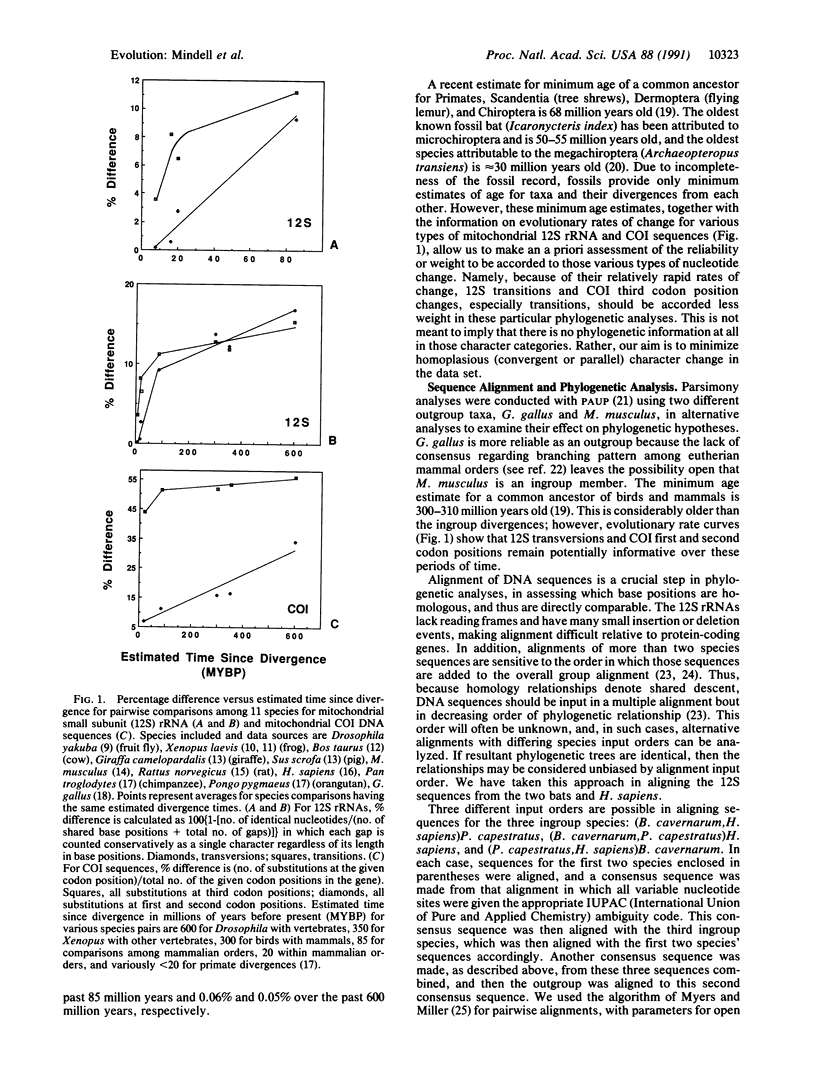
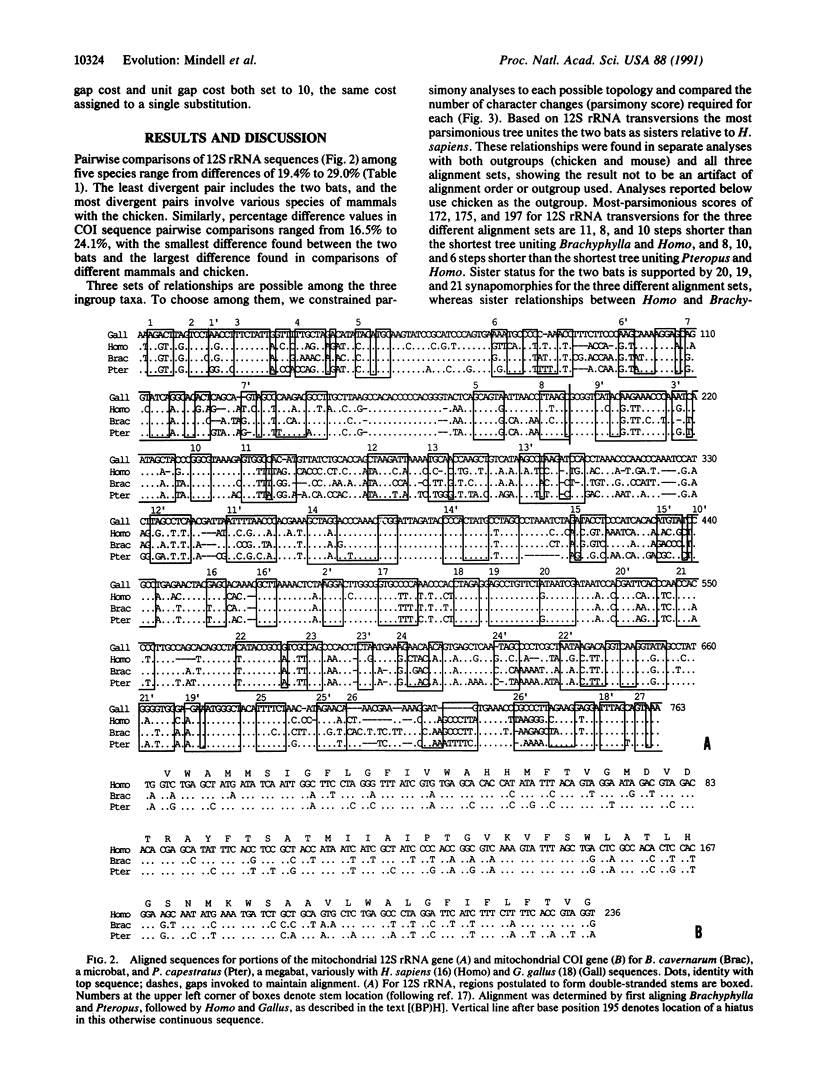
Abstract
We present 744 nucleotide base positions from the mitochondrial 12S rRNA gene and 236 base positions from the mitochondrial cytochrome oxidase subunit I gene for a microbat, Brachyphylla cavernarum, and a megabat, Pteropus capestratus, in phylogenetic analyses with homologous DNA sequences from Homo sapiens, Mus musculus (house mouse), and Gallus gallus (chicken). We use information on evolutionary rate differences for different types of sequence change to establish phylogenetic character weights, and we consider alternative rRNA alignment strategies in finding that this mtDNA data set clearly supports bat monophyly. This result is found despite variations in outgroup used, gap coding scheme, and order of input for DNA sequences in multiple alignment bouts. These findings are congruent with morphological characters including details of wing structure as well as cladistic analyses of amino acid sequences for three globin genes and indicate that neurological similarities between megabats and primates are due to either retention of primitive characters or to convergent evolution rather than to inheritance from a common ancestor. This finding also indicates a single origin for flight among mammals.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Aquadro C. F., Greenberg B. D. Human mitochondrial DNA variation and evolution: analysis of nucleotide sequences from seven individuals. Genetics. 1983 Feb;103(2):287–312. doi: 10.1093/genetics/103.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton M. J. Phylogeny of the major tetrapod groups: morphological data and divergence dates. J Mol Evol. 1990 May;30(5):409–424. doi: 10.1007/BF02101113. [DOI] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The ribosomal RNA genes of Drosophila mitochondrial DNA. Nucleic Acids Res. 1985 Jun 11;13(11):4029–4045. doi: 10.1093/nar/13.11.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins P., Morais R. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol. 1990 Apr 20;212(4):599–634. doi: 10.1016/0022-2836(90)90225-B. [DOI] [PubMed] [Google Scholar]
- Dunon-Bluteau D., Brun G. The secondary structures of the Xenopus laevis and human mitochondrial small ribosomal subunit RNA are similar. FEBS Lett. 1986 Mar 31;198(2):333–338. doi: 10.1016/0014-5793(86)80431-8. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989 Apr;5(2):151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Hixson J. E., Brown W. M. A comparison of the small ribosomal RNA genes from the mitochondrial DNA of the great apes and humans: sequence, structure, evolution, and phylogenetic implications. Mol Biol Evol. 1986 Jan;3(1):1–18. doi: 10.1093/oxfordjournals.molbev.a040379. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Seki T., Yaginuma K., Koike K. Nucleotide sequences of small ribosomal RNA and adjacent transfer RNA genes in rat mitochondrial DNA. Gene. 1981 Dec;16(1-3):297–307. doi: 10.1016/0378-1119(81)90085-8. [DOI] [PubMed] [Google Scholar]
- Kocher T. D., Thomas W. K., Meyer A., Edwards S. V., Päbo S., Villablanca F. X., Wilson A. C. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A. A rate-independent technique for analysis of nucleic acid sequences: evolutionary parsimony. Mol Biol Evol. 1987 Mar;4(2):167–191. doi: 10.1093/oxfordjournals.molbev.a040433. [DOI] [PubMed] [Google Scholar]
- Lake J. A. The order of sequence alignment can bias the selection of tree topology. Mol Biol Evol. 1991 May;8(3):378–385. doi: 10.1093/oxfordjournals.molbev.a040654. [DOI] [PubMed] [Google Scholar]
- Li W. H., Wolfe K. H., Sourdis J., Sharp P. M. Reconstruction of phylogenetic trees and estimation of divergence times under nonconstant rates of evolution. Cold Spring Harb Symp Quant Biol. 1987;52:847–856. doi: 10.1101/sqb.1987.052.01.092. [DOI] [PubMed] [Google Scholar]
- Maly P., Brimacombe R. Refined secondary structure models for the 16S and 23S ribosomal RNA of Escherichia coli. Nucleic Acids Res. 1983 Nov 11;11(21):7263–7286. doi: 10.1093/nar/11.21.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E. W., Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988 Mar;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Novacek M. J. Evidence for echolocation in the oldest known bats. Nature. 1985 May 9;315(6015):140–141. doi: 10.1038/315140a0. [DOI] [PubMed] [Google Scholar]
- Pettigrew J. D. Flying primates? Megabats have the advanced pathway from eye to midbrain. Science. 1986 Mar 14;231(4743):1304–1306. doi: 10.1126/science.3945827. [DOI] [PubMed] [Google Scholar]
- Pettigrew J. D., Jamieson B. G., Robson S. K., Hall L. S., McAnally K. I., Cooper H. M. Phylogenetic relations between microbats, megabats and primates (Mammalia: Chiroptera and Primates). Philos Trans R Soc Lond B Biol Sci. 1989 Nov 30;325(1229):489–559. doi: 10.1098/rstb.1989.0102. [DOI] [PubMed] [Google Scholar]
- Roe B. A., Ma D. P., Wilson R. K., Wong J. F. The complete nucleotide sequence of the Xenopus laevis mitochondrial genome. J Biol Chem. 1985 Aug 15;260(17):9759–9774. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten R. A., Walberg M. W., Clayton D. A. Precise localization and nucleotide sequence of the two mouse mitochondrial rRNA genes and three immediately adjacent novel tRNA genes. Cell. 1980 Nov;22(1 Pt 1):157–170. doi: 10.1016/0092-8674(80)90164-6. [DOI] [PubMed] [Google Scholar]


