Abstract
Aims
One of six heparin biosynthetic enzymes, cloned and expressed in Escherichia coli as a soluble fusion protein, requires large-scale preparation for use in the chemoenzymatic synthesis of heparin, an important anticoagulant drug.
Methods and Results
The 6-O-sulfotransferase isoform-3 (6-OST-3) can be conveniently prepared at mg/L levels in the laboratory by culturing E. coli on Luria–Bertani medium in shake flasks and inducing with isopropyl β-D-1-thiogalactopyranoside at an optical density of 0·6–0·8. The production of larger amounts of 6-OST-3 required fed-batch cultivation of E. coli in a stirred tank fermenter on medium containing an inexpensive carbon source, such as glucose or glycerol. The cultivation of E. coli on various carbon sources under different feeding schedules and induction strategies was examined. Conditions were established giving yields (5–20 mg g-cell-dry weight−1) of active 6-OST-3 with excellent productivity (2–5 mg l−1 h−1).
Conclusions
The production of 6-OST-3 in a fed-batch fermentation on an inexpensive carbon source has been demonstrated.
Significance and Impact of the Study
The ability to scale-up the production of heparin biosynthetic enzymes, such as 6-OST-3, is critical for scaling-up the chemoenzymatic synthesis of heparin. The success of this project may someday lead to a commercially viable bioengineered heparin to replace the animal-sourced anticoagulant product currently on the market.
Keywords: biosynthetic enzyme expression, fed-batch fermentation, heparin, high cell density cultivation, optimization of induction, sulfotransferase
Introduction
Heparin, an important anticoagulant drug, is a sulfated polysaccharide that is biosynthesized in the Golgi compartment of certain animal cells (Linhardt 2003). Heparin biosynthesis involves the consecutive action of a number of different types of Golgi enzymes including glycosyl transferases, N-deacetylases and N-sulfotransferases, C5 epimerase, and 2-, 3-, and 6-O-sulfotransferases (Esko and Selleck 2002). The drug heparin is currently prepared in metric ton scale from animal tissues, primarily pig intestine (Linhardt 2003). In 2007–2008, there was a contamination crisis in which a significant portion of the heparin on the commercial market was adulterated with oversulfated chondroitin sulfate (Guerrini et al. 2008). The death of over 100 patients in the US receiving contaminated heparin has been attributed to this adulteration (Liu et al. 2009). As part of an ongoing effort to separate the food chain from the drug chain and reduce the risks associated with the use of heparin (Bhaskar et al. 2012), our laboratory has been actively investigating the production of a nonanimal derived bioengineered heparin (Wang et al. 2011). The chemoenzymatic process, used to prepare bioengineered heparin, involves fermentative production of heparosan, chemical de-N-acetylation N-sulfation to prepare the intermediate N-sulfoheparosan, and enzymatic treatment with C5-epimerase, 2-O-sulfotransferase, 6-O-sulfotransferase isoform-1 and 6-O-sulfotransferase isoform-3, and 3-O-sulfotransferase isoform-1 (Wang et al. 2011). The enzymes used in these enzymatic steps are part of the biosynthetic pathway in the Golgi of animal cells (Esko and Selleck 2002). Truncated Golgi proteins, missing their transmembrane domain, are expressed from Escherichia coli as maltose-binding protein (MBP)-fusion proteins (Chen et al. 2005). As part of an effort to reduce process costs and to scale-up, these recombinant enzymes must be prepared in large-scale fed-batch fermentations. We recently reported the fedbatch production of 6-O-sulfotransferase isoform-1 (Restaino et al. 2013). In the current manuscript, we now report the fed-batch production of 6-O-sulfotransferase isoform-3 (6-OST-3).
Materials and methods
Materials
All media components, fermentation reagents, chemicals used for preparation of biomass extraction buffer, and chemicals and standards used in HPLC analysis, were from Sigma-Aldrich (St. Louis, MO). LB broth powder was purchased from BD Biosciences (San Jose, CA). Kanamycin, Ampicillin, tetracycline and IPTG were from Gold Biotechnology (St. Louis, MO).
Bacterial strain and media
The recombinant E. coli Rosetta-gami B (DE3) cells (Novagen, Cambridge, MA) strain with the plasmid pMalc2x, and the catalytic domain of mouse 6-OST-3 (Pro121–Pro450) was used to express 6-OST-3 maltose-binding protein fusion product (Chen et al. 2005). Shake flask experiments were performed in LB medium, LB plus 43·5 mmol l−1 glycerol-medium, LB plus 22·2 mmol l−1 glucose-medium and LB plus 22·2 mmol l−1 fructose-medium. Ampicillin (143 μmol l−1), tetracycline (28 μmol l−1) and kanamycin (103 μmol l−1)) were always added to the media to select for the plasmid and prevent the growth of contaminating microbes, after being filtered with a syringe through a 0·22-μm membrane.
Shake flask experiments
Transformed cells were incubated at 37°C on shaker with 220 rev min−1 overnight to prepare cultures from stock bacteria. A 1-l media flask was inoculated with 10% (v/v) of stock culture. When the fermentation reached an optical density at 600 nm (OD600) of 0·6–0·8, the temperature was decreased to 22°C for 30 min. A 200-μl aliquot of 1 mol l−1 isopropyl-thiogalactopyranoside (IPTG) was added to each 1 l of fermentation media for induction. The culture was shaken for 16–18 h at 220 rev min−1 at the reduced temperature of 22°C.
Fed-batch experiments
A 20-l bioreactor (Bio-Flo 4500, New Brunswick Scientific, Enfield, CT), in situ sterilizable and equipped with pH and pO2 probes (Mettler, Toledo, Switzerland), was used for fed-batch experiments. During growth, the fermentation parameters were controlled and data were collected by Biocommand A4 software (Eppendorf, Inc., Enfield, CT).
For the 5-l fed-batch fermentation, 200 μl of glycerol stock bacteria was inoculated into a 250-ml shake flask containing 50 ml LB medium. After 12 h of cultivation in a shaker incubator (220 rev min−1) at 37° C, the entire preculture was aseptically transferred into a 2·5-l shake flask containing 500 ml LB medium and incubated under the same condition for 10 h. This was used to inoculate the 5-l fed-batch fermentation that took place in the 20-l bioreactor. Fed-batch experiments were performed at 37°C and pH of 7·0 in 5-l LB containing 43·5 mmol l−1 glycerol. The DO was set to 20% and cascade controlled by inlet airflow rate and agitation speed. The pH was set to 7·0 and cascade controlled by acid pump for hydrochloric acid and base pump for ammonia water. After 4–10 h of batch phase, the culture was fed with a concentrated solution (435 mmol l−1 glycerol, 150 g l−1 yeast extract and 48·7 mmol l−1 MgSO4·7H2O). In a first fed-batch experiment (FB A), the culture was fed after 6 h at a constant feeding rate of 1 ml min−1. IPTG was added to a final concentration of 1 mmol l−1 as soon as the OD600 value reached about 7·5. In the second fed-batch experiment (FB B), the culture was constantly fed after 10 h with a feeding rate of 3 ml min−1 over the first 4 h and then at 1·7 ml min−1 until the end of fermentation. FB B was induced at an OD600 value of 9·5. In a third fed-batch experiment (FB C), the culture was fed after 4 h at a constant feeding rate of 1·8 ml min−1 over the first 8 h and then at a constant feeding rate of 1·0 ml min−1 until 22 h into the fermentation. FB C was induced at an OD600 value of 3·5.
Bacterial growth was monitored at various time points by measuring OD600 with an UVmini-1240 spectrophotometer (Shimadzu, Kyoto, Japan). The culture was diluted into the linear range to make these measurements. The cell pellet was collected by centrifugation at 3501 g for 20 min at 4°C and frozen for storage at −80°C.
Biomass extraction and enzyme purification
The pellet (20 g) was suspended in 100 ml of ice-cold loading buffer A (25 mmol l−1 Tris-HCl, pH 7·4, containing 500 mmol l−1 NaCl) by vortexing, and cells were lysed on ice using a Q700 sonicator (Qsonica, Newtown, CT) at power level 4·5 for 1 min (30 strokes, 1s on and 1s off). Sonication of the re-suspended cells was performed three times according to the programme, taking 30 s break between each cycle. Cell debris was spun down at 19 868 g at 4°C for 30 min, and the supernatant was filtered through a 0·22-μm filter using vacuum system into 50-ml tube cooled in an ice bath. The sample containing the expressed 6-OST was maintained on ice briefly prior to FPLC purification.
The FPLC system was manually washed, without an attached column, for 5 min at a flow rate 5 ml min−1 with eluting buffer B (25 mmol l−1 Tris-HCl, pH 7·5, containing 500 mmol l−1 NaCl and 40 mmol l−1 maltose monohydrate) and then washed for 5 min at flow rate 5 ml min−1 with loading buffer A. The amylose column was connected to the FPLC, the sample was injected, and the column was washed with buffer A for 10–15 min at a flow rate 2 ml min−1 and then washed with elution buffer B at a flow rate 2 ml min−1 for 5 min by manual operation. Purified enzyme was collected on a fraction collector with UV280 detection, and fractions containing the 6-OST peak were pooled together in a 50-ml tube. Glycerol (555 m mol l−1) was added to the enzyme before it was stored at −80°C.
SDS-PAGE and protein concentration analysis
Protein concentration was determined with Nanodrop 3300 (Thermo Fisher Scientific Inc., Wilmington, DE). SDS-PAGE protein analysis was performed in a Mini- Protean Tetra system (BIORAD, Hercules, CA), by loading 30 μl of 1: 1 (v:v) boiled protein solution and dye buffer on a 4–20% precast gel (Mini protean TGX gels, BIORAD) and running in a Tris, glycine and SDS buffer (10× Tris/glycine/SDS buffer, BIORAD) at 100 V for 45 min. A 250 kDa to 10-kDa protein standard (Precision Plus Protein Kaleidoscope, BIORAD) was used as ladder. Gels were washed with tap water for 30 min and then stained by soaking for 5 h in Coomassie blue solution (Gel Code Blue Safe Protein Stain, Thermo Fisher Scientific).
Activity assay and mass analysis
The 6-OST activity assay was performed as previously described with some modification (Restaino et al. 2013). The activity was analysed by incubating 50 μg (0·6 nmol) of purified 6-OST-3 with 25 μg (approx. 25 nmol) of N-sulfoheparosan decasaccharide, 25 μg (0·7 nmol) of arylsulfotransferase, 0·5 μmol l−1 para-nitrophenylsulfate (PNPS) and 4 nmol of 3′-phosphoadenosine-5′-phosphosulfate (PAPS) in 250 μl system with supplement of 50 mmol l−1 MES buffer pH 7·0. The assays were conducted in transparent, 96-well plates purchased from Greiner Bio-One (Monroe, NC). The plates were incubated at 37°C in SpectraMax plate reader (Molecular Devices, Sunnyvale, CA). A kinetic model was used to measure the absorbance at 400 nm min−1 over 1 h. The enzyme activity was calculated using the following equation:
Abst2 and Abst1 are the absorbance at 400 nm at t2 min and t1 min, respectively. The time range was chosen over a linear segment of the plot. The value is the extinction coefficient is 10·5 × 10−3 mol l−1, RT is the reaction time (min), and [6-OST-3] is the concentration of 6-OST-3. One unit of the activity represents the amount of 6-OST-3 enzyme required to produce 1 nmol PNP product per min.
HPLC analysis
Concentrations of glycerol in the medium during the fermentation were measured by HPLC as previously described (Restaino et al. 2013).
Results
Optimization of culture conditions of shake flask experiments
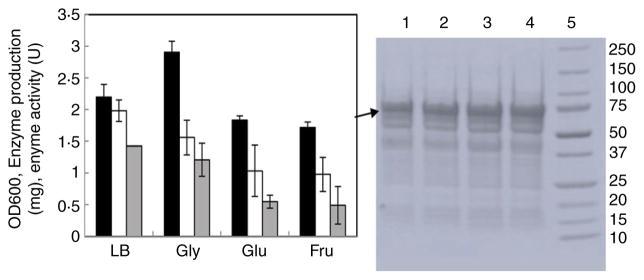
Shake flask experiments were preformed to investigate the medium composition, culturing and induction conditions that might be suitable for fed-batch expression of 6-OST-3. Different carbon sources have been reported that are suitable for recombinant protein expression in E. coli, including glucose, acetate, glycerol, galactose, mannitol, fructose, xylose and succinate (Maheswaran and Forchhammer 2003; Andersson et al. 2007; Carvalho et al. 2012). For 6-OST-3 expression, fructose (22·2 mmol l−1), glucose (22·2 mmol l−1) and glycerol (43·5 mmol l−1) were finally selected for the media studies. Based on the growth rate, enzyme production and enzyme-specific activity, glycerol was selected over the other sugars, although both the enzyme amount (1·56 mg l−1) and specific activity (1·21 U mg−1) were lower than the values obtained with growth on LB media, 1·98 mg l−1 and 1·43 U mg−1, respectively (Fig. 1).
Figure 1.
Expression of 6-OST-3 with LB and LB plus 0·4% of different carbon sources. Left panel:OD600, enzyme production production and activity of 6-OST-3 expressed from LB media, LB plus 43·5 mmol l−1 glycerol, LB plus 22·2 mmol l−1 glucose and LB plus 22·2 mmol l−1 fructose. Black columns:OD600, White columns: enzyme protein production, Grey columns: enzyme activity. Right panel: SDS-PAGE analysis of purified protein solution; lanes 1, LB plus 22·2 mmol l−1 fructose; 2, LB plus 22·2 mmol l−1 glucose; 3, LB plus 43·5 mmol l−1 glycerol; 4, LB; and 5, 250–10 kDa molecular weight ladder. The 6-OST-3 bands are indicated by arrows.
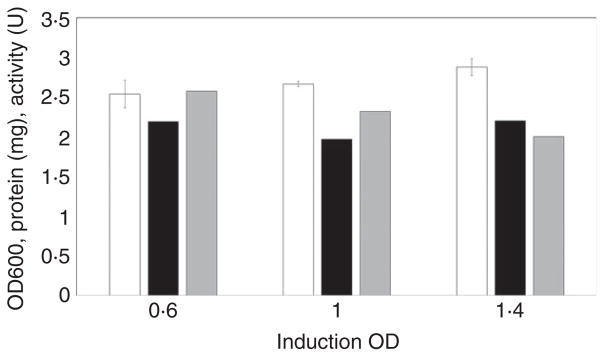
LB medium with a 2% inoculum is generally used to produce 6-OST-3 at small scale. In such cases, we have observed that induction conditions have a significant influence on the activity of 6-OST-3. Therefore, we investigated the effect of inducing at different OD600 on the resulting 6-OST-3 activity (Fig. 2). The results showed that although the activities were decreased slightly when induction takes place at higher OD600 values, there was no significant difference in induced protein production.
Figure 2.
Expression of 6-OST-3 with LB plus 43·5 mmol l−1 glycerol of different induction OD. The inoculation amount was 10%, and the culture temperature was 37°C. The induction OD values were 0·6, 1·0 and 1·4. Black columns:OD600, White columns: enzyme protein production, Grey columns: enzyme activity.
High cell density cultivation
The high cell density culture of recombinant E. coli was used to obtain recombinant proteins with high yield and high productivities. This is required to meet the high demand for enzymes in the large-scale production of bioengineered heparin. High cell density culture also has the advantage of increasing the effectiveness of the process, reducing culture and wastewater volumes, production costs, and upfront investment for fermentation equipment.
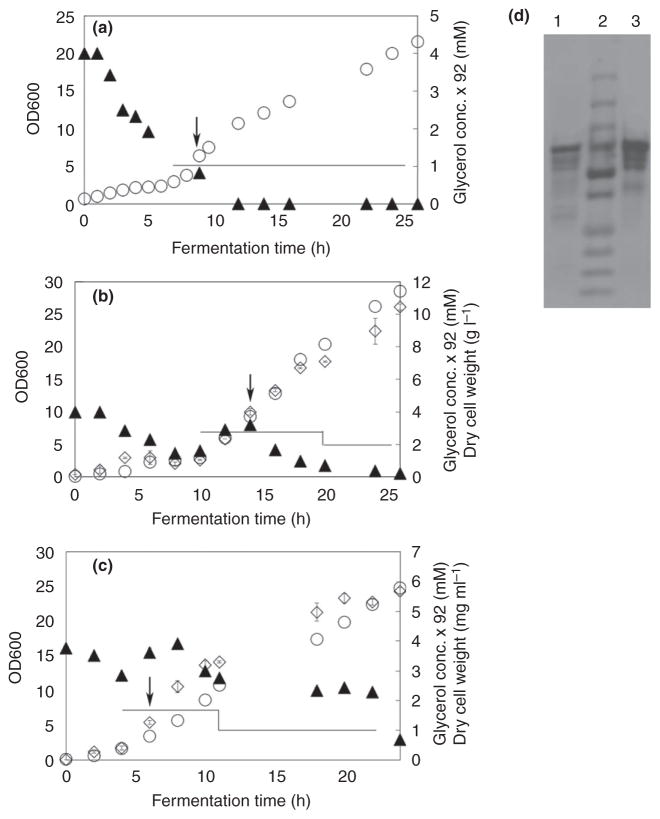
Three experiments were performed to examine the effect of feeding time, feeding rate and induction time on the yield and activity of 6-OST-3. The results of these studies are presented in Figs 3 and 4 and Table 1.
Figure 3.
Escherichia coli 6-OST-3 fed-batch fermentation in glycerol and LB medium. Panels a, b and c show fed-batch fermentation profiles from batch a, b and c, respectively. Bacterial growth (empty circles), glycerol concentration (filled triangles), and dry weight of biomass (empty diamonds), feeding profile (lines). Induction was performed with 0·1 mmol l−1 IPTG at 7·5, 9·5 and 3·5 Abs600nm, respectively, as indicated by the arrows. d: SDS-PAGE analysis of purified enzyme; lane M, ladder as reported in Fig. 1; lane 1, purified enzyme from batch a; lane 2, purified enzyme from batch b; and lane 3, purified enzyme from batch c.
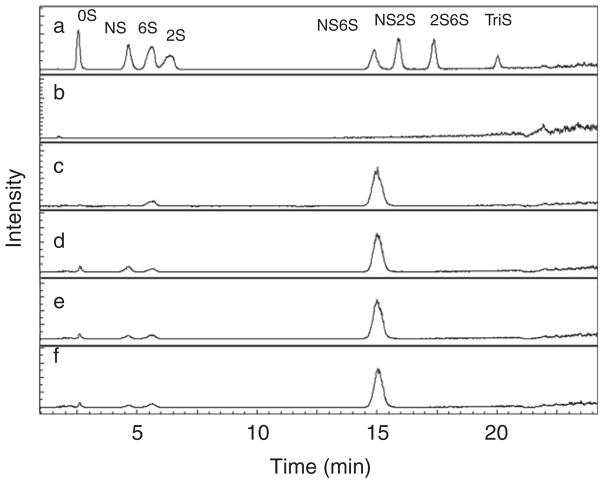
Figure 4.
Extracted ion chromatographs (EICs) of 6OST treated products analysed by LC-MS. a. 8 HP/HS disaccharide standards; b. control; c. product from LB shake flask 6OST treatment; d. product from batch A 6OST treatment; e. product from batch B 6OST treatment; and f, product from batch C 6OST treatment. Unsaturated disaccharides standards of heparan sulfate (0S, ΔUA-GlcNAc; NS, ΔUA-GlcNS; 6S, ΔUA-GlcNAc6S; 2S, ΔUA2S-GlcNAc; 2SNS, ΔUA2S-GlcNS; NS6S, ΔUA-GlcNS6S; 2S6S, ΔUA2S-GlcNAc6S; and TriS, ΔUA2S-GlcNS6S, where GlcNAc is N-acetyl-D-glucosamine and ΔUA 4-deoxy-α-L-threo-hexenopyranosyluronic acid).
Table 1.
Comparison of flask fermentation with LB medium and three fed-batch fermentation experiments with different induction time points and feeding rates
| FB A | FB B | FB C | LB | |
|---|---|---|---|---|
| OD600 at induction time | 7·5 | 9·5 | 3·5 | 0·6 |
| Cell density at induction (gcdw l−1) | 2·6 | 3·5 | 0·8 | |
| Feeding start time (h) | 6 | 10 | 4 | |
| Fermentation time (h) | 26 | 26 | 24 | |
| Maximum cell density (gcdw l−1) | 7·6 | 10·5 | 5·7 | |
| Purified 6-OST-3 concentration (mg l−1) | 61·5 | 52·9 | 120·7 | 6·1 |
| Yield 6-OST-3/biomass (mg gcdw−1) | 8·1 | 5·0 | 21·2 | |
| Productivity 6-OST-3 (mg l−1 h−1) | 2·4 | 2·0 | 5·0 | 0·29 |
| Final specific activity (nmol min−1 mg−1) | 1·36 ± 0·185 | 1·55 ± 0·144 | 1·67 ± 0·172 | 2·55 |
| Productivity 6-OST-3 (U l−1 h−1) | 3·26 | 3·10 | 8·35 | 0·74 |
| NS6S content (% ± SD) | 79·93 ± 2·45 | 82·94 ± 0·58 | 83·18 ± 3·07 | 93·68 |
For the first batch, we controlled the feeding rate at 0·6 ml min−1 from 6 h, so that the glycerol concentration was almost zero (Fig. 3). At this situation, the growth rate of the cells should be lower than when nutrition was enough. However, because it was induced when the OD600 reached 7·52, the maximum cell density was still up to 7·6 gcdwl−1. The productivity of 6-OST-3 was 2·4 mg l−1 h−1), and the final specific activity was 1·36 ± 0·185 nmol min−1 mg−1. The mass analysis showed that the proportion of ΔUA-N-sulfo, 6-O-sulfoglucosamine (NS6S) was up to 79·9 ± 2·5% after overnight reaction with 50 μg of 6-OST-3 in a 250-μl system.
For the second batch, the expression was induced when the OD600 reached 9·3 (Fig. 3), and the feeding rates were 3 ml ml−1 from 9 h and 1 ml min−1 after induction at 14 h. During the expression period, the glycerol concentration was lower than 21·8 mmol l−1, but the nutrition was enough for cells growth, so that a high maximum cell density of 10·5 gcdwl−1 was obtained (Table 1). However, at this situation, the productivity of 6-OST-3 was 2·0 mg l−1 h−1, which was lower than the first batch. The final specific activity was 1·55 ± 0·144 nmol min−1 mg−1.
For the third batch, the expression was induced when the OD600 reached 3·46 (Fig. 3), the feeding rates were 1·8 ml ml−1 from 4 h and 1 ml min−1 after 8 h, and the glycerol concentration was between 21·8 mmol l−1 and 32·7 mmol l−1. Because the induction time was early, a low maximum cell density of 5·7 gcdwl−1 was obtained (Table 1). However, at this situation, the productivity of 6-OST-3 was 5·0 mg l−1 h−1, which was much higher than the first and second batch. The final specific activity was 1·67 ± 0·172 nmol min−1 mg−1.
The SDS-PAGE showed that 6-OST-3 was the main component of the purified proteins from all three batches.
The polysaccharide substrate used in this reaction is N-sulfo, N-acetyl heparosan. This substrate contains 89% N-sulfoglucosamine residues and 11% N-acetylglucosamine residues. The complete conversion of this substrate by 6-OST-3, followed by heparin lyase conversion to unsaturated uronic acid (ΔUA) disaccharide-containing products, could afford up to 89% NS6S product. Liquid chromatography-mass spectral analysis showed that the NS6S percentage conversion of 6-OST-3 from batch 2 and batch 3 were up to 82·9 ± 0·6% and 83·2 ± 3·1, respectively. They were close to the quantitative conversion (88·7%) observed for enzyme obtained from LB shake flask fermentation.
Discussion
The enzyme of 6-OST-3 is a transmembrane protein, and only the catalytic domain of mouse 6-OST-3 was cloned into the plasmid; therefore, it is easy to form inclusion bodies. We tried different media, such as M9 plus 2% glucose and 0·5% peptone, and M9 plus 2% glycerol and 0·5% peptone, to express this enzyme, however, we got some soluble proteins without activity. Even when we tried LB plus 2% or 1% different carbon sources, the activities were very low. The soluble but inactive enzyme that was formed may be the result of incorrect protein folding or soluble aggregates (Dobson 2003). But when we lowered the concentration of the carbon sources, the activities were increased. The growth of E. coli was greatly inhibited by acetate, and the results showed that galactose was a good carbon source for 6-OST-3 expression, but compared with glycerol, it is not an economic source.
The cultivation of E. coli under different feeding schedules and induction strategies was examined. For batch A, we control the cell growth rate during the expression period, to make sure that we can get active enzymes on batch fermentation. For batch B, we tried to get high productivity active enzymes by getting a high cell density, but it was not so successful. For batch C, the enzyme expression was induced earlier, and we obtained the highest productivity levels, 8·35 U l−1 h−1, although the maximum cell density was only 5·7 gcdwl−1. Despite the lower cell density, productivity was over 10-fold higher than the flask fermentation using LB medium, which was 0·74 U l−1 h−1.
The lack of appropriate folding-assistant proteins and a post-translational modification system in E. coli often causes aggregation and low productivity of recombinant eukaryotic proteins (Song et al. 2012). The engineered strain was used to express the catalytic domain of mouse 6-OST-3 (Pro121-Pro450), a transmembrane protein. Although maltose-binding protein was used as a fusion partner in the expressing host E. coli strain, inclusion bodies were still formed during the expression process (data not shown). Assuming there is a degree of leaky expression, a higher induction OD could mean more preexisting inclusion bodies or soluble aggregates before induction, leading to easier incorporation of the enzyme into these inactive forms after induction. This may be the reason that there is a correlation between high induction OD and low enzyme productivity.
In summary, the cultivation of E. coli on various carbon sources under different feeding schedules and induction strategies was examined, LB plus 0·4% of glycerol (v/v) were finally selected for the expression media, and a concentrated solution (435 mmol l−1 glycerol, 150 g l−1 yeast extract and 48·7 mmol l−1 MgSO4·7H2O) was chosen as feeding media. When induction OD600 was 3·46 and the feeding rates were 1·8 ml min−1 from 4 h and 1 ml min−1 after 8 h, the productivity of 6-OST-3 was 5·0 mg l−1 h−1, and a maximum cell density of 5·7 gcdwl−1 was obtained. The final specific activity was 1·67 nmol min−1 mg−1.
Acknowledgments
The authors gratefully acknowledge funding from the NIH in the form of grant HL096972, from the National Natural Science Foundation of China under Grant No. 31171737 and the PRC grant 210208310507 supporting the visit of Jianhua Zhang, and from the Heparin Consortium.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Andersson C, Hodge D, Berglund KA, Rova U. Effect of different carbon sources on the production of succinic acid using metabolically engineered Escherichia coli. Biotechnol Prog. 2007;23:381–388. doi: 10.1021/bp060301y. [DOI] [PubMed] [Google Scholar]
- Bhaskar U, Sterner E, Hickey AM, Onishi A, Zhang F, Dordick JS, Linhardt RJ. Engineering of routes to heparin and related polysaccharides. Appl Microbiol Biotechnol. 2012;93:1–16. doi: 10.1007/s00253-011-3641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho RJ, Cabrera-Crespo J, Tanizaki MM, Gonçalves VM. Development of production and purification processes of recombinant fragment of pneumococcal surface protein A in Escherichia coli using different carbon sources and chromatography sequences. Appl Microbiol Biotechnol. 2012;94:683–694. doi: 10.1007/s00253-011-3649-9. [DOI] [PubMed] [Google Scholar]
- Chen J, Avci FY, Muñoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, et al. Enzymatic redesigning of biologically active heparan sulfate. J Biol Chem. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Guerrini M, Beccati D, Shriver Z, Naggi A, Viswanathan K, Bisio A, Capila I, Lansing JC, et al. Oversulfated chondroitin sulfate is a contaminant in heparin associated with adverse clinical events. Nat Biotechnol. 2008;26:669–675. doi: 10.1038/nbt1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhardt RJ. Heparin: structure and activity. J Med Chem. 2003;46:2551–2554. doi: 10.1021/jm030176m. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang Z, Linhardt RJ. Lessons learned from the contamination of heparin. Nat Prod Rep. 2009;26:313–321. doi: 10.1039/b819896a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswaran M, Forchhammer K. Carbon-source-dependent nitrogen regulation in Escherichia coli is mediated through glutamine-dependent GlnB signalling. Microbiology. 2003;149:2163–2172. doi: 10.1099/mic.0.26449-0. [DOI] [PubMed] [Google Scholar]
- Restaino OF, Bhaskar U, Paul P, Li L, De Rosa M, Dordick JS, Linhardt RJ. High cell density cultivation of a recombinant E. coli strain expressing a key enzyme in bioengineered heparin production. Appl Microbiol Biotechnol. 2013;97:3893–3900. doi: 10.1007/s00253-012-4682-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JA, Lee DS, Park JS, Han KY, Lee J. The N-domain of Escherichia coli phosphoglycerate kinase is a novel fusion partner to express aggregation-prone heterologous proteins. Biotechnol Bioeng. 2012;109:325–335. doi: 10.1002/bit.23320. [DOI] [PubMed] [Google Scholar]
- Wang Z, Li J, Cheong S, Bhaskar U, Akihiro O, Zhang F, Dordick JS, Linhardt RJ. Response surface optimization of the heparosan N-deacetylation in producing bioengineered heparin. J Biotechnol. 2011;156:188–196. doi: 10.1016/j.jbiotec.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]