Abstract
Compelling clinical, social, and economic reasons exist to innovate in the process of drug discovery for neuropsychiatric disorders. The use of patient-specific, induced pluripotent stem cells (iPSCs) now affords the ability to generate neuronal cell-based models that recapitulate key aspects of human disease. In the context of neuropsychiatric disorders, where access to physiologically active and relevant cell types of the central nervous system for research is extremely limiting, iPSC-derived in vitro culture of human neurons and glial cells is transformative. Potential applications relevant to early stage drug discovery, include support of quantitative biochemistry, functional genomics, proteomics, and perhaps most notably, high-throughput and high-content chemical screening. While many phenotypes in human iPSC-derived culture systems may prove adaptable to screening formats, addressing the question of which in vitro phenotypes are ultimately relevant to disease pathophysiology and therefore more likely to yield effective pharmacological agents that are disease-modifying treatments requires careful consideration. Here, we review recent examples of studies of neuropsychiatric disorders using human stem cell models where cellular phenotypes linked to disease and functional assays have been reported. We also highlight technical advances using genome-editing technologies in iPSCs to support drug discovery efforts, including the interpretation of the functional significance of rare genetic variants of unknown significance and for the purpose of creating cell type- and pathway-selective functional reporter assays. Additionally, we evaluate the potential of in vitro stem cell models to investigate early events of disease pathogenesis, in an effort to understand the underlying molecular mechanism, including the basis of selective cell-type vulnerability, and the potential to create new cell-based diagnostics to aid in the classification of patients and subsequent selection for clinical trials. A number of key challenges remain, including the scaling of iPSC models to larger cohorts and integration with rich clinicopathological information and translation of phenotypes. Still, the overall use of iPSC-based human cell models with functional cellular and biochemical assays holds promise for supporting the discovery of next-generation neuropharmacological agents for the treatment and ultimately prevention of a range of severe mental illnesses.
Keywords: Human stem cells, iPSC models, drug discovery, CRISPR-Cas9, high-throughput screening, high-content imaging, neuropharmacology, bipolar disorder, schizophrenia, autism spectrum disorders, dementia
1. Phenotypic assays in human iPSC models of neuropsychiatric disorders
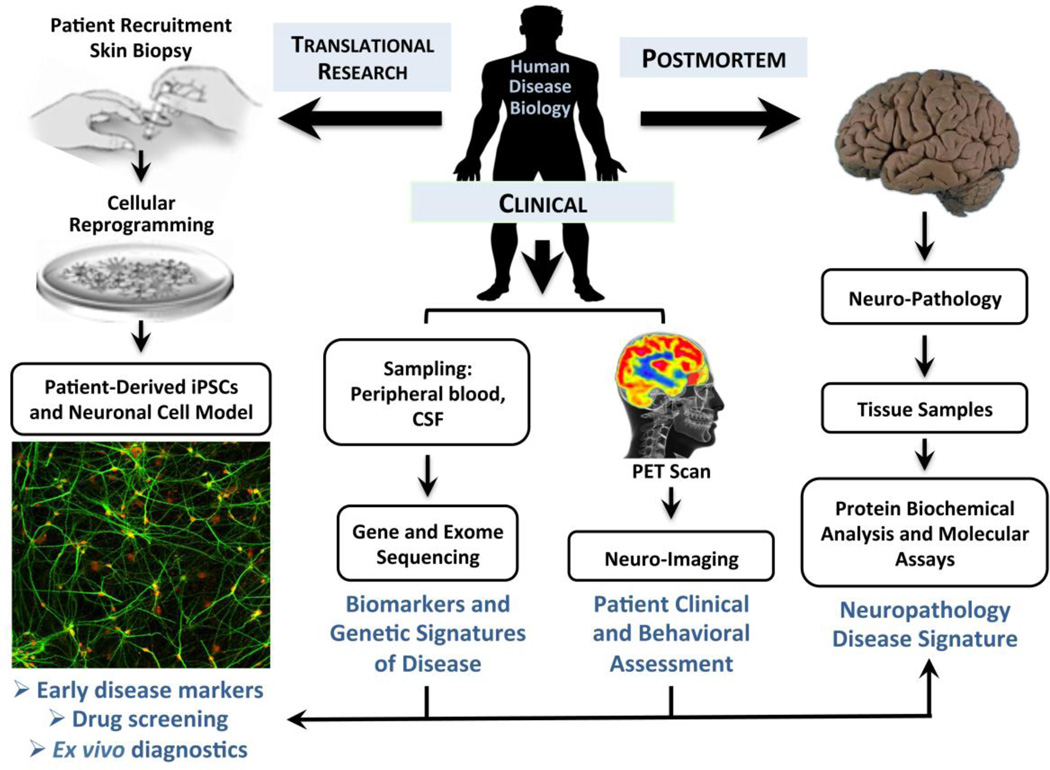
Identifying disease-causing genetic variation is only the first step in understanding the process by which a gene contributes to the pathophysiology of disease1. Indeed, several causal variants identified decades ago— for Huntington’s disease and Cystic fibrosis, for example— are only now beginning to yield targeted therapeutics. While common neuropsychiatric disorders are highly polygenic in nature2,3, there are also numerous highly-penetrant, single-gene disorders with relevant neurologic or psychiatric symptomology. The generation of induced pluripotent stem cell (iPSC) models of monogenic or oligogenic neuropsychiatric disorders increasingly provides a powerful means to dissect the underlying disease biology at the level of cells and molecules and to create scalable systems that can support functional genomic and pharmacological screens4. This is particularly true for disorders like amyotrophic lateral sclerosis (ALS) and Alzheimer’s disease where cellular phenotypes and cellular context are more well understand and thus able to help define disease-relevant cellular phenotypes5. Here we review recent advances in models of both monogenic and genetically complex, polygenic neuropsychiatric disorders using human stem cell technology. We focus, in particular, on the development of functional and phenotypic cellular assays and their additional potential for characterization of pharmacological agents in an integrated manner guided by complementary information emerging from biomarker, clinical and post-mortem studies of human disease biology (Figure 1).
Figure 1. Strategies for integrative modeling of human disease pathophysiology and drug discovery using patient-specific iPSC models.
Somatic cell sources other than fibroblasts from skin biopises can also be used. Molecular assays on iPSC-derived neuronal cells and glia can range from single cell imaging to whole transcriptomes via RNA-seq or and proteomes using quantiative mass spectrometry.
1.1 Autism spectrum disorders: Timothy Syndrome
Timothy syndrome is a rare autosomal dominant neurodevelopmental disorder, with characteristic cardiac abnormalities, caused by mutations in the gene coding for the alpha 1C subunit of the L-type calcium channel (CACNA1C) associated with gain-of-function channel activity due to a delay in channel closing. Studies by the Dolmetsch laboratory have demonstrated that Timothy syndrome patients’ iPSC-derived neurons with a characteristic CACNA1C G406R substitution have aberrant action potential firing and intracellular calcium signaling after depolarization, leading to alteration of activity-dependent gene expression6,7, when compared to wild-type neurons generated in the same manner. Besides electrophysiological and signaling defects, Timothy syndrome iPSC-derived neural progenitor cells also exhibited aberrant neuronal differentiation patterns, as measured by elevated number of tyrosine hydroxylase positive neurons, along with decreased expression of SATB2 from lower cortical layer neurons and callosal projection neurons6. A comparable decrease in SATB2 expression was also observed, in the same study, in brain sections from transgenic mice expressing a Cacna1c mutant allele in forebrain neurons. Remarkably, aberrant intracellular calcium signaling after depolarization in these neurons was reversible and could blocked by treatment with the voltage-dependent L-type calcium channel antagonist nimodipine6. Though nimodipine was effective at normalizing the defects in calcium signaling, it was ineffective at restoring proper levels of tyrosine hydroxylase expression. In contrast, the cyclin-dependent kinase inhibitor roscovitine, which enhances voltage-dependent inactivation of the L-type calcium channel, was able to decrease the number of tyrosine hydroxylase neurons6. While neither of these pharmacological agents was identified by an unbiased screen, but rather on the basis of an a priori mechanism, the potential for screening for such agents as demonstrated. Finally, as another example of convergence between human and mouse neuronal models of Timothy syndrome, both patient iPSC-derived neurons and mouse model neurons exhibited activity-dependent dendritic retraction due to ectopic activation of the GTPase RhoA, but in a calcium-independent manner7. Taken together, the cellular phenotypes observed with a Timothy syndrome human iPSC-derived model provide insight into the role of CACNA1C in regulating diverse aspects of neuroplasticity. Assuming relevance to disease pathophysiology, these findings may stir the development of novel therapeutics that can restore proper L-type calcium channel function and rescue downstream signaling at the level of neuronal gene expression and neurodevelopment. Whether or not human-specific phenotypes can be identified using the Timothy syndrome iPSC model that could not be observed with the corresponding mouse model remains to be demonstrated, and will be an excellent test case of the merits of human iPSC model contributions. A critical if subtle point is that the investigators identified drugs of distinct mechanism associated with short-term normalization of function versus normalization of neurodevelopmental phenotypes: this distinction may be particularly relevant to the neuropsychiatric diseases discussed below, where different strategies may be required to identify symptom-controlling versus disease-modifying agents. More generally, these results illustrate perhaps the canonical example of iPSC-derived neurons being applied to identify in vitro phenotypes associated with rare genetic variation, and then to identify drugs that rescue these phenotypes. These findings also illustrate a key challenge in the development of iPSC-derived models for the identification of molecular mechanism and potential drug targets. It is often the case that several phenotypic changes can be defined in these model systems, the challenge is to determine the most relevant phenotype(s) to use for pharmacological investigation or phenotypic screening.
1.2 Autism spectrum disorders: Fragile X syndrome
Fragile X syndrome is the most prevalent genetic cause of an autism spectrum disorder, as well as intellectual disability in males. Fragile X syndrome has been shown to be caused predominantly by an expanded CGG trinucleotide repeat in the 5’ untranslated region (5’UTR) of the Fragile X Mental Retardation (FMR1) gene on the X-chromosome, leading to epigenetic silencing and loss of expression of the Fragile X Mental Retardation Protein (FMRP)8–10. Despite the known relationship between the CGG-repeat expansion in the 5’ untranslated region of the Fragile X Mental Retardation 1 gene (FMR1) that leads to FMR1 silencing and the loss of expression of the Fragile X Mental Retardation Protein (FMRP), the consequences of the loss of FMRP still remain poorly understood10. As reviewed in more detail by Bhattacharyya and Zhao in this special issue11, multiple human embryonic stem cell (hESC)12,13 and in iPSC models of Fragile X syndrome have now been generated14,15. For example, Sheridan et al.14 reported on the characterization of iPSC models of Fragile X syndrome from multiple subjects with CGG repeat sizes in the full mutation range (>200), and demonstrated that the FMR1 locus remained epigenetically silenced in iPSC-derived CNS neural progenitor cells and neurons. A neurodevelopmental time course analysis also revealed aberrant neuronal differentiation in terms of axonal and dendritic complexity and elevated expression of a glial marker, indicative of a fate-specification defect14. They also isolated both iPSCs with a full mutation (~700 CGG repeats) sized repeats and an otherwise isogenic iPSC line from the same individual with pre-mutation sized repeats (~142 CGG repeats). This shortening of the repeat was sufficient to restore normal levels of DNA methylation at the locus, allow FMRP expression, and rescued the neural progenitor cells in terms of neurodevelopmental deficits14. These observations point to the importance of repeat length in epigenetic silencing of FMRP and highlight the need to consider somatic mosaicism (or repeat instability) along with the need to characterize multiple clones from subjects if single cell clones are selected16. Importantly in terms of replication of findings in different FMR1 mutation lines, similar neurodevelopmental deficits have been reported in both human embryonic stem cell (hESC) models12,13 and in other iPSC models15. Taken together, these results suggest the possibility that adult neurogenesis pathways are dysregulated in Fragile X syndrome patients. This is further supported by recent analysis of Fmr1 deletion mouse models17–19, as well as post-mortem studies that have shown increased morphological alternations of the CA1 region and dentate gyrus regions of the hippocampus20. Importantly, recent studies on a larger collection (n=9) of Fragile X syndrome hESC lines have demonstrated FMR1 epigenetic gene silencing occurs in 66% of the lines analyzed. This contrasts to the conclusions made concerning differences between iPSC models versus hESC models since the FMR1 gene was initially found to be expressed in a hESC line from a full mutation individual and only silenced upon differentiation. This discrepancy, as the authors point out, serves to illustrate the critical importance of examining multiple cell models of disease.
Since the majority of the existing mouse models of Fragile X syndrome involve deletion of the gene rather than epigenetic silencing of the locus in a CGG-repeat dependent manner, studies of the actual molecular mechanisms that are most tightly linked to the causally mutation for the disorder are not readily possible. This makes patient-derived iPSC models well suited for modeling Fragile X syndrome in vitro and for therapeutic screening to target the fundamental cause of the disorder namely the epigenetic silencing of FMRP. To this end, building on the findings of silenced FMR1 expression in Fragile X syndrome iPSC-derived neural progenitor cells14, two groups have now reported on high-throughput screens for compounds that can rescue expression of FMRP in these cells. Kumari et al.21 developed a homogenous, time-resolved fluorescence resonance energy transfer (FRET) assay for FMRP detection in a 1,536-well plate format and screened 5,000 bioactive and tool compounds. This screen yielded a total of six compounds that modestly increased FMR1 mRNA levels, but not at sufficient levels to be therapeutically relevant. Similarly, Kaufmann et al.22 employed a high-content, image-based assay for FMRP expression to screen 50,000 compounds resulting in a small number of active compounds that also modestly restored expression of FMRP. That more effective compounds were not identified likely reflects a combination of the relatively modest size of the compound libraries screened and the complex nature of repeat-mediated epigenetic dysregulation at the locus23. Nonetheless, these proof-of-concept studies pave the way for larger-scale therapeutic screening for Fragile X syndrome, including high-content screening assays that in addition to FMRP expression in post-mitotic neurons, and not just progenitor cells, quantitatively assess diverse morphological properties of FXS patient neuron (e.g.24; S.J.H., personal communication).
Additional applications of epigenetic and functional assays in Fragile X syndrome iPSC models comes from recent clinical findings with the mGluR5 (Metabotropic glutamate receptor 5) antagonist AFQ056 in Fragile X syndrome25. These studies suggest that efficacy of AFQ056 may be restricted to a subset of patients with full FMR1 promoter methylation since many patients with only partial methylation showed no response. Conceivably, iPSC-derived neuronal cells from responder and non-responder Fragile X subjects could be used to dissect the relationship between FMR1 methylation status and mGluR5 antagonist response. Moreover, assuming faithful recapitulation of the epigenetic status of the FMR1 gene in vitro and in vivo in brain, pre-testing of compounds in patient iPSC-derived neuronal cells may help improve prediction of clinical response better than methylation and repeat testing alone.
1.3 Rett syndrome, CDKL5 disorder, and beyond
Rett syndrome is a neurodevelopmental X-linked disorder that occurs primarily due to mutations in the gene encoding methyl-CpG binding protein 2 (MECP2) in young girls. The encoded protein MECP2 plays a key role in the recruitment of co-repressor complexes and other forms of epigenetic regulation of the genome. Multiple groups have now generated Rett syndrome iPSC-derived neuronal models that have collectively revealed a number of potentially disease-relevant phenotypes, including changes in soma size, glutamatergic synaptic deficits, and altered gene transcription26–32. Since these Rett syndrome iPSC models were generated from multiple different patients and with a range of methodologies, the observation of similar phenotypes by different research groups is promising. Moreover, since MECP2 is located on the X-chromosome, which is subject to X-inactivation, it has been possible for investigators to identify otherwise isogenic iPSC lines, from the same Rett syndrome patient, that only vary in which chromosome is X-inactivated. Such isogenicity allows a single patient-derived line to essentially act as its own control for functional studies. Through these iPSC-derived neuronal models, it has been possible to identify a number of phenotypes that are dependent upon the loss of the healthy copy of the MECP2 gene, providing strong genetic evidence for MECP2-linked phenotypes.
Besides demonstrating the feasibility of genetic rescue of the MECP2-dependent phenotypes with neuronal cells neurons expressing a functional copy of MECP2, Marchetto and colleagues tested whether the insulin-like growth factor 1 (IGF-1), a candidate therapeutic agent for Rett syndrome previously shown to ameliorate synaptic and behavioral symptoms in the Rett syndrome mouse model33, was also able to rescue the decreased number of glutamatergic synapses in human cultured neurons30. As expected form the studies with the Mecp2 knockout mouse model, IGF-1 treatment was able to restore proper numbers of glutamatergic synapses, as measured by quantifying VGLUT positive synaptic punctae30. In terms of translation, on-going clinical studies in Rett syndrome subjects described by Pini et al. indicate IGF-1 administrations is safe and well tolerated34. In an open label study in a single patient with repeated treatment, modest improvement in Rett symptoms (cognitive, social, and autonomic); however, the apparent benefits were short term since they did not persist between the two cycles of treatment. Understanding the mechanistic basis for why prolonged treatment may be necessary, or whether it is due to exposure levels, maybe assisted using Rett patient iPSC-derived neuronal models. Additionally, since one of the Rett syndrome iPSC models was generated from a patient with a nonsense mutation, that introduces a stop codon in the MECP2 gene, the authors were able to test another drug, gentamicin, an aminoglycoside shown previously to enhance the degree to which the ribosomes are capable of reading through a stop codon30. As for the IGF-1, gentamicin was also capable of increasing VGLUT-positive synaptic puncta, providing a potentially distinct mechanism for treatment of Rett syndrome patients30. However, with only imaging of VLGUT puncta, it remains to be demonstrated if either IGF-1 or read-through agents are capable of restoring functional glutamatergic synapses. Such functional synaptic assays remain difficult to scale, but may be critical if the goal is to demonstrate normalization of function, not just morphology. Moreover, whether these same agents are capable or rescuing other cellular phenotypes in Rett syndrome iPSC-derived neurons has not yet been investigated. Nonetheless, these results provide an important proof-of-concept for rescuing loss-of-function variants in neurodevelopment disorders. In this context, the in vitro demonstration of the rescuing effect of IGF-1 is particular encouraging given the implication of downstream PI3K/AKT/MAPK signaling pathways whose investigation may lead to the identification of alternative and possibly improved therapeutic targets.
Often classified as an “early-onset seizure” variant of Rett syndrome with developmental delay, cognitive deficits, and autistic features35–38, mutations in the CDKL5 (cyclin-dependent kinase-like 5) gene on the X-chromosome are also associated with a severe neurodevelopment disorder. While CDKL5 has been shown to be localized within the nucleus, and at excitatory synapses where it regulates synaptic plasticity and dendritic spine morphology, the physiological functions of CDKL5, in particular those relevant for disease pathogenesis, remain poorly understood. To gain insight into these questions, Amenduni et al.39 reported on the characterization of iPSCs from a female with the CDKL5 p.Q347X mutation (early onset seizure variant) and one male with p.T288I mutation (X-linked epileptic encephalopathy), associated with different clinical presentations. As with the Rett syndrome studies described above, exploiting the fact that X-chromosome inactivation occurs randomly, the authors were able to isolate clonal isogenic lines that expressed either wild-type CDKL5 or the mutant allele. Expression profiling of these neurons revealed elevated expression of a GRID1 (orphan glutamate receptor δ-1 subunit) encoding a cell adhesion molecule that in cortical neurons plays a role in controlling inhibitory presynaptic differentiation39. Intriguingly, the same GRID1 expression difference was also found in MECP2 mutant iPSC models39,40,as well as in iPSC models from an atypical Rett syndrome patient with a FOXG1 mutation41. In addition to gene expression differences, imaging of neurons from different CDKL5 mutant iPSCs by Ricciardi et al.42 revealed deficits in dendritic spine morphology and excitatory synapse stability at least in part though interaction and phosphorylation of the cell adhesion molecule LRRC4C (Leucine Rich Repeat Containing 4C; NGL-1). Taken together these findings point to a potential common molecular mechanisms affecting excitatory/inhibitory synapse imbalance in these neurodevelopment disorders, and suggest that the development of high-content image-based assays focused on GRID1 expression, dendritic spine density, and balanced of excitatory and inhibitory synaptic markers as therapeutic targets could yield novel therapeutics for Rett syndrome, CDKL5 mutations, and related neurodevelopmental disorders.
Beyond mutations in MECP2 in Rett syndrome, CDKL5 disorder, or FMR1 in Fragile X syndrome where a single gene is investigated, other examples where transcriptional dysregulation is seen in human iPSC models of disorders with cognitive and behavioral symptoms are that of Williams-Beuren deletion syndrome and 7q–microduplication syndrome, where dosage imbalance at 7q11.23 occurs leading to shared but symmetrically opposite phenotypes43. Using integrative genome-wide chromatin immunoprecipitation and sequencing (ChIP-seq) and functional genomic approaches, the authors implicate GTF2I (General Transcription Factor IIi) as the key mediator of these reciprocal changes in transcription even in the pluripotent state43. These findings providing a powerful example of how the relevant genes and cellular consequences of large copy number variants can be dissected using human iPSC models.
1.4 Schizophrenia
Schizophrenia is a severe neurodevelopmental disorder characterized by alterations in cognition, affect, and behavior that affects ~1% of the population and has been shown to be highly heritable, exhibiting a complex genetic architecture that is only now beginning to be unraveled. In particular, while more than 100 loci harboring common schizophrenia-associated variants have been identified, many of them span multiple genes of unclear function, and the effect sizes of each are modest. Conversely, aggregating over thousands of variants can explain up to 15% of schizophrenia liability44. The first report neuronal phenotypes of iPSCs from schizophrenia subjects comes by the Gage laboratory45. Analysis of long-term (1–3 months) differentiated iPSC-derived neurons, co-cultured with human cerebellar astrocytes, revealed that schizophrenia patient-derived neurons exhibited a significant decrease in neuronal connectivity (measured by a rabies virus tracing assay), decreased PSD-95 protein levels and glutamate receptor expression (GRIK1, GRIK4, GRM7, GRIN2A)45. These neuronal cultures also exhibited altered gene expression profiles with evidence for dysregulation of cAMP and WNT signaling pathways45. Remarkably, treatment of these schizophrenia patient iPSC-derived neurons with the tricyclic dibenzoxazepine antipsychotic loxapine (10 µM) - but not multiple other antipsychotics -for the last 3 weeks of the total 3 month differentiation time course, reversed the gene expression abnormalities and connectivity differences45.
This landmark study highlights important methodological considerations for investigation of neuropsychiatric disorders with iPSCs in particular as we consider these cells for high-throughput screens where the assays need to be relevant, robust and reproducible to enable the more classical design-test-iterate process of early drug discovery. First, a major challenge arises from the polygenicity of schizophrenia44, such that the disorder is likely to be influenced by multiple genetic factors that can vary significantly between individuals. Since one of the iPSC models in this study was reportedly diagnosed with early onset schizophrenia at 6 years of age (a very unusual presentation of unclear relationship to adult-onset disease), and a second patient had schizoaffective disorder, where the etiological relationship to schizophrenia is also unclear, a range of phenotypic variation might be expected. Second, like many studies published to date, the neuronal cultures analyzed represented a mixture of neuronal subtypes generated under a limited number of differentiation conditions, leaving open the possibility that differentiation into more defined neural subtypes would reveal different phenotypes. Third, given the puzzling inactivity of related antipsychotic agents (thioridazine, clozapine, olanzapine, risperidone) in the same connectivity and expression assay, the precise mechanism through which loxapine treatment was able to reverse phenotypes is an important unresolved question; the inactivity of the structurally similar compound clozapine is particularly surprising. Moreover, many of the known targets of these agents are differentially expressed across neuronal subtypes, introducing the additional challenge of understanding the specific cellular targets in the test system. Therefore, it will be crucial to reproduce these results with other dibenzoxazepines along with concentration responses to define the minimal effective concentration and time period of loxapine treatment as well as additional pharmacological dissection of the mechanism of action. As a broader point, this effect also underscores the importance of selecting phenotypes and positive control conditions corresponding to an appropriate aspect of disease. Where targeting neurodevelopment may be obvious in early-onset illnesses such as Rett syndrome, the relevant phenotypes in most neuropsychiatric diseases remain unclear; in particular, one might expect distinct phenotypes and therapeutic effects corresponding to normalization of developmental versus functional phenotypes. Apart from these cautionary considerations regarding the data interpretation, the findings provide proof of principle for the ability to pharmacologically reverse certain complex cellular and molecular phenotypes in iPSC-derived neurons from schizophrenic subjects. This finding undoubtedly will prompt efforts to both reproduce these observations with other schizophrenia iPSCs, in order to determine reproducibility among cell lines and clinical subtypes, and to better understand to what extent, if any, the deficits observed are unique to the specific culture conditions and differentiation methods employed.
Other phenotypes have also been described in additional patient-derived cell studies. A set of iPSC models was developed from three schizophrenia patients by Pedrosa and colleagues, including one individual with the 22q11.2 deletion that causes velocardiofacial syndrome46, a multisystem disorder that may present with affective and/or psychotic symptoms. Studies on these iPSC-derived neuron identified alterations in expression of the pluripotency genes OCT4 and NANOG, although the significance of this observation to disease pathogenesis remains unknown, as well as dysregulated miRNA expression47. Moreover, analysis of iPSC-derived neural progenitor cells from two clones of a single schizophrenia patient identified elevated levels of extra-mitochondrial oxygen consumption, which were not detected in the fibroblasts or iPSCs from the same patient48. These iPSC-derived neural progenitor cells exhibited elevated levels of reactive oxygen species. These results were consistent with proteomic analysis of human brain tissue from schizophrenia patients, that showed that more than half of affected proteins in disease were associated with mitochondrial function and oxidative stress responses49. Interestingly, the elevation of reactive oxygen species in schizophrenia neural progenitor cells could be reversed by the mood stabilizer valproate48, although of note, valproate has not demonstrated efficacy in schizophrenia, suggesting that not all of these phenotypes may be disease-relevant.
Finally, as an example of investigating a potentially high-penetrant rare variant strategy to gain insight into schizophrenia pathogenesis, iPSC models have been generated from an American family with a 4 base pairs deletion in the DISC1 (Disrupted-in-Schizophrenia 1) gene that segregates, although not full penetrant, with mental illness in a an American pedigree50. As described below in the section of genome editing, analysis of forebrain, cortical-like neurons from isogenic lines with and without this mutation has provided evidence for neurodevelopmental abnormalities, synaptic deficits, and transcriptional dysregulation caused by the truncation of DISC151. Taken together, the current studies with schizophrenia subjects’ iPSC-derived neural progenitors and neuronal cells, although promising, highlight the need to further dissect specific disease molecular mechanisms and be critical on our assessment of phenotypes and their relevance to the clinical diagnosis, integrate information with human brain-based studies, and take advantage of only relevant and robust phenotypic assays for therapeutic screening.
1.5 Bipolar disorder
Bipolar disorder is a severe neuropsychiatric disorder of mood and cognition for which the relationship between etiology and the underlying pathophysiology is poorly understood. Like schizophrenia, genetic evidence points to the highly polygenic nature of susceptibility factors52,53, with multiple risk loci having been identified by recent genome-wide association studies. However, the connection of these genetic factors to the molecular and cellular underpinnings of disease pathophysiology remains largely elusive. One hypothesis, reviewed in greater detail by O’Shea and McInnis in this special issue54, is for bipolar disorder to have a neurodevelopmental component. Recent findings with bipolar disorder patient iPSC-derived neural progenitor and neuronal cell models provide growing support for this notion. This includes studies by Madison et al.55 employing multiplexed mRNA expression assays and high-content imaging to phenotype iPSC-derived neural progenitors and neurons from bipolar disorder subjects relative to controls. Following a family based design, and a strategy designed to isolate a specific subpopulation of CXCR4-positive neural progenitor cells by multi-chromatic, fluorescence-activated cell sorting, the authors observed differential expression of key for neurodevelopmental genes in progenitor cells from the two bipolar disorder subjects not present in their unaffected parents55. Consistent with these expression differences, deficits in neurogenesis were also observed in the two bipolar disorder subjects compared to their unaffected parents using imaging and neural lineage-specific markers as functional read outs55. Interestingly, given the known pharmacological effects of the bipolar disorder medication lithium on GSK3 (Glycogen synthase kinase 3)56, the proliferation deficits of the bipolar disorder subjects could be rescued to the level of their unaffected parental controls by treatment with the highly selective and potent GSK3 inhibitor (CHIR-99021). Overall, while multiple independent iPSC clones from each subject were characterized, a clear short-coming of this study is the limited number of subjects characterized—a limitation now overcome by the creation of a large cohorts of bipolar disorder subjects that have been generated (S.J.H., R.H.P., personal communication).
Additionally, Chen et al.57 have reported phenotypes in bipolar disorder iPSC-derived neurons that included alteration of transcription of genes critical for neurodevelopmental patterning, in particular downregulation of dorsal telencephalic maker genes such as PAX6, TBR2, FEZF2, EMX2, and TCF354. Kim et al.58 have also observed transcriptional differences in iPSC-derived neurons from bipolar disorder subjects from an Old Order Amish pedigree, although not the same transcripts seen in other studies perhaps due to methodological differences in the culture conditions. Together with the observations from Madison et al.55, these findings provided evidence for unique developmental phenotypes of bipolar disorder neurons that suggest there may be changes in cell type and identity. With advances in single-cell genomics, using highly parallel genome-wide expression profiling methodology such as Drop-seq that allows transcriptome analysis in thousands of individually identifiable cells59, it will be possible to more precisely define that nature of these transcriptional changes in defined cell populations over a neurodevelopmental time course. Such expression differences could conceivably be exploited in high-throughout gene expression signature-based screens or in high-content imaging assays with lineage specific markers.
Beyond protein-coding mRNAs, additional characterization of a subset of the bipolar disorder family iPSCs described by Madison et al.55 enabled Bavamian et al.60 to discover elevated expression of the noncoding microRNA miR-34a in induced neurons (iNs) from bipolar disorder subjects, as well as in and post-mortem brain tissue, in an orthogonal but still patient-derived neuronal culture system. Using biochemical methods and high-resolution imaging assays, overexpression of miR-34a was found to decrease dendritic complexity and impair the expression of synaptic proteins, whereas reducing endogenous miR-34a expression was found to promote dendritic complexity60. miR-34a was also shown to target the bipolar disorder risk genes ANK3 (Ankyrin-3) and CACNB3 (Voltage-dependent L-type calcium channel subunit beta-3). Given this ability to target multiple risk genes implicated by large-scale genetics, miRNAs may play an important role connecting together different risk factors and pathways. For this reason, the potential of noncoding RNAs as therapeutic agents/targets warrants further investigation, particularly as innovative strategies for delivery of nucleic acids to defined tissues and cells improves61. Alternatively, small molecule pharmacological approaches targeting coding or non-coding RNAs may be an attractive and feasible approach62. Conceivably, in the future, imaging-based phenotypic assays of dendritic complexity could be scaled to a high-content, automated microscopy screen to identify pharmacological agents that rescue neurodevelopment abnormalities in the context of bipolar disorder iPSC models.
As the most recent example of the application of functional assays to characterize bipolar disorder iPSC models, Mertens et al.63 reported the generation of VGLUT1-positive, hippocampal dentate gyrus (DG) granule cell-like glutamatergic neurons that were identified on the basis of their expression of a lentivirally delivered, Prox1 promoter-driven eGFP reporter (Prox1::eGFP). Using a related mitochondrial dye, JC-1, which reports on mitochondrial membrane potential, in a low-throughput, flow cytometry assay, analysis of these bipolar disorder DG-like neurons (N=4) revealed elevated mitochondrial membrane potential relative to control controls (N=6) DG-like neurons63. Additionally, imaging of mitochondrial size with DsRed2-mito genetically encoded fluorescent reporter revealed that the mitochondria in bipolar disorder cells were smaller in size. With evidence for mitochondrial abnormalities, as well as mRNA expression differences, in this DG-like neuron population, the authors went on to test disease-dependent neural network dysfunction with the calcium indicator dye Fluo-4-AM in the background of a Synapsin promoter-driven DsRed lentiviral reporter. Bipolar disorder neurons exhibited significantly hyperexcitability at a younger age, but not older age, as measured by a higher frequency of spontaneous Ca2+ events in comparison to healthy control neural networks63. Remarkably, the hyperexcitability of bipolar disorder DG-like neurons was silenced by lithium treatment, but only for the subset corresponding to clinically lithium-responsive subjects63. This finding is particularly notable given the challenge in clearly distinguishing responsive from resistant subjects- i.e., the phenotype itself is difficult to reliably characterize in some patients, inasmuch as lithium resistance can reflect differences in tolerability and adherence (and disease course) as well as in efficacy. Also important is the finding that the hyperexcitability phenotype was only identified in a particular phase of development (i.e., younger cells), suggesting the importance of careful consideration of developmental time course in assay development. These findings, using genetically encoded fluorescent reporters suggests that defined cell populations should be used for further pharmacological investigation of the pathogenesis of bipolar disorder, and for dissecting the mechanisms of action of lithium in order to identifying lithium-like drugs that may have improved efficacy or safety. Future studies will be required to test the generalizability of these observations in other bipolar disorder subjects, and in neuronal subtypes other than DG-like neurons, to understand the biological significance of the age-dependency of the observations of hyperexcitability.
2. Genome engineering in human stem cell models for drug discovery
Genome engineering approaches through the use of64 zinc finger nucleases (ZFNs)65, transcription activator-like effector nucleases (TALENs)64, and the increasingly powerful and flexible clustered regularly interspaced short palindromic repeats-CRISPR-associated protein-9 nuclease (CRISPR-Cas9) system66, provide powerful strategies for creating new types of cellular models and tools for use in therapeutic screening efforts66. This includes providing insight into pathways involved in disease pathogenesis by increasing confidence in genotype-phenotype relationships, and allowing a number of innovative methods to create reporters of the underlying molecular pathways involved in disease etiopathogenesis. The applicability of genome-engineering technologies is particularly relevant to the field of neuropsychiatric diseases, allowing to probe gene-by-gene effects, individually and in combination, in a isogenic background that allows to determine the specificity of phenotypic consequences of each gene and mutation.
2.1 Using genome editing for the study of genetic variants of unknown significance
As large-scale exome and whole genome sequencing in human genetics continue to expand in years to come, a burning question that emerges is how to interpret the relevance of variants of unknown significance (VUS) to disease. While in the case of the insertion of a premature stop codon, frameshift mutations, or splice site mutations that in many cases (but not always) can be assumed to cause loss-of-function, many nonsynonymous, single nucleotide substitutions that alter the amino acid sequence of a protein that have unknown functional effects. How to ascertain whether there are functional consequences from such variants remains a critical question. Rather than the traditional approaches of cloning and mutating each gene of interest in order to test in overexpression or biochemical assays the consequence of genetic variation, with the advent of genome-editing technologies it is now feasible to introduce the specific mutation of interest in the context of the human where genome it was identified66,67. Such precise genome modifications have the advantage of allowing the study of genetic variants in the cell types of interest, which can be derived from patient iPSCs, expressed at physiologically relevant levels (natural promoters, splicing, post-translational modifications), which is predicted to account for more accurate phenotypes than the conditions of overexpression.
A leading example of this strategy applied to a neuropsychiatric-disease relevant gene are studies using TALENs to both correct a 4 base pair deletion in the DISC1 gene that segregates, although not full penetrant, with mental illness in a an American pedigree51. Characterization of forebrain, cortical-like neurons from multiple individuals with this 4 base pair deletion revealed deficits in morphology and pre-synaptic development linked to SV2+ and transcription of synaptic genes and DISC1 interacting proteins that remarkably could be rescued by correcting the mutation51. To demonstrate sufficiency of this mutation to cause the same phenotypes, introduction of the mutation on a healthy control background recapitulated the defects. From a therapeutic discovery perspective, the authors went on to show that a PDE4 (Phosphodiesterase 4) inhibitor rolipram was also able to rescue the synaptic deficits, providing further evidence for a critical role of cAMP-mediated signaling in response to disrupted DISC1 function51,68. Extending this type of work will undoubtedly aid in the identification of pathways and networks affected by human genetic variation an in doing so provide new targets for therapeutic intervention.
Outside of monogenic scenarios, a key challenge in employing genome editing to study neuropsychiatric disease will be determining how to model polygenic disorders, for two reasons. First, while loss-of-function variants - modeled through knockout or knockdown strategies - might be expected to have large and readily-detectable effects (as some of the Rett and Fragile X studies noted above would suggest), the individual variants associated with diseases such as schizophrenia are rarely if ever of this type -although studying syndromes such as velocardiofacial syndrome that frequently present with schizophrenia or bipolar-like features may provide a more tractable alternative. CRISPR-Cas9 and related methods do allow introduction of (for example) SNPs in a manner agnostic to their effect, but whether these variants can be expected to cause measurable phenotypes remains to be seen. Second, the genetic background on which such variants should be introduced is difficult to establish. In a healthy control line, it seems highly unlikely that a single risk variant should be sufficient to produce disease; in a schizophrenia line, it seems equally unlikely that correcting a single risk variant should be sufficient to rescue the phenotype. While techniques for multiplexed CRISPR-Cas9 can be developed, introducing many variants simultaneously may be technically challenging and the interpretation of such models compared to, for example, patient-derived disease lines would be difficult.
2.2 Leveraging genome engineering for cell-based screening assays
In addition to generating monoallelic and biallelic reporters by targeting ‘safe harbor sites’ (that are refractory to silencing) such as the AAVS1 site on Chr19, Pei et al.69 has recently described the generation of iPSC lineage-specific reporters by targeting a NanoLuc-Halotag construct to the 3’-prime end of genes of interest to allow normal levels of expression of the endogenous genes. For example, optimized ZFN pairs targeting the MAP2 (neural lineage specific) or GFAP (astrocyte lineage specific) genes were designed to allow the insertion of a reporter cassette consisting of a 2A peptide (to allow self cleavage from the endogenous protein), followed by a NanoLuc luciferase gene fused with a HaloTag, and a neomycin resistance gene in frame with the C-terminal of the targeted gene69. Upon differentiation of these iPSCs into neurons expression MAP2-NanoLuc-KI reporter, or astrocytes expression he GFAP-NanoLuc-KI reporter, lineage specific identity can be monitored by secreted luciferase or detection of the HaloTag69. In the absence of the P2A cleavage peptide, the use of the HaloTag or other fluorescent reporters would allow the study of protein localization and trafficking in both live and fixed cells, which could be highly advantageous for investigating the kinetics of response of a tagged protein to pharmacological agents. Strategies focused on lineage-specific reporters and targeted reporter genes open up multiple new ways to developed improved assays for therapeutic screening and human disease modeling.
As an example of this strategy, although not yet in an iPSC model system, Ingles et al. recently reported on the used TALEN-mediated genome editing to precisely embed a secreted bioluminescent transcription reporter within the genetic locus harboring the Peripheral Myelin Protein 22 (PMP22) gene implicated in Charcot-Marie-tooth disease that causes peripheral neuropathy70. Using a disease-relevant Schwann cell line that naturally expresses high levels of PMP22 in a 1536-well plate format, quantitative high-throughput screening (qHTS) identified novel chemotypes not previously observed to regulate PMP22 transcription when a randomly inserted reporter gene was used.
The application of a locus targeting strategy can be extended not only to transcriptional reporters, but also for tagging of proteins with widely used epitope tags such as the human influenza hemagglutinin (HA-tag), DYKDDDDK (FLAG-tag), or polyhistidine tag (His-tag). The introduction of these tags into the endogenous locus allows for immunodetection of protein localization in differentiated cells, or in a developmental time-dependent manner, with potential for use in high-content image-based screens or for purification of proteins for use in biochemical assays. Overall, genome-editing technologies allow unprecedented opportunities to address questions of genetic variation, dysregulation and biochemical imbalance in neuropsychiatric diseases, through direct testing of the phenotypic contribution of each gene, and the molecular pathways affected, in patient-derived iPSC models. Also, by engineering reporters of gene and protein expression, that inform on endogenous gene regulation, cellular function and lineage specificity, drug discovery screening efforts have higher probabilities of identifying effective disease-modifying and preventive therapeutics.
3. High-content and functional imaging assays for pharmacological screening
In contrast to target or mechanism-based screening, human iPSC models hold great potential for phenotypic screening, where the target is unknown and the assay is more pathophysiologically relevant. Phenotypic screening offers the potential of identifying compounds acting through novel molecular mechanisms or unprecedented targets and as such these assays circumvent our incomplete understanding of pathophysiology and disease biology. However, given the complexity and diversity of biological processes involved in cellular phenotypic assays, it is unlikely that traditional approaches involving iterative improvements in compound properties (the design-make- test cycle) will be successful. In this case, phenotypic assays need to replicate the disease phenotype to such an extent that it is possible to confidently predict the translation of a given compound to clinically relevant features of illness.
There are still relatively few cases in which even small scale compound screens have been performed using human iPSC-derived model systems, but with the advances described above in the identification of cellular phenotypes linked to disease and new tools like CRISPR-Cas9 this should begin to change rapidly. For example, Zhao et al.71 recently described a stably integrated, luciferase-based WNT pathway reporter system in human iPSC-derived neural progenitor cells. The WNT signaling pathway is a key regulator of nervous system development, including axon guidance, dendrite development, and synapse formation, and as such, WNT dysregulation has been associated with neurodevelopment and psychiatric diseases72. After development and optimization for a 384-well plate format, this assay was used to screen a set of known bioactive compounds and FDA-approved drugs, leading to the identification of a number of both known and novel small molecule regulators of WNT signaling in human neural progenitors. To add another dimension to the screen, compounds were tested either under basal conditions, by culturing cells in WNT3A–conditioned media, or with the addition of lithium to the culture media, a known WNT pathway activator and drug used to treat bipolar disorder. This strategy led to the identification of compounds that specifically enhanced WNT signaling under each of these conditions71, which included selective mGLUR1 (Metabotropic glutamate receptor 1) antagonists and the FDA-approved drug riluzole (Rilutek). The approach taken is a demonstration of the ability of human iPSC models to yield novel targets for modulating WNT signaling in the brain, and potentially already-approved drugs for repurposing in combination with lithium in studies of neuropsychiatric disorders71. Recently, this reporter assay has been scaled to a fully automated format for large-scale compound screening as part of the NIH-supported Molecular Libraries Program (MLPCN). It has now been successfully used to screen over 300,000 compounds, to identify activators and inhibitors of WNT signaling, yielding a collection of novel pharmacological probes of this pathway that is critical for neurogenesis and neuroplasticity (S.J.H., manuscript in preparation). Further testing of these compounds using functional assays in human disease-specific iPSC models where deficits of WNT signaling have been observed, such as bipolar disorder55,56 and frontotemporal dementia73,74, holds promise to yield new probes of human disease mechanisms and potentially leads for therapeutic development.
Moving beyond reporter genes, Efthymiou et al.75 have described an efficient human stem cell-derived neural progenitor-based culture system for use in high-throughput screens. Here, directed differentiation from Nestin-positive progenitors cells originated lineage-specific neurons or astrocytes that were cultured separately or together and used in proof-of-concept high-throughput viability screens in a 96-well plate format75. For the viability assay, reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) to formazan was used as an inexpensive surrogate measure. For high-content imaging in live cells, the authors used the MitoTracker Green FM dye to track mitochondria, and a baculovirus-derived nuclear targeted GFP for labeling of post-mitotic nuclei75,76. In a comparative profiling of a set of 80 compounds, performed in human iPSC-derived neural stem cells using the MTT assay, only four compounds were found to be cytotoxic in all four cell types, demonstrating the advantages of lineage-specific assessment of effects of pharmacological agents76.
Live cell assays such as the ones mentioned here, in combination with time-lapse automated high-resolution imaging, establishes a means of conducting functional screens of organelle trafficking and function in response to pharmacological agents or as phenotyping tools in the context of disease models. Advances in automated microscopy and high-resolution imaging technologies now allow single cell and subcellular level analysis of phenotypes with increased throughput in image acquisition77–79. Additionally, increased sophistication and speed of computational methods for analysis of thousands of images and parameters enables automated quantification of a diverse range of cellular features77,80.
4. Key questions for stem cell modeling and therapeutic discovery
In order to improve usefulness for drug discovery, iPSC modeling efforts will benefit from standardization of the production of iPSCs themselves and their extensive quality control. While a number of steps in the production of iPSC cells can be automated to allow population-level modeling81, there is still a need to develop robust, scalable, functional assays for phenotypic assays and for quantitative biochemistry that are ultimately disease relevant4. Related to this issue, one driving question for human iPSC-derived models and therapeutic screening efforts is whether they can be effectively used to model and understand the mechanisms of selective cellular and circuit vulnerability and dysfunction that occur in many neurological and psychiatric disorders82. For example, in frontotemporal dementia only specific neuronal subtypes and defined regions of the human brain undergo pathological changes associated with accumulation of aggregated proteins such as tau (microtubule-associated protein tau) or TDP-43 (transactive response DNA binding protein 43 kDa). The consequences of protein accumulation include aberrant proteostasis and selective neuronal loss in specific regions of the brain, while neurons immediately proximal remain unaffected at the level of such pathological aggregates83. Here, the ability to develop functional assays and pharmacological screens in defined neuronal subtypes, representative of the affected and non-affected cell types in disease-relevant circuits, holds promise in the identification of therapies targeting core aspects of disease pathogenesis, which is much more complex that solving genetically-linked phenomena84.
A related goal, and opportunity offered by iPSC-derived models of disease, is to investigate the early steps of disease pathogenesis that are predicted to occur much earlier than the clinical manifestation or the downstream neuropathology seen in the context of post-mortem human brain studies85. Exciting examples include efforts, guided by recent exome sequencing studies implicating disruption of actin cytoskeleton dynamics in schizophrenia86,87, to investigate developmentally-dependent dendritic spine morphology and density on glutamatergic cortical-like neurons, which correspond to cortical layer III pyramidal (S.J.H., personal communication). Similarly with undifferentiated neural progenitor cells from schizophrenia subjects, Brennad et al.88 have reported differences in cell migration properties that may reflect early alterations of cytoskeletal and cell adhesion pathways, leading to morphological or connectivity differences later in development. Additionally, in the context of human iPSC-derived neuronal models of tauopathy, multiple research groups have now reported aberrant tau expression and post-translational changes as early markers of pathogenesis associated with late onset of neurodegeneration89–94. The observation of differences in tau by biochemical and imaging analysis has set the stage to investigate the underlying molecular and cellular mechanisms of disease pathogenesis. Similarly, exciting phenotypic differences have been reported in iPSC models of other genetic forms of neurodegeneration, including Alzheimer’s disease (e.g. 95–98) and Parkinson’s disease (e.g. 99–101). Taken together, these findings provide guidance for developing a battery of high-throughput screens to identify novel pharmacological agents and leads for therapeutic development.
If robust cellular phenotypes that are disease relevant can routinely be assessed in patient-derived in vitro stem cell models, might it become feasible to use these biochemical, imaging, and physiological assays as new diagnostic tools or biomarkers to classify subjects and to guide the selection of subjects for clinical trials? Such notions are clearly at a nascent stage for the field of neuropsychiatry (as compared to the cancer field for example), but given the state-of-the-art the potential for such integrative, cellular systems-levels diagnostic assays is hinted at in a number of studies. These assays could conceivably include base line, homeostatic conditions, as well as conditions in which neuronal and glial cells are perturbed with pharmacological agents or functional genomic probes, such as gene-specific using CRISPR-Cas9 activation or silencing66, to measure cells response to specific stimuli or stressors, as an indirect measure of “cell health”. This question is particularly pertinent to polygenic disorders where the risk is divided between hundreds or even thousands of common variants, each of small effect size. Whilst it may be possible to identify treatment responders based on a composite polygene risk, the ability to empirically predict response based on iPSC-based tests would be an attractive alternative.
Since human stem cell-derived neurons allow access to physiologically relevant protein complexes without the need for overexpression in heterologous systems or assays in non-human systems, they may also provide more meaningful systems to characterize and select drug candidates. There are numerous examples of a disconnect between the pharmacological properties of drugs in an isolated recombinant system compared to a ‘native’ system or in phenotypic assays102. This translational disconnect could potentially be overcome using iPSC-based screens. This may also provide important new avenues to discover and investigate pharmacological agents with allosteric binding models or functional selectivity, including classical GPCRs targeted by classical antipsychotics103, and other emerging CNS targets, including HDAC (Histone deacetylase) complexes (104–106; S.J.H., personal communication),where differences in the mode of antagonism or agonism of complexes can result in different functional response.
While the use of human stem cell technology provides a number of advantages and promising avenues for novel pharmacological probe and therapeutic agent discovery, there are still a number of limitations and roadblocks. One general limitation is the fact that post-mitotic cell types obtained through the directed differentiation of iPSCs into neural or glial lineages are still relatively immature, based on a number of gene expression markers and physiological readouts. Possible strategies to circumvent this aspect, and promote faster maturation and “aging” of neural cells in culture, include the expression of progerin, a shortened form of lamin A that has been shown to elevate levels of multiple aging-related markers107, and the creation of 3-D organoid culture systems108,109. Certainly assays relying on longer time in culture are likely to better capture features present only in mature neurons, but at the cost of diminished feasibility and scalability: even with modern culture techniques and robotics, maintaining neurons in culture for 8 weeks or more can be labor intensive. In part for this reason, a parallel line of work is investigating ‘shortcuts’ for rapid generation of neural cells without requiring iPSC culture, so-called induced neurons, has promise and has begun to be used to reveal disease phenotypes 110,111.
Another limitation in the iPSC field is the relative high cost of certain cell culture media and supplements like growth factors (e.g. brain-derived neurotrophic factor (BDNF); glial cell line–derived neurotrophic factor (GDNF), and Sonic hedgehog (SHH)), that are commonly used to direct the differentiation to specific lineages, as well as coatings reagents (e.g. laminin) utilized to mimic appropriate extracellular environments. In terms of factors utilized for directed differentiation, one promising direction is the development of substituting pharmacological agents, that once synthesized on a larger scale can reduce costs and reduce variability inherent to protein purification, a number of which are already routinely used, including SB431542 (Activin receptor-like kinase receptor inhibitor), cyclopamine (Hedgehog signaling pathway inhibitor), CHIR-99021 (GSK3 inhibitor), and dibutryl-cAMP (cAMP analog) (112–114). Still, the cost and expertise required to reliably generate homogeneous neurons, particularly on a large scale, remain prohibitive for many laboratories and requires further standardization of protocols and optimization.
A further concern remains the high risk for type I error associated with initial iPSC modeling reports: given the labor required to establish and phenotype these lines, many publications describe only one or a small number of case and control lines, characterized in terms of multiple phenotypes. These results may be highly susceptible to artifacts introduced by subtle differences in cell line generation at any step in the process. The current state of the field might be compared to the early days of genetic association studies: given a lack of understanding about effect sizes to expect, it seemed plausible that psychiatric diseases might be attributable to one or a few genes of large effect. As such, small case-control association studies could report that the combination of (for example) SLC6A4, HTR2A, and BDNF might explain depression risk. While with the benefit of hindsight it is easy to be critical of such reports, it must be recognized that their limitations became apparent only much later as the problems of publication bias and multiple comparisons became dogmatic115.
Practically, then, it will be critical for initial findings in small numbers of lines to be replicated in larger cohorts and in multiple lines established from individual subjects. Here directly addressing cell variability within a culture, between clones of a patient, and between patients with the same disorder must be done rigorously so robust and relevant phenotypes can be identified. The number of cellular phenotypes examined must also be made clear, so that investigators can consider the risk for false positives; given the work required to make and characterize these cells, temptation will remain strong to chalk negative findings up to failed assays that are not mentioned in print. The availability of large neuropsychiatric patient-derived iPSC repositories generated with a single highly-automated methodology from our labs (R.H.P., S.J.H.) and others116 may be useful in allowing effects to be estimated with greater precision and reliability, and reducing the publication of false positives. Further, these larger biobanks may enable investigation and identification of more clinically-relevant phenotypes, by selecting patient subgroups to model on the basis of treatment response or individual neurocognitive phenotypes.
A final caveat in the application of these cell lines to modeling is the importance of distinguishing phenotypes related to neurodevelopment and those related to neural function, recognizing that there is likely complex overlap between these categories. For psychiatric disease, there are no known disease-modifying agents, only those that control symptoms. (This observation does not mean that such agents do not exist, only that their ability to modify underlying illness course has not been established). As a result, it may be difficult to establish positive controls for chemical screening if the goal is disease modifiers. In the near term, screens are likely to rely on comparisons with control drugs known to be efficacious in psychiatric disease - but such screens should not necessarily be expected to reveal neurodevelopmental modifiers. On the other hand, as we have noted, studies of monogenic diseases are already beginning to reveal such modifiers.
5. Future potential of stem cell models for drug discovery efforts
Recent examples of successful drug discovery leading to F.D.A. approval of first-in-class, new chemical entities in disease areas where animal models were deemed inadequate to accurately test efficacy (e.g. Kalydeco in cystic fibrosis;117), combined with the uncertainties of modeling neuropsychiatric disorders in animals118, provides an example of a new paradigm for neuropsychiatric drug discovery. Given the potential of human iPSC derived neuronal assays to both discover and systematically test experimental therapeutic agents to generate a more relevant translational pharmacology, these ex-vivo systems may be the only requirement to determine efficacy prior to commencing clinical trials.
Along these lines, while not representing the discovery of a new chemical entity, studies in ALS patient iPSC-derived motor neuron subtypes revealed a hyperexcitable phenotype that could be silenced by the existing anti-epileptic drug retigabine (ezogabine; Potiga)119. On the basis of these pre-clinical findings, a large multi-site clinical trial to evaluate the efficacy of retigabine in ALS is being conducted (ClinicalTrials.gov Identifier: NCT02450552). In parallel to this efficacy trial, investigators are also generating iPSC from ALS patients enrolled in the trial that will be used to predict which patients are likely to exhibit a beneficial clinical response prior to their actual use of the drug in the trials. As generation of patient-derived models becomes more routine and more scalable, the generation of cellular models may facilitate a new generation of biomarkers to understand and predict neuropsychiatric treatment response120. Conceptually, the advance testing of experimental therapeutics using patient-specific iPSC models enrolled in a trial provides a powerful proof-of-concept for linking physiologically relevant models of human disease to drug development efforts. In doing so, this strategy closes the loop of discovery in a powerful manner driven by human disease biology at each step of the process (Figure 1). With time, such innovative has great potential to contribute to the development of clinically effective drugs.
Acknowledgments
Members of the Haggarty and Perlis laboratories are thanked for their continued dedication to advancing patient-specific iPSC models of neuropsychiatric disorders. We apologize for the inability to cite a number of other important papers in the field due to space limitations. Research in the Haggarty laboratory on human stem cell models of neuropsychiatric disease has been supported in part by the FRAXA Research Foundation, Harvard Stem Cell Institute Seed Grant, Marigold Foundation, Stanley Medical Research Institute, Tau Consortium, Association for Frontotemporal Degeneration, Bluefield Project to Cure FTD, Pitt-Hopkins Research Foundation, Loulou Foundation, Defense Medical Research and Development Program, National Institute of Mental Health (R33MH087896; R01MH091115; R01MH095088), National Institute of Aging (RF1AG042978), National Institute of Neurological Disorders & Stroke (R21NS085487), NHGRI (P50MH106933), National Center for Complementary and Integrative Health (R01AT009144). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial Disclosures
Dr. Haggarty has served on scientific advisory boards for Rodin Therapeutics and PsyBrain; and has received speaker honoraria and/or consulting fees from Sunovion Pharmaceuticals, Biogen-Idec, and AstraZeneca. Dr. Perlis has served on scientific advisory boards or received consulting fees from Genomind, Nestle Health, Perfect Health, Pfizer, Proteus Biomedical, PsyBrain, and RID Ventures. He receives royalties from Bracket (a Medco subsidiary). Dr. Cross and Dr. Brandon are employees and shareholders of AstraZeneca, PLC.
REFERENCES
- 1.Gusella JF, MacDonald ME. Huntington’s disease: seeing the pathogenic process through a genetic lens. Trends Biochem Sci. 2006;31:533–540. doi: 10.1016/j.tibs.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neale BM, Sklar P. Genetic analysis of schizophrenia and bipolar disorder reveals polygenicity but also suggests new directions for molecular interrogation. Curr Opin Neurobiol. 2015;30:131–138. doi: 10.1016/j.conb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Haggarty SJ, Perlis RH. Translation: screening for novel therapeutics with disease-relevant cell types derived from human stem cell models. Biol Psychiatry. 2014;75:952–960. doi: 10.1016/j.biopsych.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khurana V, Tardiff DF, Chung CY, Lindquist S. Toward stem cell-based phenotypic screens for neurodegenerative diseases. Nat Rev Neurol. 2015;11:339–350. doi: 10.1038/nrneurol.2015.79. [DOI] [PubMed] [Google Scholar]
- 6.Pasca SP, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krey JF, et al. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2012;16:201–209. doi: 10.1038/nn.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin JP, Bell J. A Pedigree of Mental Defect Showing Sex-Linkage. J Neurol Psychiatry. 1943;6:154–157. doi: 10.1136/jnnp.6.3-4.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinton VJ, Brown WT, Wisniewski K, Rudelli RD. Analysis of neocortex in three males with the fragile X syndrome. Am J Med Genet. 1991;41:289–294. doi: 10.1002/ajmg.1320410306. [DOI] [PubMed] [Google Scholar]
- 10.Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharyya A, Zhao X. Human pluripotent stem cell models of Fragile X Syndrome. Mol Cell Neurosci. 2015 doi: 10.1016/j.mcn.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Telias M, Segal M, Ben-Yosef D. Neural differentiation of fragile X human embryonic stem cells reveals abnormal patterns of development despite successful neurogenesis. Dev Biol. 2013;374:32–45. doi: 10.1016/j.ydbio.2012.11.031. [DOI] [PubMed] [Google Scholar]
- 13.Telias M, Kuznitsov-Yanovsky L, Segal M, Ben-Yosef D. Functional Deficiencies in Fragile X Neurons Derived from Human Embryonic Stem Cells. J Neurosci. 2015;35:15295–15306. doi: 10.1523/JNEUROSCI.0317-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheridan SD, et al. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doers ME, et al. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014;23:1777–1787. doi: 10.1089/scd.2014.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willmann CA, et al. To clone or not to clone? Induced pluripotent stem cells can be generated in bulk culture. PLoS One. 2013;8:e65324. doi: 10.1371/journal.pone.0065324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazarov O, et al. Impaired survival of neural progenitor cells in dentate gyrus of adult mice lacking FMRP. Hippocampus. 2012;22:1220–1224. doi: 10.1002/hipo.20989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo W, et al. Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nat Med. 2011;17:559–565. doi: 10.1038/nm.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo W, et al. Inhibition of GSK3beta improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21:681–691. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greco CM, et al. Neuropathologic features in the hippocampus and cerebellum of three older men with fragile X syndrome. Mol Autism. 2011;2:2. doi: 10.1186/2040-2392-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumari D, et al. High-Throughput Screening to Identify Compounds That Increase Fragile X Mental Retardation Protein Expression in Neural Stem Cells Differentiated From Fragile X Syndrome Patient-Derived Induced Pluripotent Stem Cells. Stem Cells Transl Med. 2015;4:800–808. doi: 10.5966/sctm.2014-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufmann M, et al. High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. J Biomol Screen. 2015;20:1101–1111. doi: 10.1177/1087057115588287. [DOI] [PubMed] [Google Scholar]
- 23.Usdin K, Kumari D. Repeat-mediated epigenetic dysregulation of the FMR1 gene in the fragile X-related disorders. Front Genet. 2015;6:192. doi: 10.3389/fgene.2015.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuai L, et al. Chemical genetics identifies small-molecule modulators of neuritogenesis involving neuregulin-1/ErbB4 signaling. ACS Chem Neurosci. 2010;1:325–342. doi: 10.1021/cn900046a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacquemont S, et al. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci Transl Med. 2011;3:64ra1. doi: 10.1126/scitranslmed.3001708. [DOI] [PubMed] [Google Scholar]
- 26.Ananiev G, Williams EC, Li H, Chang Q. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS One. 2011;6:e25255. doi: 10.1371/journal.pone.0025255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farra N, et al. Rett syndrome induced pluripotent stem cell-derived neurons reveal novel neurophysiological alterations. Mol Psychiatry. 2012;17:1261–1271. doi: 10.1038/mp.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheung AY, Horvath LM, Carrel L, Ellis J. X-chromosome inactivation in rett syndrome human induced pluripotent stem cells. Front Psychiatry. 2012;3:24. doi: 10.3389/fpsyt.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung AY, et al. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchetto MC, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci U S A. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Squillaro T, et al. Reduced expression of MECP2 affects cell commitment and maintenance in neurons by triggering senescence: new perspective for Rett syndrome. Mol Biol Cell. 2012;23:1435–1445. doi: 10.1091/mbc.E11-09-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tropea D, et al. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci U S A. 2009;106:2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pini G, et al. Repeated insulin-like growth factor 1 treatment in a patient with rett syndrome: a single case study. Front Pediatr. 2014;2:52. doi: 10.3389/fped.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalscheuer VM, et al. Disruption of the serine/threonine kinase 9 gene causes severe X-linked infantile spasms and mental retardation. Am J Hum Genet. 2003;72:1401–1411. doi: 10.1086/375538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaving LS, et al. Mutations of CDKL5 cause a severe neurodevelopmental disorder with infantile spasms and mental retardation. Am J Hum Genet. 2004;75:1079–1093. doi: 10.1086/426462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tao J, et al. Mutations in the X-linked cyclin-dependent kinase-like 5 (CDKL5/STK9) gene are associated with severe neurodevelopmental retardation. Am J Hum Genet. 2004;75:1149–1154. doi: 10.1086/426460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mari F, et al. CDKL5 belongs to the same molecular pathway of MeCP2 and it is responsible for the early-onset seizure variant of Rett syndrome. Hum Mol Genet. 2005;14:1935–1946. doi: 10.1093/hmg/ddi198. [DOI] [PubMed] [Google Scholar]
- 39.Amenduni M, et al. iPS cells to model CDKL5-related disorders. Eur J Hum Genet. 2011;19:1246–1255. doi: 10.1038/ejhg.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Livide G, et al. GluD1 is a common altered player in neuronal differentiation from both MECP2-mutated and CDKL5-mutated iPS cells. Eur J Hum Genet. 2015;23:195–201. doi: 10.1038/ejhg.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patriarchi T, et al. Imbalance of excitatory/inhibitory synaptic protein expression in iPSC-derived neurons from FOXG1 patients and in foxg1 mice. Eur J Hum Genet. 2015 doi: 10.1038/ejhg.2015.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricciardi S, et al. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol. 2012;14:911–923. doi: 10.1038/ncb2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adamo A, et al. 7q11.23 dosage-dependent dysregulation in human pluripotent stem cells affects transcriptional programs in disease-relevant lineages. Nat Genet. 2015;47:132–141. doi: 10.1038/ng.3169. [DOI] [PubMed] [Google Scholar]
- 44.Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedrosa E, et al. Development of patient-specific neurons in schizophrenia using induced pluripotent stem cells. J Neurogenet. 2011;25:88–103. doi: 10.3109/01677063.2011.597908. [DOI] [PubMed] [Google Scholar]
- 47.Zhao D, et al. MicroRNA Profiling of Neurons Generated Using Induced Pluripotent Stem Cells Derived from Patients with Schizophrenia and Schizoaffective Disorder, and 22q11.2 Del. PLoS One. 2015;10:e0132387. doi: 10.1371/journal.pone.0132387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.da Silveira Paulsen B, Souza da Silveira M, Galina A, Kastrup Rehen S. Pluripotent stem cells as a model to study oxygen metabolism in neurogenesis and neurodevelopmental disorders. Arch Biochem Biophys. 2012 doi: 10.1016/j.abb.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 49.Prabakaran S, et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. doi: 10.1038/sj.mp.4001511. 643. [DOI] [PubMed] [Google Scholar]
- 50.Chiang CH, et al. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011;16:358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wen Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Green EK, et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry. 2013;18:1302–1307. doi: 10.1038/mp.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muhleisen TW, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. 2014;5:3339. doi: 10.1038/ncomms4339. [DOI] [PubMed] [Google Scholar]
- 54.Sue O’Shea K, McInnis MG. Neurodevelopmental Origins of Bipolar Disorder: iPSC Models. Mol Cell Neurosci. 2015 doi: 10.1016/j.mcn.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 55.Madison JM, et al. Characterization of bipolar disorder patient-specific induced pluripotent stem cells from a family reveals neurodevelopmental and mRNA expression abnormalities. Mol Psychiatry. 2015;20:703–717. doi: 10.1038/mp.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valvezan AJ, Klein PS. GSK-3 and Wnt Signaling in Neurogenesis and Bipolar Disorder. Front Mol Neurosci. 2012;5:1. doi: 10.3389/fnmol.2012.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen HM, et al. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl Psychiatry. 2014;4:e375. doi: 10.1038/tp.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim KH, et al. Transcriptomic Analysis of Induced Pluripotent Stem Cells Derived from Patients with Bipolar Disorder from an Old Order Amish Pedigree. PLoS One. 2015;10:e0142693. doi: 10.1371/journal.pone.0142693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bavamian S, et al. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol Psychiatry. 2015;20:573–584. doi: 10.1038/mp.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumar L, Verma S, Vaidya B, Gupta V. Exosomes: Natural Carriers for siRNA Delivery. Curr Pharm Des. 2015;21:4556–4565. doi: 10.2174/138161282131151013190112. [DOI] [PubMed] [Google Scholar]
- 62.Bernat V, Disney MD. RNA Structures as Mediators of Neurological Diseases and as Drug Targets. Neuron. 2015;87:28–46. doi: 10.1016/j.neuron.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mertens J, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hockemeyer D, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 66.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Dis Model Mech. 2013;6:896–904. doi: 10.1242/dmm.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brandon NJ. Uncovering the function of Disrupted in Schizophrenia 1 through interactions with the cAMP phosphodiesterase PDE4: Contributions of the Houslay lab to molecular psychiatry. Cell Signal. 2015 doi: 10.1016/j.cellsig.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 69.Pei Y, et al. A platform for rapid generation of single and multiplexed reporters in human iPSC lines. Sci Rep. 2015;5:9205. doi: 10.1038/srep09205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Inglese J, et al. Genome editing-enabled HTS assays expand drug target pathways for Charcot-Marie-tooth disease. ACS Chem Biol. 2014;9:2594–2602. doi: 10.1021/cb5005492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao WN, et al. A high-throughput screen for Wnt/beta-catenin signaling pathway modulators in human iPSC-derived neural progenitors. J Biomol Screen. 2012;17:1252–1263. doi: 10.1177/1087057112456876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okerlund ND, Cheyette BN. Synaptic Wnt signaling-a contributor to major psychiatric disorders? J Neurodev Disord. 2011;3:162–174. doi: 10.1007/s11689-011-9083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosen EY, et al. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71:1030–1042. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raitano S, et al. Restoration of progranulin expression rescues cortical neuron generation in an induced pluripotent stem cell model of frontotemporal dementia. Stem Cell Reports. 2015;4:16–24. doi: 10.1016/j.stemcr.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Efthymiou A, et al. Functional screening assays with neurons generated from pluripotent stem cell-derived neural stem cells. J Biomol Screen. 2014;19:32–43. doi: 10.1177/1087057113501869. [DOI] [PubMed] [Google Scholar]
- 76.Pei Y, et al. Comparative neurotoxicity screening in human iPSC-derived neural stem cells, neurons and astrocytes. Brain Res. 2015 doi: 10.1016/j.brainres.2015.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boutros M, Heigwer F, Laufer C. Microscopy-Based High-Content Screening. Cell. 2015;163:1314–1325. doi: 10.1016/j.cell.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 78.Wheeler HE, Wing C, Delaney SM, Komatsu M, Dolan ME. Modeling chemotherapeutic neurotoxicity with human induced pluripotent stem cell-derived neuronal cells. PLoS One. 2015;10:e0118020. doi: 10.1371/journal.pone.0118020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sirenko O, Hesley J, Rusyn I, Cromwell EF. High-content high-throughput assays for characterizing the viability and morphology of human iPSC-derived neuronal cultures. Assay Drug Dev Technol. 2014;12:536–547. doi: 10.1089/adt.2014.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Finkbeiner S, Frumkin M, Kassner PD. Cell-based screening: extracting meaning from complex data. Neuron. 2015;86:160–174. doi: 10.1016/j.neuron.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paull D, et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat Methods. 2015;12:885–892. doi: 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- 82.Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71:35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 83.Bang J, Spina S, Miller BL. Frontotemporal dementia. Lancet. 2015;386:1672–1682. doi: 10.1016/S0140-6736(15)00461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou J, Seeley WW. Network dysfunction in Alzheimer’s disease and frontotemporal dementia: implications for psychiatry. Biol Psychiatry. 2014;75:565–573. doi: 10.1016/j.biopsych.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 85.Jack CR, Jr, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brennand K, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015;20:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fong H, et al. Genetic correction of tauopathy phenotypes in neurons derived from human induced pluripotent stem cells. Stem Cell Reports. 2013;1:226–234. doi: 10.1016/j.stemcr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Usenovic M, et al. Internalized Tau Oligomers Cause Neurodegeneration by Inducing Accumulation of Pathogenic Tau in Human Neurons Derived from Induced Pluripotent Stem Cells. J Neurosci. 2015;35:14234–14250. doi: 10.1523/JNEUROSCI.1523-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wren MC, et al. Frontotemporal dementia-associated N279K tau mutant disrupts subcellular vesicle trafficking and induces cellular stress in iPSC-derived neural stem cells. Mol Neurodegener. 2015;10:46. doi: 10.1186/s13024-015-0042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iovino M, et al. Early maturation and distinct tau pathology in induced pluripotent stem cell-derived neurons from patients with MAPT mutations. Brain. 2015;138:3345–3359. doi: 10.1093/brain/awv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ehrlich M, et al. Distinct Neurodegenerative Changes in an Induced Pluripotent Stem Cell Model of Frontotemporal Dementia Linked to Mutant TAU Protein. Stem Cell Reports. 2015;5:83–96. doi: 10.1016/j.stemcr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Silva MC, et al. Society for Neuroscience, Annual Meeting. Chicago, IL: 2015. Tauopathy: Discovery of Small Molecule Modulators of Tau Phenotypes in Human iPSC-Derived Neuronal Models of Frontotemporal Degeneration. [Google Scholar]
- 95.Shi Y, et al. A human stem cell model of early Alzheimer’s disease pathology in Down syndrome. Sci Transl Med. 2012;4:124ra29. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muratore CR, et al. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–3536. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kondo T, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell Stem Cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 98.Israel MA, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cooper O, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci Transl Med. 2012;4:141ra90. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Woodard CM, et al. iPSC-derived dopamine neurons reveal differences between monozygotic twins discordant for Parkinson’s disease. Cell Rep. 2014;9:1173–1182. doi: 10.1016/j.celrep.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Flierl A, et al. Higher vulnerability and stress sensitivity of neuronal precursor cells carrying an alpha-synuclein gene triplication. PLoS One. 2014;9:e112413. doi: 10.1371/journal.pone.0112413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fallahi-Sichani M, Honarnejad S, Heiser LM, Gray JW, Sorger PK. Metrics other than potency reveal systematic variation in responses to cancer drugs. Nat Chem Biol. 2013;9:708–714. doi: 10.1038/nchembio.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mailman RB, Murthy V. Third generation antipsychotic drugs: partial agonism or receptor functional selectivity? Curr Pharm Des. 2010;16:488–501. doi: 10.2174/138161210790361461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fass DM, Schroeder FA, Perlis RH, Haggarty SJ. Epigenetic mechanisms in mood disorders: targeting neuroplasticity. Neuroscience. 2014;264:112–130. doi: 10.1016/j.neuroscience.2013.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Becher I, et al. Chemoproteomics reveals time-dependent binding of histone deacetylase inhibitors to endogenous repressor complexes. ACS Chem Biol. 2014;9:1736–1746. doi: 10.1021/cb500235n. [DOI] [PubMed] [Google Scholar]
- 106.Soragni E, Chou CJ, Rusche JR, Gottesfeld JM. Mechanism of Action of 2-Aminobenzamide HDAC Inhibitors in Reversing Gene Silencing in Friedreich’s Ataxia. Front Neurol. 2015;6:44. doi: 10.3389/fneur.2015.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Miller JD, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pasca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang JL, et al. Label-free, live optical imaging of reprogrammed bipolar disorder patient-derived cells reveals a functional correlate of lithium responsiveness. Transl Psychiatry. 2014;4:e428. doi: 10.1038/tp.2014.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vadodaria KC, et al. Generation of functional human serotonergic neurons from fibroblasts. Mol Psychiatry. 2015 doi: 10.1038/mp.2015.161. [DOI] [PubMed] [Google Scholar]
- 112.Li W, et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci U S A. 2011;108:8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ladewig J, et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods. 2012;9:575–578. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- 114.Hu W, et al. Direct Conversion of Normal and Alzheimer’s Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell. 2015;17:204–212. doi: 10.1016/j.stem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 115.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Morrison M, et al. StemBANCC: Governing Access to Material and Data in a Large Stem Cell Research Consortium. Stem Cell Rev. 2015;11:681–687. doi: 10.1007/s12015-015-9599-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hadida S, et al. Discovery of N-(2,4-di-tert-butyl-5-hydroxyphenyl)-4-oxo-1,4-dihydroquinoline-3-carboxamide (VX-770, ivacaftor), a potent and orally bioavailable CFTR potentiator. J Med Chem. 2014;57:9776–9795. doi: 10.1021/jm5012808. [DOI] [PubMed] [Google Scholar]
- 118.Kaiser T, Feng G. Modeling psychiatric disorders for developing effective treatments. Nat Med. 2015;21:979–988. doi: 10.1038/nm.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wainger BJ, et al. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Perlis RH. Translating biomarkers to clinical practice. Mol Psychiatry. 2011;16:1076–1087. doi: 10.1038/mp.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]