μg/ml for the combination imipenem-EDTA). Spectrophotometric assays demonstrated carbapenemase activity against carbapenems (hydrolyzing activity against neropenem between 0.0113 and 0.0287 absorbance units/min), which was 85% EDTA inhibited in all five P. aeruginosa isolates.
Preliminary PCR amplification experiments failed to detect previously described MβL genes, whereas controls with blaVIM-, blaIMP-, and blaSPM-like genes yielded PCR products of the expected sizes.
Considering that mobile MβL genes were found in class 1 integron structures which also contain an aacA4 cassette (6, 17, 32), a PCR was performed with specific primers for these elements. Amplicons of 900 bp were obtained from PCR with 3F1 and aacA4FR primer pair (Table 1). Sequence analysis of these fragments revealed an open reading frame of 753 bp coding for a protein of 250 amino acids with significant similarities to some class B MβLs. The new MβL gene was named blaGIM-1 (German imipenemase).
Independent PCRs with primers targeting different regions of the integrase and the aacA4 cassette and reactions with primers specifically designed for the 753-bp open reading frame (Table 1) confirmed the presence of a novel MβL gene.
Sequence analysis and its deduced protein sequence.
The nucleotide sequence of blaGIM-1 showed a GC content of 42.1% and encoded a mature protein of 25,501 Da with a theoretical pI of 5.3. Analytical IEF of P. aeruginosa revealed three β-lactamase bands, one with a pI of 5.4 and that likely corresponded to GIM-1 and two additional bands focused in pI 6.7 and 8.4. The additional band in pI 8.4 was probably the chromosomally encoded AmpC. The results obtained in the IEF showed that the pI value was 5.4, in agreement with the theoretical value. The identity of the β-lactamase focused in pI 6.7 could not be determined.
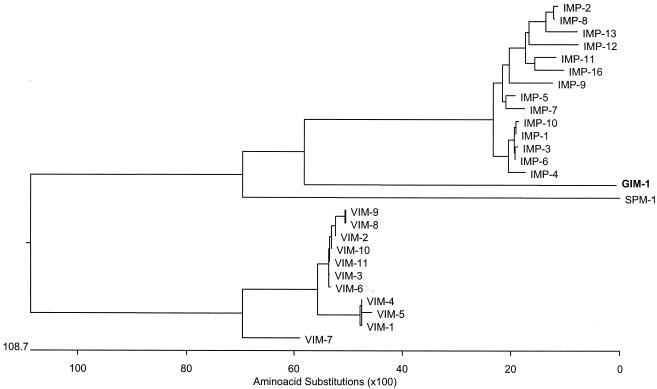
The amino acid sequence of GIM-1 had low identity with other clinically significant MβL genes. The amino acid sequence displayed most identity with IMP variants IMP-6, IMP-1, and IMP-4 (43.5, 43.1, and 43.1%, respectively), followed by VIM variants ranging from a high of 31.2% with VIM-7 to 28.8% with VIM-1, VIM-4, and VIM-5 and only 28.0% similarity with SPM-1. A phylogenetic analysis placed the new enzyme in a new subclass of class B β-lactamases (Fig. 1).
FIG. 1.
Phylogenetic tree obtained for most of the mobile MβLs. The alignment used was prepared with CLUSTALW.
The predicted protein sequence showed that in the active site, GIM-1 has amino acid motifs that are conserved in MβL enzymes (4, 10, 11), namely, the consensus zinc binding motif HXHXD (residues 116 to 120) and the other residues involved in the coordination of the Zn2+ ions (Hys196, Cys221, and Hys293, according to the BBL numbering system). However, the zinc-binding motif of GIM-1 was unique in that there was a serine at position 117 and a glutamic acid residue at position 119, amino acid residues that were not found at these positions in other MβL enzymes (Fig. 2).
FIG. 2.
Alignment of the amino acid sequence of GIM-1 with three representatives of the MβL groups IMP-1, VIM-1, and SPM-1. Differences in the amino acid sequences are noted by a single letter representing the amino acid change within a particular sequence. Residues involved in the coordination of the zinc iron are underlined and numbered according to the BBL system (in italics).
Genetic environment harboring blaGIM-1.
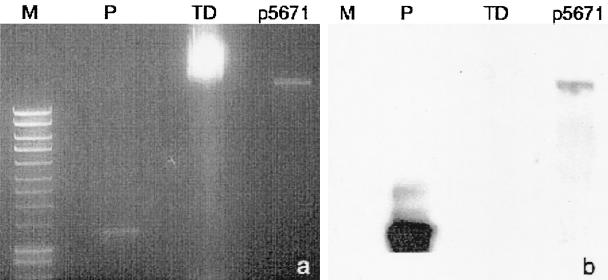
Plasmid DNA analysis showed that the five P. aeruginosa clinical isolates harbored plasmids of the same size. The plasmid obtained from strain 73-5671 was estimated to be ∼22 kb in size, as determined from DNA fragment profiles generated with different restriction enzymes. Southern hybridization experiments showed that blaGIM-1 is in the plasmid (Fig. 3), although transfer of the β-lactam resistance by electroporation and conjugation could not be demonstrated.
FIG. 3.
(a) Agarose gel electrophoresis. Lanes: M, 1-kb plus ladder; P, probe; TD, genomic DNA; p5671, plasmid from strain 73-5671. (b) Results of Southern blot analysis showing the hybridization of intl1F/GIMR marked probe with the probe fragment and the 22-kb plasmid from the index isolate harboring blaGIM-1.
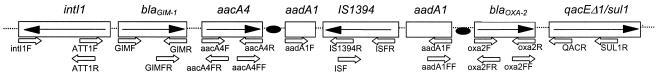
Extended analysis of the genetic environment of blaGIM-1 revealed key genetic components found in class 1 integrons: the 5′-CS, containing the intI1 integrase gene with its own promoter and the attI1 recombination site, and the 3′-CS accommodating the fused structure qacEΔ1/sul1 (2). The MβL gene is located in the first gene cassette position of this 6-kb class 1 integron, which was named In77. Two putative promoters (P1 and P2) precede the blaGIM-1 start codon (ATG), both of which lie within the integrase structural gene. The primary promoter (P1) had all of the features of an intermediate strength promoter: the −35 box TGGACA and the −10 box TAAACT being separated by 17 bp (5). An insertion of three guanosine residues has been reported to activate the second promoter (P2) by spacing the −35 and −10 hexamers to 17 bp (4). This feature was not present in this integron, signaling that only the P1 drives the expression of the gene cassettes embedded in the integron.
The MβL gene has been inserted at the attI1 recombination site and, although blaGIM-1 is preceded by the expected core site (GTTAGAA), it is not immediately followed by an inverse core site and other elements of a 59-be (2L and 2R regions). The second cassette in In77 accommodates the aacA4 gene; however, the expected 59-be between blaGIM-1 and aacA4 was absent, even within the MβL gene. The aacA4 allele in this integron encodes an AAC(6′)-Ib aminoglycoside acetyltransferase that confers resistance to netilmicin, gentamicin, and tobramycin. The 3′ end of the aacA4 gene cassette leads to a 59-be that is 72 bp long, which is in turn followed by a second aminoglycoside-resistant determinant, aadA1. However, this gene is interrupted at nucleotide position 135 by a copy of IS1394 (GenBank accession no. U37284) (36), a 1,100-bp insertion sequence (IS) previously described in a strain of Pseudomonas alcaligenes. The IS1394 encodes a transposase that shows identity with those of the IS30 family of elements. As shown in Fig. 4, the transcriptional orientation of the IS1394 copy in In77 was opposite that of the genes acquired as cassettes. After the IS1394 was the remainder of the aadA1 gene cassette. The 59-be of the aadA1 gene cassette was 60 bp long and shows all consensus features of the recombination site structures (core site GTTRRRY, inverse core site RYYYAAC, and the internal domains 2L and 2R). Finally, the aadA1 gene cassette is followed by another β-lactamase gene cassette accommodating blaOXA-2. This was followed by the fused gene cassette qacEΔ1/sul1.
FIG. 4.
Schematic representation of integron ln77 carrying blaGIM-1 (the arrows in the gene boxes indicating the direction of transcription). The black dots indicate 59-bes. In the third cassette position the aadA1 is interrupted by a copy of the IS1394. Block arrows beneath the gene map indicate the positions of the primers used for PCRs and sequence analyses.
Patterns of β-lactam susceptibility of E. coli producing the GIM-1 enzyme.
The substrate specificity of GIM-1 and its contribution to resistance were investigated by testing the susceptibility to β-lactams of E. coli DH5α carrying the recombinant plasmid containing the integron borne blaGIM-1 (pGIM-1) and that produces GIM-1 enzyme in contrast to DH5α, which carries an empty vector (pSCRIPT). The GIM-1 production in the host strain was confirmed by measuring the hydrolysis rate of the E. coli DH5α carrying pGIM-1 compared to DH5α carrying the empty vector (0.0097 and 0.0011 Abs/min, respectively).
As shown in Table 2, GIM-1 production was associated with a decrease in the in vitro susceptibility of the E. coli host to ceftazidime, cefotaxime, cefepime, imipenem, meropenem, and ampicillin, indicating that the enzyme can contribute to broad-spectrum resistance in the microbial host.
Kinetic properties of GIM-1.
Analysis of the purified preparation of GIM-1 by sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed a single 26-kDa band and was estimated to be >95% pure (data not shown). In addition, the pI of the protein was determined and, after nitrocefin staining, only one pI 5.4 band was observed in the gel.
The kinetic parameters of GIM-1, including kcat, Km, and the kcat/Km ratio, were determined for several different β-lactam compounds, as presented in Table 3. Under the experimental conditions adopted, GIM-1 hydrolyzed most tested compounds, with the exception of aztreonam, azlocillin, and the serine β-lactamase inhibitors, clavulanic acid and tazobactam. Even after prolonged incubation (1 to 3 h) of the enzyme with these agents, enzyme activity was undetectable.
TABLE 3.
Steady-state kinetic parameters of purified GIM-1 compared to those of IMP-1, VIM-1, VIM-2, and SPM-1
| Antibiotic | Kinetic parametersa of:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GIM-1b
|
IMP-1c
|
VIM-1d
|
VIM-2e
|
SPM-1f
|
|||||||||||
| kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) | |
| Penicillin | 6.6 | 46 | 0.14 | 320 | 520 | 0.62 | 29 | 841 | 0.034 | 55.8 | 49 | 1.14 | 108 | 38 | 2.8 |
| Ampicillin | 3.3 | 20 | 0.16 | 950 | 200 | 4.8 | 37 | 917 | 0.04 | - | - | - | 117 | 72 | 1.6 |
| Carbenicillin | 4.1 | 170 | 0.02 | ND | ND | 0.02 | 167 | 75 | 2.2 | - | - | - | 74 | 814 | 0.09 |
| Azlocillin | ND | ND | ND | - | - | - | 1,525 | 123 | 12 | - | - | - | 53 | 147 | 0.35 |
| Piperacillin | 6.9 | 69 | 0.10 | ND | ND | 0.72 | 1,860 | 3,500 | 0.53 | 32.7 | 72 | 0.45 | 117 | 59 | 2 |
| Ticarcillin | 2.3 | 57 | 0.04 | 1.1 | 740 | 0.0015 | 452 | 1,117 | 0.41 | 31.7 | 46 | 0.69 | ND | <0.35 | ND |
| Nitrocefin | 5.8 | 12 | 0.47 | 63 | 27 | 2.3 | 95 | 17 | 5.6 | - | - | - | 0.53 | 4 | 0.12 |
| Cephalothin | 16 | 22 | 0.72 | 48 | 21 | 2.4 | 281 | 53 | 5.3 | 56.2 | 44 | 1.28 | 43 | 4 | 11.7 |
| Cefuroxime | 5.9 | 7 | 0.80 | 8 | 37 | 0.22 | 324 | 42 | 7.7 | 12.1 | 22 | 0.55 | 37 | 4 | 8.8 |
| Cefoxitin | 8.3 | 206 | 0.04 | 16 | 8* | 2 | 26 | 131 | 0.2 | 3 | 24 | 0.12 | 8 | 2 | 4 |
| Ceftazidime | 18 | 31 | 0.58 | 8 | 44 | 0.18 | 60 | 794 | 0.076 | 89 | 98 | 0.90 | 28 | 46 | 0.6 |
| Cefotaxime | 1.1 | 4 | 0.24 | 1.3 | 4* | 0.35 | 169 | 247 | 0.68 | 27.5 | 32 | 0.86 | 16 | 9 | 1.9 |
| Cefepime | 17 | 431 | 0.04 | 7 | 11* | 0.66 | 549 | 145 | 3.8 | 4.7 | 184 | 0.03 | 18 | 18 | 1 |
| Imipenem | 27 | 287 | 0.09 | 46 | 39 | 1.2 | 2.0 | 1.5 | 1.3 | 9.9 | 10 | 0.99 | 33 | 37 | 1 |
| Meropenem | 2.7 | 25 | 0.11 | 50 | 10 | 0.12 | 13 | 48 | 0.27 | 1.4 | 5 | 0.28 | 63 | 281 | 0.22 |
| Moxalactam | 14 | 1,035 | 0.01 | 88 | 10* | 8.8 | - | - | - | 14.8 | 80 | 0.18 | 13 | 97 | 0.13 |
| Aztreonam | ND | ND | ND | >0.01 | >1,000 | <10−5 | <0.01 | >1,000 | <10−5 | <0.5 | ND | ND | ND | <0.3 | ND |
| Clavulanic acid | ND | ND | ND | - | - | - | - | - | - | - | - | - | ND | >0.1 | ND |
| Tazobactam | ND | ND | ND | >1,000 | >3.98 | 0.0039 | 5.3 | 337 | 0.016 | - | - | - | 0.6 | 3 | 0.2 |
The kinetic parameters of the purified GIM-1 reveal a broad substrate profile but no clear preferences for any of the β-lactam subfamilies tested. Individual kinetic parameters for GIM-1 for different β-lactam agents differed considerably. The highest kcat/Km ratios were observed with cefuroxime, cephalothin, and ceftazidime (0.802, 0.718, and 0.577 μM−1 s−1, respectively). The lowest values were demonstrated with moxalactam, carbenicillin, and ticarcillin (0.014, 0.024, and 0.039 μM−1 s−1, respectively).
For the carbapenems, GIM-1 showed 10 times greater turnover of imipenem than meropenem (kcat values of 27.1 s−1 and 2.7 s−1, respectively). However, the affinity of the enzyme for imipenem is 10 times higher than for meropenem (Km of 287.5 and 25.4 μM, respectively), which makes the kcat/Km ratios for the two compounds very similar (0.094 for imipenem and 0.106 μM−1 s−1 for meropenem). This finding contrasts those for other clinically important class B β-lactamases, which show larger kcat/Km ratios for imipenem than for meropenem (Table 3).
The comparison of GIM-1 kinetic values with the parameters reported for other clinically relevant MβL (Table 3) showed that, in general, the kcat/Km ratios obtained for GIM-1 are lower than those of other MβLs, specifically IMP-1, VIM-1, VIM-2, and SPM-1.
DISCUSSION
The novel class B β-lactamase, GIM-1, is divergent from other class B β-lactamase enzymes and is the fourth subclass of mobile MβL thus far identified. The enzymes most closely related to GIM-1 are the IMP variants, although GIM-1 shares ca. 40% amino acid identity with this group. GIM-1 possesses the major consensus features of the MβL family (8), such as the zinc-binding motif (HXHXD), with amino acids not previously reported at the variable positions in this motif likely to be relevant to the particular activity of the protein.
The kinetic properties of GIM-1 demonstrate that the enzyme has similar activity to other members of the class B β-lactamases (7, 14, 18, 21), although the kinetic parameters for GIM-1 show that these values are closer to those of IMP-1 than to those of VIM-1, VIM-2, and SPM-1. GIM-1, like IMP-1, prefers penicillin and ampicillin (kcat/Km = 0.142 and 0.157 μM−1 s−1) over other penam antimicrobial agents, such as carbenicillin and ticarcillin (kcat/Km 0.024 and 0.039 μM−1 s−1, respectively). Although GIM-1 and IMP-1 exhibit similar kinetic properties when tested against penicillin and ampicillin, there are differences in the kinetic activities of these enzymes against cephalothin and cefoxitin as substrates (kcat/Km = 0.718 and 0.040 μM−1 s−1 for GIM-1 compared to 2.4 and 2.0 μM−1 s−1 for IMP-1). Unlike the other class B β-lactamases, which show more significant activity against imipenem than against meropenem, GIM-1 demonstrates similar kinetic ratios with imipenem and meropenem (kcat/Km = 0.094 and 0.106 μM−1 s−1, respectively). In conclusion, the low kcat/Km ratios determined for GIM-1 with most β-lactam antimicrobial agents reflect high substrate affinities (Km) and low substrate turnover rates (kcat).
Like the majority of MβL genes, blaGIM-1 was found on a class 1 integron, In77, that is carried on a 22-kb plasmid in P. aeruginosa 73-5671. In addition to a novel MβL gene, this integron harbors three other resistance genes, aacA4, aadA1, and blaOXA-2, in that order (Fig. 4). The blaGIM-1 and aacA4 genes appeared to be accommodated in a single gene cassette that has probably been generated from individual cassettes by deletion of most of the intervening 59-be. The aadA1 cassette is bisected by an insertion element, IS1394, that inactivates this particular gene. In addition, the IS1394 is oriented so that its transposase gene is transcribed toward the integrase gene of the In77. Since gene cassettes in class 1 integrons do not carry their own promoters (3) and thus are under the control of the integron promoter embedded in the intI1, in this integron structure, gene cassettes located downstream from the IS1394, such as blaOXA-2, are unlikely to be expressed.
The finding of blaGIM-1 in what appears to be five clonal isolates of P. aeruginosa from different patients from the same hospital is evidence of a nosocomial outbreak. The production of a novel MβL by these isolates could be a warning of future clinical problems with these and similar strains. However, in its present form, the GIM-1 gene may not be as mobile as those encoding IMP or VIM MβLs because the 22-kb plasmid carrying In77 is nonconjugative and appears to have a restricted host range. Only time will tell whether the two-gene cassette accommodating blaGIM-1 and aacA4 will move to another integron on a more easily transmissible plasmid, as happened with other cassette-borne MβL genes.
Several important points arise from this and other studies of MβL found in clinical isolates. (i) Of the four subclasses of mobile MβL gene identified to date, two were originally found in Europe, perhaps signaling future difficulties for empirical treatment with carbapenems in this region. (ii) Thus, it would be desirable that the incidence of MβL production in clinical isolates of key gram-negative bacteria, such as P. aeruginosa, Enterobacter cloacae, Serratia marcescens, and Klebsiella pneumoniae, be carefully monitored. (iii) The results reported in this communication attest to the importance of global resistance surveillance programs such as the SENTRY Program in identifying the emergence and epidemic spread of new threats to the efficacy of antimicrobial therapy and beginning into genetic risk of further spread.
Acknowledgments
We are grateful to Alan M. Simm and Tanya A. Murphy for their important advice during the purification of GIM-1 and to Peter M. Bennett, Ayman Hawrani, and Helio S. Sader for critically reviewing the manuscript.
This study was funded in part by EU grant LSHM-CT-2003-503335.
REFERENCES
- 1.Avison, M. B., C. S. Higgins, C. J. von Heldreich, P. M. Bennett, and T. R. Walsh. 2001. Plasmid location and molecular heterogeneity of the L1 and L2 beta-lactamase genes of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 45:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahar, G., A. Mazzariol, R. Koncan, A. Mert, R. Fontana, G. M. Rossolini, and G. Cornaglia. 2004. Detection of VIM-5 metallo-beta-lactamase in a Pseudomonas aeruginosa clinical isolate from Turkey. J. Antimicrob. Chemother. 54:282-283. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, P. M. 1999. Integrons and gene cassettes: a genetic construction kit for bacteria. J. Antimicrob. Chemother. [PubMed]
- 4.Bush, K. 1998. Metallo-β-lactamases: a class apart. Clin. Infect. Dis. 27(Suppl. 1):S48-S53. [DOI] [PubMed] [Google Scholar]
- 5.Collis, C. M., and R. M. Hall. 1995. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob. Agents Chemother. 39:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Silva, G. J., M. Correia, C. Vital, G. Ribeiro, J. C. Sousa, R. Leitao, L. Peixe, and A. Duarte. 2002. Molecular characterization of blaIMP-5, a new integron-borne metallo-beta-lactamase gene from an Acinetobacter baumannii nosocomial isolate in Portugal. FEMS Microbiol. Lett. 215:33-39. [DOI] [PubMed] [Google Scholar]
- 7.Franceschini, N., B. Caravelli, J. D. Docquier, M. Galleni, J. M. Frere, G. Amicosante, and G. M. Rossolini. 2000. Purification and biochemical characterization of the VIM-1 metallo-beta-lactamase. Antimicrob. Agents Chemother. 44:3003-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gales, A. C., R. N. Jones, J. Turnidge, R. Rennie, and R. Ramphal. 2001. Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S146-S155. [DOI] [PubMed] [Google Scholar]
- 9.Gales, A. C., M. C. Tognim, A. O. Reis, R. N. Jones, and H. S. Sader. 2003. Emergence of an IMP-like metallo-enzyme in an Acinetobacter baumannii clinical strain from a Brazilian teaching hospital. Diagn. Microbiol. Infect. Dis. 45:77-79. [DOI] [PubMed] [Google Scholar]
- 10.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, and J. M. Frere. 2001. Standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garau, G., I. Garcia-Saez, C. Bebrone, C. Anne, P. Mercuri, M. Galleni, J. M. Frere, and O. Dideberg. 2004. Update of the standard numbering scheme for class B beta-lactamases. Antimicrob. Agents Chemother. 48:2347-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibb, A. P., C. Tribuddharat, R. A. Moore, T. J. Louie, W. Krulicki, D. M. Livermore, M. F. Palepou, and N. Woodford. 2002. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new bla(IMP) allele, bla(IMP-7). Antimicrob. Agents Chemother. 46:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koh, T. H., G. C. Wang, and L. H. Sng. 2004. IMP-1 and a novel metallo-beta-lactamase, VIM-6, in fluorescent pseudomonads isolated in Singapore. Antimicrob. Agents Chemother. 48:2334-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laraki, N., N. Franceschini, G. M. Rossolini, P. Santucci, C. Meunier, E. de Pauw, G. Amicosante, J. M. Frere, and M. Galleni. 1999. Biochemical characterization of the Pseudomonas aeruginosa 101/1477 metallo-β-lactamase IMP-1 produced by Escherichia coli. Antimicrob. Agents Chemother. 43:902-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livermore, D. M., and N. Woodford. 2000. Carbapenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 17.Mendes, R. E., M. Castanheira, P. Garcia, M. Guzman, M. A. Toleman, T. R. Walsh, and R. N. Jones. 2004. First isolation of bla(VIM-2) in Latin America: report from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 48:1433-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy, T. A., A. M. Simm, M. A. Toleman, R. N. Jones, and T. R. Walsh. 2003. Biochemical characterization of the acquired metallo-beta-lactamase SPM-1 from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 47:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.Patzer, J., M. A. Toleman, L. M. Deshpande, W. Kaminska, D. Dzierzanowska, P. M. Bennett, R. N. Jones, and T. R. Walsh. 2004. Pseudomonas aeruginosa strains harbouring an unusual blaVIM-4 gene cassette isolated from hospitalized children in Poland (1998-2001). J. Antimicrob. Chemother. 53:451-456. [DOI] [PubMed] [Google Scholar]
- 21.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J. D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quinn, J. P., A. E. Studemeister, C. A. DiVincenzo, and S. A. Lerner. 1988. Resistance to imipenem in Pseudomonas aeruginosa: clinical experience and biochemical mechanisms. Rev. Infect. Dis. 10:892-898. [DOI] [PubMed] [Google Scholar]
- 24.Riccio, M. L., N. Franceschini, L. Boschi, B. Caravelli, G. Cornaglia, R. Fontana, G. Amicosante, and G. M. Rossolini. 2000. Characterization of the metallo-beta-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla(IMP) allelic variants carried by gene cassettes of different phylogeny. Antimicrob. Agents Chemother. 44:1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., P. MacCallum, and D. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toleman, M. A., D. Biedenbach, D. Bennett, R. N. Jones, and T. R. Walsh. 2003. Genetic characterization of a novel metallo-β-lactamase gene, blaIMP-13, harboured by a novel Tn5051-type transposon disseminating carbapenemase genes in Europe: report from the SENTRY worldwide antimicrobial surveillance programme. J. Antimicrob. Chemother. 52:583-590. [DOI] [PubMed] [Google Scholar]
- 28.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2003. Molecular and biochemical characterization of OXA-45, an extended-spectrum class 2d' beta-lactamase in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 47:2859-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, and T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-beta-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 30.Tsakris, A., S. Pournaras, N. Woodford, M. F. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh, T. R., D. J. Payne, A. P. MacGowan, and P. M. Bennett. 1995. A clinical isolate of Aeromonas sobria with three chromosomally mediated inducible beta-lactamases: a cephalosporinase, a penicillinase and a third enzyme, displaying carbapenemase activity. J. Antimicrob. Chemother. 35:271-279. [DOI] [PubMed] [Google Scholar]
- 32.Walsh, T. R., M. A. Toleman, W. Hryniewicz, P. M. Bennett, and R. N. Jones. 2003. Evolution of an integron carrying blaVIM-2 in Eastern Europe: report from the SENTRY Antimicrobial Surveillance Program. J. Antimicrob. Chemother. 52:116-119. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe, M., S. Iyobe, M. Inoue, and S. Mitsuhashi. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 35:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan, J. J., P. R. Hsueh, W. C. Ko, K. T. Luh, S. H. Tsai, H. M. Wu, and J. J. Wu. 2001. Metallo-β-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob. Agents Chemother. 45:2224-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yatsuyanagi, J., S. Saito, Y. Ito, K. Ohta, J. Kato, S. Harata, N. Suzuki, and K. Amano. 2004. Identification of Pseudomonas aeruginosa clinical strains harboring the bla(VIM-2) metallo-β-lactamase gene in Akita prefecture, Japan. Jpn. J. Infect. Dis. 57:130-132. [PubMed] [Google Scholar]
- 36.Yeo, C. C., and C. L. Poh. 1996. IS1394 from Pseudomonas alcaligenes N.C.I.B. 9867: identification and characterization of a member of the IS30 family of insertion elements. Gene 175:109-113. [DOI] [PubMed] [Google Scholar]
- 37.Ziha-Zarifi, I., C. Llanes, T. Kohler, J. C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]