Abstract
Single nucleotide polymorphisms (SNPs) are replacing microsatellites for population genetic analyses, but it is not apparent how many SNPs are needed or how well SNPs correlate with microsatellites. We used data from the gopher tortoise, Gopherus polyphemus – a species with small populations, to compare SNPs and microsatellites to estimate population genetic parameters. Specifically, we compared one SNP dataset (16 tortoises from 4 populations sequenced at 17,901 SNPs) to two microsatellite datasets, a full dataset of 101 tortoises and a partial dataset of 16 tortoises previously genotyped at 10 microsatellites. For the full microsatellite dataset, observed heterozygosity, expected heterozygosity, and FST were correlated between SNPs and microsatellites; however, allelic richness was not. The same was true for the partial microsatellite dataset, except that allelic richness, but not observed heterozygosity, was correlated. The number of clusters estimated by Structure differed for each dataset (SNPs = 2; partial microsatellite = 3; full microsatellite = 4). PCA showed four clusters for all datasets. More than 800 SNPs were needed to correlate with allelic richness, observed heterozygosity, and expected heterozygosity, but only 100 were needed for FST. The number of SNPs typically obtained from NGS far exceeds the number needed to correlate with microsatellite parameter estimates. Our study illustrates that diversity, FST, and PCA results from microsatellites can mirror those obtained with SNPs. These results may be generally applicable to small populations, a defining feature of endangered and threatened species, because theory predicts that genetic drift will tend to outweigh selection in small populations.
Keywords: microsatellites, target enrichment, sequence capture, next-generation sequencing, immunogenetics, population genomics
Introduction
Molecular markers vary in their utility and application to population genetic studies, and geneticists use available markers suited to answering questions at hand. Initially, geneticists only had allozymes and used them to infer nucleotide changes underlying differences in protein migration during electrophoresis. Later, variable mitochondrial DNA markers were used because of the availability of conserved primers and the high copy number of mitochondria, but mitochondrial markers mostly provided information on broad-scale genetic patterns (Moritz, 1994). Presently, markers such as microsatellites are commonly used in population genetics because most are presumed neutral, are found throughout genomes, and can elucidate fine-scale spatial genetic patterns (e.g., Clostio et al., 2012).
Genomic resources, hybridization arrays, fluorescent probes, and next-generation sequencing (NGS) have allowed researchers to access other types of genomic markers, and recently large arrays of single nucleotide polymorphisms (SNPs) have become particularly popular in population genetic studies of not only model but also non-model organisms (Allendorf et al., 2010). SNPs are one of the most numerous molecular markers (Gupta et al., 2001), and thousands to millions of them can be examined simultaneously using NGS techniques compared to dozens observed in traditional Sanger sequencing-based approaches. However, as the preferred tool shifts from microsatellites to genome-wide SNPs, it is important to understand new results in the context of previous research.
Prior research has shown that microsatellite-derived population genetic parameters generally correlate with parameters derived from SNPs. Most data from pre-NGS SNP methods find correlations between microsatellites and SNPs (e.g., Ryynanen et al., 2007; Narum et al., 2008; Coates et al., 2009; Glover et al., 2010; Garke et al., 2012), but there are some exceptions (e.g., Vali et al., 2008; DeFaveri et. al., 2013). Considerably fewer studies have compared genetic inferences derived from microsatellites to inferences from thousands of NGS generated SNPs, but there are some examples from restriction site-associated DNA sequencing (RADseq) studies where correlations are present (Jeffries et al., 2016) between the two types of markers for population genetic parameters or not (Lozier, 2014). As more and more studies use NGS data, a better understanding of this relationship is imperative because many current management and recovery plans currently in effect are based on genetic data from microsatellites, and these plans may change if results from microsatellites and NGS data are consistently and substantively different.
Although microsatellites are frequently presumed to be neutral because they are not transcribed or translated, they can be linked to functional genes under selection (e.g., Vasemägi et al., 2005; Li et al., 2014) or may be involved in DNA folding (Li et al., 2002). SNPs can be influenced by either neutral or adaptive genetic processes and can represent functional, coding regions of the genome, which on the one hand are under purifying selection to avoid deleterious changes and on the other under positive selection for advantageous changes. For example, SNPs present in genes that influence immune response are likely to be under strong positive selection as such changes could provide resilience to infectious disease (Bernatchez & Landry 2003; Sommer 2005). Additionally, SNPs in immune genes may be under balancing selection to maintain polymorphisms in populations (e.g., Niskanen et al., 2014) by types of balancing selection such as heterozygote advantage, frequency-dependent selection, and variable selection in time and space (Hedrick, 1999).
Although genes such as immune genes are predicted to be under strong selective pressure, neutral genetic processes affect the entire genome, including genes under selection, even when selection is the main evolutionary force (Kuo et al., 2009; Lynch et al. 2011). However, when effective population sizes (Ne) are small, genes influenced by selection may behave like effectively neutral loci because genetic drift tends to outweigh selection in small populations (e.g., Grueber et al., 2013; Miller et al., 2004). In particular, loci under selection may be effectively neutral if their selection coefficient (s) is less than or equal to (1/(2Ne)) (Wright 1931). For example, for alleles of immune response genes such as those of the major histocompatibility complex (MHC), which can have high selection coefficients of 1%, such alleles could behave like effectively neutral loci if effective population sizes are less than 50 individuals (Frankham et al., 2010). Empirical studies support these conclusions as MHC loci behave like effectively neutral loci for a variety of threatened vertebrates with small, bottlenecked populations (Weber et al., 2004; Miller et al., 2008; Taylor et al. 2012).
We recently applied genomic approaches to the threatened gopher tortoise (Gopherus polyphemus) by isolating genes involved in immune responses to better understand susceptibility to a chronic and occasionally fatal infectious upper respiratory tract disease (Elbers & Taylor, 2015). These samples were also previously genotyped at 10 microsatellites by Clostio et al., (2012) providing an excellent opportunity to compare population genetic parameters derived from presumably neutrally evolving microsatellites and presumably drift and/or selection-influenced immune gene SNPs from an organism with generally small population sizes.
We leveraged the NGS (Elbers & Taylor 2015) and microsatellite (Clostio et al., 2012) data already collected for G. polyphemus to compare estimates of population genetic diversity, differentiation, and admixture derived from immune gene SNPs and microsatellites using samples from the same populations to better understand how NGS SNP inferences relate to those from microsatellites. We also subsample our SNPs to determine how many are needed to replace a given number of microsatellites for estimating genetic diversity and differentiation. Although immune gene SNPs are putatively under selection and microsatellites are presumably neutral, we predict inferences from immune gene SNPs will mostly correlate with microsatellite inferences as there will be a preponderance of selectively neutral immune gene SNPs due to the generally small population sizes of G. polyphemus. We also predict that not all of the discovered SNPs will be needed to replace microsatellites for estimating diversity and differentiation.
Methods
Samples
Because SNP analyses are often costly, smaller sample sizes than those used in microsatellite studies are typical. In this study we were interested in how a smaller sample size but a larger number of SNP markers would compare to a typical microsatellite dataset. We were limited to analyzing SNPs from 16 tortoises, so we randomly chose 16 G. polyphemus from 4 sample populations (4 per population, Fig. 1). These 4 sample populations were chosen out of the 24 used by Clostio et al., (2012) because they were distributed along an east to west gradient and were likely representative of the genetic variability for the species. We compared the SNP dataset to two microsatellite datasets: (1) the full microsatellite dataset of 101 tortoises sampled by Clostio et al., 2012 (Table 1); and, (2) a partial microsatellite dataset of 16 tortoises. We used two microsatellite datasets to: 1) equalize sample size (partial), and; 2) use a sample size representative of a typical microsatellite study (full). Only 1 GA tortoise in the SNP dataset had been previously genotyped at all 10 microsatellite loci by Clostio et al., (2012), so for the partial microsatellite dataset, we randomly chose 3 additional tortoises from the GA population that had been genotyped at all 10 microsatellites. Thus, the SNP dataset and the partial microsatellite dataset only differed by 3 samples from the GA population.
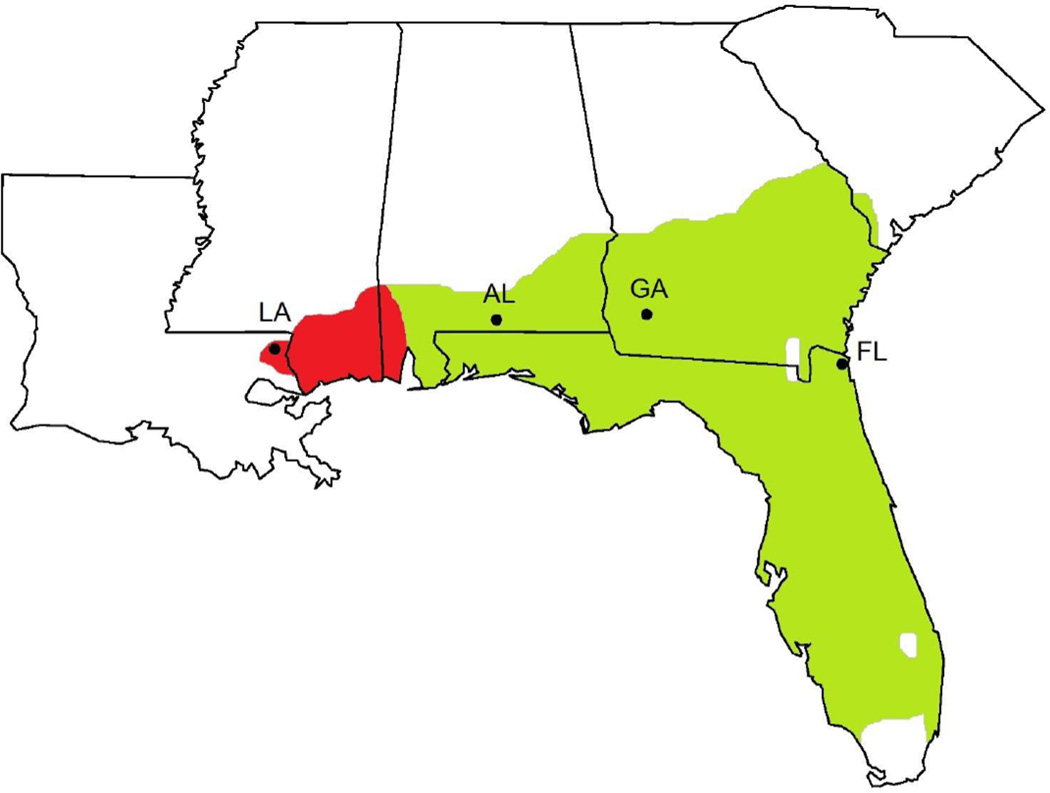
Fig. 1.
Gopherus polyphemus range map and sampling sites used in this study. Range of western G. polyphemus populations darkly shaded on the left with eastern populations lightly shaded on the right. LA for Florida Gas Pipeline, Washington Parish, Louisiana, USA (latitude, longitude, sample size for full microsatellite dataset = 30.78, −90.00; N = 36). AL for Solon Dixon, Andalusia, Alabama, USA (31.16, −86.70; N = 20). GA for Jones Ecological Research Center, Georgia, USA. (31.23, −84.47; N = 26). FL for Private Site, Nassau County, Florida, USA (30.59, −81.56; N = 19)
Table 1.
Comparisons of full (101 individuals) and partial (16 individuals) microsatellite datasets with SNP dataset (16 individuals) for Gopherus polyphemus. Values with decimals represent mean population genetic parameter values. AR for allelic richness, Ho for observed heterozygosity, HE for expected heterozygosity, No. pops for number of optimum populations determined with STRUCTURE HARVESTER for STRUCTURE or visually for PCA.
| Variable | SNP dataset | Full Microsatellite Dataset |
Partial Microsatellite Dataset |
|---|---|---|---|
| AR | 1.541 | 5.487 | 2.900 |
| Correlation with SNPs | not significant | not significant | |
| Ho | 0.267 | 0.495 | 0.469 |
| Correlation with SNPs | significant | not significant | |
| HE | 0.228 | 0.543 | 0.531 |
| Correlation with SNPs | significant | significant | |
| FST | 0.282 | 0.336 | 0.320 |
| Correlation with SNPs | significant | significant | |
| No. pops STRUCTURE | 2 | 4 | 3 |
| No. pops PCA | 4 | 4 | 4 |
Target region for sequencing SNPs
The methods for acquiring SNP data are presented in Elbers & Taylor (2015). Briefly, we created a target region to capture the immunome (i.e., genes involved in immune response, sensu amplo Ortutay & Vihinen (2006)) of Chrysemys picta bellii (western painted turtle) using the GO2TR workflow (Elbers & Taylor, 2015). The workflow filtered the C. p. bellii 3.0.1 genome assembly (Shaffer et al., 2013) annotated by the NCBI Eukaryotic Genome Annotation Pipeline (annotation release 100) using the gene ontology term “immune response” (i.e., genes that function in the immune system's response to internal or invasive threats). Jean-Marie Rouillard of MYcroarray Inc. (Ann Arbor, MI, USA) generated 120-bp bait sequences with 60-bp overlap to capture our 1.4Mbp target region.
Library preparation and sequence capture
We used biotinylated RNA baits from MYcroarray in an in-solution hybridization experiment to capture the immunomes of 16 G. polyphemus. We created 16 Illumina adaptor-ligated libraries using Agilent Sure-Select XT2 Reagent Kits for the Illumina MiSeq (Agilent Technologies, Santa Clara, CA, USA), pooled 16 prepared libraries per capture reaction, and used MYcroarray reagents and protocols for sequence capture. We then sequenced post-capture amplification libraries on two Illumina MiSeq sequencer flow cells (i.e., all individuals sequenced twice) using MiSeq version 3 chemistry and 75-bp paired-end reads at Pennington Biomedical Research Center (Baton Rouge, LA, USA).
Read quality control and mapping
We demultiplexed reads for each MiSeq run, allowing for up to one mismatch in the 8-bp barcode using MiSeq Reporter software. We used TRIMMOMATIC v0.32 (Bolger et al., 2014) default settings for adapter trimming, and for base quality filtering, we trimmed leading and trailing bases with quality scores less than 5 and 15, respectively. We also used sliding window scans to remove the 3' end of reads when average quality dropped below 15, and discarded reads with less than 40 bases. We next merged overlapping paired-ends reads with BBMerge v5.4 from the BBMap suite (https://sourceforge.net/projects/bbmap/) and then combined unpaired single reads (n=9.08 million) and merged paired reads for downstream analysis. Paired and single plus merged reads were first mapped separately to the C. p. bellii 3.0.3 genome using the BWA-MEM algorithm (Li, 2013) implemented in BWA v0.7.12 (Li & Durbin, 2009), and then less stringently using STAMPY v1.0.23 (Lunter & Goodson, 2011). We used SAMTOOLS v1.1 (Li et al. 2009) to merge binary alignment map (BAM) files from paired reads and single plus merged reads. NCBI remap (http://www.ncbi.nlm.nih.gov/genome/tools/remap) was used to convert our bait intervals from C. p. bellii 3.0.1 to C. p. bellii 3.0.3 coordinates.
Variant and genotype calling
Mapped reads were then processed using the Genome Analysis Toolkit v3.3.0 (McKenna et al., 2010, GATK), adhering to GATK best practices for exome sequencing and calling variants such as SNPs with GATK's Haplotype Caller and Unified Genotyper. We then filtered variants to remove those with bad validation, low quality, low read depth, or low genotype quality to produce a high quality set of SNPs called by the Unified Genotyper. Next, we called variants from base-recalibrated BAM files using the Haplotype Caller and filtered variants in the same manner as before. We then looked for concordance between the two variant callers and used concordant SNPs for variant quality filtering of the Haplotype Caller's call set. Finally, we used BEAGLE v4.0 r1398 (Browning & Browning, 2007) for genotype imputation on the variant-recalibrated SNP set. Following variant calling, we used PICARD's v1.128 (http://broadinstitute.github.io/picard/) Calculate HSMetrics to estimate sequencing metrics, and featureCounts (Liao et al., 2014) to estimate the number of genes and exons covered by each sample.
Population genomic analyses
For all population genomic analyses, we analyzed only di-allelic polymorphic SNP loci, as the tri- (n=758) and tetra-allelic (n=7) loci we obtained would influence SNP heterozygosity estimates. We used VCFTOOLS v0.1.12b (Danecek et al., 2011) to recalculate allele frequencies from our Beagle-imputated SNPs and then removed loci with allele frequencies of one. We then pruned SNP loci that were out of Hardy-Weinberg Equilibrium (HWE) or in Linkage Disequilibrium (LD) within each population using default settings in VCFTOOLS. We used the p.adjust function in R (R Core Team, 2015) to correct P values for HWE and LD tests using a false discovery rate (Benjamini & Hochberg, 1995) of 0.05.
We examined what polymorphic SNPs might be under selection with BayeScan v2.1 (Foll & Gaggiotti, 2008) with the intent of pruning those SNPs that were putatively under selection. We used the make_bayescan_input.py script to convert variant call format (VCF) to BayeScan input format (De Wit et al., 2012) and a false discovery rate of 0.05. In order for a given SNP to be included in the analysis, we required at least four good quality genotypes from each population and at least one copy of the minor allele for a locus.
For genetic diversity analyses and all subsequent file conversions, we used PGDSpider v2.0.7.4 (Lischer & Excoffier, 2012) and the R package hierfstat v0.04-10 (Goudet, 2005) to assess observed and expected heterozygosity and allelic richness. For population genomic differentiation, we estimated FST values with hierfstat. For estimating admixture, we performed principle component analyses (PCA) with hierfstat. We also assessed population admixture using STRUCTURE v2.3.4 (Hubisz et al., 2009; Pritchard et al., 2000). We ran STRUCTURE with 100,000 burnins and 100,000,000 replicates using correlated allele frequency and the admixture ancestry models from K = 1–5 with 20 replicates per K value. We used STRUCTURE HARVESTER web v0.6.94 (Earl & vonHoldt, 2012) to select the best K value and CLUMPAK web server (Kopelman et al., 2015) to average data from multiple runs and to visualize population assignments.
Microsatellite analyses
We assessed HWE and LD for the full and partial microsatellite datasets using ARLEQUIN v3.5 (Excoffier & Lischer, 2010). All 10 loci for both datasets were in HWE and linkage equilibrium. Genetic diversity, differentiation, and admixture were estimated in the same manner as SNPs using hierfstat and STRUCTURE.
Random sampling of SNPs for subsampling analysis
We examined how many SNP loci would be needed to obtain P values < 0.05 for Pearson’s r correlation coefficient with the full and partial microsatellite datasets for allelic richness, heterozygosities, and FST values by randomly subsampling our 17,901 SNPs. We did not include allelic richness when comparing the SNP and full microsatellite datasets because they were not correlated at the 0.05 level, and we did not include observed heterozygosity when comparing the SNP and partial microsatellite datasets because they were not correlated. We randomly chose SNPs among the following sample sizes using a custom R script: 10, 20, 40, 100, 200, 400, 800, 1,600, 3,200, 6,400, or 13,200 SNPs and calculated the P value of the Pearson’s correlation coefficient using the cor.test function in R for each sample size of SNP loci for allelic richness, observed heterozygosity, expected heterozygosity, and FST. We repeated the process and chose 10 replicates for each sample size for both the full and partial microsatellite datasets.
Effective population size
We estimated effective population size using the full microsatellite and SNP datasets with the program NeEstimator v2.01 (Do et al., 2014) and employed one single-sample estimator of Ne (i.e., the linkage disequilibrium method of Waples & Do (2008)), and two single-sample estimators of the number of effective breeders per year (i.e., Nb using the heterozygote-excess method of Zhdanova & Pudovkin (2008) and the molecular coancestry method of Nomura (2008)). We converted Nb to Ne by multiplying Nb by the generation time of 31 years for the gopher tortoise (Enge et al., 2006).
Results
From two Illumina MiSeq sequencer runs, we obtained 47.5 million reads that passed quality control and were assignable to individuals. Each tortoise had 3 ± 0.7 (mean ± standard deviation) million reads of which 47.9 ± 3.2 % were unique (i.e., were not PCR duplicates), and 98.8 ± 0.1 % of these unique reads could be aligned to our target region (Table S1, Supporting information). Mean sample coverage over the entire target region was 65.4 ± 13 reads, and each sample had 69.3 ± 3.6 % target bases with coverage greater than 20 reads (Fig. S2, Fig. S3, Supporting information). Only 4.7 % (66.3 Kbp) of the 1.4 Mbp target region had coverage of less than 2 reads. Although our target region contained a total of 632 immune genes and 5,425 exons, only 611 genes and 4,837 exons were represented by usable reads. Each sample had reads for 592.1 ± 4.2 genes and 4,106.2 ± 98.1 exons (mean ± standard deviation).
There were 17,901 di-allelic polymorphic SNP loci after filtering and imputation. None of these loci were out of HWE or in LD, but the lack of LD is unlikely given the close proximity of loci within the same exon. This may have occurred because we had to correct P values to account for thousands of multiple tests. Polymorphic SNPs were present in 491 immune genes (Table S2, Supporting information) and included broad classes such as major histocompatibility and Toll-like receptor genes (Table 2).
Table 2.
Histocompatibility and Toll-like Receptor Loci with di-allelic, polymorphic SNPs in the Gopherus polyphemus SNP dataset (16 G. polyphemus sequenced at 17,901 immune gene SNPs).
| Histocompatibility Loci |
|---|
| CD74 molecule, major histocompatibility complex, class II invariant chain |
| Class I histocompatibility antigen, F10 alpha chain-like |
| Class II histocompatibility antigen, M alpha chain |
| Class II, major histocompatibility complex, transactivator |
| DLA class II histocompatibility antigen, DR-1 beta chain-like |
| H-2 class II histocompatibility antigen, A-R alpha chain-like |
| H-2 class II histocompatibility antigen, E-S beta chain-like |
| HLA class II histocompatibility antigen, DP alpha 1 chain-like |
| HLA class II histocompatibility antigen, DR alpha chain-like |
| HLA class II histocompatibility antigen, DR beta 5 chain-like |
| HLA class II histocompatibility antigen, DRB1-15 beta chain-like |
| Major histocompatibility complex class I-related gene protein-like |
| Rano class II histocompatibility antigen, A beta chain-like |
| Toll-like Receptor Loci |
| Toll-like Receptor 13 |
| Toll-like Receptor 2 |
| Toll-like Receptor 7 |
| Toll-like Receptor 8 |
| Toll-like Receptor adaptor molecule 1 |
| Toll-like Receptor adaptor molecule 2 |
There were 66 SNP loci that may have been under selection, which represented 31 genes. Pruning these SNPs did not significantly influence results, so we chose to analyze the full SNP dataset when comparing genetic diversity, differentiation, or admixture between SNPs and microsatellites.
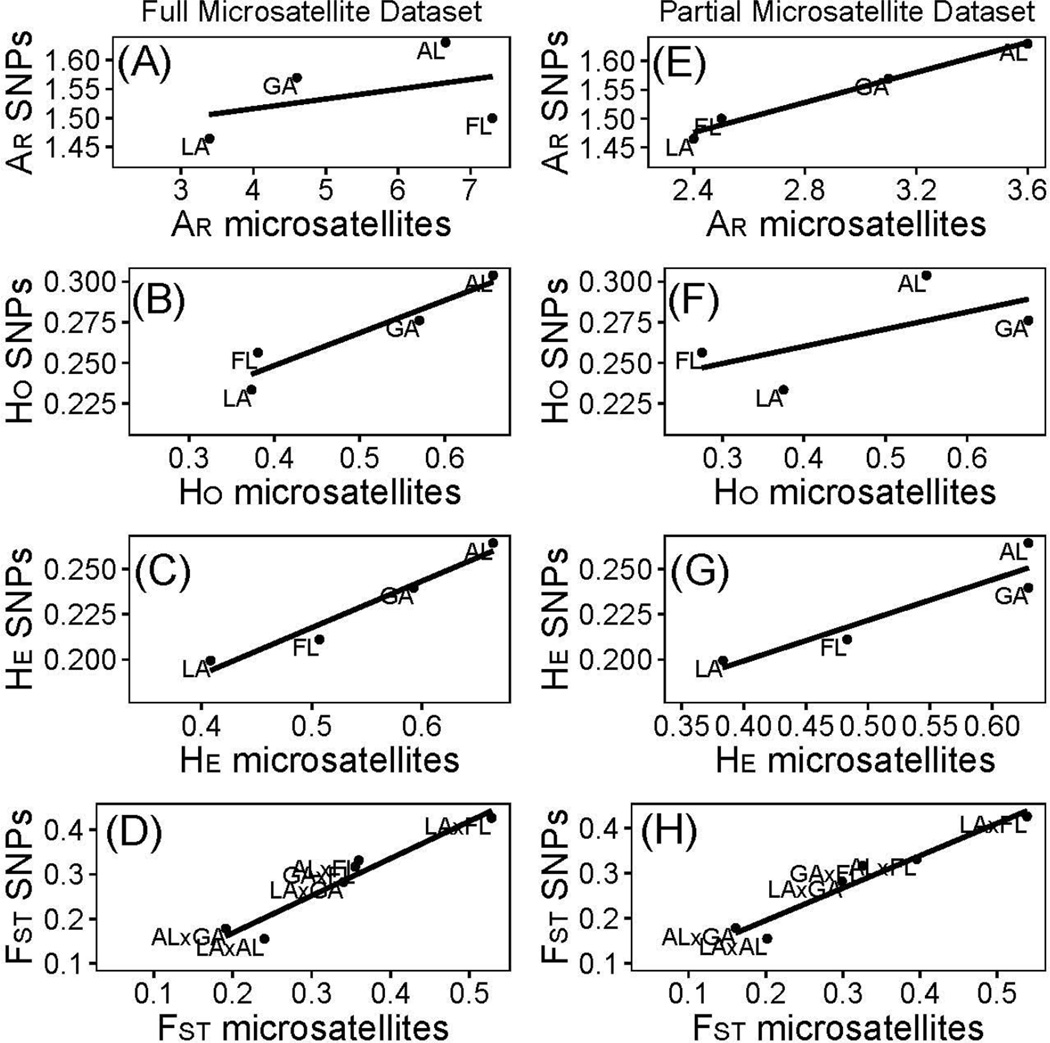
SNP allelic richness was not positively correlated with values derived from the full microsatellite dataset (Fig. 2A, Pearson's r = 0.411, P = 0.294); however, SNP and microsatellite observed (Fig. 2B, Pearson's r = 0.945, P = 0.028) and expected heterozygosities (Fig. 2C, Pearson's r = 0.976, P = 0.012) were highly correlated. Allelic richness was correlated between the SNP and partial microsatellite datasets (Fig. 2E, Pearson's r = 0.992, P = 0.004). Observed heterozygosity was not correlated (Fig. 2F, Pearson's r = 0.630, P = 0.185), but expected heterozygosity was (Fig. 2G, Pearson's r = 0.924, P = 0.038). The LA population followed by FL then GA and AL populations had the lowest to highest heterozygosity and allelic richness for SNPs. This suggests lower genetic diversity in the western LA population versus eastern FL, GA, and AL populations based on SNPs, a similar result to that obtained with both microsatellite datasets.
Fig. 2.
Correlations between 10 microsatellites and 17,901 immune gene SNPs for Gopherus polyphemus samples. Left column for full microsatellite dataset (101 G. polyphemus genotyped at 10 microsatellites) for (A) allelic richness, Pearson's r = 0.411, P = 0.294; (B) observed heterozygosity, Pearson's r = 0.945, P = 0.028; (C) expected heterozygosity, Pearson's r = 0.976, P = 0.012; and (D) FST, Pearson's r = 0.96, P = 0.001. Right column for partial microsatellite dataset (16 G. polyphemus genotyped at 10 microsatellites) for (E) allelic richness, Pearson's r = 0.992, P = 0.004; (F) observed heterozygosity, Pearson's r = 0.63, P = 0.185; (G) expected heterozygosity, Pearson's r = 0.924, P = 0.038; and (H) FST, Pearson's r = 0.968, P < 0.001. AR for allelic richness, Ho for observed heterozygosity, HE for expected heterozygosity.
Pairwise FST values were also positively correlated for SNP and the full (Fig. 2D, Pearson's r = 0.96, P = 0.001) and partial (Fig. 2H, Pearson's r = 0.968, P < 0.001) microsatellite datasets. However, LA and AL had the lowest differentiation for SNPs compared to second lowest for microsatellites.
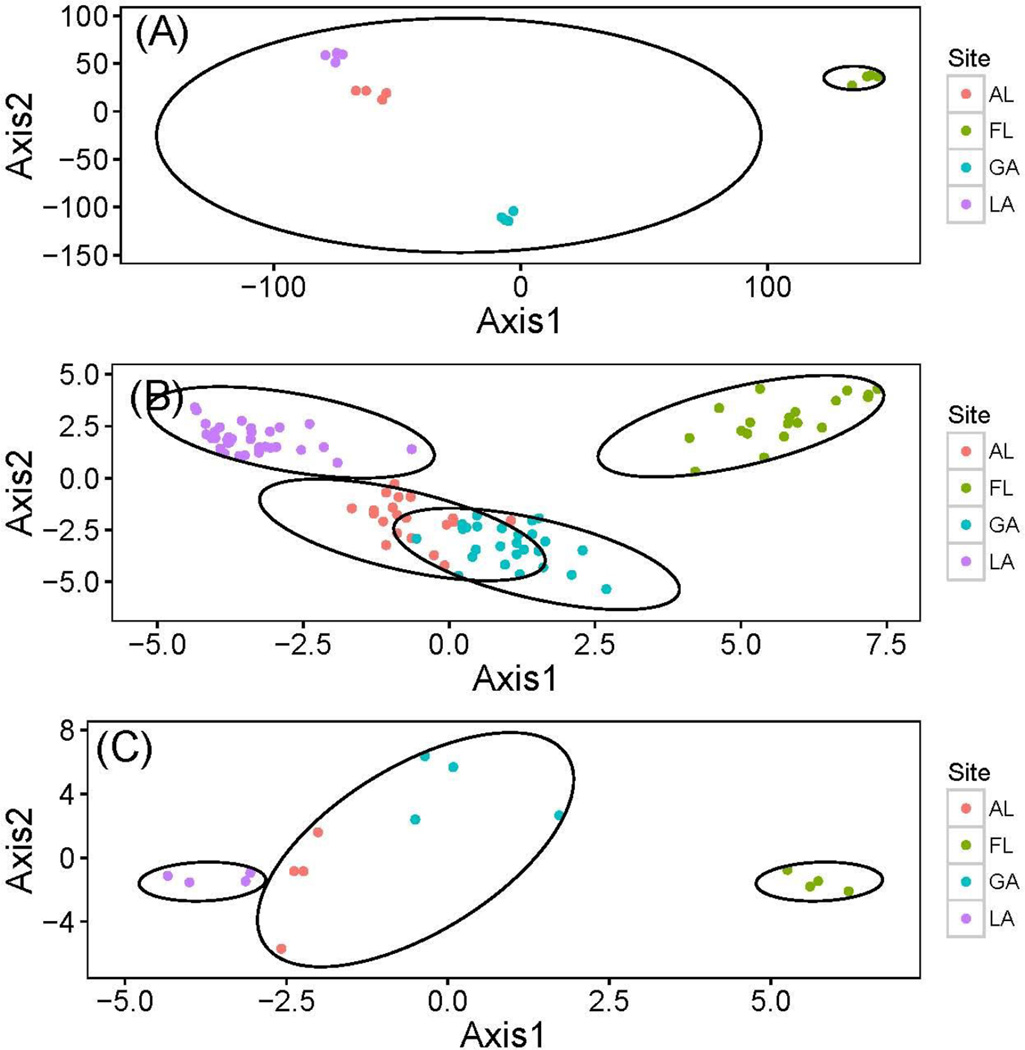
Population admixture inferred using SNPs suggested an optimum number of two clusters with STRUCTURE, the first consisting of AL, GA, and LA; the second with FL by itself (Fig. S3, Supporting information). For the full microsatellite dataset, there was an optimum of four clusters: one for each population examined (Fig. S4, Supporting information). The partial microsatellite dataset had three optimum clusters: the first with LA; the second with AL and GA; and the third with FL (Fig. S5, Supporting information). PCA analysis produced four clusters for SNPs and both microsatellite datasets (one for each population, Fig. 3A–3C).
Fig. 3.
Principle component analysis for Gopherus polyphemus datasets: (A) the SNP dataset (16 G. polyphemus sequenced at 17,901 immune gene SNPs); (B) full microsatellite dataset (101 G. polyphemus genotyped at 10 microsatellites); and (C) partial microsatellite dataset (16 G. polyphemus genotyped at 10 microsatellites). Circles indicate optimum clusters identified using STRUCTURE and STRUCTURE HARVESTER.
Random sampling of SNP loci showed that at least 1,600 SNPs were needed to obtain a significant correlation between SNP- and the full microsatellite dataset for allelic richness (Fig. S6A, Supporting information). Nearly 800 SNPs were needed for expected heterozygosity (Fig. S6B, Supporting information), but only 100 SNPs were needed for SNP- and microsatellite-derived FST values to be correlated (Fig. S6C). There was a similar pattern for the partial microsatellite dataset for allelic richness, expected heterozygosity, and FST, where at least 800, 800, and 100 SNPs were needed for significant correlations, respectively (Fig. S7A–7C, Supporting information). Parameter variability decreased as the number of randomly chosen SNPs increased, especially after 200, 100, 40, and 40 SNPs for allelic richness, observed and expected heterozygosity, and FST values respectively (Fig. S6, Fig. S7, Supporting information).
Effective population sizes estimated using the full microsatellite dataset were not particularly informative, especially the estimates of infinite population sizes from the heterozygous-excess and linkage disequilibrium methods (Fig. S8A, Supporting information). Minus the FL population’s estimate of infinite effective population size, the molecular coancestry method suggested more reasonable estimates of effective population sizes between 34–589 individuals per population. Effective population sizes estimated using immune gene SNPs were more realistic with the heterozygous-excess method suggesting between 133–186 tortoises, and the molecular coancestry method suggesting between 319–427 tortoises per population (Fig. S8B, Supporting information). The linkage disequilibrium method was not informative as all effective population sizes were estimated to be infinite.
The Ne estimates that ranged between 34–589 individuals (microsatellite and SNP molecular coancestry and SNP heterozygous-excess approaches) suggest that selection coefficients for SNPs would need to be less than 0.1% for genetic drift to outweigh selection.
Discussion
Estimates of genetic diversity derived from gopher tortoise immunome SNPs and both microsatellite datasets were typically correlated. Given that most gopher tortoise populations are small, immune gene SNPs may be behaving like effectively neutral loci. Thus, these correlations are theoretically reasonable and may hold true for other small populations, for example, endangered and threatened species generally.
Other studies have observed similar and contrasting correlations between SNP and microsatellite-derived estimates of genetic diversity. For example, previous work using 7 SNPs/indels and 14 microsatellites found that expected heterozygosity and allelic richness are positively correlated between the two types of markers in Atlantic salmon populations (Ryynanen et al., 2007). On the contrary, SNP (n=1–46) and microsatellite (n=10–27) heterozygosities are not correlated for European and North American wolf populations (Vali et al., 2008). Likewise, microsatellite-estimated diversity is different between Bombus bumble bee species, but similar when using RADseq loci (Lozier, 2014), thus diversity estimates from these two markers are not correlated.
In gopher tortoises, the rank order for allelic richness and observed heterozygosity was similar but not the same for immune gene SNPs and the full and partial microsatellite datasets, respectively. Similar observations have been made by other studies including those comparing SNPs and microsatellites in Atlantic salmon (Ryynanen et al., 2007). Rank order may be skewed between the markers because microsatellites are poly-allelic while SNPs are di-allelic. In particular, for a microsatellite or SNP marker, there are n ((n − 1)/2) combinations that result in a heterozygote where n is the number of alleles. Thus, for a di-allelic marker, there is only one combination of alleles that results in a heterozygote, and for a microsatellite that has at least 5 alleles (i.e., the average allelic richness for our 10 microsatellites in the full microsatellite dataset), there are 10 combinations of alleles that are heterozygous. This could explain why observed heterozygosity was not correlated between SNPs and microsatellites for the partial microsatellite dataset.
Previous work with microsatellites showed that genetic variation was lower in western versus eastern G. polyphemus populations (Ennen et al., 2010), and our results with the SNP and re-analysis of the full microsatellite datasets support this finding. For the partial microsatellite dataset, the FL and not LA population had the lowest observed heterozygosity, but in the full microsatellite dataset, the LA population had the lowest observed heterozygosity. The full microsatellite dataset probably provides better estimates as 36 and 19 tortoises were analyzed for the LA and FL populations, respectively as compared to just four tortoises in the partial microsatellite dataset, therefore observed heterozygosity is likely lower in the LA than FL population. Because we only sampled a single western population (Fig. 1), it is not appropriate to generalize all western populations as genetically depauperate. Ultimately, additional sampling and immunome sequencing from other western G. polyphemus populations is warranted.
Genetic differentiation
We also observed strong correlations between SNP and microsatellite-derived genetic differentiation, albeit the order of least to most differentiated comparisons varied. The same was observed for SNP- and microsatellite-derived FST estimates from four populations of western corn rootworms (Coates et al., 2009). The incongruence in rank order may have occurred in both scenarios because of homoplasy issues with microsatellites, where high mutation rates can cause repeat number to revert to a particular allele size, which can then inflate estimates of gene flow (Coates et al., 2009).
Genetic admixture
Population admixture assessments had few inconsistencies between SNPs and microsatellites. PCAs suggested four clusters using either marker. We did observe differences in STRUCTURE admixture results with the optimum number of clusters being 2 for SNPs and 4 and 3 for the full and partial microsatellite datasets. Morin et al. (2012) compared 42 SNPs versus 22 microsatellites in bowhead whales and also found that the optimum number of clusters is different when using STRUCTURE. SNPs and microsatellites may have suggested different estimates of the optimum number of clusters because some of the SNPs may represent functional rather than neutral genetic variation like the microsatellites, with both types of markers differing to what extent they have been influenced by selection and/or genetic drift..
Experimental design considerations
So far, we have discussed how population genetic parameters estimated from immune gene SNPs mirror patterns estimated from microsatellite loci, but marker choice also depends on additional considerations such as cost, number of loci, computational issues with NGS generated SNPs, and neutral versus selective processes. First, although sequencing costs are decreasing, NGS techniques can be more expensive than microsatellites on a per sample basis depending on availability of equipment. In particular, the NGS technique used in this paper, in-solution hybridization, requires synthesis of expensive RNA baits/probes, in the order of several thousand dollars (USD). Although tagged microsatellite primers are not trivial in cost, they are far cheaper than biotinylated RNA baits. Further, most genetics labs are not equipped for NGS workflows that require specialized equipment, so lab work must either be outsourced to commercial or non-commercial core facilities.
The number of loci required to adequately address the genetic question at hand is also an important consideration when choosing between SNPs and microsatellites and will vary depending on the question being asked. In general, simulations suggest many more SNPs are needed than microsatellite loci when trying to achieve similar statistical power or parameter estimates. For example, between 60–100 SNP loci are needed for accurate parentage assignment (Anderson & Garza, 2006), and empirical data from sockeye salmon suggest 80 SNPs have higher assignment success and are more accurate for parentage assignment than 11 microsatellites (Hauser et al., 2011). Furthermore, 80 or more SNPs are needed for detecting low levels of divergence (i.e., FST < 0.005) (Morin et al., 2009). Ryynanen et al. (2007) observed significant correlations between 7 SNPs/indels and 14 microsatellite loci when estimating FST. Our data subsampling results suggest at least 100 SNP loci are needed for correlating SNP and microsatellite-derived FST. For allelic richness and heterozygosities, our data suggest more than 800 SNP loci are needed to correlate with 10 microsatellite loci in G. polyphemus, but Ryynanen et al. (2007) only needed 7 SNP/indel loci to obtain similar correlations, possibly because they analyzed 21 populations. Acquiring data from a large number of SNPs is not a problem with NGS approaches. Not all SNP loci are equally informative, and smaller SNP panels may occasionally perform well in comparison to much larger SNP arrays.
Computational issues with NGS are also not trivial, as our own NGS analysis relied on high performance computing resources and required many gigabytes of data storage. This does not include the time or expertise required to write code and scripts to analyze the gigabytes of raw data.
Neutral versus selective processes are also important to consider when deciding between SNPs and microsatellites. Markers such as microsatellites may be neutrally evolving unless linked to functional genes while SNPs could represent both functional and neutral markers and be influenced by both neutral and adaptive processes. Our SNP data had very few SNPs that were putatively under selection (less than 1%), which is in line with previous NGS studies (e.g., Hohenlohe et al., 2010; Lemay & Russello 2015; Blanco-Bercial & Bucklin 2016). This together with the observed correlations between SNPs and microsatellites suggests that most of our SNPs were effectively neutral. The gopher tortoise populations we surveyed appear to have small effective population sizes, likely less than 500 individuals per population, so perhaps the selection coefficients of many of the immune gene SNPs were small enough (i.e., less than 0.1%) that they behaved as effectively neutral loci.
Conclusion
As more and more population genetic studies are publishing NGS generated SNPs as opposed to microsatellites, it would be useful to identify patterns between microsatellites and NGS derived SNPs and to appreciate the additional functional information commonly provided by SNPs. One apparent pattern is that high variation observed at microsatellites can translate into high SNP-estimates of genetic diversity (Ryynanen et al., 2007) and vice versa. Further, genetic diversity estimated by allelic richness between microsatellites and SNPs may be a less stable metric than diversity estimated by observed and/or expected heterozygosity as more alleles are present in microsatellites than SNPs. This does not mean allelic richness should be ignored especially for conservation purposes because some traits including disease resistance are associated with particular alleles (e.g., Langefors et al., 2001), which is not accounted for by heterozygosity. Another important pattern that may be observed between microsatellites and SNP studies is presence/absence of genetic structure, with any potential inconsistencies resulting from different evolutionary forces acting on the markers. The addition of adaptive processes acting on SNPs can result in similar but disparate structure patterns between the two marker types. Finally, even SNPs that are putatively influenced by selection may behave as effectively neutral loci when effective population sizes are small, thus we recommend researchers consider this when comparing population genetic results derived from potentially functional and neutral markers in small populations such as those of threatened and endangered species.
Supplementary Material
Acknowledgments
This material is based upon work that is supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture, McIntire Stennis project LAB04066 and LAB94169. The Lucius Gilbert Foundation provided support for sequencing and for J.P.E. We are grateful to Richard Carmouche of Pennington Biomedical Research Center's Genomic Core Facility for performing next-generation sequencing laboratory work. This project/work used genomics core facilities that are supported in part by COBRE (NIH 8 P20 GM103528) and NORC (NIH 2P30DK072476) center grants from the National Institutes of Health. We appreciate the access we had to the LSU High Performance Computing resources, which we used to analyze next-generation sequencing data.
Footnotes
Data Accessibility
Raw sequencing data are available from the Sequence Read Archive (accession: SRP061247). BAM and VCF files are available from Dryad repository (doi: 10.5061/dryad.40c7c). Detailed analytical methods and scripts to create Tables and Figures are available from https://github.com/jelber2/immunome_2014
Author Contributions
J.P.E. designed the study and performed SNP analyses. R.W.C. performed microsatellite analyses. J.P.E. and S.S.T. wrote the paper.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1 Sequencing metrics for Gopherus polyphemus samples. Percent UR for percent of total reads that were unique, Percent URA for percent of unique reads that were alignable, Mean coverage for mean number of reads across the target region, Percent 20× for percent of bases in target region with greater than 20× coverage, No. genes for number of genes, and No. exons for number of exons.
Table S2 All genes with di-allelic, polymorphic SNPs from 16 Gopherus polyphemus samples.
Fig. S1 Coverage plots for first eight Gopherus polyphemus samples showing number of sequencing reads at or above specified proportions. A value at 100 Depth and 0.5 fraction means 50 percent of bases were at or above 100× coverage.
Fig. S2 Coverage plots for last eight Gopherus polyphemus samples showing number of sequencing reads at or above specified proportions.
Fig. S3 STRUCTURE plot for 16 Gopherus polyphemus sequenced at 17,901 immune gene SNPs with optimum number of clusters K = 2 determined by STRUCTURE HARVESTER.
Fig. S4 STRUCTURE plot for the full microsatellite dataset (101 Gopherus polyphemus genotyped at 10 microsatellite loci) with optimum number of clusters K = 4 determined by STRUCTURE HARVESTER.
Fig. S5 STRUCTURE plot for the partial microsatellite dataset (16 Gopherus polyphemus genotyped at 10 microsatellite loci) with optimum number of clusters K = 3 determined by STRUCTURE HARVESTER.
Fig. S6 Subsampling analysis showing how many randomly sampled SNP loci out of the total of 17,901 are needed in comparison to the full microsatellite dataset (101 Gopherus polyphemus genotyped at 10 microsatellite loci) for Pearson's r correlation coefficient to be significant at 0.05 level (dotted line) for (A) observed heterozygosity; (B) expected heterozygosity; and (C) FST. There were 10 simulations for each size class of SNPs. Ho for observed heterozygosity, HE for expected heterozygosity.
Fig. S7 Subsampling analysis showing how many randomly sampled SNP loci out of the total of 17,901 are needed in comparison to the partial microsatellite dataset (16 Gopherus polyphemus genotyped at 10 microsatellite loci) for Pearson's r correlation coefficient to be significant at 0.05 level (dotted line) for (A) allelic richness; (B) expected heterozygosity; and (C) FST. There were 10 simulations for each size class of SNPs. AR for allelic richness, HE for expected heterozygosity.
Fig. S8 Effective population sizes per generation (Ne) along with 95 % confidence intervals for Gopherus polyphemus samples estimated with the program NeEstimator using (A) the full microsatellite dataset (101 G. polyphemus genotyped at 10 microsatellite loci) or (B) the SNP dataset (16 G. polyphemus sequenced at 17,901 immune gene SNPs). Dots that are on the top of the graph represent Ne estimates of infinity, and lines that extend to the top of the graph represent upper 95 % confidence limits of infinity. LD for linkage disequilibrium method of Waples & Do (2008), HET for heterozygote-excess method of Zhdanova & Pudovkin (2008), and MOL for the molecular coancestry method of Nomura (2008). Note that the HET and MOL methods estimate the effective number of breeders per year (Nb), which were converted to Ne by multiplying Nb by the generation time of 31 years for G. polyphemus (Enge et al. 2006).
References
- Allendorf FW, Hohenlohe PA, Luikart G. Genomics and the future of conservation genetics. Nature Reviews Genetics. 2010;11:697–709. doi: 10.1038/nrg2844. [DOI] [PubMed] [Google Scholar]
- Anderson EC, Garza JC. The power of single-nucleotide polymorphisms for large-scale parentage inference. Genetics. 2006;172:2567–2582. doi: 10.1534/genetics.105.048074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? Journal of Evolutionary Biology. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- Blanco-Bercial L, Bucklin A. New view of population genetics of zooplankton: RAD-seq analysis reveals population structure of the North Atlantic planktonic copepod Centropages typicus. Molecular Ecology. 2016;25:1566–1580. doi: 10.1111/mec.13581. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. TRIMMOMATIC: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. American Journal of Human Genetics. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clostio RW, Martinez AM, LeBlanc KE, Anthony NM. Population genetic structure of a threatened tortoise across the south-eastern United States: implications for conservation management. Animal Conservation. 2012;15:613–625. [Google Scholar]
- Coates BS, Sumerford DV, Miller NJ, et al. Comparative performance of single nucleotide polymorphism and microsatellite markers for population genetic analysis. Journal of Heredity. 2009;100:556–564. doi: 10.1093/jhered/esp028. [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, et al. The Variant Call Format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit P, Pespeni MH, Ladner JT, et al. The simple fool's guide to population genomics via RNA-Seq: an introduction to high-throughput sequencing data analysis. Molecular Ecology Resources. 2012;12:1058–1067. doi: 10.1111/1755-0998.12003. [DOI] [PubMed] [Google Scholar]
- DeFaveri J, Viitaniemi H, Leder E, Merila J. Characterizing genic and nongenic molecular markers: comparison of microsatellites and SNPs. Molecular Ecology Resources. 2013;13:377–392. doi: 10.1111/1755-0998.12071. [DOI] [PubMed] [Google Scholar]
- Do C, Waples RS, Peel D, Macbeth G, Tillett BJ, Ovenden JR. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources. 2014;14:209–214. doi: 10.1111/1755-0998.12157. [DOI] [PubMed] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Elbers JP, Taylor SS. GO2TR: a gene ontology-based workflow to generate target regions for target enrichment experiments. Conservation Genetics Resources. 2015;7:851–857. [Google Scholar]
- Enge K, Berish J, Bolt R, Dziergowski A, Musinsky H. Biological status report gopher tortoise. Report, Florida Fish and Wildlife Conservation Commission. 2006 [Google Scholar]
- Ennen JR, Kreiser BR, Qualls CP. Low genetic diversity in several gopher tortoise (Gopherus polyphemus) populations in the Desoto National Forest, Mississippi. Herpetologica. 2010;66:31–38. [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. 2nd. Cambridge: Cambridge University Press; 2010. [Google Scholar]
- Garke C, Ytournel F, Bedhom B, et al. Comparison of SNPs and microsatellites for assessing the genetic structure of chicken populations. Animal Genetics. 2012;43:419–428. doi: 10.1111/j.1365-2052.2011.02284.x. [DOI] [PubMed] [Google Scholar]
- Glover KA, Hansen MM, Lien S, Als TD, Hoyheim B, Skaala O. A comparison of SNP and STR loci for delineating population structure and performing individual genetic assignment. BMC Genetics. 2010;11:1–12. doi: 10.1186/1471-2156-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Molecular Ecology Notes. 2005;5:184–186. [Google Scholar]
- Grueber CE, Wallis GP, Jamieson IG. Genetic drift outweighs natural selection at toll-like receptor (TLR) immunity loci in a re-introduced population of a threatened species. Molecular Ecology. 2013;22:4470–4482. doi: 10.1111/mec.12404. [DOI] [PubMed] [Google Scholar]
- Gupta P, Roy J, Prasad M. Single nucleotide polymorphisms SNPs: a new paradigm in molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Current Science. 2001;80:524–535. [Google Scholar]
- Hauser L, Baird M, Hilborn RAY, Seeb LW, Seeb JE. An empirical comparison of SNPs and microsatellites for parentage and kinship assignment in a wild sockeye salmon (Oncorhynchus nerka) population. Molecular Ecology Resources. 2011;11:150–161. doi: 10.1111/j.1755-0998.2010.02961.x. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. Balancing selection and MHC. Genetica. 1999;104:207–214. doi: 10.1023/a:1026494212540. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stifer N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genetics. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Molecular Ecology Resources. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries DL, Copp GH, Lawson Handley L, Olsen KH, Sayer CD, Hanfing B. Comparing RADseq and microsatellites to infer complex phylogeographic patterns, an empirical perspective in the Crucian carp, Carassius carassius, L. Molecular Ecology. 2016;25:2997–3018. doi: 10.1111/mec.13613. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CH, Moran NA, Ochman H. The consequences of genetic drift for bacterial genome complexity. Genome Research. 2009;19:1450–1454. doi: 10.1101/gr.091785.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langefors A, Lohm J, Grahn M, Andersen O, von Schantz T. Association between major histocompatibility complex class IIb alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proceedings of the Royal Society of London B. 2001;268:479–485. doi: 10.1098/rspb.2000.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay MA, Russello MA. Genetic evidence for ecological divergence in kokanee salmon. Molecular Ecology. 2015;24:798–811. doi: 10.1111/mec.13066. [DOI] [PubMed] [Google Scholar]
- Li C, Sun Y, Huang HW, Cannon CH. Footprints of divergent selection in natural populations of Castanopsis fargesii (Fagaceae) Heredity. 2014;113:533–541. doi: 10.1038/hdy.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013;1303.3997 [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Korol AB, Fahima T, Beiles A, Nevo E. Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Molecular Ecology. 2002;11:2453–2465. doi: 10.1046/j.1365-294x.2002.01643.x. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lischer H, Excofer L. PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics. 2012;28:298–299. doi: 10.1093/bioinformatics/btr642. [DOI] [PubMed] [Google Scholar]
- Lozier JD. Revisiting comparisons of genetic diversity in stable and declining species: assessing genome-wide polymorphism in North American bumble bees using RAD sequencing. Molecular Ecology. 2014;23:788–801. doi: 10.1111/mec.12636. [DOI] [PubMed] [Google Scholar]
- Lunter G, Goodson M. STAMPY: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Research. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Bobay LM, Catania F, Gout JF, Rho M. The repatterning of eukaryotic genomes by random genetic drift. Annual Review of Genomics and Human Genetics. 2011;12:347–366. doi: 10.1146/annurev-genom-082410-101412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller HC, Lambert DM. Genetic drift outweighs balancing selection in shaping post-bottleneck major histocompatibility complex variation in New Zealand robins (Petroicidae) Molecular Ecology. 2004;13:3709–3721. doi: 10.1111/j.1365-294X.2004.02368.x. [DOI] [PubMed] [Google Scholar]
- Miller HC, Miller KA, Daugherty CH. Reduced MHC variation in a threatened tuatara species. Animal Conservation. 2008;11:206–214. [Google Scholar]
- Morin PA, Archer FI, Pease VL, et al. An empirical comparison of SNPs and microsatellites for population structure, assignment, and demographic analyses of bowhead whale populations. Endangered Species Research. 2012;19:129–147. [Google Scholar]
- Morin PA, Martien KK, Taylor BL. Assessing statistical power of SNPs for population structure and conservation studies. Molecular Ecology Resources. 2009;9:66–73. doi: 10.1111/j.1755-0998.2008.02392.x. [DOI] [PubMed] [Google Scholar]
- Moritz C. Applications of mitochondrial DNA analysis in conservation: a critical review. Molecular Ecology. 1994;3:401–411. [Google Scholar]
- Narum SR, Banks M, Beacham TD, et al. Differentiating salmon populations at broad and fine geographical scales with microsatellites and single nucleotide polymorphisms. Molecular Ecology. 2008;17:3464–3477. doi: 10.1111/j.1365-294x.2008.03851.x. [DOI] [PubMed] [Google Scholar]
- Niskanen AK, Kennedy LJ, Ruokonen M, et al. Balancing selection and heterozygote advantage in major histocompatibility complex loci of the bottlenecked Finnish wolf population. Molecular Ecology. 2014;23:875–889. doi: 10.1111/mec.12647. [DOI] [PubMed] [Google Scholar]
- Nomura T. Estimation of effective number of breeders from molecular coancestry of single cohort sample. Evolutionary Applications. 2008;1:462–474. doi: 10.1111/j.1752-4571.2008.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortutay C, Vihinen M. Immunome: a reference set of genes and proteins for systems biology of the human immune system. Cellular Immunology. 2006;244:87–89. doi: 10.1016/j.cellimm.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- Ryynanen HJ, Tonteri A, Vasemagi A, Primmer CR. A comparison of biallelic markers and microsatellites for the estimation of population and conservation genetic parameters in Atlantic salmon (Salmo salar) Journal of Heredity. 2007;98:692–704. doi: 10.1093/jhered/esm093. [DOI] [PubMed] [Google Scholar]
- Shaffer HB, Minx P, Warren DE, et al. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biology. 2013;14:R28. doi: 10.1186/gb-2013-14-3-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Frontiers in Zoology. 2005;2:16–16. doi: 10.1186/1742-9994-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Jenkins DA, Arcese P. Loss of MHC and neutral variation in Peary caribou: genetic drift is not mitigated by balancing selection or exacerbated by MHC allele distributions. PLoS One. 2012;7:e36748. doi: 10.1371/journal.pone.0036748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vali U, Einarsson A, Waits L, Ellegren H. To what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations? Molecular Ecology. 2008;17:3808–3817. doi: 10.1111/j.1365-294X.2008.03876.x. [DOI] [PubMed] [Google Scholar]
- Vasemagi A, Gross R, Paaver T, Koljonen ML, Saisa M, Nilsson J. Analysis of gene associated tandem repeat markers in Atlantic salmon (Salmo salar L.) populations: implications for restoration and conservation in the Baltic Sea. Conservation Genetics. 2005;6:385–397. [Google Scholar]
- Waples RS, Do C. LDNE: a program for estimating effective population size from data on linkage disequilibrium. Molecular Ecology Resources. 2008;8:753–756. doi: 10.1111/j.1755-0998.2007.02061.x. [DOI] [PubMed] [Google Scholar]
- Weber DS, Stewart BS, Schienman J, Lehman N. Major histocompatibility complex variation at three class II loci in the northern elephant seal. Molecular Ecology. 2004;13:711–718. doi: 10.1111/j.1365-294x.2004.02095.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhdanova OL, Pudovkin AI. Nb HetEx: a program to estimate the effective number of breeders. Journal of Heredity. 2008;99:694–695. doi: 10.1093/jhered/esn061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.