Abstract
The present standard of care for hepatitis C virus (HCV) infection is pegylated alpha interferon (IFN-α) in combination with ribavirin. However, specific antivirals such as HCV NS3-NS4A protease inhibitors are now in clinical development, and these agents can potentially be used in combination with the present treatments. Therefore, it is important to investigate the potential benefits or adverse effects of these new combinations by using available in vitro HCV culture systems first. In the present study we demonstrate that the combination of a specific HCV NS3-NS4A protease inhibitor and IFN-α synergistically inhibits HCV RNA replication in replicon cells, with little or no increase in cytotoxicity. Furthermore, the benefit of the combination was sustained over time, such that a greater than 3-log reduction in HCV RNA levels was achieved following 9 days of treatment. The viral RNA appeared to be cleared from the replicon cells after 14 days of treatment, and no viral RNA rebound was observed upon withdrawal of the inhibitors. In each case, the antiviral effects obtained with higher concentrations of either the protease inhibitor alone or IFN-α alone can be achieved by a combination of both agents at lower concentrations, which may potentially reduce the risk of possible adverse effects associated with high doses of either agent.
Despite the recent improvement in hepatitis C treatment with the introduction of pegylated interferon alpha (IFN-α) plus ribavirin, chronic hepatitis C continues to present a serious health challenge that affects 170 million people worldwide, including 4 million people in the United States and 8 million people in Europe and Japan (30). Treatment with pegylated IFN-α plus ribavirin results in a sustained viral response in less than 50% of the patients infected with hepatitis C virus (HCV) genotype 1, which is more difficult to treat; and the majority of patients in the United States, Europe, and Japan are infected with genotype 1. Therefore, new and better treatments for hepatitis C are much needed.
HCV has a 9.6-kb plus-strand RNA genome that encodes a polyprotein precursor of about 3,000 amino acids which is processed proteolytically upon translation by both cellular and viral proteases to 10 individual proteins: C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (for a review, see reference 23). Recently, an additional protein, F/ARFP, of unknown function was identified. F/ARFP is encoded by an alternative open reading frame overlapping with the core (C) protein-coding sequence (34-36). Two of the nonstructural (NS) proteins, NS5B and NS3, are the most characterized. NS5B is the viral RNA-dependent RNA polymerase. NS3 comprises an N-terminal protease domain of 181 amino acids and a C-terminal helicase domain. The serine protease activity of NS3 in complex with the NS4A cofactor is responsible for the proteolytic cleavage at four junctions of the HCV polyprotein precursor: NS3-NS4A (self-cleavage), NS4A-NS4B, NS4B-NS5A, and NS5A-NS5B (2, 9, 10, 13, 22, 31, 33). The NS3-NS4A protease has been an attractive target in the development of new antivirals with activities against HCV (for a review, see references 5 and 28) since it is essential for viral replication (16). Recently, a proof-of-concept clinical study (18) demonstrated the antiviral efficacy of a potent HCV NS3-NS4A protease inhibitor in hepatitis C patients.
With new, specific antivirals on the horizon, the treatment options for HCV are likely to expand. The use of combinations of antiviral agents with different mechanisms of action is likely to be an important strategy to increase antiviral potency, to reduce the toxicities associated with individual agents, and to suppress viral resistance. Therefore, it is important to explore the feasibility and potential benefit of combination therapy with new anti-HCV agents. Although the ultimate test for any new treatment is the demonstration of clinical efficacy and safety, given the high cost and limited number of clinical studies that can be performed, it would be very helpful to investigate potential drug-drug combinations in vitro beforehand. Although a robust, reliable infectious cell culture system is not available for HCV, the discovery of a subgenomic HCV replicon (25) and the subsequent optimization of the system (3, 17, 24) have greatly facilitated the evaluation of antiviral activities of new anti-HCV drug candidates. It has been reported that HCV RNA replication in replicon cells can be inhibited by either IFN-α (3, 8, 12) or NS3-NS4A protease inhibitors (21, 27). Here we report on a quantitative analysis of the effects of the combination of a specific HCV NS3-NS4A protease inhibitor and IFN-α on the HCV replicon in replicon cells. We demonstrate that these two agents act synergistically to inhibit the replication of the HCV replicon RNA. The benefit of the combination treatment was sustained over time, reduced HCV replicon RNA levels by more than 4 orders of magnitude after up to 14 days of treatment, and prevented a rebound of the HCV replicon in cells.
MATERIALS AND METHODS
Compounds.
Protease inhibitor 1 (PI-1) (Fig. 1) was synthesized by Vertex Pharmaceuticals Inc. (Cambridge, Mass.), dissolved in dimethyl sulfoxide (DMSO) as a 20 mM solution, and stored at −20°C. Human recombinant IFN-α was purchased from Calbiochem (La Jolla, Calif.) and was stored at −70°C. Ribavirin was obtained from Sigma (St. Louis, Mo.), dissolved in DMSO as a 500 mM solution, and stored at −20°C.
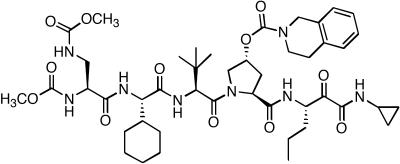
FIG. 1.
Chemical structure of PI-1, a specific HCV NS3-NS4A protease inhibitor.
Generation of HCV replicon cells.
Parental Huh-7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; JRH Biosciences, Lenexa, Kans.) containing 10% heat-inactivated fetal bovine serum (ΔFBS; JRH Biosciences), 2 mM l-glutamine, and nonessential amino acids (JRH Biosciences). The cells were transfected with an in vitro-transcribed subgenomic HCV replicon RNA whose sequence was identical to that of the I377neo/NS3-3′/wt replicon described by Lohmann et al. (25). Stable cells containing the self-replicating HCV replicon were selected and maintained in the presence of 250 μg of G418 (Invitrogen, Carlsbad, Calif.) per ml and were used for HCV replicon assays.
Two-day HCV replicon assay.
HCV replicon cells were plated in a 96-well plate at a density of 104 cells per well in DMEM with 10% ΔFBS. On the following day (∼16 h later), the culture medium was replaced with DMEM containing either no compound as a control or compounds serially diluted in the presence of 2% ΔFBS and 0.5% DMSO. After the cells were incubated with the compounds for 48 h, the intracellular RNA was extracted with an RNeasy 96 kit (Qiagen, Valencia, Calif.). The level of HCV RNA, both the positive strand and the negative strand, was determined by a real-time multiplex quantitative reverse transcription-PCR (RT-PCR) assay (the Taqman assay) with a pair of HCV-specific primers (5′-CCA TGA ATC ACT CCC CTG TG-3′ and 5′-CCG GTC GTC CTG GCA ATT C-3′), an HCV-specific probe (5′-6-FAM-CCT GGA GGC TGC ACG ACA CTC A-TAMRA-3′, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine), and an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, Calif.). In each experiment, each datum point represents the average of five replicates in cell culture. The IC50 was defined as the concentration of compound at which the HCV RNA level in the replicon cells was reduced by 50%. To monitor any cytotoxic effect, the viabilities of the replicon cells following 48 h of treatment with compound were determined by using a tetrazolium compound (3-[4,5-dimethylthiazol-2-yl]-5-[3-carboxymethoxyphenyl]-2-[4-sulfophenyl]-2H-tetrazolium, inner salt [MTS])-based assay (CellTiter 96 AQueous One Solution Cell Proliferation Assay; Promega, Madison, Wis.). The CC50 was defined as the concentration of the compound at which cell viability was reduced by 50%.
Synergy and antagonism analysis.
The effects of drug-drug combinations were evaluated in two different mathematical models, the Bliss independence model and the Loewe additivity model, which have distinct definitions of a theoretical additive effect, synergy (a greater than additive effect), or antagonism (a less than additive effect) (11).
For the Bliss independence model, the experimental data were analyzed by using MacSynergy, a three-dimensional analytical method developed by Prichard and Shipman (29). In this model, the theoretical additive effect is calculated from the dose-response curves of individual compounds by the equation Z = X + Y(1 − X), where X and Y represent the inhibition produced by drug 1 alone and drug 2 alone, respectively, and Z represents the effect produced by the combination of drug 1 and drug 2. The theoretical additive surface is subtracted from the actual experimental surface, resulting in a surface that would appear as a horizontal plane at 0% inhibition if the combination were merely additive. Any peak above this plane would indicate synergy, whereas any depression below it would indicate antagonism. The 95% confidence intervals for the experimental dose-response surface are used to evaluate the data statistically. The volume of the peak or depression is calculated to quantify the overall synergy or antagonism produced.
For the Loewe additivity model, the experimental data were analyzed by using CalcuSyn (Biosoft, Ferguson, Mo.), a computer program based on the method of Chou and Talalay (4). Briefly, the dose-effect curves for each drug or drug combination were converted to median-effect plots with the program. Then, a combination index (CI) value for each experimental combination was calculated on the basis of the following equation: [(D)1/(Dx)1] + [(D)2/(Dx)2] + [(D)1(D)2/(Dx)1(Dx)2], where (Dx)1 and (Dx)2 are the doses of drug 1 and drug 2 that have x effect when each drug is used alone, respectively, and (D)1 and (D)2 are the doses of drug 1 and drug 2 that have the same x effect when they are used in combination, respectively. CI values of <1, 1, and >1 indicate synergy, an additive effect, and antagonism, respectively.
Nine-day HCV replicon assay.
HCV replicon cells were plated at a very low density of 500 cells per well in a 96-well plate so that the cells would not reach confluence during 9 days in culture. Compounds were serially diluted in DMEM containing 10% ΔFBS and 0.2% DMSO in the absence of G418. Fresh medium and compounds were added to the cells every 3 days. After the cells were treated with antiviral compounds for 3, 6, or 9 days, the number of cells in each well was determined by the tetrazolium compound (MTS)-based assay with an established standard curve. The level of HCV RNA in the cells was determined by quantitative RT-PCR (Taqman), as described above for the 2-day replicon assay. The copy number of HCV replicon RNA per cell was calculated for cells treated with compound and compared to that for control cells treated with 0.2% DMSO in medium.
HCV replicon clearance and rebound study.
HCV replicon cells were plated in a six-well tissue culture plate at a density of 2 × 105 cells per well. Compounds were serially diluted in DMEM containing 10% ΔFBS and 0.2% DMSO in the absence of G418. The cells were split every 3 to 4 days and placed into fresh medium with compound or 0.2% DMSO (no-compound control) before they reached confluence, while a cell sample was harvested. After 14 days of treatment, the medium with compound was removed and the cells were washed three times before they were split and placed into fresh medium containing 10% ΔFBS and 250 μg of G418 per ml but no antiviral compound. G418 was added to the culture to enrich the cells from which the HCV replicon had not been completely cleared. During the 3 weeks after the antiviral compound was removed, cell samples were taken whenever the cells reached about 80% confluence and were split and placed into fresh medium containing 250 μg of G418 per ml. For all the samples taken, the number of live cells in each sample was determined by a ViaCount assay (Guava Technologies, Hayward, Calif.). The level of HCV RNA in the cells was determined by the quantitative RT-PCR (Taqman), as described above for the 2-day replicon assay, and then the copy number of HCV replicon RNA per cell in each sample was calculated.
RESULTS
PI-1, a potent HCV NS3-NS4A protease inhibitor, reduces the level of HCV replicon RNA in a dose-dependent manner.
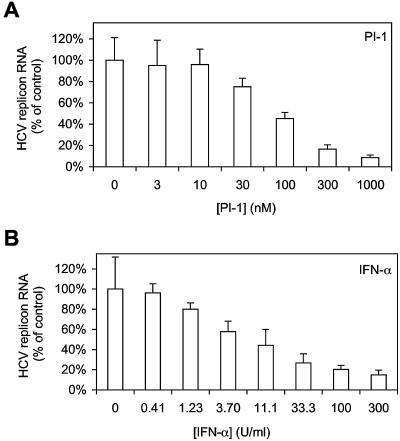
PI-1 (Fig. 1) is a competitive, reversible, peptidomimetic inhibitor of HCV NS3-NS4A protease with a Ki of 8 nM against the NS3 serine protease domain in the presence of an NS4A peptide (1, 6). The antiviral activity of PI-1 was examined in HCV replicon cells, in which inhibition of NS3-NS4A protease is expected to lead to inhibition of viral RNA replication and, subsequently, a reduction in the total level of HCV replicon RNA in the cells. As shown in Fig. 2A, there was a dose-dependent reduction of replicon RNA in cells treated with PI-1. The average IC50 of PI-1 following 48 h of treatment was calculated to be 68 nM from three independent experiments. No significant cytotoxicity, as determined by the MTS-based cell viability assay, was observed with up to 10 μM PI-1 after 48 h of treatment, and the CC50 for HCV replicon cells was greater than 50 μM (data not shown). As shown in Fig. 2B, IFN-α also inhibited the HCV replicon in a dose-dependent manner, with an IC50 of 6.55 U/ml, and no significant cytotoxicity was observed with up to 1,000 U of IFN-α per ml. The typical IC50 of IFN-α in this assay ranged from 1 to 10 U/ml in several independent experiments (data not shown).
FIG. 2.
Dose-dependent inhibition of HCV replicon by PI-1 or IFN-α alone. HCV replicon cells were treated with PI-1 (A) or IFN-α (B) for 48 h. At the end of the 48-h treatment, total cellular RNA was extracted with the RNeasy-96 kit. The levels of HCV replicon RNA remaining were then determined by quantitative RT-PCR, as described in Materials and Methods, and are shown as a percentage of the levels of replicon RNA in cells treated with 0.5% DMSO (control). Each bar represents the average of five cell culture replicates with the standard deviation.
Combination of an NS3-NS4A protease inhibitor with IFN-α results in a synergistic reduction in HCV replicon RNA levels.
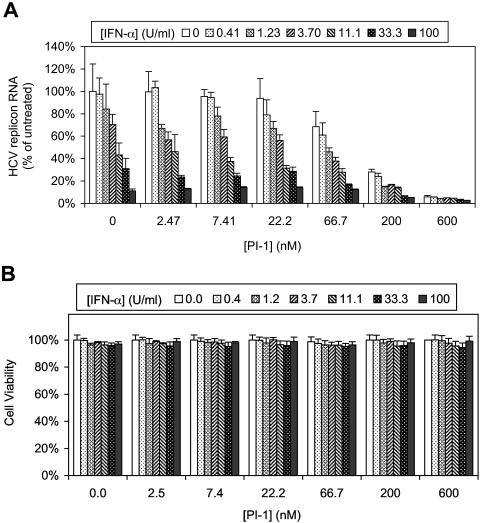
The present standard of care for the treatment of HCV infection is the combination of pegylated IFN-α and ribavirin, which significantly improves the sustained viral response rate compared to that achieved with IFN-α monotherapy. However, ribavirin itself does not exhibit a strong antiviral effect when it is administered as monotherapy, and its potency in the HCV replicon assay is poor (IC50 > 20 μM; data not shown). Therefore, in this study we chose to investigate the combination of IFN-α and an HCV NS3-NS4A protease inhibitor for its direct antiviral activity. HCV replicon cells were treated for 48 h with various concentrations of PI-1, IFN-α, or the two in combination. As shown in Fig. 3A, at any given concentration of PI-1, the addition of IFN-α led to a further decrease in HCV replicon RNA levels in a dose-dependent manner. Similarly, at any given concentration of IFN-α, there was a dose-dependent decrease in HCV replicon RNA levels when PI-1 was added. In other words, the combination of the HCV NS3-NS4A protease inhibitor and IFN-α always resulted in a greater reduction in the level of HCV RNA compared to that achieved by each agent alone. It is important that there was little or no increase in cytotoxicity when PI-1 and IFN-α were used together. In fact, none of the treatments reduced cell viability by more than 6% (Fig. 3B).
FIG. 3.
Combination of PI-1 with IFN-α in the 2-day HCV replicon cell assay. HCV replicon cells were treated with various concentrations of PI-1 (indicated on the x axis) in combination with various concentration of IFN-α (indicated at the top) for 48 h. (A) The level of HCV RNA remaining in replicon cells treated with PI-1 and IFN-α in combination was determined by quantitative RT-PCR and is shown as a percentage of the level in cells treated with 0.5% DMSO (control). Each bar represents the average of six cell culture replicates with the standard deviation. (B) The viability of HCV replicon cells following compound treatment for 48 h was determined by an MTS-based cell viability assay and is presented as a percentage of the viability of cells treated with 0.5% DMSO (control). Each bar represents the average of four cell culture replicates with the standard deviation.
Next, we wanted to address whether the activity of the combination of PI-1 and IFN-α was synergistic or simply additive. The data shown in Fig. 3A were analyzed in both the Bliss independence and the Loewe additivity models (11).
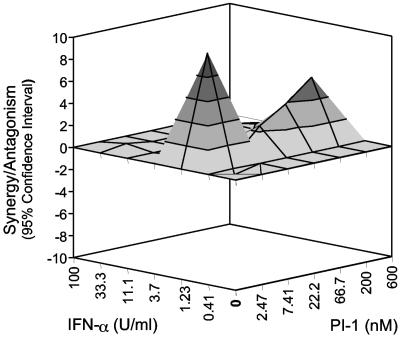
For the Bliss independence model, the experimental data were analyzed by use of a computer program, MacSynergy (29). The theoretical additive effect of any given combination of PI-1 and IFN-α was calculated from the dose-response curve of a single agent alone, either PI-1 or IFN-α, by the equation described in Materials and Methods. The calculated theoretical additive effect was subtracted from the actual experimental effect shown in Fig. 3A, and only the statistically significant (95% confidence interval) difference between the two was plotted in Fig. 4. If the combination were merely additive, the resulting surface in the plot shown in Fig. 4 would appear as a horizontal plane at the zero value. Any peak above the zero plane indicates more than an additive effect, namely, synergy, whereas any depression below the zero plane indicates a less than additive effect, namely, antagonism. As shown in Fig. 4, the combination of PI-1 and IFN-α was synergistic. The total log volume of the synergy was calculated to be 6, which is considered moderate synergy, according to the guidelines of the program.
FIG. 4.
Analysis of the combination of PI-1 and IFN-α by use of the Bliss independence model. A three-dimensional graph was used to illustrate the difference between the experimental effect and the theoretical additive effect for each combination. The predicted theoretical additive effect was calculated by using the MacSynergy program, as described in Materials and Methods, and was subtracted from the experimental effect (shown in Fig. 3A), and then the difference between the two effects was plotted in the graph shown here. The zero plane (the gray plane) across the z axis represents the theoretical additive effect, a positive value displayed as a peak above the plane (the height is correlated with increasing grayness) indicates synergy, and a negative value shown as a valley below the plane indicates antagonism. Each experimental datum point represents the average of six cell culture replicates, and 95% confidence intervals were used to evaluate the data.
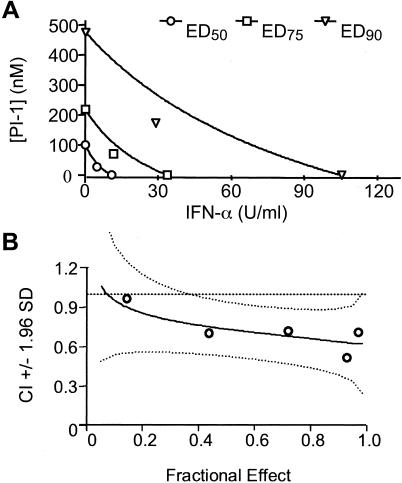
For the Loewe additivity model, the experimental data were analyzed by use of a computer program, CalcuSyn (4). In this model, the antiviral effects produced by the combination of PI-1 and IFN-α at various fixed ratios of concentrations were compared to those produced by PI-1 or IFN-α alone. As an example, the results of a detailed analysis of the combination of PI-1 (in nanomolar) and IFN-α (in units per milliliter) at a fixed ratio of 6:1 are shown in Fig. 5. The analysis can be presented as a traditional isobologram (Fig. 5A), in which the lines represent the effective doses (EDs) of the two agents that would be needed to achieve 50, 75, or 90% inhibition if the combination was simply additive. As shown by the datum points in Fig. 5A, the actual experimental doses needed to achieve these effects are less than the expected doses, indicating synergy. In order to further quantify the combination effect, the CI value was calculated for each experimental combination and plotted in Fig. 5B, in which a CI value of less than 1 indicates synergy. As shown in Table 1, the combination of PI-1 and IFN-α at three different ratios was analyzed by this method, and at all three ratios the combination showed moderate synergy, according to the guidelines of the program.
FIG. 5.
Analysis of the combination of PI-1 and IFN-α by use of the Loewe additivity model. (A) The data for the combination shown in Fig. 3A were also analyzed by using CalcuSyn program and are presented as a traditional isobologram, in which the lines represent the EDs of the two drugs required to achieve 50, 75, or 90% inhibition if the effects of the two compounds were simply additive. The dots are the actual experimental doses used to achieve those effects. (B) The CI can be calculated for each combination, as described in the Materials and Methods, and is plotted as the solid line versus the percent inhibition (i.e., the fractional effect). The 95% confidence intervals (1.96 standard deviations) of the CI are shown as two curved dotted lines. The theoretic additive effect (CI = 1) is represented by a straight dotted line.
TABLE 1.
CIs for the combination of PI-1 and IFN-α at three different ratios in Loewe additivity modela
| PI-1 (nM)/IFN-α (U/ml) ratio | CI values at:
|
||
|---|---|---|---|
| ED50 | ED75 | ED90 | |
| 2:1 | 0.49 | 0.65 | 0.91 |
| 6:1 | 0.86 | 0.79 | 0.74 |
| 18:1 | 0.86 | 0.78 | 0.73 |
ED50, ED75, and ED90 are the EDs of the compounds required to achieve 50, 75, and 90% inhibition, respectively. The CI values at various percentages of inhibition were calculated with the CalcuSyn program, as described in Materials and Methods.
Combination of an NS3-NS4A protease inhibitor and IFN-α facilitates multilog reductions in HCV replicon RNA levels.
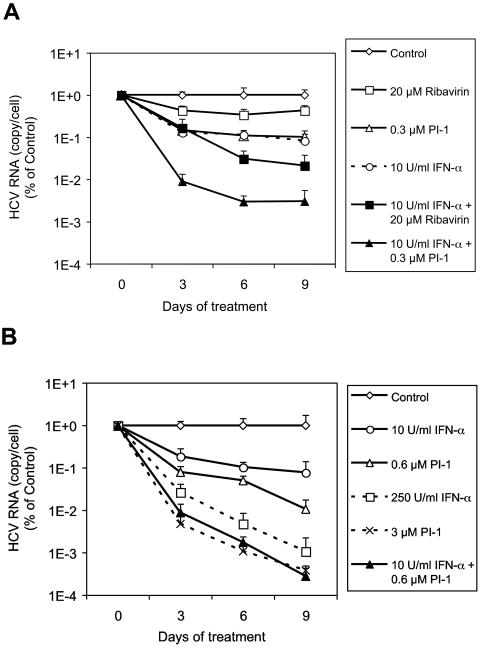
To investigate whether the enhanced antiviral effect of PI-1 and IFN-α in combination can be sustained over time and whether multiple logarithms of viral RNA reduction can be achieved, we treated the cells with PI-1, IFN-α, or ribavirin alone or with the combination of IFN-α and either PI-1 or ribavirin for 3, 6, or 9 consecutive days and measured the amount of HCV replicon RNA remaining in the cells. As shown in Fig. 6A, there was a time-dependent decrease in the level of HCV replicon RNA following treatment with the various agents. After 9 days of treatment, 0.3 μM PI-1 or 10 U of IFN-α per ml each reduced the level of HCV replicon RNA by ∼1 log, whereas the combination of the two agents together resulted in a greater reduction (2.5 logs) in the amount of viral RNA. In comparison, only a small decrease (0.4 log) in the HCV RNA level was observed with 20 μM ribavirin, which was the highest concentration of the compound that could be tested in this assay without significantly affecting cell viability. When 20 μM ribavirin was combined with 10 U of IFN-α per ml, a 1.7-log decrease in the HCV RNA level was induced, which was more than the effect achieved with IFN-α alone but less than that achieved with PI-1 and IFN-α in combination. Another comparison of combination and single treatments is shown in Fig. 6B, in which 0.6 μM PI-1 alone and 10 U of IFN-α per ml alone caused 2.0- and 1.1-log decreases in the level of HCV replicon RNA after 9 days of treatment, respectively. Combination of the two treatments resulted in a 3.6-log reduction in HCV RNA levels. Treatment with each compound alone would require higher concentrations of either PI-1 (3 μM) or IFN-α (250 U/ml) to achieve the same degree of inhibition.
FIG. 6.
Nine-day treatment of HCV replicon cells with PI-1, IFN-α, or ribavirin. The cells were treated with compound for 3, 6, or 9 days. At the end of each treatment period, cell numbers were determined by an MTS-based cell viability assay with an established standard curve, and the level of HCV RNA in the cells was determined by the quantitative RT-PCR assay. The copy number of HCV RNA per cell in each sample is plotted as the percentage (on a log scale) of the copy number in replicon cells incubated with 0.2% DMSO (control) for the same period of time. Each datum point represents the average of five cell culture replicates with the standard deviation.
Combination of an NS3-NS4A protease inhibitor and IFN-α clears HCV RNA from replicon cells and prevents viral RNA rebound after withdrawal of treatment.
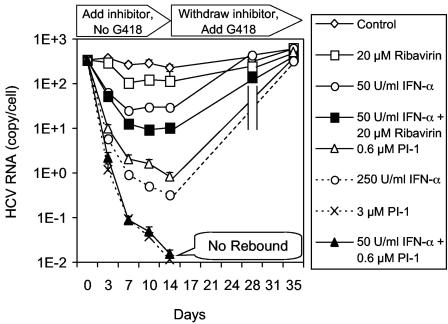
The goal of anti-HCV therapy is to completely eradicate the virus and to prevent a rebound after the termination of treatment, also known as a sustained viral response. We developed a method to model antiviral therapy in HCV replicon cells by identifying the antiviral agents and the conditions which could completely remove HCV RNA from the replicon cells (after a >4-log drop) and test for rebound after the antiviral compounds are withdrawn. As shown in Fig. 7, HCV replicon cells were treated with PI-1, IFN-α, or ribavirin alone or the combination of IFN-α and either PI-1 or ribavirin for 14 days in the absence of G418. The cells grew normally and were split every 3 or 4 days and placed into fresh medium with inhibitors before they reached confluence. A sample was taken at the same time to determine the level of HCV RNA remaining in the cells. In control cells treated with 0.2% DMSO, the HCV RNA level remained stable for 14 days in the absence of G418. After 14 days of treatment, only a 0.5-log drop in the level of HCV replicon RNA was observed with 20 μM ribavirin alone, 50 U of IFN-α per ml reduced the HCV RNA level by ∼1 log, and the combination of the two reduced the HCV RNA level by 1.5 logs. A 2.5-log reduction in the HCV RNA level was observed when cells were treated with 0.6 μM PI-1 alone for 14 days. In contrast, the combination of 0.6 μM PI-1 and 50 U of IFN-α per ml resulted in a ∼4.5-log reduction in the HCV RNA level, which was similar to that of 3 μM PI-1 alone and which was better than that of 250 U of IFN-α per ml alone (∼3-log drop).
FIG. 7.
Rebound of HCV replicon after a 14-day treatment with PI-1, IFN-α, or ribavirin. HCV replicon cells were treated with PI-1, IFN-α, or ribavirin alone or the combination of IFN-α and either PI-1 or ribavirin in the absence of G418. After 14 days of treatment, the compounds were withdrawn and 250 μg of G418 per ml was added to enrich the remaining HCV replicon-positive cells that are capable of growing in the presence of G418 (rebound). The cultures were monitored for another 21 days in the presence of G418, and cell samples were collected whenever the cell monolayer reached confluence. The cell number was determined by the Guava ViaCount assay, as described in the Materials and Methods, and the level of HCV RNA in the cells was determined by the quantitative RT-PCR assay. The absolute numbers of HCV replicon RNA copies per viable cell are shown.
The level of HCV RNA remaining after an ∼4.5-log reduction was close to the detection limit of our quantitative RT-PCR assay. The signal may simply represent a background of degraded HCV RNA rather than viable replicons capable of replicating in cells. In other words, functional HCV replicon may already have been cleared from the cells, even though an extremely small amount of residual HCV RNA was detected. In order to test this hypothesis, we performed a rebound experiment in which the inhibitors were withdrawn from the culture medium on day 14. Unlike a viral infection system, the HCV replicon is a stable cell line maintained under G418 selection, which enriches the population of replicon-positive cells over that of replicon-negative cells. Therefore, when the inhibitors were withdrawn, 250 μg G418 per ml was added back to the culture medium in order to expand any cell from which viable HCV replicon RNA had not been completely cleared. The cells were cultured for three additional weeks in the presence of G418, during which the cells were split whenever they reached 80% confluence and a cell sample was taken at the same time. Cells that had completely lost the HCV replicon died between 10 and 14 days in the presence of 250 μg of G418 per ml, just like the parental Huh-7 cells. The levels of HCV replicon RNA in cells treated with 20 μM ribavirin alone, 50 U of IFN-α per ml alone, or the combination of both compounds rebounded to the same levels as the control cells within 2 weeks after the withdrawal of the inhibitors. The levels of HCV replicon RNA also rebounded to the original level after 3 weeks in cells treated with 0.6 μM PI-1 or 250 U of IFN-α per ml. In contrast, no HCV replicon cells were recovered after the 3-week incubation with G418 from cells treated with the combination of 0.6 μM PI-1 and 50 U of IFN-α per ml or from cells treated with 3 μM PI-1, suggesting that any viable HCV replicon RNA had been completely cleared from these treated cells.
DISCUSSION
With novel, specific therapeutics against HCV such as NS3-NS4A protease inhibitors on the horizon, it is important to try to evaluate the potential benefits or adverse effects of the new agents in combination with the present standard of care, namely, pegylated IFN-α plus ribavirin. In this study, we used a specific inhibitor of HCV NS3-NS4A protease, PI-1, to investigate the antiviral effect of PI-1 alone or PI-1 in combination with IFN-α in a subgenomic HCV replicon system. We demonstrated that treatment with the combination of the two agents resulted in moderate but significant synergy in reducing the HCV RNA levels in the replicon cells, with little or no increase in cytotoxicity.
In analysis of drug-drug combinations, the definition of theoretical additivity is critical so that a statistical analysis of the actual effect of a combination can reveal synergistic (greater than additive) or antagonistic (less than additive) effects. The Bliss independence model is based on statistical probability, which assumes that the two drugs should affect viral replication independently. The Loewe additivity model is based on dose addition, which assumes that the two drugs should be indistinguishable from each other; i.e., the effect is the same as adding one drug to itself (11). It is not unusual for different or even contradictory conclusions to be reported in the literature for the same drug-drug combination because different models were used. Therefore, rather than being biased toward one theory over the other, we analyzed our data with two different programs representing the two main models. We used one analysis to corroborate the other to confirm the synergistic effects of PI-1 and IFN-α.
The goal of antiviral treatment in patients is eradication of the virus with no relapse upon withdrawal of treatment. In contrast, the most common in vitro measurement of antiviral activity is the IC50 or the IC90 assay, which measures a two- or a ninefold drop in the viral RNA level, respectively, over a short period of time. In an attempt to establish a more relevant in vitro assay, we increased the length of treatment of replicon cells to 9 days and examined the effect on HCV RNA reduction. The combination of a relatively low concentration of the PI-1 protease inhibitor and a low concentration of IFN-α resulted in a greater than 3-log reduction in HCV replicon RNA levels, which can be achieved only with higher concentrations of PI-1 or IFN-α alone. It was difficult to quantitatively assess whether the result of the combination was synergistic or additive because of the small number of combinations and the high degree of variability of the assay compared to that of the 2-day assay. Nonetheless, on the basis of the findings with the Bliss independence model, the observed effect of the PI-1 and IFN-α combination in the 9-day assay (a 3.6-log decrease in HCV RNA level) slightly exceeded the expected additive effect (3.1-log drop) of PI-1 alone (2.0-log drop) plus IFN-α alone (1.1-log drop), although this analysis is not statistically significant due to assay variability and sample size. Finally, the sustained antiviral effect and the benefit of combination treatments were further demonstrated in the HCV replicon rebound study, in which the inhibitors were withdrawn after 2 weeks of treatment. A caveat of the replicon system is that it required the addition of G418 to help amplify HCV replicon cells and facilitate the rebound process. Nonetheless, under the same experimental conditions, the advantage of the combination treatment over single treatments was apparent, in that lower concentrations of PI-1 and IFN-α in combination were able to clear the HCV replicon RNA and to prevent rebound compared to the concentration of either agent alone required to achieve the same results. Therefore, all of the studies described above present a potential strategy for the use of two agents in combination to achieve a similar or better antiviral effect while reducing the overall exposure and potential adverse effects associated with the use of a high dose of either agent alone.
A recent published study demonstrated that the combination of type I (α and β) and type II (γ) IFNs can lead to synergistic inhibition of HCV RNA in replicon cells (19). Another study suggested that the combination of an HCV NS5B polymerase inhibitor and IFN-α also results in synergy in the inhibition of viral RNA replication (14). This is the first report of a thorough and quantitative analysis that demonstrates that the combination of a specific, small-molecule inhibitor of HCV NS3-NS4A protease and IFN-α is synergistic in reducing HCV RNA levels in vitro. Our results suggest that a combination therapy consisting of an HCV NS3-NS4A protease inhibitor and IFN-α could potentially have a better antiviral effect than that achieved with the present standard of care for hepatitis C. With many new inhibitors with different mechanisms being developed for the treatment of HCV infections, we proposed that various combinations of these agents should be evaluated in vitro using a similar process, as described in this paper. There are also limitations in interpreting the results of in vitro experiments. For example, the concentrations of drugs used in this study were selected according to their relative potency in the HCV replicon system and may not reflect clinically relevant doses. Also, the effects of the drugs in vivo will be affected by many other factors, such as the physical properties and pharmacokinetics of the drugs. In addition, agents such as IFN-α and ribavirin could exert indirect antiviral activities, such as immunomodulatory effects, that are absent in the in vitro system (15, 20, 26, 32). It remains to be determined whether the synergy observed between HCV protease inhibitors and IFN-α in vitro can ultimately be translated into clinical efficacy.
Another important reason for combining antiviral agents is to suppress the emergence of viral resistance. In the case of human immunodeficiency virus, monotherapy has been shown to be ineffective, largely due to the rapid emergence of resistant variants. It was not until the introduction of cocktail therapies, i.e., the use of combinations of agents with different mechanisms of action, that sustained suppression of viral replication and resistance could be achieved. In the case of HCV, specific mutations that result in resistance to new antivirals, such as protease or polymerase inhibitors, are likely to emerge, given the high mutation rate of the virus. Therefore, it remains to be evaluated both in vitro and in vivo whether the combination of different anti-HCV drugs can help to prevent or suppress the emergence of resistance.
In summary, we have demonstrated in HCV replicon cells a synergistic antiviral effect between two agents with different mechanisms of action: an HCV protease inhibitor, PI-1, and a cytokine, IFN-α. Interestingly, a recent study revealed a specific mechanism that HCV may use to circumvent the host immune response and presented a potential link between the HCV protease and the IFN pathway (7). Foy et al. (7) reported that the HCV NS3-NS4A protease can effectively block the activation of IFN regulatory factor 3 (IRF-3), a key transcription factor that is required for type I IFN synthesis. Moreover, a specific HCV protease inhibitor was able to restore the responsiveness of the IRF-3 pathway. The synergistic effect observed here could also be related to the modulation of cytokine production and the cytokine response by PI-1 and IFN-α, the exact mechanism of which remains to be elucidated. Nonetheless, these findings all support the notion that specific HCV protease inhibitors can be used to restore or enhance the effectiveness of IFN therapy. It is hoped that the introduction of HCV protease inhibitors in the clinic, either alone or in combination with IFN-α, will provide better treatment regimens for hepatitis C patients.
Acknowledgments
We thank Kevin Cottrell, Janos Pitlik, and Robert Perni for providing the compound PI-1; Yu-Ping Luong and Cynthia Gates for determining the Ki of PI-1 against the HCV NS3-NS4A protease; and Robert Perni, Steve Lyons, Michael Briggs, and John Thomson for critically reading the manuscript.
REFERENCES
- 1.Babine, R. E., S. H. Chen, J. E. Lamar, N. J. Snyder, X. D. Sun, M. J. Tebee, F. Victor, Q. M. Wang, Y. Y. M. Yip, I. Collado, C. Garcia-Paredes, R. S. I. Parker, L. Jin, D. Guo, and J. I. Glass. 7. March 2002. Peptidomimetic protease inhibitors. WO patent 02/018369.
- 2.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1993. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J. Virol. 67:3835-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Chou, T. C., and P. Talalay. 1977. A simple generalized equation for the analysis of multiple inhibitions of Michaelis-Menten kinetic systems. J. Biol. Chem. 252:6438-6442. [PubMed] [Google Scholar]
- 5.De Francesco, R., L. Tomei, S. Altamura, V. Summa, and G. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:1-16. [DOI] [PubMed] [Google Scholar]
- 6.Farmer, L., J. Pitlik, R. Perni, L. Courtney, and J. Van Drie. 23. January 2003. Bridged bicyclic serine protease inhibitors. WO patent 03/006490.
- 7.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 8.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 9.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greco, W. R., G. Bravo, and J. C. Parsons. 1995. The search for synergy: a critical review from a response surface perspective. Pharmacol. Rev. 47:331-385. [PubMed] [Google Scholar]
- 12.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kimura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. USA 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston, V. K., D. Maley, R. C. Gagnon, C. W. Grassmann, S. E. Behrens, and R. T. Sarisky. 2003. Kinetic profile of a heterocyclic HCV replicon RNA synthesis inhibitor. Biochem. Biophys. Res. Commun. 311:672-677. [DOI] [PubMed] [Google Scholar]
- 15.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 16.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 19.Larkin, J., L. Jin, M. Farmen, D. Venable, Y. Huang, S. L. Tan, and J. I. Glass. 2003. Synergistic antiviral activity of human interferon combinations in the hepatitis C virus replicon system. J. Interferon Cytokine Res. 23:247-257. [DOI] [PubMed] [Google Scholar]
- 20.Lau, J. Y., R. C. Tam, T. J. Liang, and Z. Hong. 2002. Mechanism of action of ribavirin in the combination treatment of chronic HCV infection. Hepatology 35:1002-1009. [DOI] [PubMed] [Google Scholar]
- 21.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 22.Lin, C., J. A. Thomson, and C. M. Rice. 1995. A central region in the hepatitis C virus NS4A protein allows formation of an active NS3-NS4A serine proteinase complex in vivo and in vitro. J. Virol. 69:4373-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, vol. 1, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 26.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 27.Pause, A., G. Kukolj, M. Bailey, M. Brault, F. Do, T. Halmos, L. Lagace, R. Maurice, M. Marquis, G. McKercher, C. Pellerin, L. Pilote, D. Thibeault, and D. Lamarre. 2003. An NS3 serine protease inhibitor abrogates replication of subgenomic hepatitis C virus RNA. J. Biol. Chem. 278:20374-20380. [DOI] [PubMed] [Google Scholar]
- 28.Perni, R. B., and A. D. Kwong. 2002. Inhibitors of hepatitis C virus NS3 · 4A protease: an overdue line of therapy. Prog. Med. Chem. 39:215-255. [DOI] [PubMed] [Google Scholar]
- 29.Prichard, M. N., and C. Shipman, Jr. 1990. A three-dimensional model to analyze drug-drug interactions. Antivir. Res. 14:181-205. [DOI] [PubMed] [Google Scholar]
- 30.Seeff, L. B., and J. H. Hoofnagle. 2002. National Institutes of Health Consensus Development Conference: management of hepatitis C: 2002. Hepatology 36:S1-S2. [DOI] [PubMed] [Google Scholar]
- 31.Tanji, Y., M. Hijikata, S. Satoh, T. Kaneko, and K. Shimotohno. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas, H. C., M. E. Torok, D. M. Forton, and S. D. Taylor-Robinson. 1999. Possible mechanisms of action and reasons for failure of antiviral therapy in chronic hepatitis C. J. Hepatol. 31(Suppl. 1):152-159. [DOI] [PubMed] [Google Scholar]
- 33.Tomei, L., C. Failla, E. Santolini, R. De Francesco, and N. La Monica. 1993. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J. Virol. 67:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varaklioti, A., N. Vassilaki, U. Georgopoulou, and P. Mavromara. 2002. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 277:17713-17721. [DOI] [PubMed] [Google Scholar]
- 35.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]