Abstract
Background
Lake Malawi cichlids represent one of a growing number of vertebrate models used to uncover the genetic and developmental basis of trait diversity. Rapid evolutionary radiation has resulted in species that share similar genomes but differ markedly in phenotypes including brains and behavior, nuptial coloration and the craniofacial skeleton. Research has begun to identify the genes, as well as the molecular and developmental pathways that underlie trait divergence.
Results
We assemble a compendium of gene expression for Lake Malawi cichlids, across pharyngula (the phylotypic stage) and larval stages of development, encompassing hundreds of gene transcripts. We chart patterns of expression in Bone morphogenetic protein (BMP), Fibroblast growth factor (FGF), Hedgehog (Hh), Notch and Wingless (Wnt) signaling pathways, as well as genes involved in neurogenesis, calcium and endocrine signaling, stem cell biology, and numerous homeobox (Hox) factors—in three planes using whole-mount in situ hybridization. Because of low sequence divergence across the Malawi cichlid assemblage, the probes we employ are broadly applicable in hundreds of species. We tabulate gene expression across general tissue domains, and highlight examples of unexpected expression patterns.
Conclusions
On the heels of recently published genomes, this compendium of developmental gene expression in Lake Malawi cichlids provides a valuable resource for those interested in the relationship between evolution and development.
Keywords: Cichlid fishes, Evolution of gene expression, Lake Malawi, Developmental pathways
Background
Comparative gene expression is a hallmark of the evolution and development research program [1]. This is particularly the case among closely related vertebrate species, like hominids [2], beach mice [3], cavefishes [4], stickleback [5], and cichlid fishes [6]. In these examples and many others, diversity in key traits evolves via spatial, temporal and/or quantitative variation in gene expression. Despite the importance of changes in gene expression to the evolution of closely related species, comprehensive surveys of spatial expression patterns are typically confined to laboratory models (e.g., zebrafish, [7]).
Consequently, we have produced a compendium of spatial gene expression across early development, in Lake Malawi cichlid fishes. This resource should find broad applicability for three reasons. First, genomic surveys demonstrate extreme genetic similarity among Malawi species [8, 9], and other lineages from East Africa [10]. This means the probes we develop will be useful across hundreds of African cichlid species. Second, Malawi cichlids in particular have been used to study the genetic and developmental basis of key vertebrate phenotypes, like nuptial coloration [11–13], the cranio-dental skeleton [14–18], and the brain [19]. The developmental pathways we highlight here are relevant to continued study of these important evolutionary phenotypes. Third, recently developed means to manipulate cichlid genomes and development (e.g., transgenics, treatment with small molecules, genome editing: [17, 20, 21] are informed by observations of gene expression in time and space. We use whole mount in situ hybridization (ISH) to document spatial expression patterns for approximately 160 genes, across 12 major categories. We tabulate expression domains for each gene at pharyngula and larval stages, in three planes of view (Fig. 1). This compendium of developmental gene expression should be a valuable resource for biologists interested in the relationship between development and evolution.
Fig. 1.

Schematic of planes: a frontal, b lateral, c dorsal planes and domains of expression color coded according to legend
Methods
Fish husbandry
Lake Malawi cichlids used for this study included Metriaclima zebra [MZ] and Petrotilapia chitimba “thickbar” [PC]. These species were used owing to their availability and the fact that they belong to the ‘mbuna’ rock dwelling lineage. While Malawi cichlid species share qualitative expression domains across species, those from different ecotypes (mbuna versus ‘non-mbuna’) may exhibit heterochronic and quantitative differences in expression [6, 17]. Adult cichlids were maintained in re-circulating aquarium systems at 28 °C (Georgia Institute of Technology). Fertilized embryos were removed from the mouths of brooding females and staged in days post-fertilization (dpf), according to the Nile tilapia developmental series [22]. Embryos were raised to 4dpf or 6dpf and euthanized with sodium bicarbonate buffered anesthetic MS-222, before fixation in 4% paraformaldehyde. Pre-hatching embryos at 4dpf were dechorionated using fine forceps to achieve proper fixation and reagent penetration.
Primer and probe design
Primers were designed using recently assembled and annotated tilapia and MZ genomes [10] (accession numbers KT906433-KT906561) as well as partial genome assemblies [9] (accession numbers KC633830- KC633846, EU867210-EU867217, KT851376-- KT851399) were used to amplify cichlid cDNA. Amplified DNA was inserted into the pGEM-T Easy vector system (Promega) and transformed into JM109 competent cells (Promega). Upon amplification and purification (Qiagen, Plasmid Maxi Kit), riboprobes were prepared using the Promega Riboprobe System Sp6/T7 kit. There is minimal sequence divergence between Malawi cichlid species; the average nucleotide diversity is 0.28%, less than that among laboratory strains of zebrafish [9]. Cloned plasmid insert sequences used for probe generation have been deposited in GenBank (Accession numbers are shown in Tables 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11).
Table 1.
Expression data for the TGF-β/BMP pathway genes at pharyngeal (blue) and larval (orange) stages
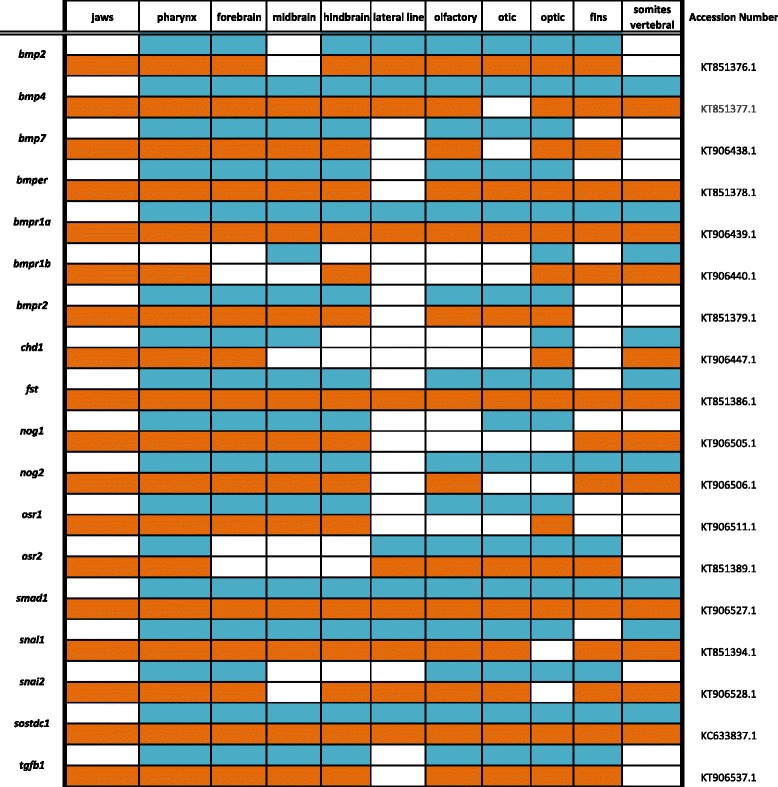
Table 2.
Expression data for FGF pathway genes at pharyngeal (blue) and larval (orange) stages
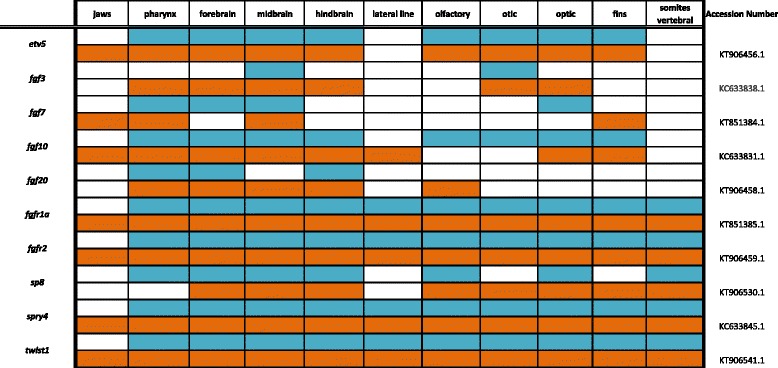
Table 3.
Expression data for Forkhead Box family genes at pharyngeal (blue) and larval (orange) stages
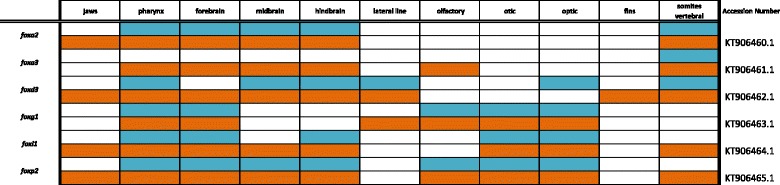
Table 4.
Expression data for Hedgehog pathway genes at pharyngeal (blue) and larval (orange) stages
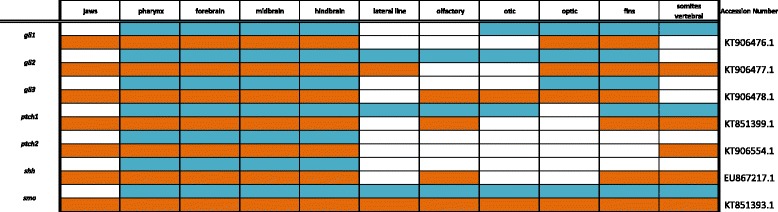
Table 5.
Expression data for Homeobox pathway genes at pharyngeal (blue) and larval (orange) stages
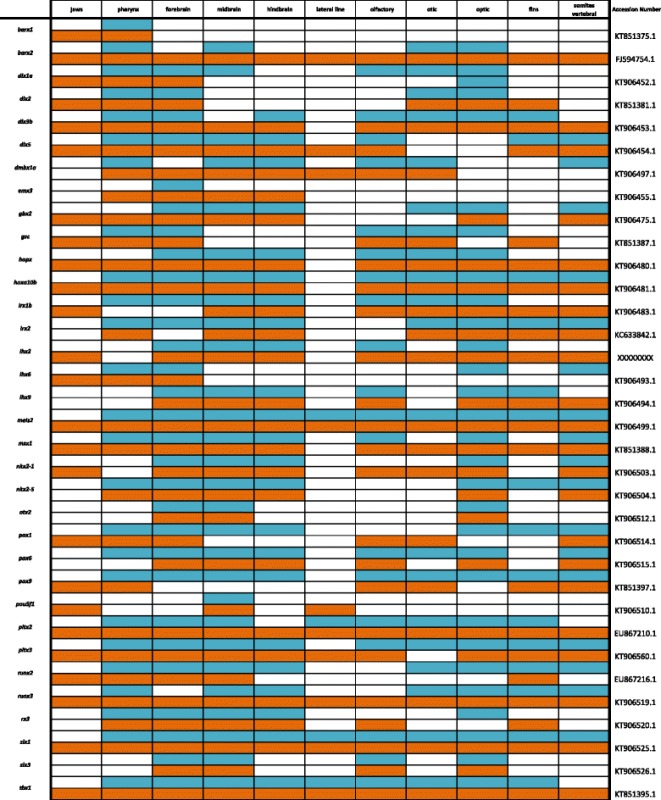
Table 6.
Expression data for Calcium, Endocrine, and Insulin signaling factors at pharyngeal (blue) and larval (orange) stages
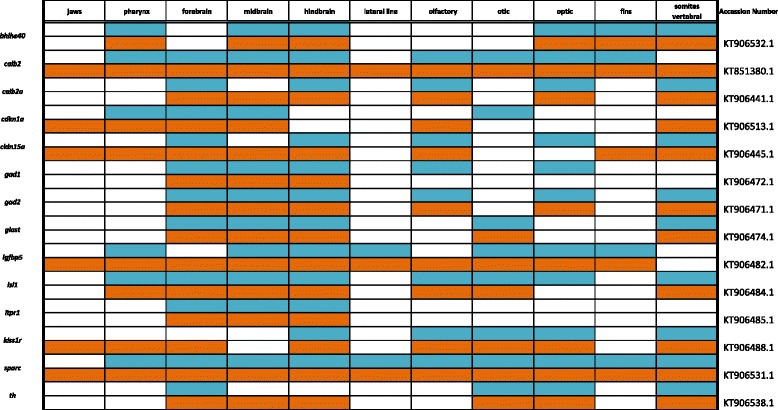
Table 7.
Expression data for Mitogens, Stem, and Tumor Suppressor factors at pharyngeal (blue) and larval (orange) stages

Table 8.
Expression data for Notch pathway genes at pharyngeal (blue) and larval (orange) stages
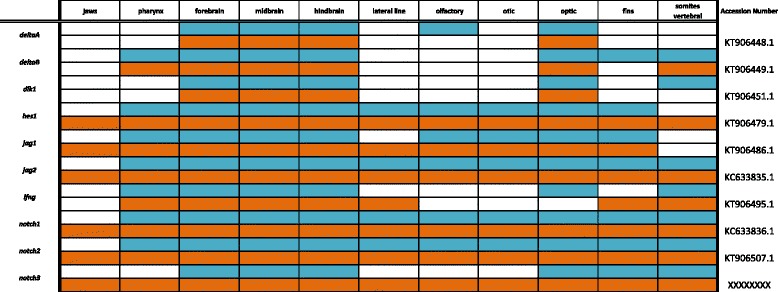
Table 9.
Expression data for brain development and neurogenesis factors at pharyngeal (blue) and larval (orange) stages
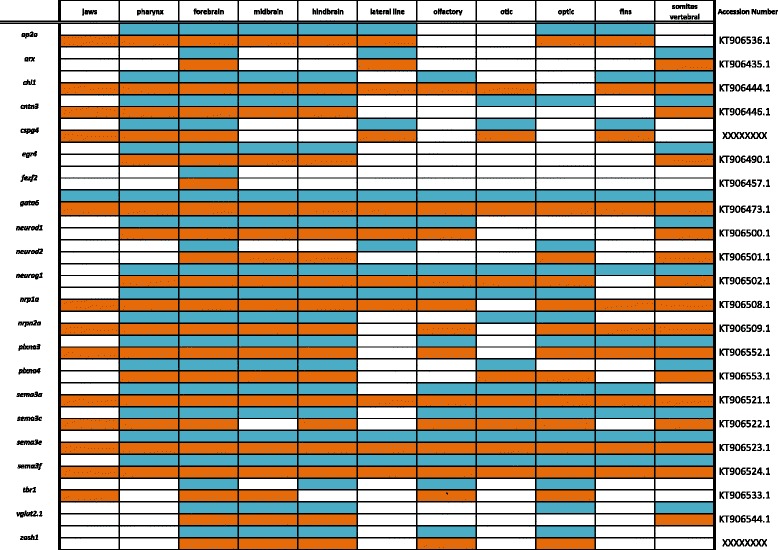
Table 10.
Expression data for brain development and neurogenesis factors at pharyngeal (blue) and larval (orange) stages

Table 11.
Expression data for developmental genes at pharyngeal (blue) and larval (orange) stages

In situ hybridization
Specimens for ISH were fixed a minimum of 48 h in 4% paraformaldehyde at 4 °C and then dehydrated into a graded series of methanol for further fixation and storage at −20 °C O/N. Embryos were rehydrated and permeabilized in 10 μg/mL proteinase K for one hour. They were then refixed in 4% PFA and incubated in prehybridization solution at 70 °C. Embryos were incubated overnight at 70 °C in digoxigenin-labeled antisense riboprobes. The following day, embryos were washed through a graded series of saline-sodium citrate buffer solutions and blocked with blocking solution (5% blocking buffer, 5% goat serum in MABT). Embryos were then hybridized with 1:3000 anti-digoxigenin-AP FAB fragments in blocking buffer overnight at 4 °C. Excess antibody was removed by washing, and color reaction with NBT/BCIP was performed on the AP-conjugated anti-dig antibodies. Gene expression was imaged in whole mount, using a LeicaDFC295 compound light microscope.
Results and discussion
Bone morphogenetic protein and transforming growth factor beta pathway
The transforming growth factor beta (TGF-β) superfamily is a class of cytokines organized into TGF-βs, bone morphogenetic proteins (BMPs), and activin/inhibins that bind to Type I and II serine/threonine kinase receptors [23]. Upon ligand activation, type II receptors phosphorylate type I receptors, leading to SMAD protein activation and ultimately gene regulation. TGF-β/BMPs play a major role in almost every aspect of vertebrate biology, from gastrulation and organization of the body plan, to the genesis of almost every organ, to renewal and adult tissue maintenance [23, 24]. Inasmuch, mutations in the TGF-β superfamily and its regulators have been demonstrated as causative for the evolution of major adaptations.
BMPs are believed to control multiple aspects of cichlid jaw shape and function [25, 26], are in part responsible for evolutionary novelty in beak shape of Darwin’s finches [27], and have a direct dose-dependent effect on the craniofacial skeleton when transgenically titrated in mice [28]. All of the BMP pathway factors we include are expressed in the jaw once it has formed in the larval stage, and many are also expressed in the pharynx, as indicated in Table 1. bmp2 and bmp4 pattern, generate, shape, and regenerate teeth in mice, squamates, and cichlids [29–35] while a large-effect QTL containing bmp6 has been reported for a doubling of pharyngeal tooth number in stickleback [36]. bmper may regulate tooth number in Malawi cichlids [17].
In mice, bmp2 and bmp7 regulate dorso-ventral patterning of the brain and in chicken, undifferentiated neural ectoderm has been induced to express dorsal-specific markers by addition of the protein products of these genes [37]. In Fig. 2 we observe expression of ligands bmp2, bmp4 and bmp7, as well as endothelial regulator bmper and receptors bmpr1a and bmpr2, along the dorsomedial telencephalon and in distinct patterns in the forebrain, similar to expression reported in mouse [38]. After mutation of bmpr1a in mouse the choroid plexus fails to form from the dorsal telencephalon, demonstrating a role of this receptor in forebrain specification [39]. While bmpr1a is expressed in all three regions of the brain, bmpr1b is localized to the developing cerebellum, preoptic region, and eyes at 4dpf; and at 6dpf is seen in the eyes and somites. chd1 is expressed in the optic nerve, pharynx, notochord, and gut.
Fig. 2.
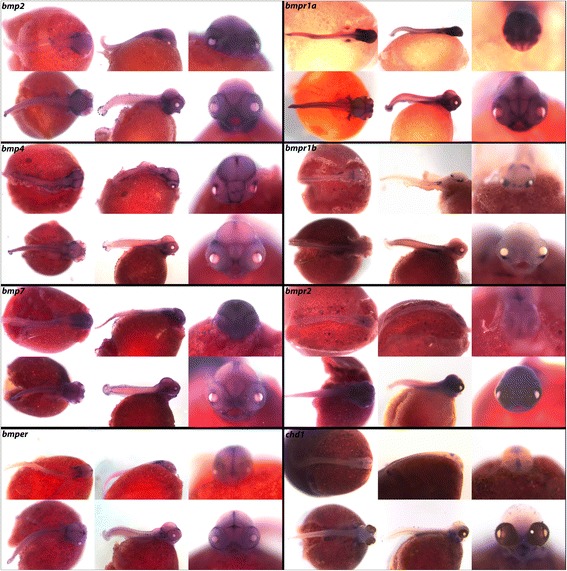
Expression of genes from the TGF-β/BMP pathway at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
In Fig. 3, BMP inhibitor fst is heavily expressed throughout the brain at 4dpf, and by 6dpf this expression sharpens. Inhibitors nog1 and nog2 are seen in the vertebrae and brain, with nog1 diffusely throughout the brain and exhibiting localized expression in the hindbrain and somites at 6dpf. nog2 demonstrates more restricted areas of expression in the preoptic region and lens at 4dpf, and additional expression in the developing dental placodes at 6dpf. Transcription factor osr1 is expressed in the optic tectum and hindbrain at both stages, and osr2 is heavily expressed in the gut and behind the eyes at 4dpf and in the retinae at 6dpf.
Fig. 3.

Expression of genes from the TGF-β/BMP pathway at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
Tumor-suppressor smad1 is phosphorylated in response to BMP pathway activation, and regulates transcription. We observe smad1 throughout the brain, fins, eyes, somites/vertebrae, jaw, and pharynx. Transcription factors snai1 and snai2 exhibit distinct expression patterns. snai1 is in all three brain regions, notably along the longitudinal fissure at 6dpf, as well as the vertebrae/somites. snai2 appears around the eyes pretectum, and pectoral fins, along with heavy expression in the pharyngeal arches and somites. We observe sostdc1 in the pharynx and cranial lateral line, and tgfb1 around the eyes, retinae, pharynx, and fins.
Fibroblast growth factor pathway
Much like the TGF-β/BMP pathway, the Fibroblast growth factor (FGF) pathway plays a part in eukaryotic development and homeostasis across ontogeny, and is particularly important for organogenesis and the generation of evolutionary novelty. FGFs and FGF receptors (FGFRs) are part of a larger family of Tyrosine Kinases and high-affinity cell surface receptors known as Receptor-Tyrosine Kinases (RTKs) that function through activation of Ras/MAP kinase and phospholipase-C gamma pathways [40]. Conserved across all metazoans, FGFs have gained redundancy in higher vertebrate genomes, presumably for the formation of complex traits. In Amphioxus, FGF’s coordinate segment reduction, perhaps permissive for the evolution of the vertebrate head [41]. In frog, FGFs work synergistically with BMPs to induce neurulation [42]. By contrast, FGFs have long been recognized as competitors of the BMP pathway in patterning limb outgrowth in mammals [43], and formation and regeneration of fish fins [44]. Genetic ablation of the FGF antagonists Spry4 and Spry2 produces mice with tusks for incisors [45], and heterochrony of fgf8 expression in the blind cavefish neural plate contributes to defective retinal morphogenesis [4]. It is apparent that FGFs are one of the key pathways exploited by nature during animal evolution [46].
In Fig. 4 and Table 2, we report expression of ligands fgf3, fgf7, fgf10 and fgf20; receptors fgfr1a and fgfr2; transcription factors etv5, sp8 and twist1; and repressor spry4. Expression of etv5 can be seen in the hindbrain, eyes, jaws, fins and pharynx. The four ligands included have unique expression patterns with fgf3 most evident in the isthmus (midbrain-hindbrain boundary, MHB), fgf7 in the jaw, fgf10 in the midbrain and around the eyes, and fgf20 in the hindbrain, olfactory placodes, and pharynx. Receptors fgfr1a and fgfr2 are both heavily expressed in the central nervous system, along the longitudinal fissure of the brain, and in the pharynx and jaw.
Fig. 4.
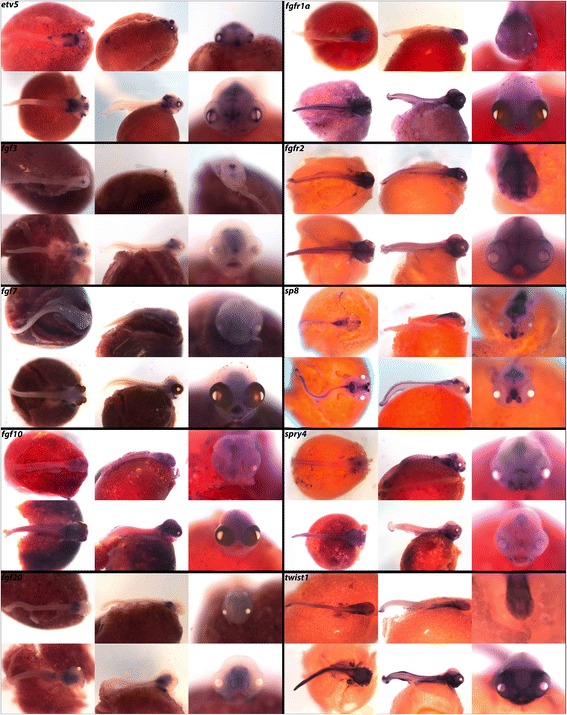
Expression of genes from the FGF pathway at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
Zinc-finger transcription factor sp8 acts under the regulation of fgf10 and Wnt/β-catenin, and has been shown to regulate fgf8 for limb formation during chicken development [47]. We observe sp8 along the spinal region and throughout the brain and olfactory placodes in a pattern similar to that seen in zebrafish [48]. Repressor spry4 and transcription factor twist1 are both expressed along the somites, as well as in the pharynx, fins, eyes, and jaws.
Forkhead box pathway
The Forkhead Box transcription factors share an evolutionary conserved “forkhead” or “winged-helix” 100 amino-acid DNA binding domain. The moniker for the Fox family was coined when the first homolog forkhead (fkh) was identified in Drosophila, with mutant flies exhibiting split heads [49]. Moreover, the helix-turn-helix motif of this domain is comprised of 3 α-helices and two large loops that resemble “wings.” To date, over 100 Fox transcription factors have been identified across eukaryotes and much work has been done to clarify their nomenclature [50]. For example, in humans there are over fifty Fox proteins categorized into 19 subgroups (FOXA to FOXS) [51].
While the characteristic protein domain of the Fox family has remained tightly conserved, individual genes have evolved greatly outside of these domains and have taken on a myriad of highly divergent and specialized functions such as tumor suppression, cell signaling, apoptosis, and DNA repair. Although functionally divergent, redundant roles exist for family members such as FoxA1 and FoxA2 in both lung and liver formation [52, 53]. In Fig. 5, we observe expression of foxa2 in the diencephalon-midbrain boundary (DMB), the oral/pharyngeal cavity and in the developing spinal cord, while foxa3 is expressed in the gut. foxg1 is expressed in the retina and telencephalon, and is differentially expressed in rock- vs. sand-dwelling Malawi cichlids [6]. As in zebrafish, foxi1 is expressed in pharynx, jaw, vertebral elements, and otic placodes [54] and in Malawi cichlids expression strongly resembles that of neural crest marker foxd3.
Fig. 5.
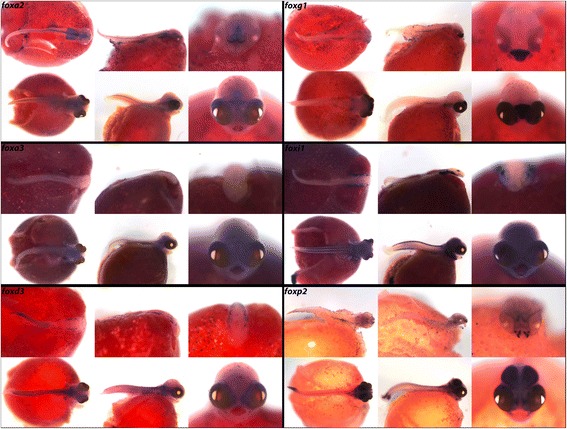
Expression of Forkhead Box genes at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
In vertebrates, FoxP2 has demonstrated roles in vocalization and the ability to learn language. Deletions in FoxP2 result in verbal dyspraxia and a collapse of the communication system at both the neural and muscular levels [55]; furthermore Foxp2 has evolved episodically in hominids [56]. In cichlids, foxp2 is expressed in distinct foci in the thalamus and telencephalon, as well as in the pharyngeal arches where sound is produced [57], in the otic placodes and tectum where sound is received and processed, and in the fins.
Hedgehog pathway
The Hedgehog pathway executes pervasive roles in embryonic development, stem cell renewal, and cancer biology. Hedgehog proteins are a group of soluble morphogens that include Indian Hedgehog, Desert Hedgehog, and perhaps the most well studied ligand in embryology, Sonic Hedgehog (SHH). These morphogens bind to the transmembrane receptor Patched (Ptch), releasing co-receptor Smoothened (Smo) and permitting activation of Hedgehog signaling. The Hh pathway is involved in the specification and morphogenesis of nearly all animal organs [58].
In Fig. 6, we report expression of transcription factors gli1, gli2 and gli3, receptors ptch1, ptch2 and smo, and the ligand shh. Similar to expression seen in zebrafish [59], we find that all of the Hh pathway genes included exhibit t-shaped expression in the ZLI boundary of the diencephalon at both stages, as well as expression in the pharynx and fins (Table 4). We observe heavy expression of gli2 and gli3 in the midbrain and dorsal telencephalon, but expression of gli1 is less prominent. In cichlids, ptch1 is responsible for adaptive variation in jaw shape [60] and function [14]. We see ptch1 and ptch2 expressed in the jaw and throughout the central nervous system and somites, and ptch1 additionally in the olfactory and otic cups. Ligand shh exhibits similar but more restricted patterns of expression in the forebrain, jaw, and somites. Similar to results found in zebrafish [61], smo appears at the midline and somites, as well as in the brain and fins at both stages.
Fig. 6.
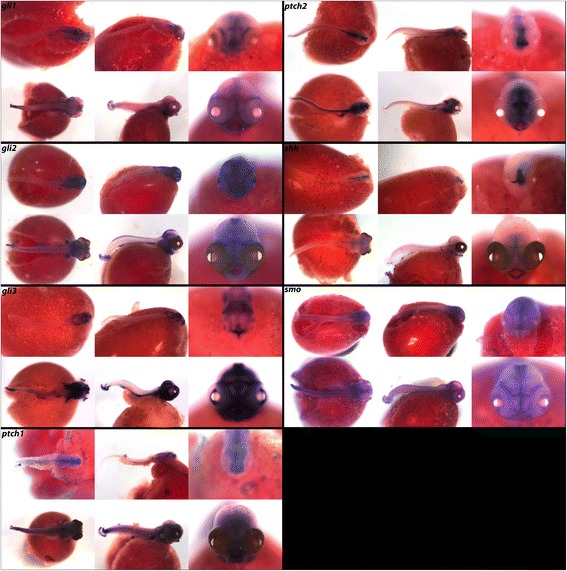
Expression of Hedgehog pathway genes at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
Homeobox pathway
Called the “Rosetta Stone of developmental biology,” the homeobox gene family is best known for its role in organizing the metazoan body plan. Hallmark to this family is the “homeobox,” a conserved homeodomain sequence approximately 60 amino acids in length that binds DNA. With an estimated 300 homeobox genes, comprised of true genes and pseudogenes, Hox transcription factors are often divided into classes (approximately eleven) and subclasses that represent their general developmental functions [62]. Belonging to the Antennapedia gene of Drosophila (ANTP) class, we report expression in cichlids of barx1 and barx2 of the NK-like (NKL) subclass (Fig. 7). barx1 expression has previously been demonstrated in cichlid pharyngeal teeth [63], and here we describe expression of barx1 and 2 in the oral jaws, pharynx, and the gut.
Fig. 7.
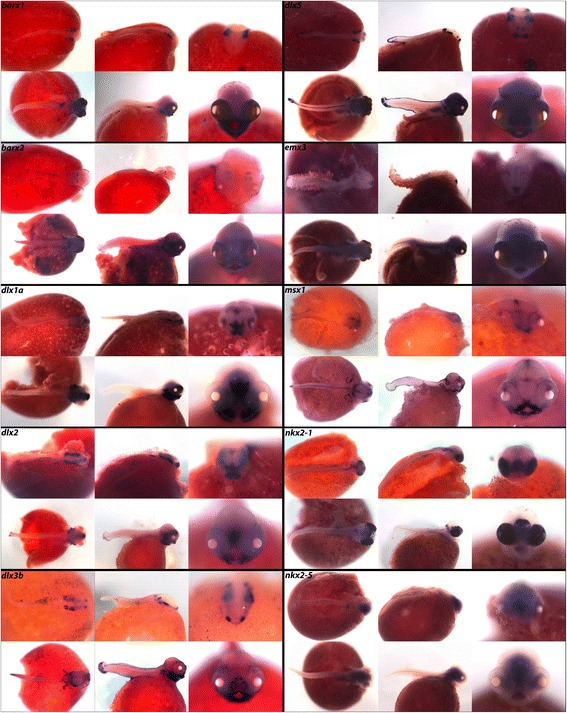
Expression of ANTP class Homeobox genes at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
dlx1a, dlx2, dlx3b, dlx5, emx3, msx1, nkx2-1 and nkx2-5 belong to the ANTP class and members have been well described as mediators of zebrafish jaw [64] and lamprey pharynx development [65], as well as important for fish brain development [66]. In Fig. 7 we note strikingly similar expression of dlx1a and dlx2, and expression of Dlx and Msx genes in the jaws and pharynx. The Dlx and Nkx genes are expressed in the ventral regions of the mid and/or forebrain. emx3 is expressed in the dorsal telencephalon early, and at later stages is seen throughout the brain and trunk.
We also present expression of Paired gene of Drosophila PRD class factors dmbx1a, gsc, hopx, otx2, pax1, pax6, pax9, pitx2, pitx3 and rx3. Many of the members of the PRD class are known to be important for eye development [67–69], and we note expression of each of these factors in either retinal or lens development at the pharyngula or larval stages. dmbx1a exhibits expression in the midbrain at 4dpf, and at 6dpf expression is visible throughout the optic tectum and hindbrain. gsc and hopx are both expressed in the pharynx and eye structures, while otx2 exhibits heavy expression in the fore-, midbrain and eyes. In Fig. 8 we note expression of the Pax genes in the somites, and expression of pax1 and pax9 in the pharynx and jaw. Paired-Like Homeodomain factors pitx2 and pitx3 demonstrate expression in the eyes, brains, and somites, while rx3 is localized to the presumptive hypothalamus/preoptic region.
Fig. 8.
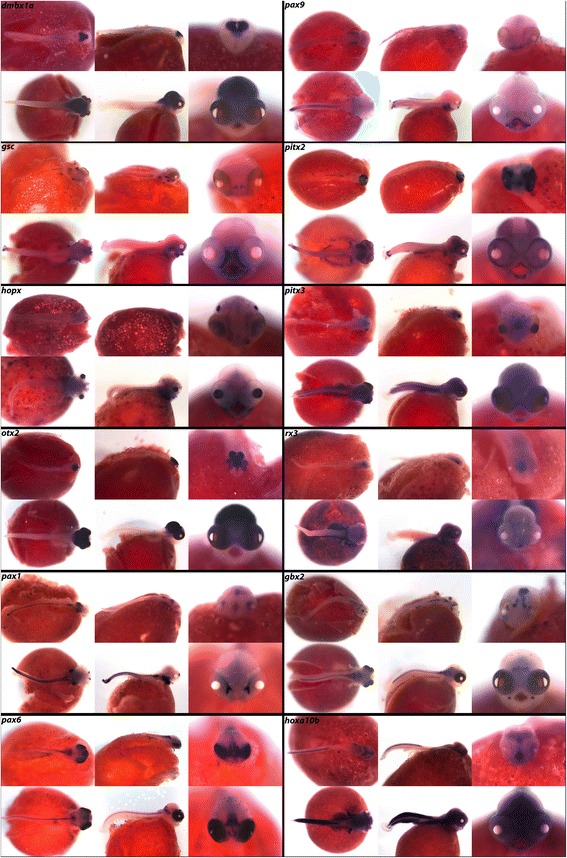
Expression of PRD and HOXL Homeobox genes at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
gbx2 and hoxa10b of the HOXL subclass are expressed in foci of the jaw joint [70] and fins respectively, the latter of which has been described as crucial for proper limb and fin patterning [71].
In the three amino acid loop extension (TALE) superclass we demonstrate expression of irx1b, irx2 and meis2, all three of which are expressed in the eyes and brain. Belonging to the LIM class we present expression of LIM homeobox 2 (lhx2), lhx6 and lhx9. lhx2 and lhx9 exhibit essentially identical expression patterns in the brain, fins, and spinal region, while lhx6 is only expressed in the jaw, pharynx, and preoptic region.
In the POU class, named for Pit, Oct, and Unc transcription factors (POU class), we document expression of pou5f1, and in the SINE class we show expression of six1 and six3. In Fig. 9 we observe expression of six1 in the brain, eyes, somites, and fins, while six3 is expressed only in the diencephalon, telencephalon, nasal placodes and eyes. Finally, we report expression of runx2 and runx3 in the jaws and pharynx, and transcription factor tbx1 throughout the brain and in the fins.
Fig. 9.

Expression of Homeobox genes at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
Calcium, endocrine, and insulin signaling
While proteins such as insulin and regulators of calcium are essential for maintaining endocrine homeostasis in adult animals, their roles in embryogenesis are pervasive, but not well known. Entire families of signaling proteins exist to precisely coordinate cellular communication and resulting developmental differentiation, often in the same places where they will signal later in ontogeny. One example is calcium signaling, which is essential for proper odorant detection and olfaction [72]. In Fig. 10, we note expression of calcium signaling regulators calb2, calb2a, cldn15a, kiss1r, and sparc in the olfactory placodes and resulting nasal epithelium. We also observe expression of calb2 in the cephalic lateral line, pharynx, gut, and somites, and calb2a in the hindbrain, as well as in the spinal region. Tight junction factor cldn15a is expressed in the three brain regions, pharynx, fins and jaw, while itpr1 expression is restricted to the forebrain and cerebellum (Table 6). Receptor kiss1r exhibits notable expression in the eyes and hindbrain at 4dpf, and by 6dpf expression also appears in the jaw and pharynx. Extracellular matrix factor sparc is expressed generally across the integument and along the cephalic lateral line and spinal region.
Fig. 10.
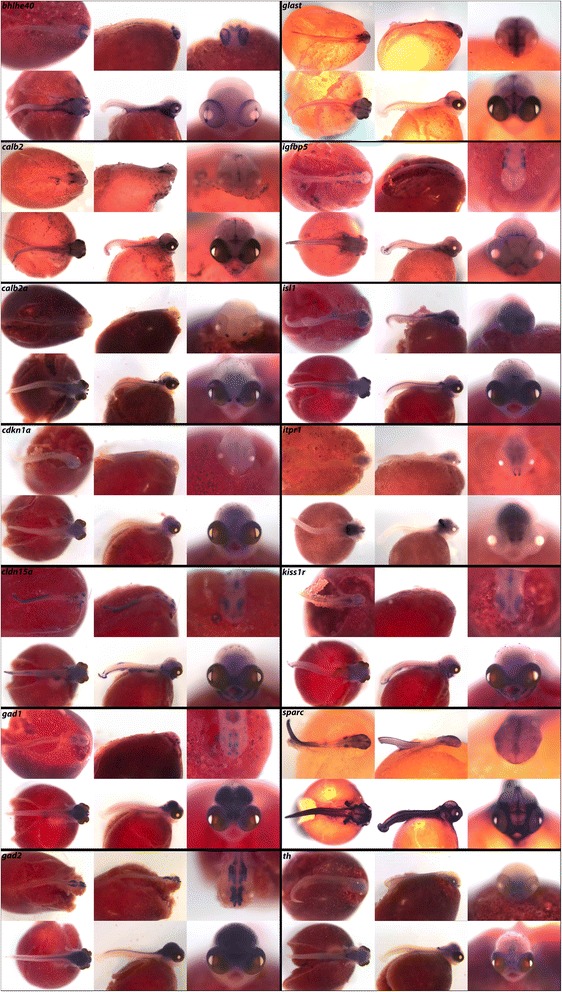
Expression of Calcium, Endocrine, and Insulin signaling factors at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientation
As another example, gad1 and gad2 (also known as gad67 and gad65, respectively) encode enzymes for the production of the neurotransmitter GABA and have known roles in schizophrenia and Parkinson’s disease. As in zebrafish [73] and other organisms, we note expression of gad1 and gad2 throughout the brain early in pharyngeal and larval stages of development. Plasma membrane transporter glast is expressed in cephalic lateral line placodes, where ion exchange will mediate sensory signaling later in the fully functional organ. In the insulin pathway, we report expression of igfbp5 in lateral line and all brachial arches. isl1, or Insulin gene enhancer protein, binds to insulin enhancer sites to regulate insulin gene expression and has known roles in diabetic disease. It is commonly used as a marker of pancreatic cells early and late in zebrafish ontogeny [74] and we show is expressed similarly in cichlids, as well as in forebrain neuronal subsets. Factors involved in endocrine signaling, such as bhlhe40, cdkn1a, and th, are all diffusely expressed in the brain. bhlhe40 exhibits notable expression in the eyes at both stages, and additional expression in the pharynx and somites at 6dpf.
Mitogens, stem cell factors and tumor suppressors
The biomedical world has greatly invested in understanding the processes of cellular renewal and division because of implications in regenerative medicine and cancer. Despite this focus, little attention has been paid to embryo-wide expression patterns of the genes involved (Fig. 11). bmi1, an epithelial stem cell marker in intestinal tissues, along with lgr5 [75], is expressed along the lateral line and in the brain, eyes and fins (see Fig. 11 for expression of lgr4 and lgr6). fut4 is a reported mitogenic factor involved in tumor suppression [76] expressed in cichlid brain structures. Arsenate resistance protein, srrt, has been shown to promote self-renewal of mouse neural stem cells by regulating sox2 expression [77]. We observe expression of srrt in the hindbrain and eyes.
Fig. 11.

Expression of WNT pathway genes at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
Mesenchymal stem cells (mSCs) can be difficult to define due their loose spatial arrangement and degrees of potency. We report expression of celsr1, mcam, pdgf, and vim, which have recently been hypothesized to maintain mSCs [78, 79] in structures including brain, eyes, lateral line, and fins.
Crucial to the dichotomy of stem cell potency is the genetic environment that houses these cells, known as the niche. For instance, a set of key genes known as the Yamanaka factors, cmyc, klf4, oct4, and sox2 are important for maintaining pluripotent stem cells (PSCs), and through retroviral induction can transfate mouse fibroblasts into induced PSCs (iPSCs) [80]. We have cloned oct4, reported as pou5f1 in the Hox panel (Fig. 9). Similar to reports in zebrafish [81] we see little whole-mount expression of oct4 past neurulation, presumably because of its defined roles in PSC maintenance. However, we report expression of klf4, noted in lateral line, fins, and brain, as well as sox2, noted even at later larval stages in adult organs capable of self-renewal, including teeth, taste buds, and the cephalic lateral line. The ability of sox2 to persist and localize to epithelial stem cell (eSC) niches has been noted before [82]. sox2 has been reported as an eSC marker in a host of adult organs [83, 84], along with bmi1 and lgr5 [75] in the intestine, and tumor suppressor lrig1 as a master regulator of eSCs [85]. We observe lrig1 throughout the brain, spinal region, fins, and eyes. Finally, we report expression of neural crest stem factor sox10 in the pharynx and somites, and mitogenic factor tp63 in the jaw, pharynx, CNS, cephalic lateral line, and fins.
Notch pathway
The intercellular Notch signaling cascade is a highly conserved pathway involved in animal cell specification and proliferation [86]. Notch signaling exhibits versatility through a gamut of posttranslational modifications that alter receptor response to ligand. Notch activation occurs primarily by juxtacrine signaling from Delta, Serrate, and Jagged class ligands, which bind the Notch receptor extracellular domain of an adjacent cell. This binding causes proteolytic cleavage of a cytosolic domain to enable it to act as a transcription factor. The Notch pathway is of particular interest in axial patterning during embryogenesis because of this characteristic signal transduction between neighboring cells. In vertebrate models, including chicken [87], and mouse [88], temporal regulation of Notch in the pre-somitic mesoderm plays an important role in segmentation. In zebrafish, segmentation can be restored in Notch-deficient embryos via delivery of artificial pulses of Notch [89]. In cichlids, the Notch pathway is involved in patterning and regeneration of teeth [32], and in the renewing mouse incisor Notch has a role in maintaining the stem niche [90].
In Fig. 12 we document expression of deltaA, deltaB, dlk1, jag1, and jag2 ligands, Notch inhibitor lnfg, transcription factor hes1, and notch1, notch2, and notch3 receptors. As indicated in Table 8, we observe deltaA throughout the brain at 4dpf, with notable expression in the mid- and hindbrain. By 6dpf, expression is concentrated along the central midline of the fore- and midbrain and around the eyes. deltaB exhibits a similar pattern in the brain, but has additional expression in the somites and pharynx at both stages.
Fig. 12.
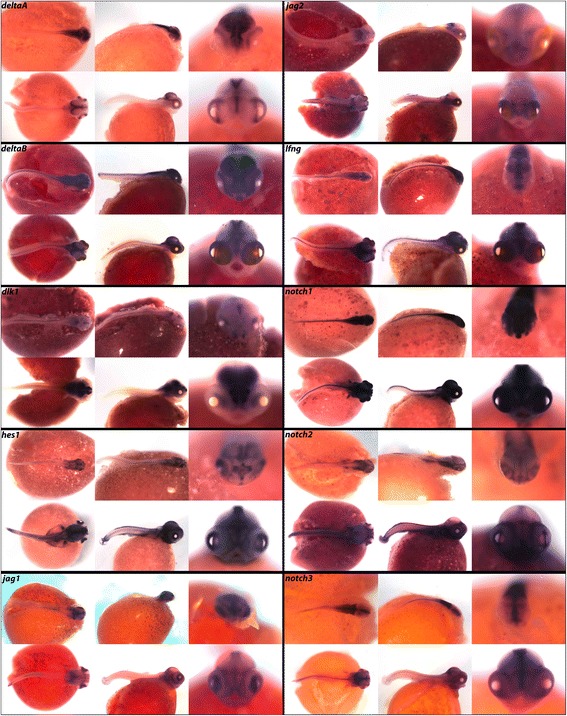
Expression of Notch pathway genes at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
In Drosophila, Notch signaling has been shown to regulate cell fate in the eye by acting at specific ommatidium photoreceptors [91]. We observe expression in the retina (see frontal view) for deltaA, hes1, jag1, notch1 and notch2, as well as expression of deltaB, dlk1, jag2, and notch3 in the eyes. At 4dpf, dlk1 expression is restricted to small areas in all three regions of the brain and dorsal side of the eyes, and by 6dpf this expression has spread to the telencephalon, optic tectum, and hindbrain. hes1 expression at 4dpf is seen in the head and lateral line, and at 6dpf expression is also evident in the vertebral somites and jaw.
Jagged1 is important for endothelial tissue development, and has been correlated with human congenital diseases of the heart [92]. We observe expression of jag1 and jag2 throughout the brain and fins at both developmental stages, and jag2 additionally in the somites and jaw. lnfg is expressed in the brain and eyes, with heavy expression along the central midline. We also observe lnfg in the somites, where it is critical for somite segmentation according to studies performed in mouse [93]. Notch receptors notch1, notch2 and notch3 all exhibit expression along the center midline of the brain and in the jaw, somites, pharynx, and lateral line.
Brain development and neurogenesis
The formation of the brain and nervous system is highly conserved and requires the integration of many, often competing, molecular signals. Cichlid brains evolve diversity via subtle modification of conserved gene regulatory networks [6, 19]. Here we show expression of transcription factors and other components of nervous system development as well as guidance cues involved in neurogenesis and axonal growth.
In vertebrates, transcription factor ap2a is required for ectodermal migration during neural tube closure and cell fate specification, and mediates regulatory networks that drive neural crest evolution [94]. In Fig. 13, ap2a is notably expressed in the eyes and brain. We observe arx, mutations of which are linked to improper CNS formation and mental retardation [95], in the somites, spinal region, and in a triangular pattern in the forebrain. Neural adhesion molecule gene chl1 is heavily expressed throughout the CNS, jaws, fins, and lateral line. We observe cntn3, a promoter of neurite outgrowth, throughout the CNS, in the eye, and in the jaw joint. Integral membrane proteoglycan cspg4 is expressed in the pharynx, gut, and dorsal fins. fezf2 expression in the telencephalon exhibits a triangular pattern similar to that of arx. We observe expression of transcription factor tbr1 in the telencephalon, olfactory bulbs and eyes, similar to the results reported in zebrafish [48]. Glutamate transporter vglut2.1 expression is only weakly in the eyes and throughout the brain. zash1, involved in body segment formation and Hox regulation [96], is seen in the brain and eyes. Disruption of highly conserved transcription factor gata6 demonstrates its role in vertebrate development [97]. We observe gata6 heavily expressed throughout the brain, somites, fins, and gut.
Fig. 13.

Expression of brain development and neurogenesis factors at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
In Fig. 13, egr4 is expressed throughout the brain at 6dpf while neuronal differentiation factor neurod1 can be seen in the brain and pharyngeal arches. neurod2 expression at 4dpf is localized to the telencephalon and eyes, but at 6dpf this expression appears throughout the brain and nerve cord. neurog1 expression is evident in the brain and the dorsal trunk.
Semaphorins are a family of secreted and membrane-bound proteins that guide the axonal growth cone during neurogenesis [98]. The semaphorin superfamily is divided into eight subclasses, all of which have a conserved 500 amino acid N-terminal sema domain [98] with variable C-terminals. In Fig. 14 we show expression of class 3 semaphorins, present in vertebrates, which are secreted proteins that act through a heterocomplex receptor of transmembrane plexins, cell adhesion molecules, and neuropilins. Specific combinations of these three receptor components allow selective binding of different Semaphorin 3 genes depending on cell type, developmental stage, and location. A model developed in the mouse molar indicates that the Wnt and Tgf-β pathways signal from the dental epithelium to sema3a in the adjacent mesenchyme, which acts to guide the growing axon via short range repulsion along the boundaries of the nerve pathway [99].
Fig. 14.
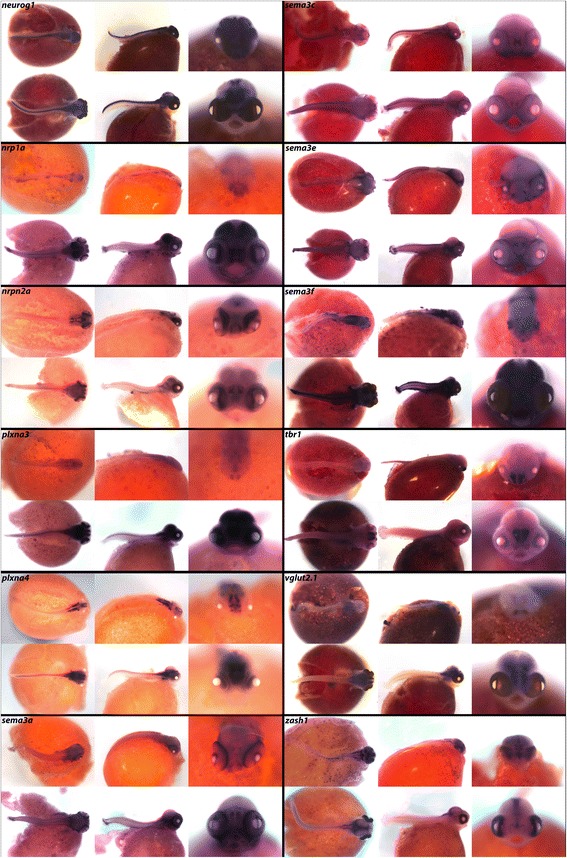
Expression of brain development and neurogenesis factors at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
We observe receptors nrp1a and nrpn2a in similar patterns in the eyes and brain, with heavier expression of nrp1a in the fore/midbrain and pharynx. plxna3 is expressed throughout the brain and eyes, while plxna4 is localized to more restricted regions of the eyes and in the dorsal region of the cerebellum. We report expression of semaphorins 3a, 3c, 3e, and 3f in the retinal tissue similar to expression reported in zebrafish [100], at both the pharyngula and larval stages. All four of these semaphorins are expressed in the early jaw, pharynx, nasal pits, somites, and presumptive optic and otic regions.
Wingless pathway
The Wingless (Wnt) signaling pathway involves many factors that alter transcription, regulate calcium levels, and affect cell polarity during embryonic development through paracrine and autocrine signal transduction. Wnt ligands initiate the pathway by binding the N-terminal extracellular domain of Frizzled family receptors, which then bind cytoplasmic Dishevelled within the cell to propagate the signal. This pathway is highly conserved across vertebrates and invertebrates, with more than 20 mammalian Wnt ligands identified [101].
The canonical Wnt pathway regulates transcription by the translocation of cytoplasmic β-catenin (ctnnb1) into the nucleus, where it co-activates Tcf/Lef family transcription factors. In the absence of Wnt activation, cytoplasmic β-catenin is ubiquitinated for proteasomal destruction by a complex containing Axin, APC, and GSK3 proteins. In Fig. 15, we observe expression of axin1, axin2 and ctnnb1 in the brain, and additional ctnnb1 expression along the cichlid trunk, pharynx, jaw, and fins. Dickkopf family inhibitor dkk3 exhibits expression in the brain, eyes, pharynx, and vertebral region. We also include four Frizzled family receptors, fzd1, fzd2, fzd7 and fzd8, which demonstrate similar expression patterns in the brain, somites, fins, jaw, and pharynx.
Fig. 15.
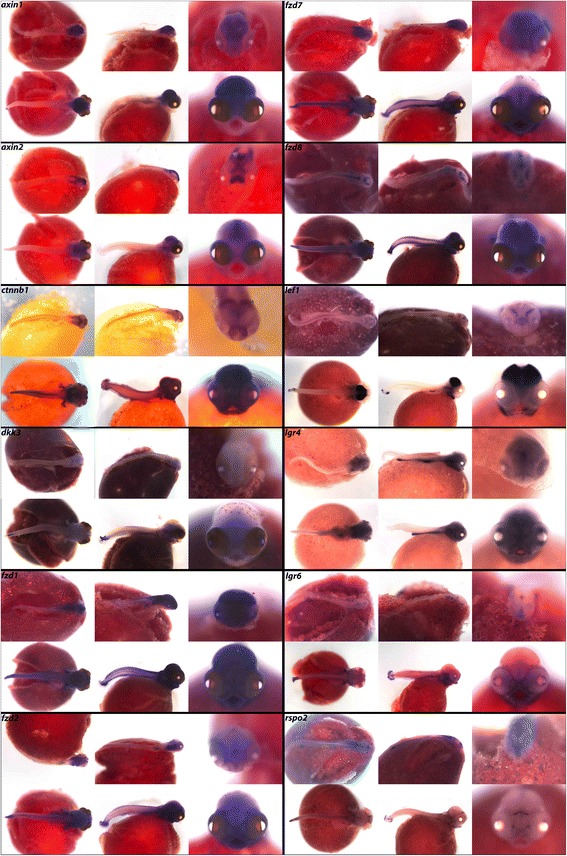
Expression of WNT pathway genes at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
Developmental roles for Wnt signaling have been demonstrated for decades, and knowledge of the effects of this pathway has continued to grow. In 1980, lethal mutations of wingless were shown to affect Drosophila larvae body segments, making boundaries between body axes indistinguishable [102]. This was further demonstrated in Xenopus embryos, which exhibited duplicated axes when injected with mouse Wnt1 RNA [103], and similar duplication was observed by injection of other Wnt related factors. This pathway is important for regulation of cell fate in self-renewing tissues, including mouse intestinal epithelium [104], and in zebrafish has been shown to be important in early neural crest development.
Nuclear β-catenin mediates transcriptional activation by transcription factors Lef1 and Tcf. We observe notable expression of lef1 in the midbrain and forebrain in a similar pattern to that of tcf712 in Fig. 15. tcf3 exhibits expression in the brain, eyes, fins, and somites. Additionally, we report R-spondin receptors lgr4 and lgr6 in distinct patterns in the brain, gut, eyes, fins, and somites, and the secreted R-spondin rspo2 in restricted regions of the fore- and hindbrain.
In Fig. 16, we show expression of Wnt antagonist sfrp1 in the hindbrain at 4dpf, and at 6dpf in the pharynx, eyes, jaw, and olfactory bulbs. sfrp5 is expressed in all three brain regions, somites, jaw, and pharynx (Table 10). In cichlids, Wnt signaling is thought to affect bone deposition to regulate phenotypic changes in craniofacial development [15]. wnt1 and wnt8 are involved in telencephalon and diencephalon development [6]. We see secreted Wnt ligands wnt1, wnt4, wnt5a, wnt7b, wnt8, wnt10a and wnt10b expressed in the hindbrain, fins and pharynx and specific Wnts, for example wnt10b and wnt5a, differentially expressed in the midbrain.
Fig. 16.
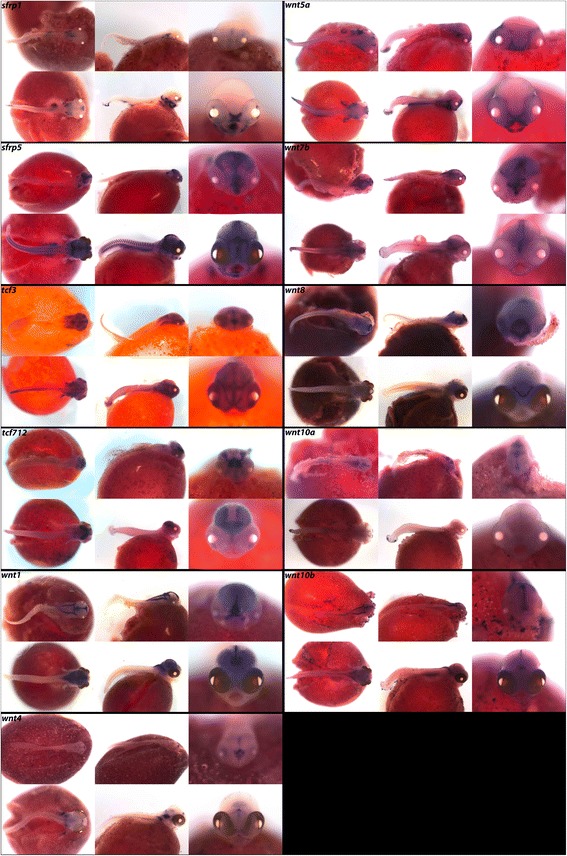
Expression of developmental genes at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
Other developmentally expressed genes
Our final expression panel (Fig. 17) includes factors not specifically involved in the above pathways and processes. We show factors involved in muscle contraction including actin gene acta2 and keratin krt8, which polymerizes into cytoplasmic filaments in epithelial cells. As in zebrafish, smooth muscle actin acta2 is expressed in the myotomes of the trunk and in the intestinal musculature [105], and we also observe expression around the eyes.
Fig. 17.
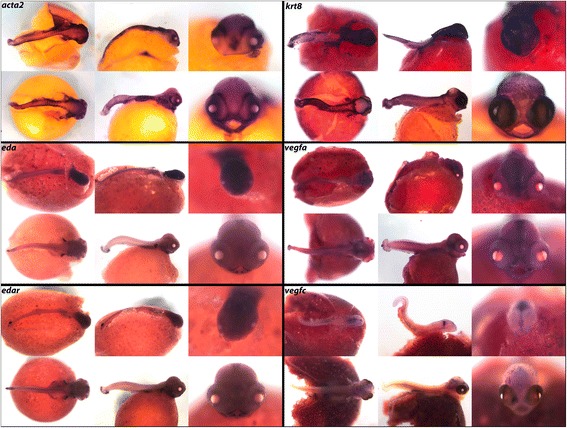
Expression of Mitogens, Stem, and Tumor Suppressor factors at pharyngeal and larval stages. Imaged whole-mount in dorsal, lateral, and frontal orientations
Transmembrane protein ectodysplasin A (eda) acts through receptor edar in ectodermal tissue development. This signaling pair helps pattern early embryonic structures including skin, hair, and teeth, from germ layers, and outlines placode derived structures such as scales and precisely patterned chicken feathers [106]. We observe eda and edar localized to the tooth placodes and fins at these stages, and note expression in and around scales later in development (not shown). Both factors appear to be expressed more heavily in pharyngula stage than at the larval stage. We see krt8 expressed generally across the entire integument at both stages of development.
Vascular endothelial growth factors vegfa and vegfc are involved in angiogenesis, vasculogenesis, and cell migration. Overexpression of this family of genes is seen in the vascularization of cancerous tumors [107], and is the target of many emerging cancer therapies. We observe expression of vegfa along the midline of the brain, as well as in the hindbrain and somites. We also observe vegfc in the somites, as well as in the olfactory, optic, and otic regions.
Conclusions
Novel expression domains
Here, we provide a set of probes for spatial analysis of gene expression, useful across hundreds of East African cichlid fishes, for studies of evolution and development. Gene expression patterns are captivating, and provide important clues to the evolution of gene regulation. Gene expression is context-dependent, dynamic in space and time. Our compendium of gene expression for early Lake Malawi cichlid development provides examples of (i) expression patterns conserved with many other animals, as well as (ii) expression patterns that can be considered novel, because they haven’t been assayed at these particular spaces and times. We highlight a few of these novel expression domains.
Calcium and endocrine signals (Fig. 10) as well as the stem cell/mitogenic factors (Fig. 17) are rarely studied at these stages, in whole mount. Particularly striking spatially delimited gene expression patterns are observed for many of these genes, including calb2, calb2a, cldn15a, kiss1r, glast, sparc, stra13, bmi1, pdgf, celsr1, klf4, trp63 and vim, suggestive of precise roles in embryonic development. We also observe new expression domains from well-studied genes. Notable from this class are osr2 (Fig. 3; expression in fins) foxp2 (Fig. 5; expression in fins and jaws), hopx (Fig. 7; expression in the pharynx), nrp1a, sema3a, sema3c and sema3e (Fig. 14; expression in fins and jaws). These novel expression domains set the stage for future exploration of function.
Acknowledgements
We thank Maya Tome and Avery Shook, along with the members of the Streelman lab and anonymous reviewers for their contributions and comments in preparation of this manuscript.
Funding
This study was funded in part by grants from the National Institutes of Health (R01GM101095, 2R01DE019637 to J.T.S.; and F30DE023013 to R.F.B.) and the National Science Foundation (IOS1146275 to J.T.S.). The funding bodies had no role in the design of the study, collection of data, analysis of data, interpretation of data or in writing the manuscript.
Availability of data and materials
Sequence data that support the findings of this study have been deposited in GenBank with the primary accession codes KT906433-KT906561, KC633830- KC633846, EU867210-EU867217, KT851376-- KT851399.
Authors’ contributions
All ISH and imaging performed by RFB, TEF, and RJM. Sequences were cloned and probes were generated by RFB, TEF, and JBS. RFB, TEF, and JTS conceptualized and composed the manuscript. All authors read and approved final versions of the article.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
All experiments conducted in relation to this publication were carried out in a humane and ethical manner in accordance with Georgia Institute of Technology policies in strict adherence to IACUC (Institutional Animal Care and Use Committee) protocols A14053 and A14055.
Abbreviations
- BMP
Bone morphogenetic pathway
- FGF
Fibroblast growth factor
- FOX
Forkhead box
- Hh
Hedgehog
- HOX
Homeobox
- ISH
In-situ hybridization
- LF
Labeotropheus fuelleborni
- MZ
Metriaclima zebra
- QTL
Quantitative trait loci
- TGF
Transforming growth factor
- Wnt
Wingless
Contributor Information
R. F. Bloomquist, Email: rbloomquist@augusta.edu
T. E. Fowler, Email: tefowler3@gmail.com
J. B. Sylvester, Email: jsylvester@gsu.edu
R. J. Miro, Email: rachelmiro315@gmail.com
J. T. Streelman, Email: todd.streelman@biology.gatech.edu, Email: streelman@biology.gatech.edu
References
- 1.Streelman JT. Advancing Evolutionary Developmental Biology. In: Advances in Evolutionary Developmental Biology (ed J. T. Streelman). Hoboken: John Wiley & Sons, Inc.; 2013.
- 2.McLean CY, Reno PL, Pollen AA, Bassan AI, Capellini TD, Guenther C, Indjeian VB, Lim X, Menke DB, Schaar BT, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471(7337):216–219. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manceau M, Domingues VS, Mallarino R, Hoekstra HE. The developmental role of Agouti in color pattern evolution. Science (New York, NY) 2011;331(6020):1062–1065. doi: 10.1126/science.1200684. [DOI] [PubMed] [Google Scholar]
- 4.Pottin K, Hinaux H, Rétaux S. Restoring eye size in Astyanax mexicanus blind cavefish embryos through modulation of the Shh and Fgf8 forebrain organising centres. Development (Cambridge, England) 2011;138(12):2467–2476. doi: 10.1242/dev.054106. [DOI] [PubMed] [Google Scholar]
- 5.Chan YF, Marks ME, Jones FC, Villarreal G, Jr, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurent deletion of a Pitx1 enhancer. Science (New York, NY) 2010;327(5963):302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sylvester JB, Rich CA, Yi C, Peres JN, Houart C, Streelman JT. Competing signals drive telencephalon diversity. Nat Commun. 2013;4:1745. doi: 10.1038/ncomms2753. [DOI] [PubMed] [Google Scholar]
- 7.Thisse B, Heyer V, Alunni V, Degrave A, Kirchner J, Parkhill J, Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/S0091-679X(04)77027-2. [DOI] [PubMed] [Google Scholar]
- 8.Loh Y, Bezault E, Muenzel F, Roberts RB, Swofford R, Barluenga M, Kidd C, Howe AE, Di Palma F, Lindblad-Toh K, et al. Origins of shared genetic variation in African cichlids. Mol Biol Evol. 2013;30(4):906–917. doi: 10.1093/molbev/mss326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loh YH, Katz LS, Mims MC, Kocher TD, Yi SV, Streelman JT. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biol. 2008;9(7):R113. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513(7518):375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albertson RC, Powder KE, Hu Y, Coyle K, Roberts RB, Parsons KJ. Genetic basis of continuous variation in the levels and modular inheritance of pigmentation in cichlid fishes. Mol Ecol. 2014;23(21):5135–5150. doi: 10.1111/mec.12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts RB, Ser J, Kocher T. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science (New York, NY) 2009;326(5955):998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parnell N, Streelman J. Genetic interactions controlling sex and color establish the potential for sexual conflict in Lake Malawi cichlid fishes. Heredity. 2012;110(3):239–246. doi: 10.1038/hdy.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Albertson RC. Hedgehog signaling mediates adaptive variation in a dynamic functional system in the cichlid feeding apparatus. Proc Natl Acad Sci. 2014;111(23):8530–4. [DOI] [PMC free article] [PubMed]
- 15.Parsons KJ, Taylor AT, Powder KE, Albertson RC. Wnt signalling underlies the evolution of new phenotypes and craniofacial variability in Lake Malawi cichlids. Nat Commun. 2014;5:3629. doi:10.1038/ncomms4629. [DOI] [PMC free article] [PubMed]
- 16.Albertson RC, Streelman JT, Kocher TD, Yelick PC. Integration and evolution of the cichlid mandible: the molecular basis of alternate feeding strategies. Proc Natl Acad Sci U S A. 2005;102(45):16287–16292. doi: 10.1073/pnas.0506649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloomquist R, Parnell N, Phillips K, Fowler T, Yu T, Sharpe P, Streelman J. Coevolutionary patterning of teeth and taste buds. Proc Natl Acad Sci U S A. 2015;112(44):E5954–5962. doi: 10.1073/pnas.1514298112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser G, Bloomquist R, Streelman J. Common developmental pathways link tooth shape to regeneration. In Submission. [DOI] [PMC free article] [PubMed]
- 19.Sylvester JB, Rich CA, Loh Y-HE, van Staaden MJ, Fraser GJ, Streelman JT. Brain diversity evolves via differences in patterning. Proc Natl Acad Sci. 2010;107(21):9718–9723. doi: 10.1073/pnas.1000395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimura K, Kocher TD. Tol2-mediated transgenesis in tilapia (Oreochromis niloticus) Aquaculture. 2011;319(3):342–346. doi: 10.1016/j.aquaculture.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juntti SA, Hilliard A, Kent K, Kumar A, Nguyen A, Jimenez M, Loveland J, Mourrain P, Fernald R. A neural basis for control of cichlid female reproductive behavior by prostaglandin F2a. Curr Biol. 2016;26(7):943–949. doi: 10.1016/j.cub.2016.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimura K, Okada N. Development of the embryo, larva and early juvenile of Nile tilapia Oreochromis niloticus (Pisces: Cichlidae). Developmental staging system. Dev Growth Differ. 2007;49(4):301–324. doi: 10.1111/j.1440-169X.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- 23.Waite KA, Eng C. From developmental disorder to heritable cancer: it’s all in the BMP/TGF-β family. Nat Rev Genet. 2003;4(10):763–773. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- 24.Hogan B. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10(13):1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 25.Albertson RC, Kocher TD. Genetic and developmental basis of cichlid trophic diversity. Heredity. 2006;97(3):211–21. [DOI] [PubMed]
- 26.Terai Y, Morikawa N, Okada N. The evolution of the pro-domain of bone morphogenetic protein 4 (Bmp4) in an explosively speciated lineage of East African cichlid fishes. Mol Biol Evol. 2002;19(9):1628–1632. doi: 10.1093/oxfordjournals.molbev.a004225. [DOI] [PubMed] [Google Scholar]
- 27.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin’s finches. Science (New York, NY) 2004;305(5689):1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 28.Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development (Cambridge, England) 2012;139(4):709–719. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5(7):499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- 30.Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science (New York, NY) 1998;282(5391):1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- 31.Fraser GJ, Bloomquist RF, Streelman JT. A periodic pattern generator for dental diversity. BMC Biol. 2008;6:32. doi: 10.1186/1741-7007-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fraser GJ, Bloomquist RF, Streelman JT. Common developmental pathways link tooth shape to regeneration. Dev Biol. 2013;377(2):399–414. doi: 10.1016/j.ydbio.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handrigan GR, Leung KJ, Richman JM. Identification of putative dental epithelial stem cells in a lizard with life-long tooth replacement. Development (Cambridge, England) 2010;137(21):3545–3549. doi: 10.1242/dev.052415. [DOI] [PubMed] [Google Scholar]
- 34.Handrigan GR, Richman JM. A network of Wnt, hedgehog and BMP signaling pathways regulates tooth replacement in snakes. Dev Biol. 2010;348(1):130–141. doi: 10.1016/j.ydbio.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 35.Streelman JT, Webb JF, Albertson RC, Kocher TD. The cusp of evolution and development: a model of cichlid tooth shape diversity. Evol Dev. 2003;5(6):600–608. doi: 10.1046/j.1525-142X.2003.03065.x. [DOI] [PubMed] [Google Scholar]
- 36.Cleves PA, Ellis NA, Jimenez MT, Nunez SM, Schluter D, Kingsley DM, Miller CT. Evolved tooth gain in sticklebacks is associated with a cis-regulatory allele of Bmp6. Proc Natl Acad Sci. 2014;111(38):13912–13917. doi: 10.1073/pnas.1407567111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liem KF, Tremml G, Roelink H, Jessell TM. Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell. 1995;82(6):969–979. doi: 10.1016/0092-8674(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 38.Furuta Y, Piston DW, Hogan B. Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development (Cambridge, England) 1997;124(11):2203–2212. doi: 10.1242/dev.124.11.2203. [DOI] [PubMed] [Google Scholar]
- 39.Hébert JM, Mishina Y, McConnell SK. BMP signaling is required locally to pattern the dorsal telencephalic midline. Neuron. 2002;35(6):1029–1041. doi: 10.1016/S0896-6273(02)00900-5. [DOI] [PubMed] [Google Scholar]
- 40.Thisse B, Thisse C. Functions and regulations of fibroblast growth factor signaling during embryonic development. Dev Biol. 2005;287(2):390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 41.Bertrand S, Camasses A, Somorjai I, Belgacem MR, Chabrol O, Escande M-L, Pontarotti P, Escriva H. Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proc Natl Acad Sci. 2011;108(22):9160–9165. doi: 10.1073/pnas.1014235108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development (Cambridge, England) 2005;132(2):299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- 43.Niswander L, Martin GR. FGF-4 and BMP-2 have opposite effects on limb growth. 1993. [DOI] [PubMed]
- 44.Knopf F, Hammond C, Chekuru A, Kurth T, Hans S, Weber CW, Mahatma G, Fisher S, Brand M, Schulte-Merker S. Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev Cell. 2011;20(5):713–724. doi: 10.1016/j.devcel.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 45.Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development (Cambridge, England) 2008;135(2):377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Popovici C, Roubin R, Coulier F, Birnbaum D. An evolutionary history of the FGF superfamily. Bioessays. 2005;27(8):849–857. doi: 10.1002/bies.20261. [DOI] [PubMed] [Google Scholar]
- 47.Kawakami Y, Esteban CR, Matsui T, Rodríguez-León J, Kato S, Belmonte JCI. Sp8 and Sp9, two closely related buttonhead-like transcription factors, regulate Fgf8 expression and limb outgrowth in vertebrate embryos. Development (Cambridge, England) 2004;131(19):4763–4774. doi: 10.1242/dev.01331. [DOI] [PubMed] [Google Scholar]
- 48.Thisse B, Thisse C. Fast release clones: A high throughput expression analysis. ZFIN direct data submission. 2004. 2015.
- 49.Weigel D, Jürgens G, Küttner F, Seifert E, Jäckle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57(4):645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- 50.Kaestner KH, Knöchel W, Martínez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14(2):142–146. [PubMed] [Google Scholar]
- 51.Lam EW-F, Brosens JJ, Gomes AR, Koo C-Y. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13(7):482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 52.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. The initiation of liver development is dependent on Foxa transcription factors. Nature. 2005;435(7044):944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 53.Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang S-L, Wert S, Stahlman MT, Whitsett JA. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280(14):13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- 54.Solomon KS, Kudoh T, Dawid IB, Fritz A. Zebrafish foxi1 mediates otic placode formation and jaw development. Development (Cambridge, England) 2003;130(5):929–940. doi: 10.1242/dev.00308. [DOI] [PubMed] [Google Scholar]
- 55.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10(4):233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enard W, Przeworski M, Fisher S, Lai C, Wiebe V, Kitano T, Monaco A, Pääbo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418(6900):869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 57.Rice AN, Lobel PS. Enzyme activities of pharyngeal jaw musculature in the cichlid Tramitichromis intermedius: implications for sound production in cichlid fishes. J Exp Biol. 2002;205(22):3519–3523. doi: 10.1242/jeb.205.22.3519. [DOI] [PubMed] [Google Scholar]
- 58.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12(6):393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 59.Thisse C, Thisse B. High throughput expression analysis of ZF-models consortium clones. ZFIN direct data submission. 2005.
- 60.Roberts RB, Hu Y, Albertson RC, Kocher TD. Craniofacial divergence and ongoing adaptation via the hedgehog pathway. Proc Natl Acad Sci. 2011;108(32):13194–13199. doi: 10.1073/pnas.1018456108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen W, Burgess S, Hopkins N. Analysis of the zebrafish smoothened mutant reveals conserved and divergent functions of hedgehog activity. Development (Cambridge, England) 2001;128(12):2385–2396. doi: 10.1242/dev.128.12.2385. [DOI] [PubMed] [Google Scholar]
- 62.Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5(1):47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, Streelman JT. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 2009;7(2):e31. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kimmel CB, Miller CT, Kruze G, Ullmann B, BreMiller RA, Larison KD, Snyder HC. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol. 1998;203(2):245–263. doi: 10.1006/dbio.1998.9016. [DOI] [PubMed] [Google Scholar]
- 65.Cerny R, Cattell M, Sauka-Spengler T, Bronner-Fraser M, Yu F, Medeiros DM. Evidence for the prepattern/cooption model of vertebrate jaw evolution. Proc Natl Acad Sci. 2010;107(40):17262–17267. doi: 10.1073/pnas.1009304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alunni A, Blin M, Deschet K, Bourrat F, Vernier P, Rétaux S. Cloning and developmental expression patterns of Dlx2, Lhx7 and Lhx9 in the medaka fish (Oryzias latipes) Mech Dev. 2004;121(7):977–983. doi: 10.1016/j.mod.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 67.Matsuo I, Kuratani S, Kimura C, Takeda N, Aizawa S. Mouse Otx2 functions in the formation and patterning of rostral head. Genes Dev. 1995;9(21):2646–2658. doi: 10.1101/gad.9.21.2646. [DOI] [PubMed] [Google Scholar]
- 68.Macdonald R, Barth KA, Xu Q, Holder N, Mikkola I, Wilson SW. Midline signalling is required for Pax gene regulation and patterning of the eyes. Development (Cambridge, England) 1995;121(10):3267–3278. doi: 10.1242/dev.121.10.3267. [DOI] [PubMed] [Google Scholar]
- 69.Pommereit D, Pieler T, Hollemann T. Xpitx3: a member of the Rieg/Pitx gene family expressed during pituitary and lens formation in Xenopus laevis. Mech Dev. 2001;102(1):255–257. doi: 10.1016/S0925-4773(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 70.Jeong J, Li X, McEvilly RJ, Rosenfeld MG, Lufkin T, Rubenstein JLR. Dlx genes pattern mammalial jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development (Cambridge, England) 2008;135:2905–2916. doi: 10.1242/dev.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woltering JM, Noordermeer D, Leleu M, Duboule D. Conservation and divergence of regulatory strategies at hox Loci and the origin of tetrapod digits. PLoS Biol. 2014;12(1):e1001773. doi:10.1371/journal.pbio.1001773. [DOI] [PMC free article] [PubMed]
- 72.Menini A. Calcium signalling and regulation in olfactory neurons. Curr Opin Neurobiol. 1999;9(4):419–426. doi: 10.1016/S0959-4388(99)80063-4. [DOI] [PubMed] [Google Scholar]
- 73.Filippi A, Mueller T, Driever W. Vglut2 and gad expression reveal distinct patterns of dual GABAergic versus glutamatergic cotransmitter phenotypes of dopaminergic and noradrenergic neurons in the zebrafish brain. J Comp Neurol. 2014;522(9):2019–2037. doi: 10.1002/cne.23524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ward AB, Warga RM, Prince VE. Origin of the zebrafish endocrine and exocrine pancreas. Dev Dyn. 2007;236(6):1558–1569. doi: 10.1002/dvdy.21168. [DOI] [PubMed] [Google Scholar]
- 75.Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci. 2012;109(2):466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taniguchi A, Suga R, Matsumoto K. Expression and transcriptional regulation of the human α1, 3-fucosyltransferase 4 (FUT4) gene in myeloid and colon adenocarcinoma cell lines. Biochem Biophys Res Commun. 2000;273(1):370–376. doi: 10.1006/bbrc.2000.2929. [DOI] [PubMed] [Google Scholar]
- 77.Andreu-Agullo C, Maurin T, Thompson CB, Lai EC. Ars2 maintains neural stem-cell identity through direct transcriptional activation of Sox2. Nature. 2012;481(7380):195–198. doi: 10.1038/nature10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liao J, Al Shahrani M, Al-Habib M, Tanaka T, Huang GT-J. Cells isolated from inflamed periapical tissue express mesenchymal stem cell markers and are highly osteogenic. J Endod. 2011;37(9):1217–1224. doi: 10.1016/j.joen.2011.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8(9):726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 80.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 81.Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier X. Expression of the zebrafish genome during embryogenesis. ZFIN direct data submission. 2001.
- 82.Driessens G, Blanpain C. Long live sox2: sox2 lasts a lifetime. Cell Stem Cell. 2011;9(4):283–284. doi: 10.1016/j.stem.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 83.Juuri E, Jussila M, Seidel K, Holmes S, Wu P, Richman J, Heikinheimo K, Chuong C-M, Arnold K, Hochedlinger K. Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development (Cambridge, England) 2013;140(7):1424–1432. doi: 10.1242/dev.089599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okubo T, Pevny LH, Hogan BL. Sox2 is required for development of taste bud sensory cells. Genes Dev. 2006;20(19):2654–2659. doi: 10.1101/gad.1457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ordóñez‐Morán P, Huelsken J. Lrig1: a new master regulator of epithelial stem cells. EMBO J. 2012;31(9):2064–2066. doi: 10.1038/emboj.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guruharsha K, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13(9):654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pourquié O. The segmentation clock: converting embryonic time into spatial pattern. Science (New York, NY) 2003;301(5631):328–330. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- 88.Niwa Y, Shimojo H, Isomura A, González A, Miyachi H, Kageyama R. Different types of oscillations in Notch and Fgf signaling regulate the spatiotemporal periodicity of somitogenesis. Genes Dev. 2011;25(11):1115–1120. doi: 10.1101/gad.2035311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soza-Ried C, Oztürk E, Ish-Horowicz D, Lewis J. Pulses of Notch activation synchronise oscillating somite cells and entrain the zebrafish segmentation clock. Development (Cambridge, England) 2014;14(8):1780–1788. doi: 10.1242/dev.102111. [DOI] [PubMed] [Google Scholar]
- 90.Harada H, Kettunen P, Jung H-S, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147(1):105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cooper MT, Bray SJ. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature. 1999;397(6719):526–530. doi: 10.1038/17395. [DOI] [PubMed] [Google Scholar]
- 92.McElhinney DB, Krantz ID, Bason L, Piccoli DA, Emerick KM, Spinner NB, Goldmuntz E. Analysis of cardiovascular phenotype and genotype-phenotype correlation in individuals with a JAG1 mutation and/or Alagille syndrome. Circulation. 2002;106(20):2567–2574. doi: 10.1161/01.CIR.0000037221.45902.69. [DOI] [PubMed] [Google Scholar]
- 93.Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL. Lunatic fringe is an essential mediator of somite segmentation and patterning. Nature. 1998;394(6691):377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- 94.Van Otterloo E, Li W, Garnett A, Cattell M, Medeiros DM, Cornell RA. Novel Tfap2-mediated control of soxE expression facilitated the evolutionary emergence of the neural crest. Development (Cambridge, England) 2012;139(4):720–730. doi: 10.1242/dev.071308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bienvenu T, Poirier K, Friocourt G, Bahi N, Beaumont D, Fauchereau F, Jeema LB, Zemni R, Vinet M-C, Francis F. ARX, a novel Prd-class-homeobox gene highly expressed in the telencephalon, is mutated in X-linked mental retardation. Hum Mol Genet. 2002;11(8):981–991. doi: 10.1093/hmg/11.8.981. [DOI] [PubMed] [Google Scholar]
- 96.Tanaka Y, Kawahashi K, Katagiri Z-I, Nakayama Y, Mahajan M, Kioussis D. Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of Hox gene expression. PLoS One. 2011;6(11):e28171. doi: 10.1371/journal.pone.0028171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol. 2005;25(7):2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75(7):1389–1399. doi: 10.1016/0092-8674(93)90625-Z. [DOI] [PubMed] [Google Scholar]
- 99.Kettunen P, Løes S, Furmanek T, Fjeld K, Kvinnsland IH, Behar O, Yagi T, Fujisawa H, Vainio S, Taniguchi M. Coordination of trigeminal axon navigation and patterning with tooth organ formation: epithelial-mesenchymal interactions, and epithelial Wnt4 and Tgfβ1 regulate semaphorin 3a expression in the dental mesenchyme. Development (Cambridge, England) 2005;132(2):323–334. doi: 10.1242/dev.01541. [DOI] [PubMed] [Google Scholar]
- 100.Callander DC, Lamont RE, Childs SJ, McFarlane S. Expression of multiple class three semaphorins in the retina and along the path of zebrafish retinal axons. Dev Dyn. 2007;236(10):2918–2924. doi: 10.1002/dvdy.21315. [DOI] [PubMed] [Google Scholar]
- 101.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 102.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 103.McMahon AP, Moon RT. Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell. 1989;58(6):1075–1084. doi: 10.1016/0092-8674(89)90506-0. [DOI] [PubMed] [Google Scholar]
- 104.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19(8):877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 105.Davuluri G, Seiler C, Abrams J, Soriano A, Pack M. Differential effects of thin and thick filament disruption on zebrafish smooth muscle regulatory proteins. Neurogastroenterol Motil. 2010;22(10):1100–e1285. doi: 10.1111/j.1365-2982.2010.01545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Houghton L, Lindon C, Morgan BA. The ectodysplasin pathway in feather tract development. Development (Cambridge, England) 2005;132(5):863–872. doi: 10.1242/dev.01651. [DOI] [PubMed] [Google Scholar]
- 107.Skobe M, Hawighorst T, Jackson DG, Prevo R, Janes L, Velasco P, Riccardi L, Alitalo K, Claffey K, Detmar M. Induction of tumor lymphangiogenesis by VEGF-C promotes breast cancer metastasis. Nat Med. 2001;7(2):192–198. doi: 10.1038/84643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequence data that support the findings of this study have been deposited in GenBank with the primary accession codes KT906433-KT906561, KC633830- KC633846, EU867210-EU867217, KT851376-- KT851399.


