Abstract
Fluoroquinolones are broad-spectrum antimicrobial agents that target type II topoisomerases. Many fluoroquinolones are highly specific for bacterial type II topoisomerases and act against both DNA gyrase and topoisomerase IV. In Escherichia coli, mutations causing quinolone resistance are often found in the gene that encodes the A subunit of DNA gyrase. One common site for resistance-conferring mutations alters Ser83, and mutations to Leu or Trp result in high levels of resistance to fluoroquinolones. In the present study we demonstrate that the mutation of Ser83 to Trp in DNA gyrase (GyrS83W) also results in sensitivity to agents that are potent inhibitors of eukaryotic topoisomerase II but that are normally inactive against prokaryotic enzymes. Epipodophyllotoxins, such as etoposide, teniposide and amino-azatoxin, inhibited the DNA supercoiling activity of GyrS83W, and the enzyme caused elevated levels of DNA cleavage in the presence of these agents. The DNA sequence preference for GyrS83W-induced cleavage sites in the presence of etoposide was similar to that seen with eukaryotic type II topoisomerases. Introduction of the GyrS83W mutation in E. coli strain RFM443-242 by site-directed mutagenesis sensitized it to epipodophyllotoxins and amino-azatoxin. Our results demonstrate that sensitivity to agents that target topoisomerase II is conserved between prokaryotic and eukaryotic enzymes, suggesting that drug interaction domains are also well conserved and likely occur in domains important for the biochemical activities of the enzymes.
The fluoroquinolones are exceptionally potent antibacterial agents that are widely used for the treatment of a wide variety of infections (14, 15, 29, 30). Fluoroquinolones interact with two related but distinct targets within the bacterial cell, DNA gyrase and DNA topoisomerase IV (22, 32). Both are type II topoisomerases, enzymes required for DNA metabolism that introduce transient double-stranded breaks in DNA and that then pass another duplex segment of DNA through the break and religate the broken ends (for reviews, see references 7, 11, 42, 46, 55, and 56).
DNA gyrase is unique among the type II enzymes, in that it can introduce negative superhelical turns into DNA (12, 22, 47). Topoisomerase IV but not DNA gyrase unlinks catenated intermediates of replicated and recombined DNA molecules, as well as unknots DNA (12, 61, 62). Fluoroquinolones interfere with the topoisomerase reaction by disrupting the DNA breakage-reunion reaction, resulting in the trapping of both DNA gyrase and topoisomerase IV at a point in the reaction cycle where the enzyme is covalently bound to DNA (21, 33). Although both enzymes carry out indispensable reactions, trapping of the covalent complex and the generation of enzyme-mediated DNA damage causes cell killing (34, 39).
The majority of known cases of quinolone resistance encountered in clinical isolates, as well as the majority of cases of laboratory selection of mutations, results from mutations in chromosomal genes that lead to alterations in the drug targets (1, 10, 15, 27, 38, 49, 50). Mutations in GyrA, the gene for which (gyrA) encodes the A subunit of DNA gyrase, are the most common mechanisms involved in quinolone resistance among gram-negative bacteria. Moreover, mutations in parC (grlA in Staphylococcus aureus), the gene encoding the homologous A subunit of topoisomerase IV, are most commonly encountered among quinolone-resistant gram-positive bacteria (1, 14, 15, 27, 50). Mutations within the genes encoding GyrA and ParC occur within conserved regions (15, 50). These hot spots for quinolone resistance have been termed quinolone resistance-determining regions and are located between amino acids 67 and 106 in GyrA, with amino acids 83 and 87 most often being involved (15, 26, 50, 57, 59). Similar hot spots were found in analogous regions of ParC (19, 20, 25, 34, 41). Mutations that change Ser83 to either leucine or tryptophan confer high levels of quinolone resistance (∼10-fold increase), whereas mutations that change Ser83 to alanine result in lower levels of quinolone resistance (∼5-fold increase) (9).
Previous studies (31, 53) examined the effects of yeast topoisomerase II mutations that change Ser740, the amino acid equivalent to Ser83 in GyrA. A mutation that results in the change Ser740Trp resulted in resistance to CP-115,953, a fluoroquinolone that is highly active against eukaryotic type II topoisomerases (16). In addition to quinolone resistance, the Ser740Trp mutation also rendered yeast topoisomerase II hypersensitive to etoposide (31, 53). The demethylepipodophyllotoxins etoposide (VP-16) and teniposide (VM-26) are nonintercalating drugs that inhibit eukaryotic type II topoisomerases by increasing the levels of DNA cleavage complexes primarily by impairing the ability of the enzyme to religate the cleaved DNA (4, 35, 36, 46). The mechanism of eukaryotic cell killing by these agents is similar to that described above for the actions of fluoroquinolones against bacteria, i.e., the generation of enzyme-mediated DNA damage. Most of the drugs, like etoposide, that act against eukaryotic topoisomerase II do not have activity against bacterial cells and have only limited activity against the purified prokaryotic enzymes (6). Because bacterial cells have efficient efflux pumps that contribute to both intrinsic and acquired resistance to inhibitors of prokaryotic type II topoisomerases (8, 44, 48, 58), investigation of the effects of epipodophyllotoxins on viable cells requires the inactivation of efflux pump activity. This can be achieved by either loss-of-function mutations or inhibition of the pumps with efflux pump inhibitors such as MC-207,110 (37).
This study demonstrates that a single point mutation, Ser83Trp, in the gyrase-encoding gene, gyrA, is associated with high-level quinolone resistance and causes enzyme and bacterial sensitivity to other antitopoisomerase drug classes, like demethylepipodophyllotoxins and amino-azatoxin [11β-(2"-N,N-dimethylaminoethyl)-amino-azatoxin], for which wild-type gyrase is refractory. Our results suggest a novel strategy of drug screening to take into account the fact that certain drugs might be active against quinolone-resistant DNA gyrases, even though the wild-type gyrase is insensitive.
MATERIALS AND METHODS
Antibiotics, enzymes, and growth medium.
Cation-adjusted Mueller-Hinton broth was from Difco (Detroit, Mich.). Standard broth no. 1 (NI agar and NI broth) was from Merck (Darmstadt, Germany). Ciprofloxacin was from Bayer AG (Wuppertal, Germany); chloramphenicol was from by Boehringer Mannheim (Mannheim, Germany); MC-207,110, Phe-Arg-β-naphthylamide, was from Sigma (Munich, Germany); and etoposide (VP-16) was from Bristol Arzneimittel GmbH (Munich, Germany). Amino-azatoxin was kindly provided by T. Macdonald, Department of Chemistry, University of Virginia, Charlottesville. All drug stock solutions were prepared in dimethyl sulfoxide at 10 mM. Further dilutions were made in distilled water immediately before use. Expand High-Fidelity polymerase (Roche, Mannheim, Germany) was used for PCR. BamHI restriction endonuclease was purchased from Life Technologies (Eggenstein, Germany). Human c-myc inserted into pBR322, T4 polynucleotide kinase, and polyacrylamide and bisacrylamide were purchased from Lofstrand Labs (Gaithersburg, Md.), Life Technologies, Inc. (Gaithersburg, Md.), or New England Biolabs (Beverly, Mass.). [γ-32P]ATP (6000 Ci/mmol) was purchased from DuPont NEN (Boston, Mass.). PCR oligonucleotide primers were obtained from GIBCO BRL (Gaithersburg, Md.). GyrA was a gift of N. A. Gormley, and gyrA bearing the Ser83Trp mutation (gyrAS83W) was a gift of C. J. R. Willmott.
Bacterial strains and plasmids.
The strains used in this study are derivatives of Escherichia coli K-12. Strain RFM443 [rpsL200 (Strr) galK2 Δlac-74 (lac mutant)] was provided by T. K. Van Dyk (DuPont Co., Wilmington, Del.). Strain RFM443-242 was constructed for this study and is isogenic to RFM443 except for the gyrAS83W mutation. Strain JM109 [recA1 hsdR17 supE44 Δ(lac-proAB) endA1 gyrA96 relA1 thi F′ (traD36 prpAB lacIq lacZΔM15)] was used for cloning procedures. E. coli clinical isolate 4917 bearing a gyrAS83W mutation was described previously (28). The plasmids used were pMAK705 (23), pMAK705-242 (this study), pBR322 from strain C600 (laboratory stock), and pBR322-242 (this study).
Site-directed mutagenesis of gyrA in E. coli.
The allelic exchange technique (23) was applied to introduce into the chromosomal gyrA gene the S83W point mutation associated with quinolone resistance. Briefly, a PCR fragment from strain 4917 carrying the mutation flanked by BamHI restriction sites was first cloned into pBR322 to give pBR322-242. The gyrA BamHI fragment was isolated from pBR322-242 and subcloned into the BamHI site within the polylinker region of pMAK705 to give pMAK705-242 (Fig. 1). This plasmid was transformed into strain RFM443 by electroporation with a Gene Pulser apparatus (Bio-Rad, Munich, Germany). The presence of the gyrAS83W mutation in pBR322-242, pMAK705-242, and strain RFM443-242 was verified by DNA sequencing.
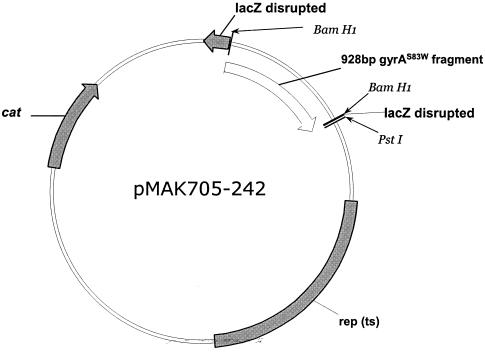
FIG. 1.
Physical map of plasmid pMAK705-242 (6.5 kbp) carrying a chloramphenicol acetyltransferase gene (cat) as a resistance marker. pMAK705-242 was constructed by inserting the 928-bp BamHI fragment carrying the truncated GyrAS83W gene from pBR322-242 into the unique BamHI site within the multiple-cloning site of pMAK705 (23) (see Materials and Methods). Replication of pMAK705 is temperature sensitive [rep (ts)].
Drug susceptibility assay.
MICs were determined by a twofold microdilution method with cation-adjusted Mueller-Hinton broth, in accordance with the recommendations of NCCLS (40). The final inoculum of 5 × 105 to 1 × 106 CFU/ml (verified by plating) was prepared by diluting suspensions harvested from overnight cultures from Luria-Bertani agar plates. The trays were incubated at 37°C for 20 h.
Time course of drug susceptibility.
Flasks (100 ml) containing 10 ml of standard broth no. 1 were inoculated with exponentially growing cells and were incubated for an additional 2 h at 37°C in a rotating shaker (200 rpm) to reach a cell density of approximately 106 CFU/ml. Drugs were then added to the culture, and 0.5-ml aliquots were taken at various times to determine the number of viable cells.
DNA supercoiling reactions.
Relaxed substrate DNA was prepared by incubating native simian virus 40 (SV40) DNA with topoisomerase I (calf thymus; GIBCO/BRL) for 2 h at 37°C. DNA supercoiling reactions were carried out in a total reaction volume of 20 μl containing 30 mM Tris-HCl (pH 7.5), 25 mM KCl, 4 mM MgCl2, 5 mM dithiothreitol, 30 μg of bovine serum albumin/ml, 9 μg of tRNA/ml, 2 mM spermidine, 1.5 mM ATP, 375 ng of relaxed SV40 DNA, and 140 ng of DNA gyrase. The reaction mixtures were incubated at 37°C for 30 min, and the reactions were stopped by the addition of 20 μl of chloroform. Samples were separated on 0.8% agarose gels. Gels were stained with ethidium bromide and viewed under UV light (300 nm). Quantitation of the amount of supercoiled DNA was performed with ImageQuant software. The amount of supercoiled DNA generated by gyrase with or without additional drug was calculated as a percentage of the total SV40 DNA (including nicked DNA, the various forms of remaining relaxed DNA, and supercoiled DNA), which was separated on agarose gels.
Preparation of end-labeled DNA fragments by PCR.
Three sets of labeled DNA fragments were prepared from the human c-myc gene by PCR: a 254-bp DNA fragment from the first intron between nucleotides (nt) 3035 and 3288 (GenBank accession no. X00364) was prepared with oligonucleotide 5′-GTAATCCAGAACTGGATCGG-3′ for the upper (coding) strand and oligonucleotide 5′-ATGCGGTCCCTACTCCAAGG-3′ for the lower (noncoding or antisense) strand (annealing temperature, 56°C). A 401-bp DNA fragment from the junction between the first intron and the first exon (between nt 2671 and 3072) was prepared with oligonucleotide 5′-TGCCGCATCCACGAAACTTT-3′ for the upper (coding) strand and oligonucleotide 5′-TTGACAAGTCACTTTACCCC-3′ for the lower (noncoding or antisense) strand (annealing temperature, 60°C). A 480-bp fragment from the first exon containing promoters P1 and P2 (between nt 2265 and 2745) was prepared with oligonucleotide 5′-GATCCTCTCTCGCTAATCTCCGCCC-3′ for the upper (coding) strand and oligonucleotide 5′-TCCTTGCTCGGGTGTTGTAAGTTCC-3′ for the lower (noncoding or antisense) strand (annealing temperature, 70°C). A 213-bp fragment from the human c-jun gene (51) was prepared with primer 5′-TGTTGACAGCGGCGGAAAGCAGS-3′ for the upper (coding) strand and primer 5′-CGTCCTTCTTCTCTTGCGTGGCTCT-3′ for the lower (noncoding or antisense) strand (annealing temperature, 64°C). Single-end labeling of these DNA fragments was achieved by 5′ end labeling of the specific primer oligonucleotide (53), and the labeled oligonucleotides were used thereafter for PCR. Approximately 0.1 μg of the c-myc DNA that had been restricted with SmaI and PvuII (the fragment from positions 2265 to 2745) and with XhoI and XbaI (the fragments from positions 2671 to 3072 and positions 3035 to 3288, respectively) was used as the template for the PCR. Ten picomoles of each oligonucleotide primer, one of which was 5′ labeled, was used in 22 temperature cycle reactions (with each cycle consisting of 94°C for 1 min, annealing for 1 min, and 72°C for 2 min).
Mapping and base sequence analysis of drug-induced DNA cleavage.
DNA fragments (5 × 104 to 8 × 104 dpm/reaction mixture) were equilibrated in cleavage buffer (10 mM Tris-HCl [pH 7.5], 50 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol, 0.1 mM disodium EDTA, 1 mM ATP, 15 μg of bovine serum albumin/ml) and drug or a 1% dimethyl sulfoxide blank was added. After 5 min of incubation, 70 ng of purified E. coli gyrase was added to give a final reaction volume of 10 μl. The reaction mixtures were incubated for 120 min at 25°C and stopped by adding EDTA and sodium dodecyl sulfate (final concentrations, 25 mM and 1%, respectively) and were further digested with proteinase K. For DNA sequence analysis, samples were separated on denaturing DNA sequencing gels and were thereafter visualized with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and ImageQuant software. The preferred bases around the gyrase DNA cleavage sites were identified as described previously (5, 45, 54).
Quantitation of drug-induced DNA cleavage.
The reaction mixtures used to quantitate drug-induced DNA cleavage included 140 ng of enzyme and 250 ng of negatively supercoiled SV40 DNA (GIBCO/BRL) in a total of 10 μl of cleavage buffer (the same buffer used for drug-induced DNA fragment cleavage). The reaction mixtures were incubated for 120 min at 25°C and stopped by adding EDTA and sodium dodecyl sulfate (final concentrations, 25 mM and 1%, respectively), and they were further digested with proteinase K. DNA fragments were separated on 0.8% agarose gels and were visualized after electrophoresis (in Tris-borate-EDTA containing 0.05 μg of ethidium bromide/ml) by UV transillumination (300 nm). The amount of linearized DNA (i.e., DNA with double-stranded breaks) was quantitated with ImageQuant and Microsoft Excel software.
RESULTS
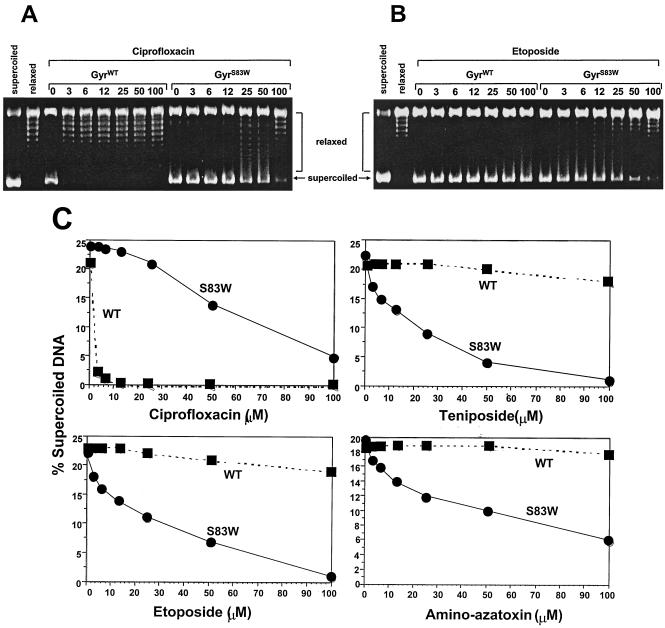
Our first experiments tested whether the Ser83→Trp mutation in DNA gyrase changed the catalytic activity of the mutant enzyme in the presence of agents active against the eukaryotic enzyme. We analyzed the effect of epipodophyllotoxins and amino-azatoxin on wild-type DNA gyrase (GyrWT)- and GyrAS83W-catalyzed supercoiling of relaxed SV40 DNA (Fig. 2). In the absence of drugs, both enzymes supercoiled relaxed SV40 DNA without detectable differences in efficacy. In the presence of ciprofloxacin (Fig. 2A), GyrWT showed complete inhibition of supercoiling at 3 μM, whereas, as expected, the mutant GyrAS83W protein retained supercoiling activity in the presence of 100 μM ciprofloxacin. We observed a different pattern of inhibition in the presence of etoposide (Fig. 2B). The GyrWT protein showed undiminished supercoiling activity in the presence of etoposide at a concentration of 100 μM, and the mutant GyrAS83W enzyme displayed reduced supercoiling activity in the presence of 3 μM etoposide and nearly complete inhibition in the presence of 100 μM etoposide. We observed similar patterns of inhibition of supercoiling with teniposide and amino-azatoxin (Fig. 2C).
FIG. 2.
Supercoiling of relaxed SV40 DNA in the presence of epipodophyllotoxins (etoposide and teniposide) and amino-azatoxin is markedly reduced with GyrAS83W. (A and B) Agarose gels after supercoiling reactions with E. coli GyrWT and GyrAS83W in the presence of the fluoroquinolone ciprofloxacin (A) and etoposide (B); (C) the supercoiled fraction in the presence of ciprofloxacin, etoposide, teniposide, and amino-azatoxin at the indicated concentrations (in micromolar) was calculated. Individual points represent the means of three to four independent experiments, with variabilities of not more than 10%.
Enhanced DNA cleavage by epipodophyllotoxins (etoposide and teniposide) and amino-azatoxin with the Ser83Trp mutant gyrase.
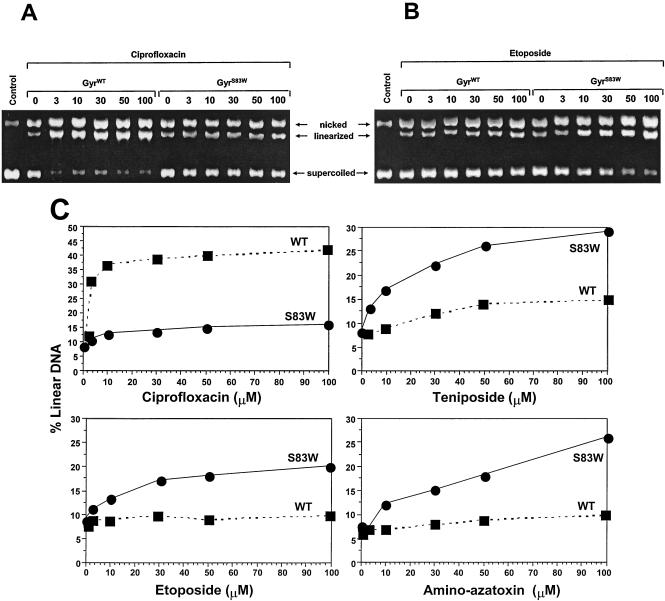
We measured DNA cleavage by purified E. coli GyrWT or GyrAS83W in the presence of either the fluoroquinolone ciprofloxacin or the epipodophyllotoxins etoposide and teniposide or amino-azatoxin (Fig. 3). The gyrase-induced DNA double-stranded breaks were measured and are illustrated in Fig. 3A for ciprofloxacin and Fig. 3B for etoposide. The levels of linearized DNA were quantitated; and plots of the levels of linearized DNA in the presence of ciprofloxacin, etoposide, teniposide, and amino-azatoxin are shown in Fig. 3C. In the absence of drug, both GyrWT and GyrAS83W enzymes generated low levels of linearized DNA. In the presence of ciprofloxacin at concentrations ranging from 3 to 100 μM, approximately 40% linearized DNA was reached with GyrWT. DNA cleavage was markedly reduced with the mutant GyrAS83W in the presence of ciprofloxacin, even at concentrations greater than 100 μM. Importantly, the opposite result was seen with etoposide, teniposide, and amino-azatoxin (Fig. 3B and C). The generation of linearized DNA by GyrWT was not significantly stimulated by etoposide, teniposide, or amino-azatoxin. In contrast, the mutant GyrAS83W protein showed at least twofold larger amounts of linearized DNA in the presence of etoposide, teniposide, or amino-azatoxin.
FIG. 3.
Cleavage of SV40 DNA in the presence of epipodophyllotoxins (etoposide and teniposide) or amino-azatoxin is enhanced with GyrAS83W. (A and B) Agarose gels after cleavage reactions with E. coli GyrWT and GyrAS83W in the presence of the fluoroquinolone ciprofloxacin (A) and etoposide (B); (C) the linearized fraction, which is proportional to the amount of gyrase-induced DNA double-stranded breaks in the presence of ciprofloxacin, etoposide, teniposide and amino-azatoxin at the indicated drug concentrations (in micromolar) was calculated. Individual points represent the means of three to four independent experiments, with variabilities of not more than 10%.
Different DNA cleavage sites for GyrAS83W compared to those for GyrWT in the presence of etoposide and amino-azatoxin.
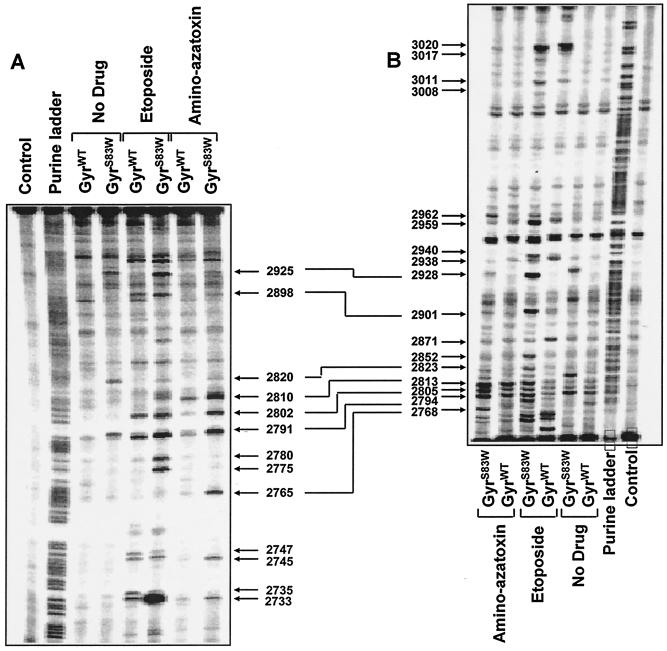
To investigate the basis for the enhanced DNA cleavage activity of GyrAS83W in the presence of specific drugs, we mapped the DNA cleavage sites induced by mutant GyrAS83W and GyrWT in the presence of etoposide and amino-azatoxin (Fig. 4). Even in the absence of drugs, differences in the cleavage sites induced by mutant GyrAS83W and GyrWT could be observed. The GyrAS83W mutant protein caused increased cleavage at specific sites in the presence of etoposide, e.g., at positions 2925, 2780, 2775, and 2928, compared to the sites of cleavage caused by GyrWT. Similarly, enhanced DNA cleavage and multiple changes in the cleavage sites were seen with the GyrAS83W protein in the presence of amino-azatoxin, another nonintercalating topoisomerase II poison.
FIG. 4.
Mapping and analysis of the DNA cleavage sites induced by mutant and wild-type gyrases in the presence of etoposide and amino-azatoxin. DNA fragments from the junction between the c-myc first intron and the first exon between positions 2671 and 3072 were prepared by PCR with one primer labeled with 32P at the 5′ terminus (51). (A) Labeling of the upper DNA strand; (B) labeling of the lower DNA strand (see Materials and Methods). The drugs (concentration, 100 μM each) are indicated above each lane. Cleavage reactions were performed at 25°C for 2 h. Purine ladders were obtained after the formic acid reaction. The control consisted of no enzyme and no drug treatment. Double-headed arrows correspond to DNA cleavage sites with a 4-bp stagger that represent potential DNA double-stranded breaks.
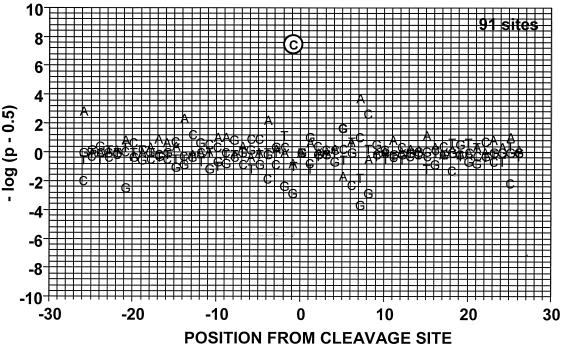
We next asked whether enhanced DNA cleavage activity and alterations in cleavage patterns were associated with an altered sequence specificity of GyrAS83W in the presence of these drugs (Fig. 5 and Table 1.) For both the GyrWT protein (data not shown) and the mutant GyrAS83W protein, etoposide preferentially stabilized sites with a C at position −1 (48 of 91 sites for GyrAS83W; Table 1). This result correlates well with previous DNA cleavage analyses of eukaryotic topoisomerase II proteins in the presence of etoposide (4, 46, 51). Taken together, the data indicate that changes in the protein-drug interaction resulting from the Ser83→Trp mutation in gyrase enhance DNA cleavage activity in the presence of etoposide. However, enhanced DNA cleavage activity with the GyrAS83W protein in the presence of drugs was not associated with increased stability (i.e., stability in the presence of heat and salt) of the covalent complexes (data not shown).
FIG. 5.
Probability of the observed deviations in base frequency at DNA cleavage sites for GyrAS83W in the presence of etoposide. Both DNA strands of three c-myc DNA fragments were analyzed for cleavage sites (see Materials and Methods). Position 0 corresponds to the cleavage site. The probability of the observed base frequency deviations from expectation is shown. On the y axis, P is the probability of observing the indicated deviation more (above the baseline) or less (below the baseline) often relative to the expected frequency of each individual base (45, 51). Cleavage sites were analyzed after treatment with 100 μM etoposide.
TABLE 1.
Base distribution at each position of etoposide-induced DNA cleavage sitesa
| Base | Total base frequency (n) at the following position from cleavage siteb:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −10 | −9 | −8 | −7 | −6 | −5 | −4 | −3 | −2 | −1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| A | 30 | 31 | 21 | 25 | 18 | 23 | 34 | 24 | 19 | 16 | 26 | 20 | 22 | 24 | 13 | 27 | 36 | 19 | 20 | 23 |
| C | 29 | 22 | 19 | 18 | 32 | 33 | 14 | 18 | 30 | 48 | 20 | 30 | 28 | 26 | 27 | 15 | 33 | 37 | 23 | 24 |
| G | 18 | 19 | 32 | 27 | 28 | 17 | 22 | 29 | 14 | 11 | 30 | 24 | 22 | 20 | 34 | 24 | 10 | 11 | 29 | 28 |
| T | 14 | 19 | 19 | 21 | 13 | 18 | 21 | 20 | 28 | 16 | 15 | 17 | 19 | 21 | 17 | 25 | 12 | 24 | 19 | 16 |
A total of 91 sites were tested.
Underlined numbers represent base frequencies significantly (P < 0.001) greater or lower than expected.
Susceptibilities of quinolone-sensible and -resistant E. coli strains.
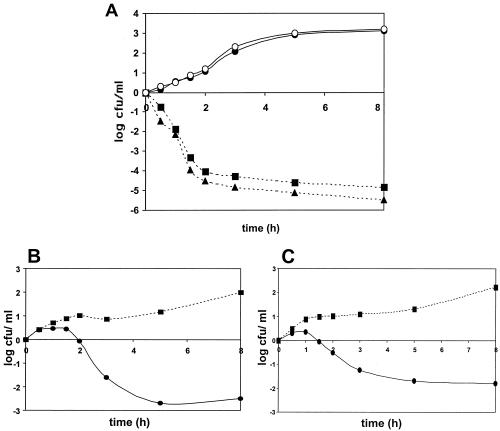
Our next goal was to demonstrate that etoposide could also act against DNA gyrase in vivo. Pilot experiments failed to demonstrate etoposide sensitivity in a variety of different strains. Since etoposide was unlikely to accumulate to significant levels in wild-type E. coli cells, we needed to be able to increase the drug levels by either increasing influx or decreasing efflux. We chose to use a chemical inhibitor of drug efflux. AcrAB is a drug efflux protein that can be inhibited by MC-207,110 (37). Therefore, we treated E. coli strain RFM443 and a GyrAS83W derivative (RFM443-242) with 40 μM MC-207,110 in the presence or absence of anticancer drugs targeting topoisomerase II (Fig. 6). Inhibition of drug efflux by MC-207,110 (final concentration, 40 μM) did not affect the ciprofloxacin sensitivity of wild-type strain RFM443 or mutant strain RFM443-242 (Fig. 6A). This result is consistent with the view that ciprofloxacin resistance in strain RFM443-242 is solely due to the Ser83Trp-mutation in GyrA. However, simultaneous treatment of cells with MC-207,110 and etoposide (Fig. 6B) resulted in the killing only of cells carrying GyrAS83W. Similar results were obtained with amino-azatoxin (Fig. 6C) and teniposide (data not shown). This experiment shows that etoposide and other topoisomerase II poisons can kill E. coli cells, provided that the cells carry a sensitizing mutation and a condition that can lead to enhanced drug accumulation. Taken together, our results show that etoposide, teniposide, and amino-azatoxin are active against E. coli GyrAS83W.
FIG. 6.
(A) Survival of E. coli strain RFM443 and its GyrAS83W mutant, strain RFM443-242, in the presence of ciprofloxacin with the efflux pump inhibitor MC-207,110 or without efflux pump inhibitor MC-207,110. For time-kill studies, ciprofloxacin (final concentration, 0.16 μM) with or without MC-207,110 (final concentration, 40 μM) was added to exponentially growing cells (cell density, approximately 106 CFU/ml), and viable cell counts were determined at the indicated times. Individual points represent the means of three independent experiments, with variabilities of not more than 0.4 log CFU. ▪, RFM443 with MC-207,110; •, RFM443-242 with MC-207,110; ▴, RFM443 without MC-207,110; ○, RFM443-242 without MC-207,110. (B and C) The Ser83Trp mutation in the GyrA subunit of strain RFM443-242 (•) is associated with reduced survival in the presence of etoposide (B) or amino-azatoxin (C) compared to that for wild-type strain RFM443 (▪). The final concentration of each drug was 110 μM. All incubations were performed in the presence of the efflux pump inhibitor MC-207,110 (final concentration, 40 μM). Viable cell counts were determined after incubation for the indicated times. Individual points represent the means of three independent experiments, with variabilities of not more than 0.4 log CFU.
DISCUSSION
In previous studies, we analyzed several point mutations within the α4 helix of the CAP homology domain of eukaryotic (yeast) type II topoisomerases (13, 31, 53). Since we observed mutations in the eukaryotic enzyme that could enhance sensitivity to some classes of inhibitors, we reasoned that homologous mutations could be used to assess whether prokaryotic topoisomerase II could be inhibited by agents thought to be specific for the eukaryotic enzyme. We carried out the analysis at the biochemical level, using purified DNA gyrase, and assessed the sensitivities of E. coli cells carrying an appropriate gyrase mutation. Our results clearly demonstrate that the Ser83 change in GyrA results in an enzyme that is sensitive to etoposide and related topoisomerase II inhibitors, whereas the wild-type enzyme is relatively insensitive to these agents.
Interestingly, the mutant with the Ser83-to-Trp mutation shares biochemical properties with the eukaryotic enzymes mutated at homologous positions. Previous studies showed that this mutant enzyme had alterations in DNA cleavage specificity even in the absence of drug (52). Etoposide-induced cleavage sites for gyrase carrying GyrAS83W showed the same preference for C at position −1 described for the wild-type yeast and human enzymes (3, 45). Not surprisingly, the properties of the prokaryotic enzyme do not exactly mimic all of those of the eukaryotic mutant enzyme. For example, the GyrAS83W protein did not demonstrate alterations in base preference, such as C at position −2 and G at position +6, which were observed for the Saccharomyces Top2pS740W and the human Top2pαS763W enzymes (53). The etoposide-induced cleavage complexes for the GyrAS83W mutant protein did not show heat or salt stability (data not shown), although these were observed with the yeast and human mutant proteins (31, 51). Nonetheless, we can clearly conclude that DNA gyrase preserves most of the determinants that are required for inhibition by etoposide and related compounds.
There are several important implications for the conservation of most determinants of drug sensitivity between prokaryotic and eukaryotic enzymes. First, drugs targeting topoisomerase II likely bind to regions of the proteins that are very highly conserved between prokaryotic and eukaryotic enzymes. This is not a surprising conclusion, given the high degree of homology found throughout many of the type II topoisomerases (56). However, the previous assumption that drugs such as etoposide acted against the eukaryotic enzyme but not the prokaryotic enzyme allowed for the possibility that such drugs interact with residues that are not highly conserved between the two kingdoms. Our results argue against this possibility.
Second, our results support the hypothesis that many topoisomerase II-targeting drugs that lead to elevated levels of covalent complexes act near the same site. The existence of mutants leading to quinolone resistance and concomitant etoposide sensitivity suggests that both drugs disrupt the same processes by a change at a particular site on the protein. This conclusion is consistent with the conclusions of previous drug competition studies performed by Osheroff and colleagues (17), who showed that quinolones inactive against the wild-type eukaryotic enzyme could block the actions of other active inhibitors.
Our studies have concentrated on sites within the gyrA homology domain that can lead to quinolone resistance. Previous work (18, 24, 26, 60) has shown that there are also important determinants of quinolone sensitivity within the gyrB domains of gyrase. Since mutations within the gyrB homology domain of eukaryotic topoisomerase II can confer resistance to etoposide and other topoisomerase II poisons (2, 43), it is likely that domains critical for the actions of both prokaryotic and eukaryotic topoisomerase II-targeting poisons occur in both gyrA and gyrB.
The antibacterial agents in clinical use that target topoisomerase II are all fluoroquinolones. The success of the fluoroquinolones as potent broad-spectrum agents is an important reason for this. However, the problems of quinolone resistance in clinical isolates will only increase as quinolone usage increases. While quinolones are the only chemical class used against prokaryotic topoisomerase II, many different chemical classes have been shown to be active against the eukaryotic enzyme. Our results suggest that other chemical classes may be exploitable as antibacterial agents. Such new chemical classes may extend the usefulness of type II topoisomerases as antibacterial targets even in the face of increasing rates of resistance to fluoroquinolones.
Acknowledgments
J.L.N. was supported by grants CA52814 and CA21765 from the National Institutes of Health and the American Lebanese Syrian Associated Charities.
Dirk Strumberg was supported by the Deutsche Forschungsgemeinschaft (grant STR 527/2-1), Bonn, Germany.
REFERENCES
- 1.Acar, J. F., and F. W. Goldstein. 1997. Trends in bacterial resistance to fluoroquinolones. Clin. Infect. Dis. 24(Suppl. 1):S67-S73. [DOI] [PubMed] [Google Scholar]
- 2.Beck, W. T., T. Khelifa, H. Kusumoto, Y. Y. Mo, Q. Rodgers, J. S. Wolverton, and Q. Wang. 1997. Novel mechanisms of resistance to inhibitors of DNA topoisomerases. Adv. Enzyme Regul. 37:17-26. [DOI] [PubMed] [Google Scholar]
- 3.Capranico, G., and M. Binaschi. 1998. DNA sequence selectivity of topoisomerases and topoisomerase poisons. Biochim. Biophys. Acta 1400:185-194. [DOI] [PubMed] [Google Scholar]
- 4.Capranico, G., G. Giaccone, F. Zunino, S. Garattini, and M. D'Incalci. 1997. DNA topoisomerase II poisons and inhibitors. Cancer Chemother. Biol. Response Modif. 17:114-131. [PubMed] [Google Scholar]
- 5.Capranico, G., F. Zunino, K. W. Kohn, and Y. Pommier. 1990. Sequence-selective topoisomerase II inhibition by anthracycline derivatives in SV40 DNA: relationship with DNA binding affinity and cytotoxicity. Biochemistry 29:562-569. [DOI] [PubMed] [Google Scholar]
- 6.Chatterji, M., S. Unniraman, S. Mahadevan, and V. Nagaraja. 2001. Effect of different classes of inhibitors on DNA gyrase from Mycobacterium smegmatis. J. Antimicrob. Chemother. 48:479-485. [DOI] [PubMed] [Google Scholar]
- 7.Chen, A. Y., and L. F. Liu. 1994. DNA topoisomerases: essential enzymes and lethal targets. Annu. Rev. Pharmacol. Toxicol. 34:191-218. [DOI] [PubMed] [Google Scholar]
- 8.Chen, F. J., and H. J. Lo. 2003. Molecular mechanisms of fluoroquinolone resistance. J. Microbiol. Immunol. Infect. 36:1-9. [PubMed] [Google Scholar]
- 9.Conrad, S., M. Oethinger, K. Kaifel, G. Klotz, R. Marre, and W. V. Kern. 1996. gyrA mutations in high-level fluoroquinolone-resistant clinical isolates of Escherichia coli. J. Antimicrob. Chemother. 38:443-455. [DOI] [PubMed] [Google Scholar]
- 10.Courvalin, P. 1990. Plasmid-mediated 4-quinolone resistance: a real or apparent absence? Antimicrob. Agents Chemother. 34:681-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cozzarelli, N. C., and J. C. Wang (ed.). 1990. DNA topology and its biological effects. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 12.Deibler, R. W., S. Rahmati, and E. L. Zechiedrich. 2001. Topoisomerase IV, alone, unknots DNA in E. coli. Genes Dev. 15:748-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, J., J. Walker, and J. L. Nitiss. 2000. A mutation in yeast topoisomerase II that confers hypersensitivity to multiple classes of topoisomerase II poisons. J. Biol. Chem. 275:7980-7987. [DOI] [PubMed] [Google Scholar]
- 14.Drlica, K. 1999. Refining the fluoroquinolones. ASM News 65:410-415. [Google Scholar]
- 15.Drlica, K., and X. Zhao. 1997. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61:377-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsea, S. H., N. Osheroff, and J. L. Nitiss. 1992. Cytotoxicity of quinolones toward eukaryotic cells. Identification of topoisomerase II as the primary cellular target for the quinolone CP-115,953 in yeast. J. Biol. Chem. 267:13150-13153. [PubMed] [Google Scholar]
- 17.Elsea, S. H., M. Westergaard, D. A. Burden, J. P. Lomenick, and N. Osheroff. 1997. Quinolones share a common interaction domain on topoisomerase II with other DNA cleavage-enhancing antineoplastic drugs. Biochemistry 36:2919-2924. [DOI] [PubMed] [Google Scholar]
- 18.Fass, D., C. E. Bogden, and J. M. Berger. 1999. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat. Struct. Biol. 6:322-326. [DOI] [PubMed] [Google Scholar]
- 19.Ferrero, L., B. Cameron, and J. Crouzet. 1995. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 39:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 21.Fournier, B., X. Zhao, T. Lu, K. Drlica, and D. C. Hooper. 2000. Selective targeting of topoisomerase IV and DNA gyrase in Staphylococcus aureus: different patterns of quinolone-induced inhibition of DNA synthesis. Antimicrob. Agents Chemother. 44:2160-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gellert, M., L. M. Fisher, and M. H. O'Dea. 1979. DNA gyrase: purification and catalytic properties of a fragment of gyrase B protein. Proc. Natl. Acad. Sci. USA 76:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Babitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heddle, J., and A. Maxwell. 2002. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob. Agents Chemother. 46:1805-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heisig, P. 1996. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 40:879-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heisig, P., B. Kratz, E. Halle, Y. Graser, M. Altwegg, W. Rabsch, and J. P. Faber. 1995. Identification of DNA gyrase A mutations in ciprofloxacin-resistant isolates of Salmonella typhimurium from men and cattle in Germany. Microb. Drug Resist. 1:211-218. [DOI] [PubMed] [Google Scholar]
- 27.Heisig, P., and B. Wiedemann. 2001. Action and reaction. Actions and resistance mechanisms of quinolone. Pharm. Unserer Zeit. 30:382-393. [DOI] [PubMed] [Google Scholar]
- 28.Heisig, P., and B. Wiedemann. 1991. Use of a broad-host-range gyrA plasmid for genetic characterization of fluoroquinolone-resistant gram-negative bacteria. Antimicrob. Agents Chemother. 35:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooper, D. C. 1998. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin. Infect. Dis. 27(Suppl. 1):S54-S63. [DOI] [PubMed] [Google Scholar]
- 30.Hooper, D. C. 1998. Clinical applications of quinolones. Biochim. Biophys. Acta 1400:45-61. [DOI] [PubMed] [Google Scholar]
- 31.Hsiung, Y., S. H. Elsea, N. Osheroff, and J. L. Nitiss. 1995. A mutation in yeast TOP2 homologous to a quinolone-resistant mutation in bacteria. Mutation of the amino acid homologous to Ser83 of Escherichia coli gyrA alters sensitivity to eukaryotic topoisomerase inhibitors. J. Biol. Chem. 270:20359-20364. [DOI] [PubMed] [Google Scholar]
- 32.Kato, J., Y. Nishimura, R. Imamura, H. Niki, S. Hiraga, and H. Suzuki. 1990. New topoisomerase essential for chromosome segregation in E. coli. Cell 63:393-404. [DOI] [PubMed] [Google Scholar]
- 33.Khodursky, A. B., and N. R. Cozzarelli. 1998. The mechanism of inhibition of topoisomerase IV by quinolone antibacterials. J. Biol. Chem. 273:27668-27677. [DOI] [PubMed] [Google Scholar]
- 34.Khodursky, A. B., E. L. Zechiedrich, and N. R. Cozzarelli. 1995. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci. USA 92:11801-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leteurtre, F., D. L. Sackett, J. Madalengoitia, G. Kohlhagen, T. MacDonald, E. Hamel, K. D. Paull, and Y. Pommier. 1995. Azatoxin derivatives with potent and selective action on topoisomerase II. Biochem. Pharmacol. 49:1283-1290. [DOI] [PubMed] [Google Scholar]
- 36.Liu, L. 1994. DNA Topoisomerases: topoisomerase-targeting drugs. Academic Press, Inc., New York, N.Y.
- 37.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez, J. L., A. Alonso, J. M. Gomez-Gomez, and F. Baquero. 1998. Quinolone resistance by mutations in chromosomal gyrase genes. Just the tip of the iceberg? J. Antimicrob. Chemother. 42:683-688. [DOI] [PubMed] [Google Scholar]
- 39.Maxwell, A. 1992. The molecular basis of quinolone action. J. Antimicrob. Chemother. 30:409-414. [DOI] [PubMed] [Google Scholar]
- 40.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 41.Ng, E. Y., M. Trucksis, and D. C. Hooper. 1996. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nitiss, J. L. 1998. Investigating the biological functions of DNA topoisomerases in eukaryotic cells. Biochim. Biophys. Acta 1400:63-81. [DOI] [PubMed] [Google Scholar]
- 43.Nitiss, J. L., and W. T. Beck. 1996. Antitopoisomerase drug action and resistance. Eur. J. Cancer 32A:958-966. [DOI] [PubMed] [Google Scholar]
- 44.Oh, H., J. Stenhoff, S. Jalal, and B. Wretlind. 2003. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb. Drug Resist. 9:323-328. [DOI] [PubMed] [Google Scholar]
- 45.Pommier, Y., G. Capranico, A. Orr, and K. W. Kohn. 1991. Local base sequence preferences for DNA cleavage by mammalian topoisomerase II in the presence of amsacrine or teniposide. Nucleic Acids Res. 19:5973-5980. (Erratum, 19:7003.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pommier, Y., F. Goldwasser, and D. Strumberg. 2001. Topoisomerase II inhibitors: epipodophyllotoxins, acridines, ellipticines, and bisdioxopiperazines, p. 538. In B. A. Chabner and D. L. Longo (ed.), Cancer chemotherapy and biotherapy: principles and practice, 3rd ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 47.Reece, R. J., and A. Maxwell. 1991. DNA gyrase: structure and function. Crit. Rev. Biochem. Mol. Biol. 26:335-375. [DOI] [PubMed] [Google Scholar]
- 48.Ricci, V., M. L. Peterson, J. C. Rotschafer, H. Wexler, and L. J. Piddock. 2004. Role of topoisomerase mutations and efflux in fluoroquinolone resistance of Bacteroides fragilis clinical isolates and laboratory mutants. Antimicrob. Agents Chemother. 48:1344-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanders, C. C. 1990. Microbiology of fluoroquinolones, p. 1-28. In W. E. Sanders, Jr., and C. C. Sanders (ed.), Fluoroquinolones in the treatment of infectious diseases. Physicians and Scientists Publishing, Glenview, Ill.
- 50.Sanders, C. C., and W. E. Sanders, Jr. 1995. Resistance to antibacterial agents, p. 15-23. In D. L. Jungkind, J. E. Mortensen, H. F. Fraimow, and G. D. Calandra (ed.), Antimicrobial resistance: a crisis in health care. Plenum Press, New York, N.Y.
- 51.Strumberg, D., J. L. Nitiss, J. Dong, K. W. Kohn, and Y. Pommier. 1999. Molecular analysis of yeast and human type II topoisomerases. Enzyme-DNA and drug interactions. J. Biol. Chem. 274:28246-28255. [DOI] [PubMed] [Google Scholar]
- 52.Strumberg, D., J. L. Nitiss, J. Dong, J. Walker, M. C. Nicklaus, K. W. Kohn, J. G. Heddle, A. Maxwell, S. Seeber, and Y. Pommier. 2002. Importance of the fourth alpha-helix within the CAP homology domain of type II topoisomerase for DNA cleavage site recognition and quinolone action. Antimicrob. Agents Chemother. 46:2735-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strumberg, D., J. L. Nitiss, A. Rose, M. C. Nicklaus, and Y. Pommier. 1999. Mutation of a conserved serine residue in a quinolone-resistant type II topoisomerase alters the enzyme-DNA and drug interactions. J. Biol. Chem. 274:7292-7301. [DOI] [PubMed] [Google Scholar]
- 54.Tanizawa, A., K. W. Kohn, and Y. Pommier. 1993. Induction of cleavage in topoisomerase I c-DNA by topoisomerase I enzymes from calf thymus and wheat germ in the presence and absence of camptothecin. Nucleic Acids Res. 21:5157-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, J. C. 1996. DNA topoisomerases. Annu. Rev. Biochem. 65:635-692. [DOI] [PubMed] [Google Scholar]
- 56.Wang, J. C. 1998. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 31:107-144. [DOI] [PubMed] [Google Scholar]
- 57.Willmott, C. J., and A. Maxwell. 1993. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolones to the gyrase-DNA complex. Antimicrob. Agents Chemother. 37:126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, S., S. R. Clayton, and E. L. Zechiedrich. 2003. Relative contributions of the AcrAB, MdfA and NorE efflux pumps to quinolone resistance in Escherichia coli. J. Antimicrob. Chemother. 51:545-556. [DOI] [PubMed] [Google Scholar]
- 59.Yoshida, H., M. Bogaki, M. Nakamura, and S. Nakamura. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida, H., M. Bogaki, M. Nakamura, L. M. Yamanaka, and S. Nakamura. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zechiedrich, E. L., and N. R. Cozzarelli. 1995. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 9:2859-2869. [DOI] [PubMed] [Google Scholar]
- 62.Zechiedrich, E. L., A. B. Khodursky, and N. R. Cozzarelli. 1997. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 11:2580-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]