Abstract
Nuclear receptors are ligand-activated transcription factors whose diverse biological functions are classically regulated by cholesterol-based small molecules. Over the past few decades, a growing body of evidence has demonstrated that phospholipids and other similar amphipathic molecules can also specifically bind and functionally regulate the activity of certain nuclear receptors, suggesting a critical role for these non-cholesterol-based molecules in transcriptional regulation. Phosphatidylcholines, phosphoinositides and sphingolipids are a few of the many phospholipid like molecules shown to quite specifically regulate nuclear receptors in mouse models, cell lines and in vitro. More recent evidence has also shown that certain nuclear receptors can “present” a bound phospholipid headgroup to key lipid signaling enzymes, which can then modify the phospholipid headgroup with very unique kinetic properties. Here, we review the broad array of phospholipid / nuclear receptor interactions, from the perspective of the chemical nature of the phospholipid, and the cellular abundance of the phospholipid. We also view the data in the light of well established paradigms for phospholipid mediated transcriptional regulation, as well as newer models of how phospholipids might effect transcription in the acute regulation of complex nuclear signaling pathways. Thus, this review provides novel insight into the new, non-membrane associated roles nuclear phospholipids play in regulating complex nuclear events, centered on the nuclear receptor superfamily of transcription factors.
Introduction
Phospholipids and Transcription
The recent discovery that phosphatidylinositols (Blind et al., 2014, 2012; Mullaney et al., 2010; Sablin et al., 2015) and short-chain phosphatidylcholines (Lee et al., 2011; Musille et al., 2012) act as hormone ligands for NR5A nuclear receptors represents the latest in a long line of phospholipid candidates that have been proposed as ligands for members of the nuclear receptor (NR) superfamily. Despite the exciting new roles these phospholipids play in nuclear receptor biology, the totality of effects phospholipids mediate on intracellular nuclear receptors remain largely uncharacterized (Irvine, 2003; Quehenberger et al., 2010). However, the potent roles phospholipids play in signaling and what is often low cellular abundance can begin to suggest real possibilities for dynamic regulation of NR function (Barlow et al., 2010; Irvine, 2003). The ability of IPMK and PTEN to directly modify phospholipids while bound to the nuclear receptor Steroidogenic Factor-1 (SF-1, NR5A1) (Blind et al., 2014, 2012), and the structure of apo-Liver Receptor Homolog-1 (LRH-1, NR5A2) (Musille et al., 2012), allow us to begin to address more mechanistic questions about how phospholipids go about regulating NRs.
The more general links between phospholipids and transcriptional regulation, independent of nuclear receptors, have been in the literature for some time. Some prominent examples include PIP3 activation of the BRG-1 component of the BAF chromatin remodeling complex (Rando et al., 2002), PIP3 dependent activation of Nucloephosmin/B23 activation of apoptotic programs (Ahn et al., 2005), and SWI/SNF activation by soluble phosphorylated inositol headgroups derived from phosphatidylinositol precursors (Shen et al., 2003; Steger et al., 2003). More recently, discoveries of PIP2 inhibition of the poly-A polymerase Star-PAP (Mellman et al., 2008), sphingosine 1-phosphate (S1P) activation of HDACs (Hait et al., 2009), inositol tetraphosphate regulation of HDAC3 (Watson et al., 2012) and PIP2 regulation of HDAC1 (Toska et al., 2013, 2012) further link lipids, phospholipids and their metabolites to transcription. Although G-protein coupled receptors (Falasca & Ferro, 2016) can mediate activation of nuclear receptors through the generation of 2nd messenger phospholipids (Lin et al., 2009), the majority of phospholipid regulation of nuclear receptors occurs through regulation of phospholipid metabolic enzymes. The most well characterized function of nuclear receptor superfamily members remains as ligand-regulated transcription factors. The past few years have revealed that many of the transcriptional effects elicited by several classes of phospholipids (Figure 1) are mediated through nuclear receptors (Musille et al., 2013; Olefsky and Glass, 2010). This review attempts to highlight the evidence that phospholipids directly regulate nuclear receptors.
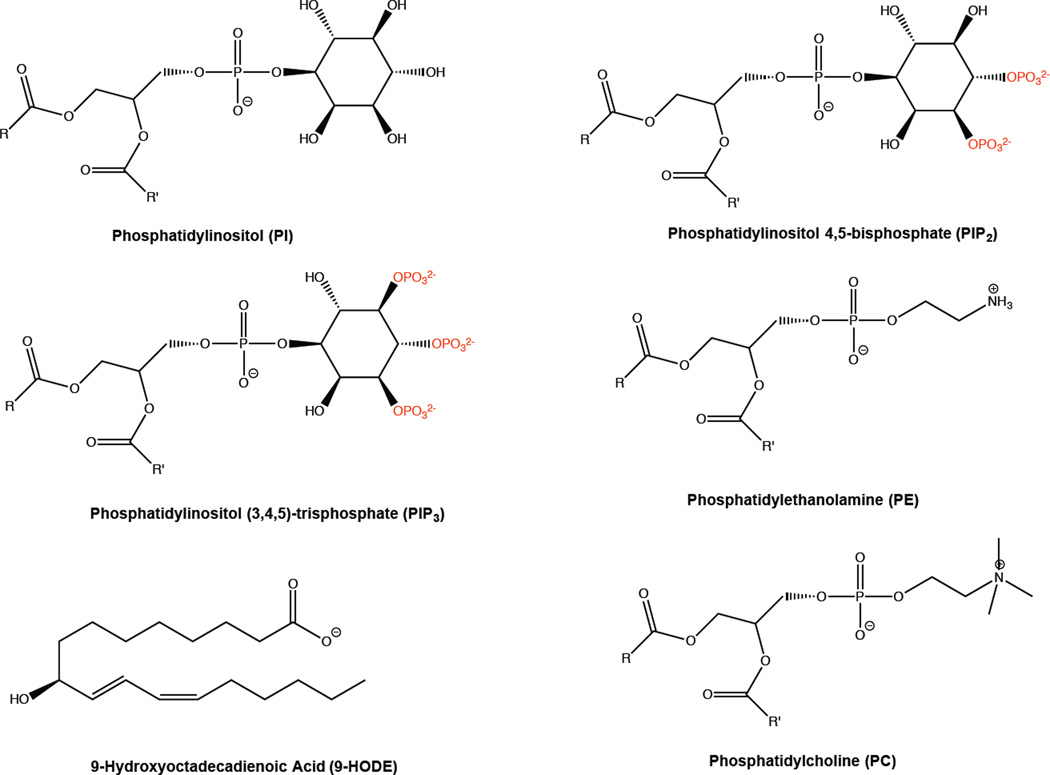
Figure 1.
Phospholipid species discussed in this review and shown in crystal structures complexed to phospholipid binding proteins. Sfh1 has been crystallized bound to phosphatidylinositol (PI), phosphatidylethanolamine (PE), and phophatidylcholine (PC). SF-1 has been crystallized bound to several lipids including phosphatidylinositol(4,5)bisphosphate (PIP2) and phosphatidylinositol(3,4,5)trisphosphate. LRH-1 has been crystallized bound to several lipids including PIP3 and dilaurylphosphatidylcholine (DLPC). PPARϒ has been crystallized bound to several lipids including 9-hydroxyoctadecadienoic Acid (9-HODE).
Topics
Abundant Endogenous Phospholipid Ligands of NRs
The phospholipid ligands of many NRs are present at relatively high levels in cells, as many of these species are metabolic intermediates of general phospholipid metabolism. At first glance, this may suggest that these abundant phospholipids may not act as bona fide ligands of NRs, simply because their high concentrations cannot fluctuate sufficiently to allow dynamic exchange with NRs. However, since most orphan NRs have relatively low affinities for their associated phospholipids, it follows that the concentration range in which a dynamic response to phospholipids would also be expected to be elevated, particularly when compared to high affinity receptor/ligand systems such as the steroid receptors. Seemingly low phospholipid concentrations also can be significantly altered by compartmentalization of these molecules into specific lipid microdomains, both in membranes (Delos Santos et al., 2015; Horejsi and Hrdinka, 2014) and non-membrane fractions within the nucleus (Barlow et al., 2010; Irvine, 2006). Further, small chemical changes to a generally abundant class of phospholipids can generate very small pools of a specific chemical species. This often occurs by altering the number or position of unsaturating double bonds in the acyl chains, or the length of the acyl chains themselves, as demonstrated for the particular POPC species associated with PPARα (Chakravarthy et al., 2009). Going far beyond simple clustering of phospholipids into phosphoinositides, cholines and ethanolamines, the Lipid MAPS consortium has demonstrated that tens of thousands of distinct lipid species are present in humans (Dennis, 2016; Quehenberger et al., 2010). Thus, the abundance of each of these unique lipid species, after fully deciphering acyl chain length (on both sn-1 and sn-2 positions), total number of saturations and their positions in the acyl chains, may lead us to realize that each species of these molecules are not as abundant as perhaps initially suspected. We begin this review with the activity of what have been considered generally abundant cellular phospholipids on nuclear receptors.
Geranylgeranyl Pyrophosphate (GGPP) on LXR
Although not technically a phospholipid, the first phosphorylated lipid species discovered to bind a nuclear receptor and regulate its transcriptional activity was geranylgeranyl pyrophosphate (GGPP), which inhibits the transcriptional activity of Liver X Receptors (LXRs) (Forman et al., 1997). GGPP functionally decreases FXR-induced luciferase reporter activity, probably by decreasing the interaction of FXR with target gene promoters, as GGPP decreases FXR DNA binding in gel shift assays (Forman et al., 1997). As an abundant and ubiquitous product of mevalonate lipid metabolism, GGPP biosynthesis can be potently inhibited by statins, which target the rate-limiting enzyme of cholesterol synthesis HMG-CoA Reductase. Indeed, statins decrease FXR transcriptional reporter activity, which can be rescued by exogenous mevalonate treatment; however, other mevalonate metabolites fail to rescue the effects of statins on FXR, or alter FXR reporter activity in any way (Forman et al., 1997). A report in 2001 also showed that GGPP decreases mRNA abundance of the LXR target gene abc1, and that GGPP decreases the physical interaction between LXR and the transcriptional co-activator SRC-1 (Gan et al., 2001). The interesting and metabolically important link between a cholesterol / GGPP / LXR signaling axis could present an opportunity for follow-up, as not much has been published since these initial discoveries.
Oxidized Phosphatidylcholine (oxPC) on PPARγ
Although most phospholipid ligands of NRs act as second messengers, never leaving the cell in which they were synthesized, several reports in 2000 and 2001 demonstrated the ability of extracellular, secreted phospholipids in lipoprotein complexes to serve as classic first messenger hormones on intracellular nuclear receptors. Oxidized phosphatidylcholine (oxPC), an important component of Low Density Lipoprotein (LDL) particles, can bind and activate intracellular Peroxisome Proliferator Activated Receptors Alpha (PPARα) (Lee et al., 2000) and Gamma (PPARγ) (Davies et al., 2001). The structure of PPARγ bound to 9-HODE is presented to demonstrate how the lipid interacts with PPARγ (Figure 2D). The latter study demonstrated that CD36, a plasma membrane scavenger receptor, enhances oxPC transport to specific intracellular compartments, and facilitates oxPC activation of PPARγ target genes. The effects these authors observed of oxPC are specific to PPARγ, as PPARαtranscriptional activation was unaffected by oxPC treatment or CD36 expression in these cells. However, oxPC was able to activate expression of pro-inflammatory genes in endothelial cells derived from wild type mice, but not in cells derived from PPARγ knockout mice (Lee et al., 2000), demonstrating some cell-line specific effects of oxPC. Further work showed that secreted phospholipase A2 (sPLA2) activity on extracellular oxLDL particles can generate intracellular ligands for PPARαand activate PPARαtarget genes (Davies et al., 2001; Delerive et al., 2000). Importantly, these studies collectively demonstrated that not unlike steroid 1st messenger hormones, phospholipids are capable of acting on nuclear receptors in an endocrine capacity, and similarly could transfer information stored in their chemical structure from one tissue to another.
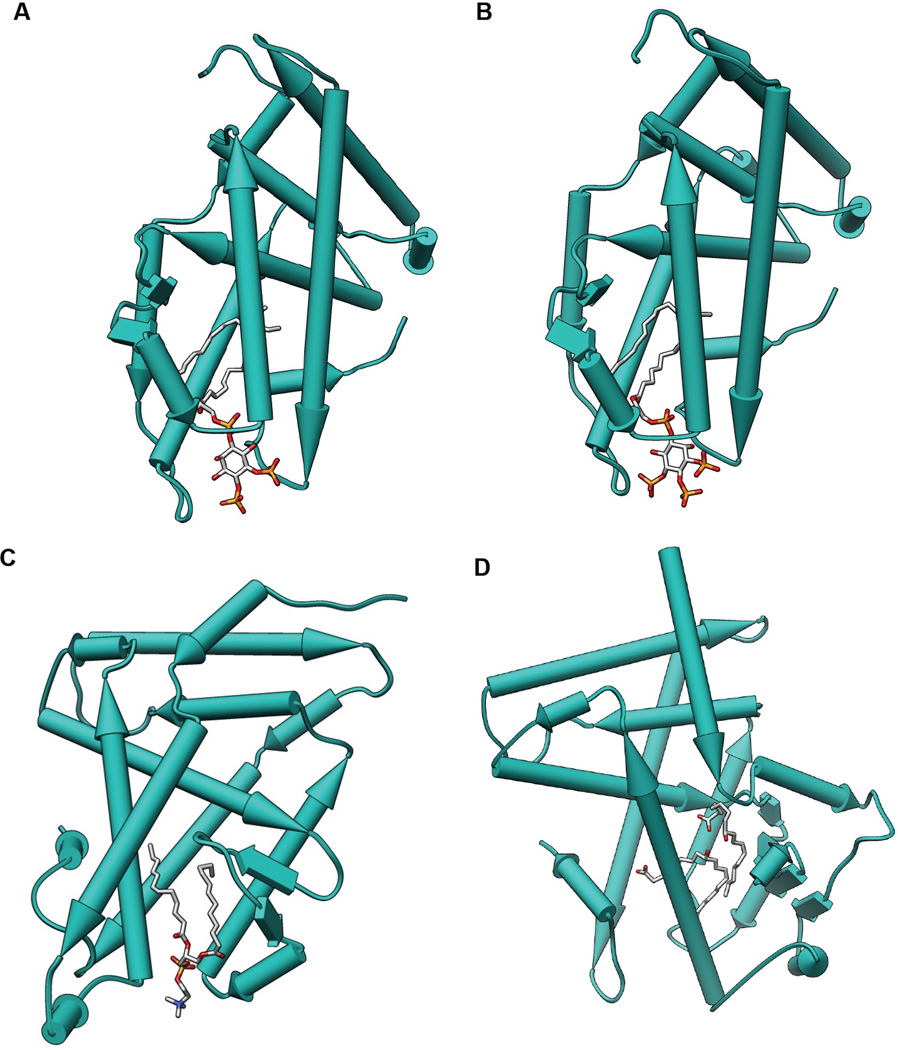
Figure 2.
Crystal structure of nuclear receptors bound to phospholipids. Proteins are depicted as pipes and planks with bound lipids shown as sticks using Chimera. A. SF-1 bound to PIP2 (PDB: 4QK4). B. SF-1 bound to PIP3 (PDB: 4QJR). C. LRH-1 bound to DLPC (PDB: 4DOS). D. PPARϒ bound to 9-HODE (PDB: 9VSR).
LysoPhosphatidic Acid (LPA) on PPARγ
PPARγ was then shown to also bind and be activated by lyso-phosphatidic acid (LPA), another high-abundance phospholipid which exists inside cells and is also present in the serum (McIntyre et al., 2003). LPA induces PPARγ-dependent changes in cellular lipid uptake, lipid metabolism and target gene activation. Importantly, the activation of PPARγ endogenous target genes by LPA occurs independently of plasma membrane LPA receptors, which are themselves important mediators of extracellular LPA effects on cellular physiology and signal transduction. Further, PPARγ transcriptional activity is unaffected by phosphatidic acid (PA), a molecule closely related to LPA that only differs in that it is acylated on only one SN position of the phospholipid glycerol backbone, while PA is acylated on both (McIntyre et al., 2003).
Phosphatidic Acid (PA) on SF-1
Although PA fails to regulate PPARγ, PA was shown to bind SF-1 and activate SF-1 transcriptional targets, as well as induce co-activator peptide recruitment to SF-1 (Li et al., 2007). PA can be generated in cells by the action of Diacylglycerol (DAG) kinases on DAG (Tu-Sekine et al., 2015), and inhibiting DAG kinase theta decreases SF-1 activation of endogenous target genes, functionally linking the metabolic enzyme that generates this phospholipid to SF-1. Interestingly, DAG kinase theta was also shown to directly bind to SF-1 protein via the classic nuclear receptor interaction motif "LXXLL", a motif that mediates the interaction between NRs and many transcriptional co-activator proteins, such as the p160s. Further, this interaction was shown to respond to cAMP signaling cascades, suggesting the interaction of a lipid metabolic enzyme (DAG kinase theta) and a nuclear receptor (SF-1), may be an important link between phospholipid metabolism and nuclear receptor regulation (Li et al., 2007).
Sphingosine on SF-1
More work from Marion Sewer’s lab showed that sphingosine, which differs chemically from a phospholipid as it contains a sphingoid base backbone rather than phospho-glycerol, binds to and inhibits the transcriptional activity and recruitment of SF-1 to the endogenous target gene cyp17a1 (Dammer et al., 2007; Urs et al., 2006). The recruitment of transcriptional co-activators to this gene is decreased upon sphingosine treatment, while co-activators and co-repressors alternate occupancy of the cyp17a1 promoter (Dammer et al., 2007). Coupled with mass spectrometry data (Li et al., 2007) and the discovery of phospholipids associated with SF-1 by crystallography (Blind et al., 2014, 2012; Krylova et al., 2005; Sablin et al., 2015), it appears likely that SF-1 is functioning as a transcriptional sensor of sphingosine metabolism, as well as phospholipid metabolism.
PIP2 and PIP3 on SF-1
The direct interaction of DAG kinase theta and SF-1 mentioned above (Li et al., 2007), coupled with the crystal structures of SF-1 bound to various phospholipids (Blind et al., 2014; Sablin et al., 2015, 2009), contributed to our recent hypothesis that a phospholipid associated with a nuclear receptor could be directly acted on by lipid metabolic enzymes (Figure 2). We showed that a non-canonical PI3- (Resnick et al., 2005) and inositol kinase called inositol polyphosphate multikinase (IPMK) (Odom et al., 2000; Mikoshiba et al., 2015) can directly phosphorylate PIP2 bound to SF-1 to generate an SF-1/PIP3 complex (Figure 2B), activating endogenous SF-1 target genes (Blind et al., 2012). We also showed that PTEN can dephosphorylate the SF-1/PIP3 product of the IPMK reaction, and that PTEN inhibits SF-1 transcriptional activity. The activity of IPMK on the SF-1/PIP2 complex (Figure 2A) is described by enzyme kinetic parameters that differ dramatically when compared to IPMK kinase activity on PIP2 in membranes, and a chemical antagonist experiment demonstrated conclusively that IPMK must be acting on the SF-1/PIP2 complex, and not the unbound phospholipid. We were also able to phosphorylate endogenous PIP2 bound to SF-1 purified from HEK cells, demonstrating that PIP2 associates with SF-1 expressed in these cells. Coupled with crystallographic and functional evidence showing several PCs of various acyl-chain lengths can regulate SF-1 and LRH-1 (Lee et al., 2011; Musille et al., 2012; Sablin et al., 2009), the association of PIP2 with SF-1 presents a strong case that both PIP2 and PCs are important to SF-1 cellular functions.
Low-abundancy Endogenous Phospholipid Ligands of NRs
Although members of the above classes of phospholipids have a relatively high abundance, some phospholipid ligands of NRs are present in cells at lower levels. The concentrations of these phospholipids suggest they may play a more potent role in regulating nuclear receptor activity. Indeed, PIP3 is often not present at detectable levels in cells, yet is one of the post potent initiators of intracellular signaling. As one of the highest abundance classes of cellular phospholipids, phosphatidylcholine (PC) might not be expected to regulate NRs. However, like all phospholipids, individual PC species vary in acyl chain length, the number of un-saturations, and the position of those oxidations in the fatty acyl chains, which gives rise to many PC species of relatively low abundance.
1-Palmitoyl 2-Oleyl PC (POPC) on PPARα
The importance of one of these lower-abundance PC species in PPARαphysiology was recently shown in Fatty Acid Synthase/PPARαliver-specific knockout mice, which were used to conclusively identify 1-palmitoyl 2-oleyl PC, (POPC) as an endogenous activator of PPARαin mouse liver (Chakravarthy et al., 2009). These authors found this lipid to compose 11.4 (+/− 0.6)% of the nuclear PC content and just 4.4 (+/− 1.2)% of all nuclear phospholipids from mouse liver. POPC associates with PPARα independent of PPARα effects on lipid metabolism, as POPC also is found associated with a DNA-binding deficient mutant of PPARα (Chakravarthy et al., 2009). The lipid metabolic enzyme responsible for generating POPC in the nucleus (CEPT1) (Henneberry et al., 2002) is required for full PPARα induction of target genes in mouse liver, while the isoform of the CEPT1 enzyme present on cytoplasmic membranes has no effect on PPARα transcriptional activity (Chakravarthy et al., 2009). Exogenous POPC directly infused into mouse liver is also able to decrease liver steatosis, and these effects are dependent on the presence of PPARα in the liver.
Cyclic PA (cPA) on PPARγ
A rare and poorly understood cellular phospholipid also regulates PPARγ, cyclic Phosphatidic Acid (CPA) (Oishi-Tanaka and Glass, 2010). Production of CPA antagonizes PPARγ in vitro, in cells and in mice, and this inhibition depends on R288 of PPARγ, an amino acid located in the ligand binding pocket of the receptor (Tsukahara et al., 2010). Again, the lipid metabolic enzyme responsible for synthesizing the phospholipid ligand also regulates this nuclear receptor. In this case, Phospholipase D2 (PLD2) cyclizes PA to cPA, and expression of this enzyme potently inhibits PPARγ transcriptional activation. Further, although both PLD1 and PLD2 isoforms are capable of generating cPA in cells, only PLD2 is capable of regulating PPARγ transcription (Tsukahara et al., 2010), suggesting that mechanisms apart from simply generating the phospholipid NR ligand in membranes must participate in phospholipid regulation of NR functions.
Exogenous Natural Phospholipids
Dilauryl PC (DLPC) on LRH-1
Non-phospholipid dietary lipids and other hydrophobic xenobiotics are well-established mediators of NRs such as PPARs, LXR, CAR and PXR, and have been reviewed in detail elsewhere (Lemberger et al., 1996; Musille et al., 2013; Reschly and Krasowski, 2006). To date, only one naturally occurring but exogenously produced phospholipid has been shown to act as a ligand for a nuclear receptor, that being the short chain phospholipid dilauryl PC (DLPC) acting on Liver Receptor Homolog 1 (LRH-1, NR5A2) to activate transcription (Figure 2C) (Lee et al., 2011). This study demonstrated that oral administration of DLPC activates LRH-1 target genes in mouse liver and increased serum levels of bile acid. Strikingly, oral administration of DLPC is able to rescue the mouse model of high fat diet-induced insulin resistance and fatty liver disease, in an LRH-1 dependent manner (Lee et al., 2011).
Although the metabolic effects of DLPC are clear, there remained some doubt as to whether DLPC could be activating LRH-1 through activation of other signaling cascades, as opposed to DLPC directly binding the receptor itself. However, the crystal structure of the complex between LRH-1 and DLPC was recently solved, demonstrating differences in both the co-activator recruitment and overall structures of DLPC vs DPPC bound LRH-1, providing structural evidence supporting direct regulation of LRH-1 by DLPC (Musille et al., 2012). Clearly, the LRH-1 dependent effects of DLPC (a natural product) on insulin resistance and metabolic disorders presents an attractive model for therapeutic intervention in these high-value human diseases.
Mechanism of Delivery: Phospholipid Transfer Proteins
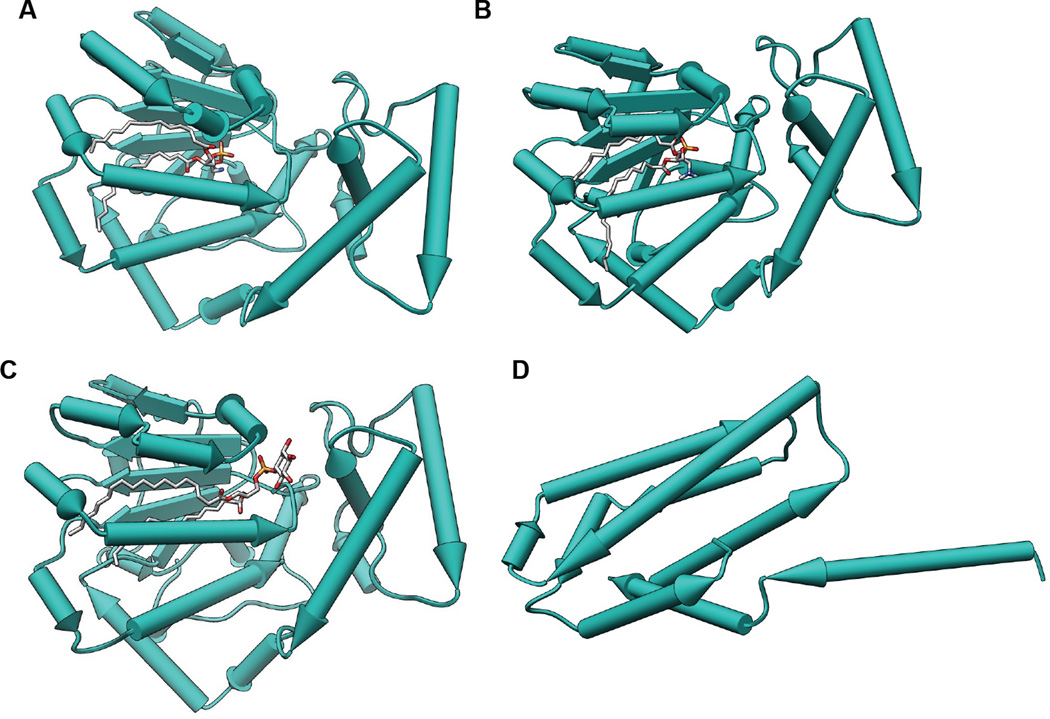
Although phospholipid effects on NR regulation lead to dramatic changes in transcriptional programs that manifest themselves at the whole organism/physiological level down to individual receptors and genes, we have almost no idea how NRs acquire these hydrophobic phospholipid ligands from cellular membrane systems. Phospholipid transfer into VLDL has been known for decades to be mediated by the phospholipid transfer proteins (PLTP1 and PLTP2), yet how/if these same proteins facilitate loading of phospholipids into nuclear receptors remains an open question (Figure 3). The Sec14 family of phospholipid transfer proteins is the best characterized family of transfer proteins (Figure 3A–C), with a crystal structure of the human homolog of yeast Sec14 informing biochemical experiments suggesting that Sec14 can act as a phospholipid "nanoreactor", facilitating the interaction of particular membrane phospholipid substrates with the enzymes that metabolize them, particularly lipid kinases and phosphatases (Figure 3C). The phospholipid transfer protein TIPE3 (Figure 3D) was also recently shown to enhance PI3-kinase activity, likely through a similar mechanism (Fayngerts et al., 2014). Notwithstanding any undiscovered effects that PLTPs may have on the exchange of phospholipids in membranes with NRs, it is clear that at least some NRs are capable of non-enzymatic, spontaneous phospholipid exchange with membrane systems in vitro, not unlike the phospholipid transfer activities of PLTPs. In fact, recombinant SF-1 LBD could technically be classified as a phospholipid transfer protein, based on its ability to exchange phospholipids between membrane systems in vitro (Bankaitis et al., 2012). It is important to clarify that whether or not nuclear receptors are capable of dynamic phospholipid exchange with membrane systems in a cellular context remains an open and very interesting mechanistic question.
Figure 3.
Crystal structures of phospholipid transfer proteins. Proteins are depicted as pipes and planks with bound lipids shown as sticks using Chimera. A. Sfh1 bound to PE (PDB: 3B74). B. Sfh1 bound to PC (PDB: 3B7Q). C. Sfh1 bound to PI (PDB: 3B7N). D. TIPE3 with no bound ligand (PDB: 4Q9V).
Phospholipid Loading by Translational Integration
Signal sequences usually direct mRNA messages that encode secretory and transmembrane proteins to the rough endoplasmic reticulum, through recognition of the signal recognition particle. Messages encoding cytoplasmic and nuclear proteins are classically thought to be translated on free ribosomes in the cytosol, independent of SRP and membranes. However, several studies have now used unbiased approaches to demonstrate that the subset of actively translating ribosomal/RNA complexes associated with membranes does not correlate with the presence of a classic signal sequence. In fact, many members of the nuclear receptor superfamily have been identified as being translated on membrane-bound ribosomes (Reid and Nicchitta, 2015). One hypothesis explaining this phenomenon is that membrane-translated proteins are co-translationally sensing the ER lipid content by acquiring lipids from the ER as the proteins fold in close proximity to this membrane system (Blind, 2014). PLTPs such as Sec14 (Figure 3C) and TIPE3 (Figure 3) are localized to ER membranes, and could facilitate this process through the same poorly understood mechanisms they use to assist in the movement of phospholipids into lipoprotein particles in the cytoplasm. Dissecting how phospholipids are loaded into NRs represents an extremely challenging problem, as these exchange reactions occur non-enzymatically in vitro, and are therefore extraordinarily difficult to measure and quantify (Bankaitis et al., 2012).
Direct Modification of Bound Phospholipids
Regardless of how NRs are able incorporate them into their structure, phospholipids are clearly found associated with several members of the NR superfamily. The classic hormone/receptor paradigm established by the activation of growth factor receptors (e.g. epidermal, fibroblast and vascular endothelial growth factor receptors), G-protein coupled receptors (rhodopsin, opioid and glutamate receptors) and steroid NRs (estrogen, glucocorticoid and mineralocorticoid receptors) has established a model regarding how hormones activate receptors. In this model, first messenger hormones synthesized far from their site of action bind to cognate receptors to activate them. This activation often leads to the enzymatic generation of intracellular second messengers, which bind and regulate cellular effector proteins. Following activation, the hormone dissociates from its receptor, helping to deactivate the signaling pathway.
There has been little recent debate over this model; however, we recently discovered that the classic second messenger PIP2 bound to SF-1 can be directly phosphorylated to PIP3 by IPMK, while PIP2 is associated with SF-1, activating SF-1 transcription (Blind et al., 2012). This discovery represents a new way that small signaling molecules can alter receptor function - small molecules statically associated with an effector can be metabolized/altered while bound to the protein, altering effector function. We posit that the direct modification of second messenger small signaling molecules bound to intracellular receptors will prove to be a general signaling paradigm, particularly in the regulation of nuclear protein/small molecule complexes (Blind, 2014). Importantly, this mechanism is capable of decoupling second messenger-bound downstream effectors from the activation of particular upstream signaling pathways.
That nature may have selected to modify a second messenger small molecule such as a phospholipid while it is bound to an effector, rather than an amino acid encoded in the genome, may reveal a new type of signaling logic that supports this mechanism. On the surface, this type of modification does not appear significantly different than serine/threonine modification of the effector. However, by allowing a small molecule bound to an effector to be modified, rather than an amino acid encoded by the genome, the cell is afforded the ability to decouple activation of the effector from second messenger signaling enzymes by replacing/displacing the small molecule associated with the effector. In the case of SF-1, this protein effector can bind PIP2, sphingosine and PC. Perhaps when the cell requires SF-1 activation to be sensitive to IPMK/PTEN signaling, the PIP2 small molecule occupies the SF-1 ligand-binding pocket. When the cell requires SF-1 to be sensitive to ceramide metabolism, sphingosine occupies the SF-1 ligand-binding pocket. If the cell wants to decouple SF-1 activity from phosphoinositide and ceramide metabolism, PC may occupy the ligand-binding pocket (Blind, 2014). It is not difficult to see the advantage that decoupling an effector from particular signaling pathways, while sensitizing it to others, may confer to complex nuclear signaling systems. In this way, the cell is able to store information that regulates protein structure outside the genome, increasing its ability to dynamically regulate these proteins. The fact that SF-1/PIP3 is robustly dephosphorylated by PTEN, yet SF-1/PIP2 is resistant to phosphorylation by p110 PI3-kinases, suggests that nuclear proteins like SF-1 can help decouple PTEN signaling in the nucleus from p110 signaling. This is particularly important given that the pharmaceutical industry has invested so heavily in p110 inhibitors, which are partly intended to compensate for loss of PTEN function.
Nuclear receptor regulation by phospholipids is only recently extending the impact of nuclear receptor biology beyond human physiology and drug design, and into far more basic science fields that seek to understand how signal transduction works. The recent structures of full-length nuclear receptors that have been solved (Chandra et al., 2013, 2008; Lou et al., 2014) will now further inform how phospholipids might play a role in these exciting ligand-activated transcription factors.
Acknowledgments
This work was supported by American Cancer Society Institutional Research Grant #IRG-58-009-56 to R.D.B.; NCI K01 CA172957 to R.D.B.; NIDDK P30 DK020593 Vanderbilt Diabetes Research and Training Center Pilot and Feasibility Award to R.D.B.; Vanderbilt Diabetes Center Discovery Award to R.D.B.; Journal of Biological Chemistry Herbert Tabor Young Investigator Award to R.D.B.; NIDDK T32 DK007563 Molecular Endocrinology Training Grant support to C.D.S.; NIGMS T32 GM007628 Training in Pharmacological Sciences support to M.K.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn J-Y, Liu X, Cheng D, Peng J, Chan P-K, Wade PA, Ye K. Nucleophosmin/B23, a nuclear PI(3,4,5)P(3) receptor, mediates the antiapoptotic actions of NGF by inhibiting CAD. Mol. Cell. 2005;18:435–445. doi: 10.1016/j.molcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Ile KE, Nile AH, Ren J, Ghosh R, Schaaf G. Thoughts on Sec14-like nanoreactors and phosphoinositide signaling. Adv. Biol. Regul. 2012;52:115–121. doi: 10.1016/j.jbior.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol. 2010;20:25–35. doi: 10.1016/j.tcb.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind RD. Disentangling biological signaling networks by dynamic coupling of signaling lipids to modifying enzymes. Adv Biol Regul. 2014;54:25–38. doi: 10.1016/j.jbior.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind RD, Sablin EP, Kuchenbecker KM, Chiu HJ, Deacon AM, Das D, Fletterick RJ, Ingraham HA. The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor NR5A1 (SF-1) Proc Natl Acad Sci U S A. 2014;111:15054–15059. doi: 10.1073/pnas.1416740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind RD, Suzawa M, Ingraham HA. Direct Modification and Activation of a Nuclear Receptor-PIP2 Complex by the Inositol Lipid Kinase IPMK. Sci Signal. 2012;5:ra44. doi: 10.1126/scisignal.2003111. doi:5/229/ra44 [pii] 10.1126/scisignal.2003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RRV, Xu HE, Turk J, Semenkovich CF. Identification of a Physiologically Relevant Endogenous Ligand for PPARa in Liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-γ–RXR-α nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Potluri N, Wu D, Kim Y, Rastinejad F. Multidomain integration in the structure of the HNF-4α nuclear receptor complex. Nature. 2013;495:394–398. doi: 10.1038/nature11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer EB, Leon A, Sewer MB. Coregulator exchange and sphingosine-sensitive cooperativity of steroidogenic factor-1, general control nonderepressed 5, p54, and p160 coactivators regulate cyclic adenosine 3’,5’-monophosphate-dependent cytochrome P450c17 transcription rate. Mol. Endocrinol. 2007;21:415–438. doi: 10.1210/me.2006-0361. [DOI] [PubMed] [Google Scholar]
- Davies SS, Pontsler AV, Marathe GK, Harrison KA, Murphy RC, Hinshaw JC, Prestwich GD, Hilaire AS, Prescott SM, Zimmerman GA, McIntyre TM. Oxidized alkyl phospholipids are specific, high affinity peroxisome proliferator-activated receptor gamma ligands and agonists. J. Biol. Chem. 2001;276:16015–16023. doi: 10.1074/jbc.M100878200. [DOI] [PubMed] [Google Scholar]
- Delerive P, Furman C, Teissier E, Fruchart J, Duriez P, Staels B. Oxidized phospholipids activate PPARalpha in a phospholipase A2-dependent manner. FEBS Lett. 2000;471:34–38. doi: 10.1016/s0014-5793(00)01364-8. [DOI] [PubMed] [Google Scholar]
- Delos Santos RC, Garay C, Antonescu CN. Charming neighborhoods on the cell surface: plasma membrane microdomains regulate receptor tyrosine kinase signaling. Cell. Signal. 2015;27:1963–1976. doi: 10.1016/j.cellsig.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Dennis EA. Lipid Cell Signaling, Enzymes, LIPID MAPS, and Mediators of Inflammation. J. Biol. Chem. 2016 [Google Scholar]
- Falasca, M and Ferro R. Role of the lysophosphatidylinositol/GPR55 axis in cancer. Adv Biol Regul. 2016;60:88–93. doi: 10.1016/j.jbior.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Fayngerts SA, Wu J, Oxley CL, Liu X, Vourekas A, Cathopoulis T, Wang Z, Cui J, Liu S, Sun H, Lemmon MA, Zhang L, Shi Y, Chen YH. TIPE3 is the transfer protein of lipid second messengers that promote cancer. Cancer Cell. 2014;26:465–478. doi: 10.1016/j.ccr.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Ruan B, Chen J, Schroepfer GJ, Evans RM. The orphan nuclear receptor LXRalpha is positively and negatively regulated by distinct products of mevalonate metabolism. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10588–10593. doi: 10.1073/pnas.94.20.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Kaplan R, Menke JG, MacNaul K, Chen Y, Sparrow CP, Zhou G, Wright SD, Cai TQ. Dual mechanisms of ABCA1 regulation by geranylgeranyl pyrophosphate. J. Biol. Chem. 2001;276:48702–48708. doi: 10.1074/jbc.M109402200. [DOI] [PubMed] [Google Scholar]
- Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberry AL, Wright MM, McMaster CR. The major sites of cellular phospholipid synthesis and molecular determinants of Fatty Acid and lipid head group specificity. Mol. Biol. Cell. 2002;13:3148–3161. doi: 10.1091/mbc.01-11-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horejsi V, Hrdinka M. Membrane microdomains in immunoreceptor signaling. FEBS Lett. 2014;588:2392–2397. doi: 10.1016/j.febslet.2014.05.047. [DOI] [PubMed] [Google Scholar]
- Irvine RF. Nuclear inositide signalling -- expansion, structures and clarification. Biochim Biophys Acta. 2006;1761:505–508. doi: 10.1016/j.bbalip.2006.02.008. doi:S1388-1981(06)00037-0. [pii]10.1016/j.bbalip.2006.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF. Nuclear lipid signalling. Nat Rev Mol Cell Biol. 2003;4:349–360. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA. Structural analyses reveal phosphatidyl inositols as ligands for the NR5A orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Lee H, Shi W, Tontonoz P, Wang S, Subbanagounder G, Hedrick CC, Hama S, Borromeo C, Evans RM, Berliner JA, Nagy L. Role for peroxisome proliferator-activated receptor alpha in oxidized phospholipid-induced synthesis of monocyte chemotactic protein-1 and interleukin-8 by endothelial cells. Circ. Res. 2000;87:516–521. doi: 10.1161/01.res.87.6.516. [DOI] [PubMed] [Google Scholar]
- Lee JM, Lee YK, Mamrosh JL, Busby SA, Griffin PR, Pathak MC, Ortlund EA, Moore DD. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011;474:506–510. doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberger T, Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: a nuclear receptor signaling pathway in lipid physiology. Annu. Rev. Cell Dev. Biol. 1996;12:335–363. doi: 10.1146/annurev.cellbio.12.1.335. [DOI] [PubMed] [Google Scholar]
- Li D, Urs AN, Allegood J, Leon A, Merrill AH, Sewer MB. Cyclic AMP-stimulated interaction between steroidogenic factor 1 and diacylglycerol kinase theta facilitates induction of CYP17. Mol. Cell. Biol. 2007;27:6669–6685. doi: 10.1128/MCB.00355-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BC, Suzawa M, Blind RD, Tobias SC, Bulun SE, Scanlan TS, Ingraham HA. Stimulating the GPR30 estrogen receptor with a novel tamoxifen analogue activates NR5A and promotes endometrial cell proliferation. Cancer Res. 2009;69:5415–5423. doi: 10.1158/0008-5472.CAN-08-1622. doi:0008-5472.CAN-08-1622 [pii] 10.1158/0008-5472.CAN-08-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou X, Toresson G, Benod C, Suh JH, Philips KJ, Webb P, Gustafsson J-A. Structure of the retinoid X receptor α-liver X receptor β (RXRα-LXRβ) heterodimer on DNA. Nat. Struct. Mol. Biol. 2014;21:277–281. doi: 10.1038/nsmb.2778. [DOI] [PubMed] [Google Scholar]
- McIntyre TM, Pontsler AV, Silva AR, St Hilaire A, Xu Y, Hinshaw JC, Zimmerman GA, Hama K, Aoki J, Arai H, Prestwich GD. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc. Natl. Acad. Sci. U. S. A. 2003;100:131–136. doi: 10.1073/pnas.0135855100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- Mikoshiba K. Role of IP3 receptor signaling in cell functions and diseases. Adv Biol Regul. 2015;57:217–227. doi: 10.1016/j.jbior.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Mullaney BC, Blind RD, Lemieux GA, Perez CL, Elle IC, Faergeman NJ, Van Gilst MR, Ingraham HA, Ashrafi K. Regulation of C. elegans fat uptake and storage by acyl-CoA synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25. Cell Metab. 2010;12:398–410. doi: 10.1016/j.cmet.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musille PM, Kohn JA, Ortlund EA. Phospholipid--driven gene regulation. FEBS Lett. 2013;587:1238–1246. doi: 10.1016/j.febslet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musille PM, Pathak MC, Lauer JL, Hudson WH, Griffin PR, Ortlund EA. Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation. Nat. Struct. Mol. Biol. 2012;19:532–537. S1–S2. doi: 10.1038/nsmb.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- Oishi-Tanaka Y, Glass CK. A new role for cyclic phosphatidic acid as a PPARgamma antagonist. Cell Metab. 2010;12:207–208. doi: 10.1016/j.cmet.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CRH, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Zhao K, Janmey P, Crabtree GR. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2824–2829. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Nicchitta CV. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2015;16:221–231. doi: 10.1038/nrm3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Krasowski MD. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr. Drug Metab. 2006;7:349–365. doi: 10.2174/138920006776873526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick AC, Snowman AM, Kang BN, Hurt KJ, Snyder SH, Saiardi A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci U S A. 2005;102:12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin EP, Blind RD, Krylova IN, Ingraham JG, Cai F, Williams JD, Fletterick RJ, Ingraham HA. Structure of NR5A1 (SF-1) bound by different phospholipids: evidence for regulatory ligands. Mol. Endocrinol. 2009;23:25–34. doi: 10.1210/me.2007-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin EP, Blind RD, Uthayaruban R, Chiu HJ, Deacon AM, Das D, Ingraham HA, Fletterick RJ. Structure of NR5A2 (LRH-1) with PIP3 hormone bound in the ligand binding pocket. J. Struct. Biol. 2015;192:342–348. doi: 10.1016/j.jsb.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Xiao H, Ranallo R, Wu W-H, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Haswell ES, Miller AL, Wente SR, O’Shea EK. Regulation of chromatin remodeling by inositol polyphosphates. Science. 2003;299:114–116. doi: 10.1126/science.1078062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toska E, Campbell HA, Shandilya J, Goodfellow SJ, Shore P, Medler KF, Roberts SG. Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Rep. 2012;2:462–469. doi: 10.1016/j.celrep.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toska E, Shandilya J, Goodfellow SJ, Medler KF, Roberts SG. Prohibitin is required for transcriptional repression by the WT1-BASP1 complex. Oncogene. 2013 doi: 10.1038/onc.2013.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukahara T, Tsukahara R, Fujiwara Y, Yue J, Cheng Y, Guo H, Bolen A, Zhang C, Balazs L, Re F, Du G, Frohman MA, Baker DL, Parrill AL, Uchiyama A, Kobayashi T, Murakami-Murofushi K, Tigyi G. Phospholipase D2-dependent inhibition of the nuclear hormone receptor PPARgamma by cyclic phosphatidic acid. Mol. Cell. 2010;39:421–432. doi: 10.1016/j.molcel.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu-Sekine B, Goldschmidt H, Raben DM. Diacylglycerol, phosphatidic acid, and their metabolic enzymes in synaptic vesicle cycling. Adv Biol Regul. 2015;57:147–152. doi: 10.1016/j.jbior.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urs AN, Dammer E, Sewer MB. Sphingosine regulates the transcription of CYP17 by binding to steroidogenic factor-1. Endocrinology. 2006;147:5249–5258. doi: 10.1210/en.2006-0355. [DOI] [PubMed] [Google Scholar]
- Watson PJ, Fairall L, Santos GM, Schwabe JWR. Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature. 2012;481:335–340. doi: 10.1038/nature10728. [DOI] [PMC free article] [PubMed] [Google Scholar]