Abstract
We report on a relatively large survey of Prader-Willi syndrome, Angelman syndrome, and control subjects with the newly described methylation polymerase chain reaction (PCR) method to determine its usefulness for molecular diagnosis. Sixty-one Prader-Willi syndrome (PWS) individuals (26 men and 35 women), 9 Angelman syndrome (AS) patients (5 men and 4 women), and 58 other individuals were studied with methylation PCR following sodium bisulfite treatment of genomic DNA. In addition, multiple tissues, including fetal tissue, were studied from several individuals to determine the effects of various tissues on methylation PCR results. The expected methylation PCR result was observed in each case. This PCR-based assay evaluates the methylation status of the CpG island of the SNRPN gene and allows for rapid molecular diagnosis of PWS or AS with less labor than Southern hybridization for methylation analysis. The PCR results were identical to those achieved by Southern hybridization in those individuals studied.
Keywords: Prader-Willi syndrome, uni-parental disomy, chromosome 15q11–q13 region, methylation, sodium bisulfite
INTRODUCTION
Herein, we report our experience with the sodium bisulfite-treated DNA methylation specific polymerase chain reaction (PCR) method previously described by Kubota et al. [1997] in a relatively large sample of previously diagnosed Prader-Willi syndrome (PWS) and Angelman syndrome (AS) subjects and control individuals in order to determine its usefulness for molecular diagnosis.
MATERIALS AND METHODS
Subjects
Sixty-one PWS subjects (26 men and 35 women), 9 AS subjects (5 men and 4 women), and 58 controls, including subjects with some PWS or AS traits but otherwise not consistent with the clinical diagnosis, were evaluated (total of 128 subjects). The previously diagnosed PWS and AS subjects were evaluated clinically by one of the authors (M.G.B.). In most cases in this study, genomic DNA was isolated from established lymphoblastoid cell lines or from peripheral blood leukocytes. However, DNA from fibroblasts was obtained from two subjects, and 21 different tissues (liver, lung, brain, fascia, adipose, large intestine, small intestine, stomach, pancreas, kidney, heart muscle, lymph node, spleen, skin, brain tumor, thymus, adrenal, ovary, placenta, blood, and skeletal muscle) were studied from three different control subjects (seven tissues from a 20-week gestation female fetus, four tissues from a 4-year-old male, and 12 tissues from a 72-year-old male) and brain tissue from two adult PWS females.
Molecular Genetics
Oligonucleotides used for the PCR analysis were commercially synthesized and represented the maternally methylated sequences at genomic position +111 and +284 of the SNRPN gene and the paternally unmethylated sequences at genomic position +140 and +239 of this gene from sequence data referenced in Genbank (Bethesda, MD) and reported previously [Sutcliffe et al., 1994; Kubota et al., 1997]. The SNRPN is a paternally expressed gene in the 15q11–q13 that has been used successfully with Southern hybridization for laboratory diagnosis of PWS and AS [Buiting et al., 1995; Kubota et al., 1996]. The two sets of primers were specifically designed to amplify the unmodified maternally methylated sequence and a second modified paternally unmethylated sequence after treatment with sodium bisulfite. DNA treated with sodium bisulfite will convert cytosine to uracil except when cytosine is methylated as found on the maternal chromosome [Kubota et al., 1997]. 5-methyl cytosine is resistant to bisulfite treatment and remains unchanged. The PCR-generated maternal fragment is 174 bp long and the paternal fragment is 100 bp [Kubota et al., 1997]. Briefly, genomic DNA (2 μg) was denatured in a 50 μl volume using freshly prepared sodium hydroxide (0.2 mol/L final concentration) for 10 min at 37°C. Thirty μl of 10 mmol/L hydroquinone and 520 μl of 3.6 mol/L sodium bisulfite (Sigma, St. Louis, MO), pH 5.0, both made fresh, were added to the denatured DNA samples and incubated at 55°C under mineral oil for 16 to 18 hr. The modified DNA was purified using the Wizard DNA Clean-Up system (Promega, Madison, WI) following the supplier’s protocol and by eluting the modified DNA into 50 μl of water. Modification was completed in final concentration of 0.3 mol/L sodium hydroxide (made fresh) at room temperature for 5 min followed by ethanol precipitation by adding 66 μl of 5 mol/L ammonium acetate, 300 μl of 99% ethanol, and 1.2 μl of 20 mg/ml glycogen (Boehringer Mannheim, Indianapolis, IN) as a carrier and the DNA resuspended in 50 μl of water and stored frozen at −20°C until used.
PCR was performed in a 30-μl volume containing 1 × PCR buffer with 1.5 mmol/L MgCl2 and 2.5 units of AmpliTaq Gold (both from Perkin-Elmer, Norwalk, CT), 250 μmol/L dNTPs, 1.0 μmol/L maternal, and 1.2 μmol/L paternal primers (sequences and protocol reported by Kubota et al., 1997) that were commercially synthesized and 3 μl of modified DNA, along with unmodified genomic DNA and negative controls containing PCR reagents except DNA. The AmpliTaq was activated at 95°C for 10 min, and DNA was amplified for 40 cycles at 94°C for 60 sec, 62°C for 30 sec, and 72°C for 30 sec, followed by a final extension at 72°C for 10 min. Fifteen μl of PCR products were subjected to electrophoresis using 2% agarose gel in 0.5 × tris boric acid-ethylenediamine tetra-acetate (TBE) buffer, stained in ethidium bromide, visualized under ultra violet illumination, and photographed.
RESULTS AND DISCUSSION
Methylation PCR data on subjects with PWS or AS and controls are summarized in Table I. In all cases, the methylation PCR analysis produced the same diagnostic results as the standard Southern hybridization and methylation analysis of the SNRPN probe. There were no differences in our study in the methylation PCR results of DNA isolated from lymphoblasts, lymphocytes, fibroblasts, or other tissues studied. Figure 1 shows representative PCR methylation results of the SNRPN gene in control, PWS, and AS subjects. The present study documents the use of methylation PCR of the promoter region of the SNRPN gene in a relatively large number of subjects with less time and with smaller quantities of DNA compared with Southern hybridization of digested DNA using the SNRPN probe. In all previously diagnosed PWS subjects, the methylation pattern was consistent with this diagnosis. In all six Angelman syndrome subjects with the 15q11–q13 deletion, the methylation PCR result was in agreement with the diagnosis. Three of the nine (33%) AS subjects with normal chromosome analysis but meeting the clinical criteria for this syndrome did not show the AS methylation PCR pattern. This percentage in Angelman syndrome is comparable with that reported by methylation testing with the Southern hybridization method [Buiting et al., 1995; Kubota et al., 1996]. Of 58 control individuals [seven parents of PWS subjects; one balanced 13;15 translocation carrier; one with ring 15 syndrome; one with a 15q25 deletion; one with a chromosome 15 marker; one with a chromosome duplication (15q11–q13); 34 individuals with some manifestations of Prader-Willi syndrome (e.g., obesity); nine patients with possible Angelman syndrome; and autopsy specimens from three subjects without evidence of PWS or AS], all showed a normal DNA pattern with methylation PCR analysis. Our data additionally support that methylation PCR analysis is accurate and relatively simple. Most AS patients and all PWS patients showed the DNA pattern consistent for each diagnosis and agreed with their Southern hybridization results. Methylation PCR provides a reliable diagnostic method for PWS and AS patients. This technique has advantages over Southern hybridization for being less time consuming and less expensive as well as requiring a smaller quantity of DNA. This technique should be incorporated into the diagnostic work up for PWS or AS subjects particularly in those with normal molecular cytogenetic studies.
TABLE I.
Methylation PCR Data in Prader-Willi Syndrome, Angelman Syndrome, and Control Subjects
| Group | Number of subjects | Methylation PCR data |
|---|---|---|
| PWS | 61 | 61: Maternal only (PWS pattern) |
| AS | 9 | 6: Paternal only (AS pattern) 3: Maternal and paternal (normal) |
| Other | 58 | 58: Maternal and paternal (normal) |
Fig. 1.
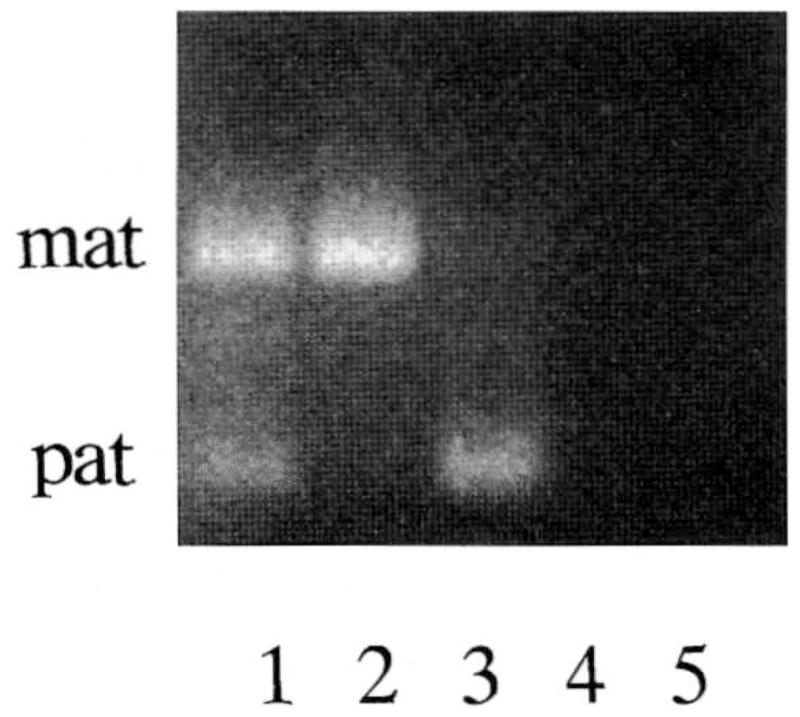
Representative methylation PCR data from three individuals. Lane 1, normal DNA pattern (maternal and paternal) in a control subject; lane 2, Prader-Willi syndrome DNA pattern (maternal only) in a PWS subject; lane 3, Angelman syndrome DNA pattern (paternal only) in an AS subject; lane 4, untreated DNA plus PCR reaction mixture; and lane 5, PCR reaction mixture but without DNA.
Acknowledgments
Contract grant sponsor: National Institute of Child Health and Human Development; Contract grant number: P01H030329.
We thank Lee Beth Kilgore for expert preparation of the manuscript and Annis Marney for technical assistance. This research was partially supported by a grant from NICHD (P01H030329).
References
- Buiting K, Saitoh S, Gross S, Dittrich B, Schwartz S, Nicholls RD, Horsthemke B. Inherited microdeletions in the Angelman and Prader-Willi syndromes define an imprinting center on human chromosome 15. Nat Genet. 1995;9:395–400. doi: 10.1038/ng0495-395. [DOI] [PubMed] [Google Scholar]
- Kubota T, Das S, Christian SL, Baylin SB, Herman JG, Ledbetter DH. Methylation-specific PCR simplifies imprinting analysis. Nat Genet. 1997;16:16–17. doi: 10.1038/ng0597-15. [DOI] [PubMed] [Google Scholar]
- Kubota T, Sutcliffe JS, Aradhya S, Gillessen-Kaesbach G, Christian SL, Horsthemke B, Beaudet AL, Ledbetter DH. Validation studies of SNRPN methylation as a diagnostic test for Prader-Willi syndrome. Am J Med Genet. 1996;66:77–80. doi: 10.1002/(SICI)1096-8628(19961202)66:1<77::AID-AJMG18>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Nakao M, Christian S, Orstavik KH, Tommerup N, Ledbetter DH, Beaudet AL. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nat Genet. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]


