Abstract
Streptococcus pneumoniae, a major cause of human disease, produces a 17-mer autoinducer peptide pheromone (competence-stimulating peptide [CSP]) for the control of competence for genetic transformation. Due to previous work linking CSP to stress phenotypes, we set up an in vivo sepsis model to assay its effect on virulence. Our data demonstrate a significant increase in the rates of survival of mice, reductions of blood S. pneumoniae counts, and prolonged times to death for mice treated with CSP. In vitro the dose of CSP used in the animal model produced a transitory inhibition of growth. When a mutant with a mutation in the CSP sensor histidine kinase was assayed, no bacteriostatic phenotype was detected in vitro and no change in disease outcome was observed in vivo. The data demonstrate that CSP, which induces in vitro a temporary growth arrest through stimulation of its cognate histidine kinase receptor, is able to block systemic disease in mice. This therapeutic effect is novel, in that the drug-like effect is obtained by stimulation, rather than inhibition, of a bacterial drug target.
Despite the availability of antimicrobial drugs, Streptococcus pneumoniae is the number one cause of bacterial pneumonia (52) and otitis media, the most frequent cause of sepsis in human immunodeficiency virus-infected patients (2), and the second most frequent cause of meningitis in all age groups (35). From the latest data, the prevalence of penicillin resistance in S. pneumoniae is 18.2% and the prevalence of macrolide resistance is 24.6% (31), yet about 3% of pneumococcal isolates in the United States may be tolerant to vancomycin and may be associated with treatment failure (51). The current treatment guidelines for pneumococcal systemic disease (sepsis and meningitis) recommend penicillin, amoxicillin, or broad-spectrum cephalosporins (if risk factors are present) and vancomycin, possibly in combination with rifampin, if penicillin resistance is suspected (12, 20). Even when appropriate antibiotic therapy is given, the overall case fatality rate for pneumococcal disease remains about 20%, and the mortality rate for pneumococcal bacteremia has remained unchanged since the 1950s (1, 2). Following pneumococcal meningitis, the mortality rate and the frequency of long-term neurological damage in survivors are very high (6, 17) and have remained unchanged for the last 40 years (50). These facts continue to foster the search for novel drug targets and drugs in order to develop novel options for the treatment of infections caused by this pathogen (40, 64, 70).
One of the major ways in which bacteria sense environmental signals is by using two-component and phosphorelay signal transduction systems (44, 63). Generally, two-component systems (TCSs) are composed of a sensor histidine protein kinase that is activated by a specific environmental signal and a response regulator that is a transcription factor. The sensor histidine kinases are commonly integral membrane proteins that, when stimulated by a specific signal, have an autokinase activity (an ATP-dependent autophosphorylation reaction on a conserved histidine residue), which in turn determines phosphorylation of a conserved aspartate residue of the cognate response regulator. The TCSs and phosphorelays have been reported in fungi and plants but not in animals (66). In fact histidine-aspartate signal transduction is distinct from the serine-threonine signaling pathways used by eukaryotes. This and several other reasons, including the finding that some TCSs are essential and that others regulate virulence in a variety of pathogens, make them attractive targets in the development of antimicrobial agents (44, 63). As for other approaches in lead discovery and drug development, the screening of combinatorial libraries has led to the identification of a variety of molecules showing in vitro inhibitory activities against TCSs (44, 63).
S. pneumoniae has 13 different TCSs (36), and a variety of reports have underlined how some of these systems are interconnected. In most cases an effect on the main multifactorial phenotypes, including virulence or competence, can be evidenced. The magnitude of the effects varies between the strains (5, 34) and the TCSs analyzed (36, 67) An example is the interconnection between CiaRH (23) (SP0798-9 in The Institute for Genomic Research [TIGR] genome) (65), TCS05 in annotation by Lange et al. (36), 494 in the work of Throup et al. (67), MicAB (SP1226-7, TCS02, 492, and VicRKX) (19, 69), and ComDE (SP2235-6, TCS12, and 498) (13, 18, 34, 43, 56). Competence for genetic transformation in S. pneumoniae is a natural process and depends on a system of coordinated gene regulation (10). In 1995 Morrison and coworkers (25) demonstrated that pneumococci produce a 17-residue peptide pheromone that induces cells to develop competence. Competence-stimulating peptide (CSP) coordinates the sudden and transient appearance of competence during the exponential growth phase in vitro. CSP is matured and exported through the ComAB ATP-binding cassette transporter (30), and cells sense CSP through a dedicated TCS composed of the sensor histidine kinase, ComD, and the response regulator, ComE (26). The genes of this TCS are cotranscribed with the CSP precursor, encoded by the comC gene. The ComE response regulator directly regulates the comAB and comCDE operons and a third gene encoding an alternative comX sigma factor, which in turn controls the rest of the pheromone quorum-sensing regulated genes (41). A variety of reports on gene expression profiling (serotype 2, 4, and 19F strains) in S. pneumoniae have focused on the specific analysis of comCDE-regulated genes (4, 13, 55, 56, 59), as this is a classical model system for quorum sensing with an identified ligand that can be produced synthetically. In addition to the well-known competence regulon (55, 59), about 180 genes (13, 56) have been found to be regulated, but few, if any, virulence genes have been found to be regulated. Some candidate genes possibly involved in virulence and induced by CSP could be the choline binding protein, CbpD (also termed Cbp3 or SP2201) (13, 56, 59), and CibAB (also termed orf62-orf51 or SP0124/5), a putative two-component bacteriocin (13, 56, 59). However, the most recent of these reports (13, 56) has indirectly linked the competence regulon to virulence by showing the down-regulation of housekeeping genes (e.g., ribosomal protein genes) and the induction of a general stress response (i.e., up-regulation of different stress response regulons). This competence-induced stress was shown to determine a temporary growth arrest and stationary-phase autolysis in a mutant with a mutation in CiaRH, a TCS involved in the exit from the competent state (13). About 30 years ago Tomasz and Zanati (68) had already described that CSP induces changes in surface properties of pneumococcal cells that lead to aggregation and temporary phenotypes, like leakage of intracellular enzymes and protoplast formation (60). These data have recently been reinforced by showing that CSP determines the release of DNA and cytoplasmic β-galactosidase from a subfraction of the pneumococcal population, a process that requires major pneumococcal autolysin (49, 62). All these reports indicate that CSP induces a competence-derived stress and that this stress, nearly undetectable as a phenotype, could, at least under some conditions, be detrimental to bacterial cells.
The ComABCDE pheromone-sensing system (11, 67) is similar to other peptide-pheromone quorum-sensing systems in gram-positive bacteria, including the second pneumococcal quorum-sensing bacteriocin-like peptide system, BlpABCRH (also denominated SpiPRH) (15, 58); the streptococcal invasion locus SilABCDE of Streptococcus pyogenes (27, 28); and the staphylococcal accessory gene regulator AgrABCD system, which controls virulence (16, 32, 33, 45). Among these autoinducible peptide quorum-sensing systems, only the staphylococcal Agr system has been linked directly to virulence by gene expression profiling, while the pneumococcal Blp system is reported to regulate only itself (15). In addition, no data have yet been published for the streptococcal Sil system (27, 28), even though both the pneumococcal BlpHR and its streptococcal homologue, SilAB (11), confer phenotypes of reduced virulence when they are mutated in a respiratory tract infection model (67) and in a local soft tissue infection model (28), respectively. As is also the case for the pneumococcal ComCD (57) and BlpCH (58), the staphylococcal AgrDC autoinducer-receptor pair shows considerable interstrain variation, which has been shown to be the molecular basis for activation of the agr response within the same group and inhibition of the agr response in strains belonging to other groups (32). This characteristic of the AgrD peptide and derivatives thereof has been exploited for the treatment of local soft tissue infections in mice (16, 32, 45). While in vivo recombinant SilCR was shown to inhibit skin disease when it was coinjected locally in mice (27), no attempt to use its pneumococcal homologue, BlpC, as a therapeutic tool has been described so far.
We hypothesized that the competence-activated stress phenotypes may make the pneumococcal cells more vulnerable to host defense factors. In the present work we report on the antibacterial activity of the 17-mer peptide CSP by means of its natural histidine kinase receptor in an in vivo model of pneumococcal sepsis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All experiments were carried out with the serotype 4 strain TIGR4 (65) and its comD-negative isogenic derivative, FP184. In order to evaluate the growth rate of the mutant in comparison to that of the wild type, bacterial strains were grown without selection in tryptic soy broth (TSB; Becton Dickinson and Company) or on tryptic soy agar supplemented with 3% blood. The growth curve was read in a single-beam spectrophotometer with digital wavelength and data displays (Spectronic 20D; Milton Roy).
In vitro mariner mutagenesis of comD.
In vitro mariner mutagenesis for the generation of mutants with mutations in comD was performed as described previously (42). The minitransposon used for mutagenesis of comD carried the kanamycin resistance determinant aphIII, and the pneumococcal recipient for the genetic construct was rough serotype 2 strain R800 (42). One of the pneumococcal transformants in which the comD gene was interrupted was named R706 and was used for further work. By using primers with sequences specific for regions external to the comCDE locus, the interrupted comD gene was amplified from strain R706 and the PCR product was directly transformed into encapsulated strain TIGR4. Transformation procedures were performed as reported previously (9). All PCR procedures were performed with the Expand High Fidelity kit (Roche) to reduce the risk of errors in the extension process. The structure of the recombinant chromosomal locus was controlled in transformants by PCR and sequencing.
Growth curve and analysis of CSP in vitro.
Bacterial strain TIGR4 and its comD mutant, FP184, from frozen stocks (mid-log-phase culture kept in 10% glycerol at −80°C) were inoculated 1/100 in 16 ml of TSB at 37°C in a water bath. When the strains reached an optical density at 590 nm (OD590) of 0.5, the culture was aliquoted into three tubes of 5 ml each, and the tubes were kept in a heat block at 37°C. For each of the two strains, one tube contained no CSP, CSP was added to the second tube at a concentration of 100 ng/ml (1.2 μl of CSP at 400 μg/ml), and CSP was added to the third tube at a concentration of 1,000 ng/ml (12.5 μl of CSP at 400 μg/ml). The ODs of the tubes were monitored every 5 min. By using 100-μl aliquots of cells from the 1 ml which remained in the original tube, the induction of competence by CSP1 and CSP2 (57) was monitored with TIGR4 and its comD mutant, FP184, by standard transformation procedures (9).
Quantitative real-time PCR.
Bacterial cells for quantitative RNA analysis were grown to early exponential phase in separate tubes inoculated in parallel. Six separate cultures of strain TIGR4 were performed. When the first culture reached an OD of 0.02, CSP1 (100 ng/ml) was added to two tubes and CSP2 (100 ng/ml) was added to two other tubes, while the last two cultures served as controls. One milliliter of cells was collected at 5 and 10 min after the addition of CSP, and the cells were immediately chilled on ice. RNA was extracted with the SV Total RNA Isolation system (Promega) by use of the standard protocol suggested by the supplier. RNA was retrotranscribed twice with random octamer primers in a final volume of 50 μl each time. The reaction mixture consisted of 10 pmol of primers, 1.25 mM deoxynucleoside triphosphates, 200 U of Moloney murine leukemia virus reverse transcriptase (Invitrogen), 40 U of RNase inhibitor (Promega), and 0.1 M dithiothreitol (Invitrogen), which were allowed to react for 30 min at 37°C. Real-time PCR was performed in a Roche LightCycler instrument with 1.5 μl of template, the LightCycler DNA Master SYBR Green I reaction mixture (Roche), 2.5 mM MgCl2, and 10 pmol of primers in a final volume of 20 μl. All PCRs were performed at 55°C as the annealing temperature. Primers for quantification were selected to amplify a fragment of less than 200 bp. Control primers were designed on the basis of the gyrB sequence (CAGATCAAGAAATCAAACTCCAA and CAGCATCATCTACAGAAACTC). The primers used for the characterization of four competence-regulated genes (13, 56) were based on the sequences of comA SP0042 (GAGACGCGAGCCATTAAGG and GGGATCTGGATCGGCAATATGA), lytA SP1937 (GGCTGGCAGAAGAATGACACTG and GCCGTCTGTGTGCTTCCT), comX1 SP0014 (AGGAAAGTCAGAAGCGTAGATA and GCGTTCTAGTTCTTCTTGTT), and dprA SP1266 (AGACTACATCGGCAATGAC and GAGACTACCTGAACGCATC). Primers specific for the capsule locus were based on cps4A SP0346 (CAAGAGTAGCCCTACCAA and AGAAAGTGAAGCGAAGTGTTAA). All PCRs were performed in duplicate. Relative quantification was performed by determining the crossing point (the point at which the fluorescence of a certain samples rises above that of the background) by the fit-to-point method indicated in the manual of the supplier (LightCycler software; Roche). Relative gene expression was analyzed by the 2−ΔΔCT method (38).
Experimental mouse model of sepsis.
Encapsulated strain TIGR4 and its isogenic comD mutant, FP184, were passaged in female CD1 mice (Charles River, Calco, Italy), as described previously (8, 9, 53). Eight-week-old animals were allowed to settle in the animal facilities at the Università di Siena, according to institutional guidelines, for 1 week before challenge. Passaged bacteria were grown to an OD590 of 0.3 in TSB at 37°C, centrifuged at 1,500 × g for 20 min, resuspended in fresh TSB with 15% glycerol, and frozen in aliquots at −70°C. Before use, the bacteria were thawed at room temperature, harvested by centrifugation, resuspended in sterile phosphate-buffered saline, and kept on ice until inoculation. Before infection, the mice were kept under an infrared lamp (200 W) for 2 to 3 min, and then the inoculum was injected intravenously (i.v.) into a lateral tail vein. The inocula were delivered in a total volume of 200 μl per mouse. Mice were regularly monitored for clinical symptoms (starry fur, hunched appearance, and lethargy), and symptoms were recorded for 10 days (240 h). During the course of the experiment the mice were humanely killed before they reached a moribund state.
Sighting study.
A preliminary toxicity study was carried out, as recommended for the fixed-dose procedure (38). Doses of 105, 106, and 107 CFU of strain TIGR4 were administered intravenously to outbred CD1 mice (n = 4). Two groups of mice were inoculated for each bacterial dose. One group was treated with CSP2, and one group served as an untreated control group. A dose of 1.3 μg of CSP2 (in 200 μl injected into a lateral tail vein) was given to each mouse at time zero and 24 h postinfection. The dose of CSP2 given at time zero was mixed on ice and administered just prior to challenge with the bacteria. The mice were regularly monitored for clinical symptoms for 10 days (240 h).
Main study.
Doses of 106 CFU of strain TIGR4 were administered i.v. to two groups of outbred CD1 mice (n = 20). One group was treated with CSP2, and one group served as an untreated control group. In a separate experiment the mice were challenged with isogenic comD-negative derivative FP184 (n = 10). As described above, a dose of 1.3 μg of CSP2 (in 200 μl injected into a lateral tail vein) was given to each mouse in the treatment group at time zero (in which the dose was mixed on ice and administered just prior to challenge with the bacteria) and 24 h postinfection. Blood was withdrawn from the cheek of each mouse infected with TIGR4 at 6 and 48 h for analysis for the presence of bacteria in the blood. The mice were regularly monitored for clinical symptoms for 10 days.
Statistical analysis.
Differences in the times to death of the mice were statistically analyzed by the Mann-Whitney-Wilcoxon test in relation to the last day at which clinical symptoms were observed (10 days). For statistical purposes, animals still alive after 10 days were assigned a time to death of 240 h. Differences in mouse survival times at the end of the experiment were statistically analyzed by Fisher's exact test. Differences in the numbers of CFU per milliliter of blood were analyzed by Student's t test. A P value of <0.05 was considered significant.
RESULTS
CSP-induced growth arrest in wild-type but not comD strains of S. pneumoniae.
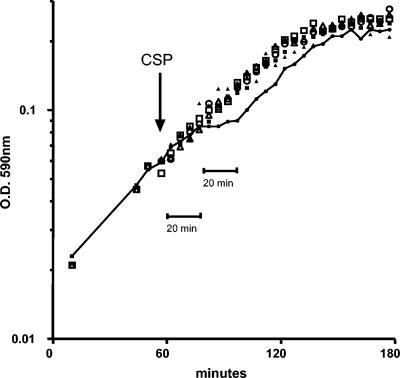
The CSP peptide was assayed for its effect on the growth of the TIGR4 strain in liquid medium at 100 ng/ml, the dose normally used for competence induction, and at 1 μg/ml, the dose used in the in vivo sepsis model (see below) (Fig. 1). Tests with aliquots of the same culture at an OD of 0.05 without a stimulus or after the addition of 100 ng of CSP/ml showed that CSP2 did not have any evident effect on growth. When CSP2 was added at 1,000 ng/ml, the strain showed a 20-min growth arrest 20 min after the addition of CSP2 (Fig. 1). After this temporary growth arrest, cells started to redivide, with the slope being comparable to those of the growth curves of all other aliquots tested (Fig. 1).
FIG. 1.
Effect of CSP2 on in vitro growth of S. pneumoniae TIGR4 and its comD mutant, FP184. Bacterial strains TIGR4 and FP184 (comD negative) were inoculated 1/100 in liquid medium, and their growth was monitored by measurement of the density of the culture. When the culture reached an OD of 0.05, 15 ml of culture was aliquoted and placed in three tubes, one without CSP2 (open squares, TIGR4; filled squares, FP184), one with 100 ng of CSP2/ml (in vitro transforming dose) (filled triangles, TIGR4; open triangles, FP184), and one with 1,000 ng of CSP2/ml (in vivo dose) (filled circles connected by a straight line, TIGR4; open circles, FP184). The time of addition of CSP2 to the cultures is shown by an arrow.
In order to assay the specificity of this effect, a mutant for the sensor histidine kinase ComD was constructed. As a first step the comD gene was inactivated in rough strain R800 by in vitro mariner mutagenesis. The minitransposon of the transformant selected for this work was found to be inserted into the TA dinucleotide at position 150 of the comD1 gene (the total size of comD is 1,323 bp). To obtain an encapsulated TIGR4 derivative for the in vivo experiments, the mutated comD1 allele was amplified from R706 by PCR and transformed into TIGR4 (which carries a comD2 allele). The comD mutant derivative of encapsulated strain TIGR4 was named FP184. Control experiments confirmed that, in contrast to TIGR4, the isogenic FP184 comD mutant was, as expected, not transformable to competence induction with either CSP1 or CSP2 (53, 57) (data not shown). No CSP-induced growth inhibition was visible in comD mutant FP184, which, otherwise, showed a growth profile identical to that of the wild-type strain (Fig. 1).
Quantitative real-time PCR was used to characterize the impact of CSP on the transcription of a few representative competence genes. The assay was performed 5 and 10 min after the addition of 100 ng of either CSP1 or CSP2 per ml to the TIGR4 strain. The data reported in Table 1 show an induction of expression of the early competence genes comA and comX1 only at 5 min, expression of the late competence gene lytA at 5 and 10 min, and a significant increase in expression of the gene for putative DNA-processing protein DprA at 5 min. No significant changes in the expression pattern of the capsule operon were detected. Irrespective of the allelic form of the peptide used, the expression patterns of the CSP-induced genes were identical (57).
TABLE 1.
Real-time PCR quantification of mRNA after addition of CSP1 and CSP2a
| Gene (strain) | Fold increase (SD) at the indicated times after addition of:
|
|||
|---|---|---|---|---|
| CSP1
|
CSP2
|
|||
| 5 min | 10 min | 5 min | 10 min | |
| comA (SP0042) | 4.25 (1.24) | 1.50 (0.28) | 3.05 (0.75) | 1.31 (0.15) |
| comX1 (SP0014) | 5.04 (0.82) | 1.52 (0.19) | 3.87 (1.46) | 1.46 (0.01) |
| lytA (SP1937) | 6.32 (1.40) | 10.63 (0.31) | 3.42 (0.03) | 10.48 (1.86) |
| dprA (SP1266) | 13.64 (5.31) | 3.72 (0.02) | 5.42 (1.34) | 2.96 (0.60) |
| cps4A (SP0346) | 1.74 (0.97) | 1.64 (0.45) | 1.22 (0.16) | 1.48 (0.74) |
All experiments were carried out in duplicate, as were the PCRs. Relative gene expression was analysed by the 2−ΔΔCT method (38).
Preliminary observations on the effectiveness of CSP against pneumococcal sepsis.
A preliminary toxicity study was carried out, as recommended for the fixed-dose procedure (38), to determine the most appropriate experimental setup and doses needed to assess the activity of the 17-mer CSP in pneumococcal sepsis. Three bacterial doses, ranging from 105 to 107 CFU of S. pneumoniae TIGR4, were given to CD1 outbred mice by the i.v. route to induce sepsis. The doses were chosen on the basis of previously published work (39, 54, 64) with mouse models of sepsis caused by other pneumococcal strains. Preliminary data showed that the mice died within 2 to 3 days when an inoculum of 107 CFU was used, within 5 days when an inoculum of 106 CFU was used, and within 6 days when an inoculum of 105 CFU was used. Detailed data are not reported, since this sighting study was designed only to provide results indicative (but not statistically significant) for a limited number of animals. In order to evaluate the possible effect of CSP2 on pneumococcal virulence, mice received 1.3 μg of CSP2 (strain TIGR4 carries the comC2 and comD2 alleles) at time zero (mixed with bacteria) and at 24 h postinfection (25, 57, 65). By considering the volume of circulating blood (about 1.2 to 1.5 ml for each 9-week-old CD1 mouse), the in vivo concentration of CSP2 should be about 1 μg/ml, which is about 10 times the concentration used to induce competence and half the calculated MICs of most known TCS-inhibiting compounds (44, 63). In this preliminary study in which mice were challenged with doses of 105 and 106 CFU, the rate of survival was increased among CSP-treated mice (six survivors among eight challenged mice) compared to that among untreated mice (two survivors among eight challenged mice).
CSP is therapeutic for pneumococcal sepsis.
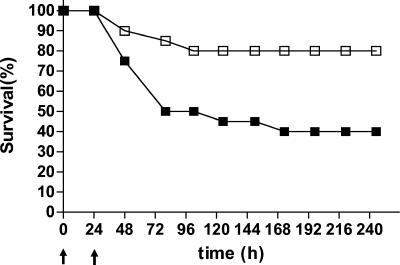
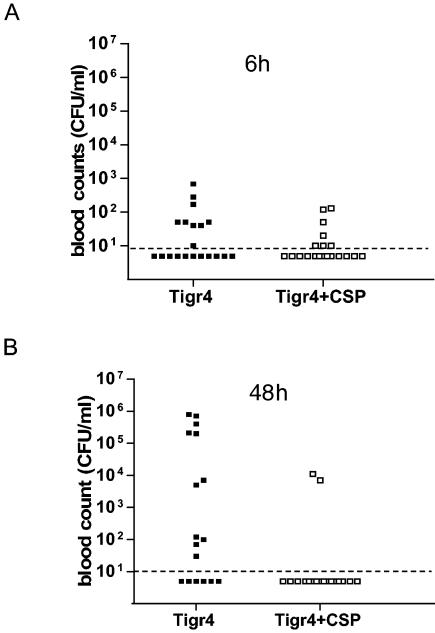
A single i.v. dose of 106 CFU of S. pneumoniae TIGR4 was found to induce sepsis in 50% of CD1 outbred mice within 3 days after challenge and in 60% of mice within 7 days after challenge (Fig. 2). To investigate the possibility that the CSP peptide could be used to inhibit pneumococcal sepsis, we administered CSP to mice at two different time points. The first dose was given at time zero, and the second was given at 24 h postinfection. In both cases, the dose of CSP was 1.3 μg per mouse, which corresponds to about 1 μg of CSP per ml of blood. This dose is 10-fold higher than the one used for the routine induction of competence (57). Figure 2 and Fig. 3 show the effects of CSP on pneumococcal sepsis in our in vivo model and report survival rates and blood counts, respectively. Only four animals in the group of CSP-treated mice developed severe sepsis, and the overall survival rate was 16 of 20 mice (80%) (Fig. 2). When the blood counts at 6 h after challenge were considered, no difference between CSP-treated mice and untreated mice could be seen. The mean bacterial count for CSP-treated mice was 19.5 CFU/ml (standard deviation [SD], 39 CFU/ml), and that for the untreated mice was 69 CFU/ml (SD, 162 CFU/ml). The situation changed at 48 h (i.e., 24 h after administration of the second dose of CSP), when the counts increased significantly in the blood of untreated mice. The majority of mice in the CSP-treated group (16 of 20 mice; 80%) were still blood culture negative, whereas only 25% (5 of 20 mice) of the mice in the untreated group were negative (mean bacterial count for CSP-treated mice, 900 CFU/ml [SD, 2,845 CFU/ml]; mean bacterial count for untreated mice, 116,000 CFU/ml [SD, 240,000 CFU/ml]) (Fig. 3). The data show that treatment with CSP results in significant differences in (i) the rate of survival at the end of the experiment (P = 0.0097; Fisher's exact test), (ii) the level of bacteremia at 48 h (P = 0.045; Student's t test), and (iii) time to death (P = 0.01; Mann-Whitney-Wilcoxon test). Three mice which received CSP without challenge were healthy until the end of the experiment.
FIG. 2.
Effect of treatment with CSP2 on pneumococcal sepsis. Two groups of CD1 outbred mice (n = 20) were injected i.v. with 106 CFU of wild-type strain TIGR4. While one group served as untreated controls (filled squares), the other group (open squares) was treated i.v. with CSP2 at 1.3 μg per mouse (corresponding to about 1 μg/ml of blood) at time zero and 24 h after challenge (both of which are indicated by arrows).
FIG. 3.
Bacterial counts in blood of mice challenged i.v. with pneumococci. The counts in the blood of individual mice were obtained by bleeding the animals from the cheek at 6 and 48 h after challenge. Data refer to the two groups of mice challenged with TIGR4, one of which received no treatment (solid squares) and one of which was treated with CSP at 0 and 24 h (open squares). (A) Samples taken at 6 h postinfection; (B) samples taken at 48 h postinfection. The detection limit of 10 CFU/ml is shown by dashed lines.
No therapeutic effect of CSP could be evidenced when the mice were challenged with comD histidine kinase sensor mutant FP184. In this challenge experiment, most mice died, with no statistically significant difference between the untreated mice and the CSP-treated mice (20 of 20 mice and 19 of 20 mice in the two groups, respectively, died). The reduced rate of survival of mice challenged with FP184 compared with the rate of survival of mice challenged with strain TIGR4 was not significant. On the other hand, when the effects of CSP treatment on the difference in the rates of survival between mice challenged with comD mutant FP184 and mice challenged with wild-type strain TIGR4 were compared, the difference was highly significant (P = 0.0001).
DISCUSSION
Despite the availability of antimicrobial drugs, treatment of infections due to S. pneumoniae remains a major problem. Even if in vitro data and work with animal models indicate that drugs are highly efficient in killing the pneumococcus, the overall mortality rate of about 20% and the frequency of long-term damage postinfection (hearing loss, cognitive impairment, learning deficiency, and other neurological sequelae) in human disease is very high and has remained unchanged for the last 40 years (1, 6, 17, 22, 46, 50, 52). The difficulty of finding efficient treatment alternatives continues to foster the search for novel narrow-spectrum or even species-specific drug targets and drugs in order to develop novel treatment options for infections caused by this pathogen. Industrial genome-based drug discovery programs generally focus on the detection of essential steps in metabolic pathways for small-molecule-drug development. An example of this strategy is the work that identified the mevalonate pathway as a potential antibacterial target (70). Other approaches report successful treatment of infections caused by pneumococci with monoclonal antibodies; for example, previous work has exploited antibodies against the surface protein PspA (64). An alternative recent approach describes the successful use of lytic phage enzymes for both topical and systemic treatments (39, 40).
Competence for genetic transformation in S. pneumoniae is the main characteristic that renders this pathogenic organism an interesting tool for scientists since the description of transformation by Griffith in 1928 and the discovery of pneumococcus DNA by Avery and coworkers in 1944 (3). Until now, most work on transformation focused on the mechanisms governing the horizontal gene transfer ability of the pneumococcus (11, 25, 26, 30, 67). Less attention was paid to the CSP-induced competence-accompanying phenomena, which is not obviously gene exchange related, and to changes in surface structures (68), temporary leakage of intracellular enzymes (60), protoplast formation (60), autolysis (49, 62), growth arrest and subsequent stationary-phase autolysis (13), and induction of a generalized stress response (13, 56). In this context the increased autolysis induced by the LytA amidase could also have a direct impact on virulence through involvement in the release of pneumolysin and other inflammatory compounds, such as teichoic and lipoteichoic acids (48). Recently, the consideration of all these phenotypes led to the formulation of the hypothesis that the competence regulatory system could be the S. pneumoniae substitute for the SOS system and that CSP could act as an alarmone (10, 13, 42). The observation that stress regulons are induced after CSP addition (13, 56) fits this interpretation. Almost 30 years ago, Seto and Tomasz (60) had already stated that the subtle damage conferred to the pneumococcus through CSP could, under appropriate conditions, be exaggerated and could become detrimental to the pneumococcal cells.
In the present work, we analyzed the effect of CSP on the virulence of S. pneumoniae in an in vivo model of acute systemic disease. All work was carried out with the serotype 4 strain TIGR4 (65), which carries a comC2 allele that encodes CSP2 (57). A model of sepsis (by the i.v. route) was set up to enable the direct inoculation of CSP into an infected organ and/or anatomical district (the blood). Other animal models of pneumococcal infection that are available, like the model of pneumonia and sepsis after intranasal inoculation or the model of sepsis after intraperitoneal inoculation, do not allow this type of study, and the involvement of host components in the meningitis model (29, 46, 47) may hide beneficial antipneumococcal therapeutic effects. Animal models of disease are dependent both on the infecting pneumococcal strain and on the genetic background of the mice (7, 14, 21, 54). We carried out a sighting study in order to identify the appropriate inoculum of strain TIGR4 that was able to induce sepsis in at least half of the outbred CD1 mice but that did not cause fulminant disease. That preliminary work indicated that an inoculum of 107 CFU per mouse generates fulminant disease with a fatal outcome within 2 to 3 days, while inocula of 106 and 105 CFU cause severe symptoms of pneumococcal sepsis after 4 to 5 days. This clinical picture was confirmed by the findings from the main study (20 mice per group), in which severe sepsis developed within 2 to 3 days in about 50% of mice upon challenge with 106 CFU of S. pneumoniae TIGR4. This appeared to be a normal picture of pneumococcal sepsis and is consistent with the low counts in blood 6 h after administration of the inoculum. The low counts are indicative of the ability of the spleen to clear or control the majority of the inoculated pneumococci.
In order to analyze the effect of CSP on S. pneumoniae in a systemic model of infection, we treated mice with CSP2 both at time zero (coinoculation of CSP2 and bacteria) and at 24 h postinfection. Both challenge and treatment were carried out by the i.v. route. The dose of 1 μg of CSP2 per ml of blood that we chose was 10 times higher than the concentration normally used for the induction of competence (25, 57), but it was lower than that used locally in the staphylococcal skin infection model (5 to 10 μg) (45) and was also lower than the MICs of most known TCS inhibitors for gram-positive bacteria that have been reported (63). The treatment of strain TIGR4 with 1 μg of CSP2 per ml in vitro induced a temporary inhibition of growth 20 min after the addition of CSP2. The timing of this CSP-induced phenomenon shows a striking similarity to the data on CSP-induced competence (from 10 to 30 min, with a sharp decline after 20 min) (56), the leakage of enzymes (which is maximal after 10 to 15 min) (60), the induction of protoplast formation (after 10 to 30 min) (60), and the temporary growth arrest (at about 18 min in ciaR mutants) (13). It is likely that all these temporary effects are directly or indirectly due to generalized stress induced by competence. Data on quantification of the expression of some selected CSP-controlled genes by real-time PCR for strain TIGR4 were similar to the data reported for D39 derivatives (13, 55), which thus provides no defined hint as to the reason for a bacteriostatic phenotype. A peculiarity of strain TIGR4 is its ability to respond to both CSP1 and CSP2 peptides, a fact already reported for other strains (57). The overall impact of this bacteriostatic effect on pneumococcal fitness in the host was assayed in a mouse sepsis model. The significant reduction in bacterial counts in blood, the significantly improved rate of survival at the end of the experiment, and the significant increase in the time to death observed in our mouse sepsis model after treatment with CSP2 are fully consistent with this interpretation. The improvements in these three parameters demonstrate that administration of CSP2 has a therapeutic effect against systemic pneumococcal disease. Abolition of this therapeutic effect in mice challenged with an isogenic comD mutant (strain FP184) demonstrated that the effect of CSP2 was due to its specific interaction with the ComD sensor, histidine kinase. This observation was consistent with the disappearance of the in vitro growth-inhibiting effect of CSP2 on this mutant strain. The difference in disease severity and outcome was even more evident (highly statistically significant) when the outcomes for CSP2-treated mice challenged with wild-type cells and mice challenged with comD mutant FP184 were compared. The fact that disease due to FP184 was more severe than disease due to wild-type pneumococci may need further attention, since this finding differs from those presented by other investigators (4, 24, 37), but it could well correlate to the finding that stimulation of this TCS by CSP2 reduced the virulence of our strain in our experimental setup. The significant impact of CSP treatment on disease outcome may be explained by the observation that under standard conditions the competence-induced growth inhibition might be essentially undetectable (13), while it may become prominent when it is combined with genetic alterations (13) or additional stress factors (60). This general competence-induced stress would thus not allow the bacteria to cope with additional loads imposed by host factors. In this case both the reduced growth and the reduced resistance to host factors could account for the more efficient clearance of bacteria by the host.
Quorum-sensing peptide pheromones have already been used in murine models of local soft tissue infection. In challenge experiments with S. pyogenes, local treatment (at the time of inoculation with bacteria) with the synthetic peptide SilCR (up to 50 μg) protected mice against necrotic skin ulcers. This effect was suggested to be due to the SilCR-dependent inhibition of proteolytic activity on the murine interleukin 8 homologue needed for macrophage recruitment (27, 28). In the more extensively studied staphylococcal model (33), the autoinducing peptides contain a thiolactone structure (45) and more than one allelic form is known, which enable investigators to define subgroups of Staphylococcus aureus (32). The peptides produced by one subgroup inhibit the expression of agr in strains belonging to other subgroups (32). Cross-inhibition of virulence gene expression by local treatment (at the time of inoculation with bacteria) with the autoinducer peptide protected mice from subcutaneous abscesses due to S. aureus (32, 45). Thus, in Staphylococcus the therapeutic effect is due to the inhibition of a virulence-inducing system, while in the pneumococcus the therapeutic effect would most likely be due to the activation of a stress-inducing TCS (i.e., up-regulation of stress genes and down-regulation of ribosomal genes, leading directly or indirectly to enzyme leakage, autolysis, and growth arrest).
TCS transduction systems have been proposed as novel targets for antimicrobial therapy for a variety of reasons, including the absence of these histidine-aspartate signal transduction systems in animals and the possibility that multiple targets could be inhibited by a single drug, which would enhance antibacterial potency and reduce the probability of resistance development (44, 63). The general strategy used to identify novel drugs and to design leads and drugs generally follows the rule which requires identification of essential targets and selection for target-inhibiting compounds. This rule was followed in all studies that have reported on novel compounds active against TCSs (44, 63). The present work with CSP2 is conceptually new and proposes the reverse mechanism for a therapeutic intervention against disease, namely, the activation of a not obviously virulence-related TCS, which by itself is not essential. It is evident that the direct clinical application of CSP2 may have some limitations, including the high degree of mutability of autoinducer loci (61) (although this has not been established for S. pneumoniae), the possible genetic instability of nonessential targets, and/or the risk of induction of increased in vivo gene exchange by transformation. However, these rather long-term risks should be weighed against the short-term therapeutic benefit. Because of the need for accessory therapeutic tools to cure pneumococcal disease, the use of CSP (or any CSP-derived product) in adjunct therapy, in addition to standard antibiotic therapy, may be one of the main directions for future development of the data reported here. In conclusion, our approach not only demonstrates the possibility that peptides can be used as therapeutic molecules for the inhibition of systemic bacterial disease but also proposes the induction of a TCS as a conceptually novel mechanism for the treatment of pneumococcal sepsis.
Acknowledgments
We thank Emanuel Hanski for presenting stimulating data on SilCR in June 2003 at the Gram-Positive Genomics meeting in Baveno, Italy. We also thank Peter Andrew for helpful discussion and Guido Memmi and Caterina Costa for help with laboratory procedures.
The work was supported in part by European Commission grant QLK2-CT2000-00543 (to G.P. and J.-P.C.), grant COFIN 2002 from MIUR (to G.P.), and grant FIRB RBAU01X9TB from MIUR (to M.R.O.).
REFERENCES
- 1.Afessa, B., W. L. Greaves, and W. R. Frederick. 1995. Pneumococcal bacteremia in adults: a 14-year experience in an inner-city hospital. Clin. Infect. Dis. 21:345-351. [DOI] [PubMed] [Google Scholar]
- 2.Afessa, B., I. Morales, and B. Weaver. 2001. Bacteremia in hospitalised patients with human immunodeficiency virus: a prospective, cohort study. BMC Infect. Dis. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J. Exp. Med. 79:137-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartilson, M., A. Marra, J. Christine, J. S. Asundi, W. P. Schneider, and A. E. Hromckyj. 2001. Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide. Mol. Microbiol. 39:126-135. [DOI] [PubMed] [Google Scholar]
- 5.Blue, C. E., and T. J. Mitchell. 2003. Contribution of a response regulator to the virulence of Streptococcus pneumoniae is strain dependent. Infect. Immun. 71:4405-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohr, V., N. Rasmussen, B. Hansen, H. Kjersem, O. Jesse, and H. S. Kristensen. 1983. 875 cases of bacterial meningitis: diagnostic procedures and the impact of preadmission antibiotic therapy. Part III of a three-part series. J. Infect. 7:193-202. [DOI] [PubMed] [Google Scholar]
- 7.Briles, D. E., M. J. Crain, B. M. Gray, C. Forman, and J. Yother. 1992. Strong association between capsular type and virulence for mice among human isolates of Streptococcus pneumoniae. Infect. Immun. 60:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canvin, J. R., A. P. Marvin, M. Sivakumaran, J. C. Paton, G. J. Boulnois, P. W. Andrew, and T. J. Mitchell. 1995. The role of pneumolysin and autolysin in the pathology of pneumonia and septicemia in mice infected with a type 2 pneumococcus. J. Infect. Dis. 172:119-123. [DOI] [PubMed] [Google Scholar]
- 9.Chiavolini, D., G. Memmi, T. Maggi, F. Iannelli, G. Pozzi, and M. R. Oggioni. 2003. The three extra-cellular zinc metalloproteinases of Streptococcus pneumoniae have a different impact on virulence in mice. BMC Microbiol. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claverys, J. P., and L. S. Havarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:1798-1814. [DOI] [PubMed] [Google Scholar]
- 11.Claverys, J. P., and B. Martin. 2003. Bacterial “competence” genes: signatures of active transformation, or only remnants? Trends Microbiol. 11:161-165. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, J. 2003. Management of bacterial meningitis in adults. BMJ 326:996-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dagkessamanskaia, A., M. Moscoso, V. Henard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. M. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 14.Denny, P., E. Hopes, N. Gingles, K. W. Broman, W. McPheat, J. Morten, J. Alexander, P. W. Andrew, and S. D. Brown. 2003. A major locus conferring susceptibility to infection by Streptococcus pneumoniae in mice. Mamm. Genome 14:448-453. [DOI] [PubMed] [Google Scholar]
- 15.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durnad, M., D. Calderwood, D. Weber, S. I. Miller, F. S. Southwick, V. S. Caviness, and M. N. Swartz. 1993. Acute bacterial meningitis in adults. A review of 493 episodes. N. Engl. J. Med. 328:21-28. [DOI] [PubMed] [Google Scholar]
- 18.Echenique, J. R., S. Chapuy-Regaud, and M. C. Trombe. 2000. Competence regulation by oxygen in Streptococcus pneumoniae: involvment of ciaRH and comCDE. Mol. Microbiol. 36:688-698. [DOI] [PubMed] [Google Scholar]
- 19.Echenique, J. R., and M. C. Trombe. 2001. Competence repression under oxygen limitation through the two-component MicAB signal-transducing system in Streptococcus pneumoniae and involvement of the PAS domain of MicB. J. Bacteriol. 183:4599-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert, D. N., R. C. Moellering, and M. A. Sande. 2003. The Sanford guide to antimicrobial therapy 2003. Antimicrobial Therapy Inc., Hyde Park, Vt.
- 21.Gingles, N. A., J. E. Alexander, A. Kadioglu, P. W. Andrew, A. Kerr, T. J. Mitchell, E. Hopes, P. Denny, S. Brown, H. B. Jones, S. Little, G. C. Booth, and W. L. McPheat. 2001. Role of genetic resistance in invasive pneumococcal infection: identification and study of susceptibility and resistance in inbred mouse strains. Infect. Immun. 69:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gransden, W. R., S. J. Eykyn, and I. Phillips. 2004. Pneumococcal bacteraemia: 325 episodes diagnosed at St Thomas's Hospital. BMJ 290:505-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guenzi, E., A. M. Gasc, M. A. Sicard, and R. Hakenbeck. 1994. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12:505-515. [DOI] [PubMed] [Google Scholar]
- 24.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1405. [PMC free article] [PubMed] [Google Scholar]
- 25.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An ummodified pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Havarstein, L. S., P. Gaustad, F. N. Ingolf, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 27.Hidalgo-Grass, C., M. Dan-Goor, A. Maly, Y. Eran, L. Kwinn, V. Nizet, M. Ravins, J. Jaffe, A. E. Moses, and E. Hanski. 2004. Effect of a bacterial pheromone peptide on host chemokine degradation in group A streptococcal necrotising soft-tissue infections. Lancet 363:696-703. [DOI] [PubMed] [Google Scholar]
- 28.Hidalgo-Grass, C., M. Ravins, M. Dan-Goor, J. Jaffe, A. E. Moses, and E. Hanski. 2002. A locus of group A streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 46:87-99. [DOI] [PubMed] [Google Scholar]
- 29.Hirst, R. A., B. Gosai, A. Rutman, P. W. Andrew, and C. O'Callaghan. 2003. Streptococcus pneumoniae damages the ciliated ependyma of the brain during meningitis. Infect. Immun. 71:6095-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hui, F., L. Zhou, and D. A. Morrison. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 153:25-31. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs, M. R., D. Felmingham, P. C. Appelbaum, R. N. Gruneberg, and The Alexander Project Group. 2003. The Alexander Project 1998-2000: susceptibility of pathogens isolated from community-acquired respiratory tract infection to commonly used antimicrobial agents. J. Antimicrob. Chemother. 52:229-246. [DOI] [PubMed] [Google Scholar]
- 32.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 33.Ji, G., R. C. Beavis, and R. P. Novick. 1995. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA 92:12055-12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadioglu, A., J. R. Echenique, S. Manco, M. C. Trombe, and P. W. Andrew. 2003. The MicAB two-component signal transduction system is involved in virulence of Streptococcus pneumoniae. Infect. Immun. 71:6676-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koedel, U., W. Scheld, and H.-W. Pfister. 2002. Pathogenesis and pathophysiology of pneumococcal meningitis. Lancet Infect. Dis. 2:721-736. [DOI] [PubMed] [Google Scholar]
- 36.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 273:223-234. [DOI] [PubMed] [Google Scholar]
- 37.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 38.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 39.Loeffler, J. M., S. Djurkovic, and V. A. Fischetti. 2003. Phage lytic enzyme Cp1-1 as a novel antimicrobial for pneumococcal bacteremia. Infect. Immun. 71:6199-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loeffler, J. M., D. Nelson, and V. A. Fischetti. 2001. Rapid killing of Streptococcus penumoniae with a bacteriophage cell wall hydrolase. Science 294:2170-2173. [DOI] [PubMed] [Google Scholar]
- 41.Luo, P., H. Li, and D. A. Morrison. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50:623-633. [DOI] [PubMed] [Google Scholar]
- 42.Martin, B., M. Prudhomme, G. Alloing, C. Granadel, and J. P. Claverys. 2000. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol. Microbiol. 38:867-878. [DOI] [PubMed] [Google Scholar]
- 43.Mascher, T., D. Zahner, M. Merai, N. Balmelle, A. de Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: ciaR target sites and transcription profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsushita, M., and K. D. Janda. 2002. Histidine kinases as targets for new antimicrobial agents. Bioorg. Med. Chem. 10:855-867. [DOI] [PubMed] [Google Scholar]
- 45.Mayville, P., J. I. Guangyong, R. Beavis, H. Yang, M. Goger, R. P. Novick, and T. W. Muir. 1999. Structure-activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Biochemistry 96:1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meli, D. N., S. Christen, S. L. Leib, and M. G. Tauber. 2002. Current concepts in the pathogenesis of meningitis caused by Streptococcus pneumoniae. Curr. Opin. Infect. Dis. 15:253-257. [DOI] [PubMed] [Google Scholar]
- 47.Mitchell, T. J. 2000. Virulence factors and the pathogenesis of disease caused by Streptococcus pneumoniae. Res. Microbiol. 151:413-419. [DOI] [PubMed] [Google Scholar]
- 48.Mortier-Barriere, I., A. de Saizieu, J. P. Claverys, and B. Martin. 1998. Competence-specific induction of recA is required for full recombination proficiency during transformation in Streptococcus pneumoniae. Mol. Microbiol. 27:159-170. [DOI] [PubMed] [Google Scholar]
- 49.Moscoso, M., and J. P. Claverys. 2004. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism, and stability of the liberated DNA. Mol. Microbiol. 54:783-794. [DOI] [PubMed] [Google Scholar]
- 50.Nau, R., and W. Bruck. 2002. Neuronal injury in bacterial meningitis: mechanisms and implications for therapy. Trends Neurosci. 25:38-45. [DOI] [PubMed] [Google Scholar]
- 51.Normark, B. H., R. Novak, A. Ortqvist, G. Kallenius, E. Tuomanen, and S. Normark. 2001. Clinical isolates of Streptococcus pneumoniae that exhibit tolerance to vancomycin. Clin. Infect. Dis. 32:552-558. [DOI] [PubMed] [Google Scholar]
- 52.Obaro, S. K., M. A. Monteil, and D. C. Henderson. 1996. The pneumococcal problem. BMJ 312:1521-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oggioni, M. R., G. Memmi, T. Maggi, D. Chiavolini, F. Iannelli, and G. Pozzi. 2003. Pneumococcal zinc metalloproteinase ZmpC cleaves human matrix metalloproteinase 9 and is a virulence factor in experimental pneumonia. Mol. Microbiol. 49:795-805. [DOI] [PubMed] [Google Scholar]
- 54.Perez-Trallero, E., M. Alkorta, M. J. Gimenez, D. Vicente, and L. Aguilar. 2001. Prediction of in-vivo efficacy by in vitro early bactericidal activity with oral β-lactams, in a dose-ranging immunocompetent mouse sepsis model, using strains of Streptococcus pneumoniae with decreasing susceptibilities to penicillin. J. Chemother. 13:118-125. [DOI] [PubMed] [Google Scholar]
- 55.Peterson, S. N., R. T. Cline, H. Tettelin, V. Sharov, and D. A. Morrison. 2000. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol. 182:6192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 57.Pozzi, G., L. Masala, F. Iannelli, R. Manganelli, L. S. Havarstein, L. Piccoli, D. Simon, and D. A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reichmann, P., and R. Hakenbeck. 2000. Allelic variation in a pepetide-inducible two-component system of Streptococcus pneumoniae. FEMS Microbiol. Lett. 190:231-236. [DOI] [PubMed] [Google Scholar]
- 59.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 60.Seto, H., and A. Tomasz. 1975. Protoplast formation and leakage of intramembrane cell components: induction by competence activator substance of pneumococci. J. Bacteriol. 121:344-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Somerville, G. A., S. B. Beres, J. R. Fitzgerald, F. R. De Leo, R. L. Cole, J. S. Hoff, and J. M. Musser. 2002. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 184:1430-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinmoen, H., E. Knutsen, and L. S. Havarstein. 2002. Induction of natural competence in Streptococcus pneumoniae triggers lysis and DNA release from a subfraction of the cell population. Proc. Natl. Acad. Sci. USA 99:7681-7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stephenson, K., and J. A. Hoch. 2002. Virulence- and antibiotic resistance-associated two-component signal transduction systems of gram-positive pathogenic bacteria as targets for antimicrobial therapy. Pharmacol. Ther. 93:293-305. [DOI] [PubMed] [Google Scholar]
- 64.Swiatlo, E., J. King, G. S. Nabors, B. Mathews, and D. E. Briles. 2003. Pneumococcal surface protein A is expressed in vivo, and antibodies to PspA are effective for therapy in a murine model of pneumococcal sepsis. Infect. Immun. 71:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umajama, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 66.Thomason, P., and R. Kay. 2000. Eukaryotic signal transduction via histidine-aspartate phosphorelay. J. Cell Sci. 113:3141-3150. [DOI] [PubMed] [Google Scholar]
- 67.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. R. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 68.Tomasz, A., and E. Zanati. 1971. Appearance of a protein “agglutinin” on the spheroplast membrane of pneumococci during induction of competence. J. Bacteriol. 105:1213-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wagner, C., A. de Saizieu, H. J. Schonfeld, M. Kamber, R. Lange, C. J. Thompson, and M. G. Page. 2002. Genetic analysis and functional characterization of the Streptococcus pneumoniae vic operon. Infect. Immun. 70:6121-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilding, E. I., J. R. Brown, A. P. Bryant, A. F. Chalker, D. J. Holmes, K. A. Ingraham, S. Iordanescu, C. Y. So, M. Rosenberg, and M. N. Gwynn. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J. Bacteriol. 182:4319-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]