Summary
The scale and scope of the global epidemic, coupled to challenges with traditional vaccine development approaches, point towards a need for novel methodologies for HIV vaccine research. While the development of vaccines able to induce broadly neutralizing antibodies remains the ultimate goal, to date vaccines continue to fail to induce these rare humoral immune responses. Conversely, growing evidence across vaccine platforms in both non-human primates and humans, point to a role for polyclonal vaccine-induced antibody responses in protection from infection. These candidate vaccines, despite employing disparate viral vectors and immunization strategies, consistently point to a role for functional or non-traditional antibody activities as correlates of immunity. However, the precise mechanism(s) of action of these “binding” antibodies, their specific characteristics, and ability to be selectively induced and/or potentiated to induce complete protection merits parallel investigation to neutralizing antibody-based vaccine design approaches. Ultimately while neutralizing and functional antibody-based vaccine strategies need not be mutually exclusive, defining the specific characteristics of “protective” functional antibodies may provide a target immune profile to potentially induce more robust immunity against HIV. Specifically, one approach to guide the development of functional antibody based vaccine strategies, termed “systems serology”, offers an unbiased and comprehensive approach to systematically survey humoral immune responses, capturing the array of functions and humoral response characteristics that may be induced following vaccination with high resolution. Coupled to machine learning tools, large datasets that explore the “antibody-ome” offer a means to step back from anticipated correlates and mechanisms of protection and toward a more fundamental understanding of coordinated aspects of humoral immune responses, their ability to more globally differentiate among vaccine candidates, and most critically, to identify the features of humoral immunity that distinguish protective from non-protective responses. Overall, the systematic serological approach described here aimed at broadly capturing the enormous biodiversity in antibody profiles that may emerge following vaccination complements existing cutting edge tools in the cellular immunology space that capture vaccine-induced polyfunctional cellular activity by flow cytometry, transcriptional profiling, epigenetic, and metabolomic analysis to offer a means to develop both a more nuanced and more complete understanding of correlates of protection to support the design of functional vaccine strategies.
Keywords: antibody, Fc Receptor, effector function, machine learning, vaccine
Introduction
Antibody responses are often thought to be mechanistic correlates of vaccine-mediated protection (1). Yet, many vaccines that induce robust antibody responses but do not provide protection indicate that beyond presence and prevalence, there are specific qualitative antibody features and in vivo antibody activities that are associated with immunity; that is, that not all antibodies are created equal. Efforts to develop a protective HIV vaccine may represent the setting in which the discrepancy between the generation of a robust (antibody quantity) versus protective (antibody quality) response has been most apparent. Likewise, tuberculosis, Ebola, and other vaccine development efforts continue to face similar challenges, highlighting the critical need to define signatures and mechanistic correlates of protection to rationally guide the design of protective vaccine strategies. The scope of this knowledge gap has led to a shift away from empirical vaccine design approaches that have been successful against less variant pathogens with simpler life cycles and less robust immune evasion tactics, to rational strategies that more systematically consider pathogen biology, dynamics within the host, and structural information on specific key antigenic determinants (2).
Rational strategies rooted in pathogen structure and life cycle have lead to a revolution in vaccine design, for example in the setting of RSV and MenB infection (3–5); and recent advances in cellular profiling approaches have enabled the robust and systematic analysis of host responses, providing insights into how immune cells respond to antigenic stimuli (5–8). However, while cellular technologies, such as those enabling robust transcriptional and multiparameter cytometric profiling have experienced considerable advances, humoral profiling efforts have been considerably more limited, most often focusing on antibody titer and neutralization activity toward select antigens or viral variants. However, comparable advances in humoral profiling strategies have the potential to more broadly identify robust correlates of protection, which are key parameters in successful translational vaccine development. In the absence of this information, vaccine candidates are advanced based on measures of response magnitude regardless of the importance of these features as mechanistic correlates of protective immunity, and despite known insufficiency. Indeed, response magnitude and even neutralization activity alone do not mechanistically drive protective immunity for most clinically approved vaccines (9, 10). Instead, evidence accumulated across a range of pathogens has suggested a critical role for numerous functional Ab responses, including Ab-dependent cellular cytotoxicity (ADCC), Ab-dependent cellular phagocytosis (ADCP), and Ab-dependent complement deposition (ADCD), in both protection from and post-infection control against HIV (11–14), influenza (15, 16), HSV (17, 18), Ebola virus (19), and malaria (20, 21). Collectively, these and other studies suggest that protective immunity in vivo can be attained in the absence and enhanced in the presence of in vitro neutralization activity.
Thus, to address the known insufficiency of titer and neutralization, and the wide spectrum of other possible mechanisms of antibody-based immunity, an analytical framework combining experimental and computational analysis has been developed to broadly characterize polyclonal Ab profiles in vaccine trials, designated “Systems Serology”. A systematic approach is necessary to identify key aspects of humoral immune responses, which are dynamic and highly polyclonal, with multiple somatic antibody variants directed to multiple epitopes on multiple antigens. Coupled to this diversity in antigen recognition, diversity in Fc domain subclass and glycosylation result in differences in ability to interact with innate immune receptors, such as Fcg receptors (FcgR) that are variably expressed on innate immune effector cells poised for the active elimination of pathogens via mechanisms beyond neutralization. The activity of polyclonal antibody pools is thus derived from the cumulative effect of specificities and activities present. Techniques capable of parsing this milieu into components that can be associated with other relevant clinical, genetic, or functional characteristics are beginning to demonstrate utility in affording enabling insights into humoral immunity via statistically principled analysis of humoral response profiles and relationships between antibody features, antibody functions, and clinical outcomes (12, 22–35). The resulting integrated analytical approach to objectively and broadly characterize diverse aspects of the humoral immune response in vaccine efficacy studies or by evaluation of natural disease resistance offers an enhanced ability to identify unanticipated correlates of protection, direct the selection of promising vaccines/immunogens, and define mechanisms of protective immunity (22).
AntibodyOMICS
The extensive diversity of the billions of antibodies produced during an immune response give rise to a remarkably flexible humoral immune response. The simultaneous optimization of antigen recognition via somatic hypermutation of the variable domain (Fv) and competition in germinal centers, and innate immune recognition via class-switch recombination and post-translationally-regulated activity cues of the constant domain (Fc), plays out over diverse antibody lineages. Thus antibodies may target a wide array or narrow distribution of antigens and epitopes and collectively form immune complexes that direct a wide or narrow spectrum of antiviral functions. As a result, antibodies act at the nexus between innate and adaptive immunity; they provide an adaptive means to engage a variety of innate immune effector cells to clear opsonized viral particles and infected cells. Significantly, despite the complexity of this landscape, considerable evidence indicates that Fc domain characteristics are not homogenized by polyclonality and avidity. Rather, there is strong evidence that antibodies can synergize to control pathogens. Just as serum neutralization profiles have been used to go beyond titer to robustly indicate the contributions of multiple specificities of neutralizing Abs that may be present in a polyclonal mixture (36), qualitative features such as the subclass and glycosylation profile of the antigen-reactive pool of multiple antibody types have been observed to support differential effector function potency, such NK cell cytotoxicity and monocyte phagocytosis in acute, infected, and vaccinated cohorts (26, 28, 37–40). For example, though a minor component of the overall polyclonal virus-specific antibody pool, IgG3 responses have been a marker of potent effector function across diverse activities and in diverse cohorts (22, 23, 35). Moreover, waves of antibodies may compete or complement each other’s activities, and these collaborative or conflicting properties appear to dynamically tune the overall activity of polyclonal pools (35, 41–45). Importantly, these and other studies provide evidence that the composite distribution of qualitative humoral response features present across polyclonal populations of Abs can exhibit key mechanistic relationships with polyclonal antibody activity, and that paradigms from reductionist passive transfer experiments may not strictly apply to polyclonal vaccine-induced responses.
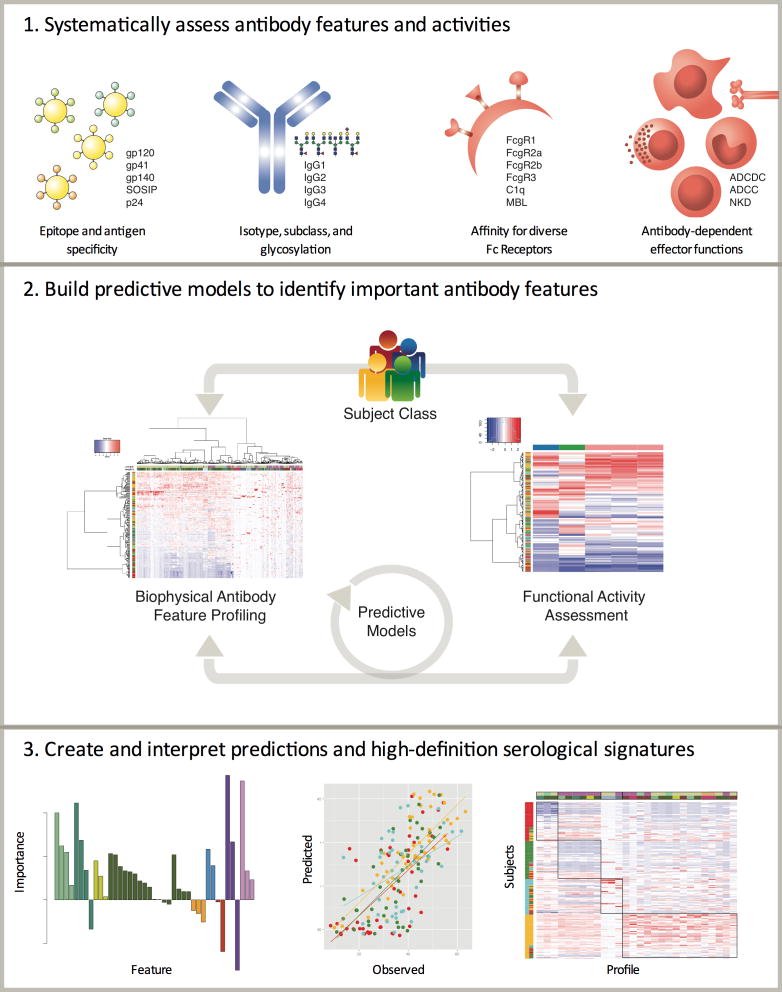
Unlike genomic or proteomic technologies that provide meaningful biological information through the direct interrogation of quantified bioanalyte levels, antibodies work as swarms to mediate their unique effector functions, and are controlled by amino acid sequence and post-translational changes at both ends of the molecule. Thus millions of individual antibodies, which may each differ in Fv and Fc character, can lead to a remarkable combinatorial diversity that may reveal critical insights into immune mechanisms of infection control. Distinct tools are required to fully interrogate the antibody-ome, related to the ability of B cells to simultaneously tune both ends of the antibody – the variable domain (Fv) that is responsible for binding to antigens and the constant domain (Fc) that directs immune activity. Remarkably, both domains are functionally tuned by a common enzyme, activation-induced deaminase (AID), which can direct DNA remodeling. On the Fv, AID drives DNA conversions that are then rapidly repaired by an array of DNA repair machinery resulting in substitution of single amino acid changes and/or larger insertions/deletions. On the Fc end, AID drives the deletion of larger segments of DNA to drive subclass/isotype changes—each with a distinct effector profile. This genetic evolutionary activity enables the humoral immune response to co-evolve to respond rapidly to simultaneously improve targeting (Fv) and destruction (Fc) of invading pathogens. While the revolution in genomic sequencing technologies have greatly advanced our ability to track the evolutionary pathways exploited by B cells to induce protective antibodies, these tools do not capture information related to the linkage between post-translational functional tuning and genomic changes. Thus, understanding antibody function requires technologies that can link the Fv and Fc variation directly at the protein level to define precise correlates of immune protection. Integrated B cell clonotypic analysis with functional profiling that captures interactions with the low affinity soluble or surface expressed FcRs, expressed on all innate immune effector cells could dramatically enhance understanding of humoral immunity. Tools that can capture humoral polyclonal and post-translational diversity, the resulting avidity effects, and subsequent innate immune cell interactions and outcomes may be critical to gaining deep and enabling insights into humoral immunity (Figure 1). A wealth of evidence, including studies ranging from anti-cancer monoclonal antibody therapeutic modes of action to the efficacy of glycoconjugate vaccines, suggests that effector function can be sufficient for efficacy, and that it significantly contributes to the in vivo activity of even the most potent neutralizing mAbs. Collectively, our emerging understanding of the remarkable breadth of antibody bioactivities, potential modes of action, and mechanisms of biodiversity provide key insights pointing toward the potential utility of systematic assays and analysis tools to provide insights into the specific features of humoral responses that are associated with potent activity and protective efficacy.
Figure 1.
Overview of the systems serology approach.
Learning from clinical efficacy studies
While empirical vaccine design approaches have shown only moderate success, rational vaccine design approaches and prioritization efforts among HIV vaccine candidates have been limited by an inadequate understanding of the correlates of protection against infection. However, large-scale efficacy trials offer critical insights into the characteristics of responses that have or have not influenced infection rates and may guide prioritization and development. Several trials, including HVTN 505, 502, and 503 evaluated HIV genes vectored by recombinant adenoviruses, and induced HIV-1 specific CD4+ and CD8+ responses, but were not efficacious. Similarly, the bivalent Envelope protein in alum vaccine evaluated in VAX003 and VAX004 raised antibody responses, but was not protective. To date, a single human HIV vaccine trial, RV144, has demonstrated efficacy, showing a significant but modest reduction in the risk of infection (46). Perplexingly, this vaccine did not induce immune responses that are typically associated with protective immunity— including neutralizing Abs, cytotoxic T cell responses, and high Ab titers. Moreover, immune profiles that were anticipated to potentially provide protection, such as tier 2 neutralization or cytotoxic T cell responses did not demonstrate statistically significant differences among cases and controls. In contrast, unanticipated correlates were observed (44), strongly motivating a more comprehensive analysis of the humoral response elicited.
Because of these unexpected results, a remarkable effort, bringing together numerous labs to explore numerous response parameters, was initiated to define the immune correlates of protection in the RV144 vaccine trial. Ultimately, the analysis pointed to elevated IgG binding to the V1V2 hypervariable loops and lower levels of Envelope-specific IgA among individuals who remained uninfected (44). Moreover, secondary analyses of breakthrough virus sequences demonstrated evidence of vaccine-mediated pressure within the V1V2 (47), supporting the potential mechanistic significance of this humoral correlate. Furthermore, in the absence of high IgA, ADCC responses were also inversely associated with risk. Additional genetic associations related to Fc receptor and HLA polymorphisms (48, 49). Collectively, and in comparison to responses in VAX003, this network of humoral response characteristics pointed toward humoral response quality as key to efficacy. Further support for this hypothesis was gained when Envelope and V-loop specific IgG3s were found to be correlated with reduced risk of infection, and associated with multiple antibody effector functions (43, 50). Thus this exhaustive humoral correlates analysis demonstrated an unexpected role of unanticipated humoral correlates, related to functional binding antibodies, to achieve protection. This collective effort pointed to the critical importance of looking beyond response magnitudes, or titers, which typically correlate with protection, as surrogate endpoints, and toward the more specific qualitative features of the humoral response that were suggested to be key indicators of protective efficacy.
Limitations of traditional correlates analysis
Traditional means of conducting correlates analysis suffer from seeking to balance competing objectives; namely, there exists a tradeoff between statistical power and likelihood of identifying insights into responses that may be associated with efficacy. As our understanding of the various modes of action of antibodies expands, additional potential correlates are considered. With expanded tools more response characteristics can be assessed, and it becomes more likely that a meaningful correlate will happen to be evaluated, but less likely that this correlate will meet the increasing standard of rigorous statistical significance tests. Thus there is a conflict between traditional vaccine efficacy analysis approaches and advancing immune-omic assessment platforms. While statistical rigor is necessary for high confidence in the identification of correlates and sets the gold standard for trial design, it can have unintended consequences on future development activities.
When only a limited number of immune assays are selected, an incomplete representation of the response elicited is developed- which may miss critical correlates of protective immunity. For example, six response measures were considered in the primary RV144 correlates analysis. This panel of measurements was restricted to include only highly robust and reproducible response measures, some of which were selected partially based on anticipated mechanistic roles in efficacy and ability to compare to previous studies, and others partially based on the desire to assess orthogonal response characteristics in order to capture the most scientific bang for each statistical buck. While necessary and rational, such downselection introduces inherent biases that can have substantial effects on subsequent development activities. In the case of RV144, of the six response measures, two unanticipated correlates were identified in primary analyses, with additional correlates observed in secondary and follow up analyses. Thus, the downselection of responses for assessment was highly successful. Multiple, robust correlates were identified, and they included unanticipated responses that were broadly reflective of a number of similar or related measurements (43, 51). However, given the observation of these similar and additional distinct correlates in follow up studies, it is clear that if additional or different measurements had been considered, different and/or additional correlates would likely have been identified.
Importantly, correlates become key road posts in advancing vaccine research. A complete picture of protective immunity can only be developed upon inclusive measurement and analytical dissection of the many features that may impact the antiviral profile of the humoral immune response. Because protection may be achievable through different mechanisms, correlates have the potential to be narrow differentiators that pertain only to specific regimens and populations, or to reflect a response with broad protective efficacy across different regimens and candidate vaccines, it is important to the trajectory of the success of ongoing efforts to ensure that an expanded and systematic analysis of responses in candidate and protective studies is performed, enabling robust meta-analyses that may be essential in the cross-evaluation of distinct vaccine strategies.
Defining humoral signatures
Unanticipated immune correlates can be defined in large datasets using robust, statistically sound, computational tools that have emerged over the past two decades to define “signatures” in big-data (Figure 1). One such analytical framework, broadly referred to as machine learning (ML), is rooted in pattern recognition, artificial intelligence, and computational statistics and learning theory (reviewed in (52)). ML approaches have become integral to data-intensive biological studies, contributing to tasks as widely ranging as predictions of protein secondary structure (53), correction of experimental bias in cDNA microarrays (54), and classification of patients into clinical groups (55), among countless others. Broadly speaking, ML approaches seek to automate the identification of “signatures” or patterns that reproducibly appear in particular populations of subjects, or among variably potent antibody pools, or differentially protected individuals, and thus may point toward key aspects of biological variation and mechanisms.
To this end, a number of unsupervised analysis methods aim to provide means to effectively distill the information value of “big data”. Clustering and dimensionality reduction techniques provide insights into natural groupings of response characteristics across study subjects or among subjects over humoral response characteristics offering insights into classes of responses, or similarly, into families of responders. Notably, these unsupervised analytic strategies are particularly amenable to combination with traditional approaches. For example, minimal signatures that identify key aspects of variation can be identified by mining comprehensive data using reduction methods that eliminate data that track together. Understanding relationships within immune response data is also valuable in and of itself, as such co-regulation suggests shared mechanisms of induction (35, 40, 51). Similarly, understanding correlated responses between humoral immunity and transcriptional programs may likewise point to linked regulatory pathways and response characteristics (7, 56–58).
Supervised approaches, in contrast, seek to use response data to build predictive models—explicitly modeling how well and which measurements can make classification or regression predictions. They extrapolate from known data to make predictions of unknowns—seeking to unveil meaningful biological relationships by learning historical associations and trends. Examples of this approach include the using features from a humoral response profile to characterize clinical class, such as the identification of response characteristics that differentiate between rapid or slow HIV progressors; models of expected infection risk, such as the humoral features that predict time to infection in an repeated exposure NHP challenge study; as well as to learn predictive relationships among features, such as estimations of effector function developed from antibody profiles. Each of these models seeks to produce reliable results based on generalization of previously observed examples.
While there are reasons to be skeptical of complex analytical approaches, there are robust means to identify whether data sets may be “too wide”, or method “too powerful” to be confident that it results from meaningful relationships rather than relying on chance in the setting of many chances. Like more traditional statistical tools, mathematical tools have evolved along with big data analytical approaches to define the robustness of observed associations. First, the quality of a model can be rigorously assessed using repeated cross validation, in which a model is developed on a subset of the data, referred to as the training set, and the models performance is then evaluated by application to the held out, or test set. Second, permutation tests, in which data labels are randomly exchanged prior to model building, can serve as an in silico negative control, setting a baseline benchmark for performance. Third, the ultimate test of model utility is application to an independent set of samples, or validation cohort, as has become a standard practice in genomic studies.
The ability of these approaches to value generalization can have advantages over traditional approaches. In our experience, diverse approaches can achieve consistent performance using a consistent set of measurements (32). In contrast, in the context of small studies that compare responses between study arms, t-tests can be very sensitive to inclusion of all subjects—that is, a response measurement difference may meet a given significance threshold across all subjects, but fall short of this cutoff if a subset or even a single subject is excluded. Immune features with this type of sensitivity are unlikely to contribute to cross-validated models, as they are not sufficiently strong predictive indicators of group differences.
Overall, these computational approaches offer a robust, data agnostic, and systematic means to sift through large datasets to find minimal humoral signatures that robustly track with subject groups, vaccine types, and efficacy outcomes—identifying critical and unanticipated landmarks in the complex landscape of humoral immunity. Application of these analytical tools, which have evolved remarkably to keep up with big data science, in combination with expanded experimental profiling, promises to provide potentially key support to advancing vaccine design.
Data-driven discovery, a complement to reductionist approaches
As technologies have rapidly evolved, large datasets are now easily captured with respect to cellular, transcriptional, and metabolomic features that collectively have illuminated mechanisms of vaccine action (6, 7). However, despite their importance, less is known about how to evaluate antibodies at similar resolution. Additionally, the need to develop vaccines for pathogens for which correlates of protection are not known, or that require accelerated development timelines to prevent epidemics may require an analytical paradigm shift. These needs can be addressed by development of new tools and new analytical strategies.
However, novel experimental analytical methods for antibodies have not advanced as quickly as those for cells. Yet, over the past two decades, advances in monoclonal antibody therapeutics have greatly expanded our understanding of antibody function, and pointed toward a diversity of potential modes of activity, and numerous structure-function relationships that impact effector potency. Moreover, the multiple, varied, and unanticipated correlates observed in RV144 (44) indicate the potential value of utilizing a broad, unbiased approach to assessing the types of responses that may be hallmarks of protective HIV-1 vaccines. Beyond neutralization breadth and potency, substantial evidence from natural infection, vaccination, and studies in animal models points to a critical role for antibody FcR engagement in reducing risk of infection (59).
With the objective to assess as many of the humoral attributes that may influence outcomes as possible, investigation of vaccine-induced antibody responses has moved beyond titer and neutralization activity to incorporate subclass, glycosylation, epitope specificity, affinity for antigen, affinity for antibody Fc receptors, immune complex valency and composition, and activity in in vitro and cell-based assays with diverse effectors, such as neutrophils, monocytes, dendritic cells, natural killer cells, and diverse effector mechanisms such as complement deposition, cytokine release, degranulation, netosis, and oxidative burst (Figure 1). Such systematic approaches offer a means to reduce investigator and confirmation bias that can be inherent in down selection efforts for traditional correlates analysis. More importantly, they simultaneously offer the ability to identify novel and facilitate a more comprehensive understanding of correlates, expand upon how response characteristics are interrelated, and provide clues as to conserved pathways in responses to vaccination. This appreciation from RV144 led to the creation of comprehensive antibody testing platforms, that together with other growing –omic technologies, hold the promise to reveal unexpected insights into correlates of protective immunity that may accelerate the development of HIV vaccines and beyond.
Systems Serology
Several recent studies have attempted to use systems serology approaches to comprehensively profile and ultimately distill the features of protective or highly functional humoral immune responses. One such study sought to identify whether different antibody types or activities were present among HIV controllers lacking as compared to those with protective HLA alleles (26). While differences in humoral immunity among genetically variant subject groups were lacking, clear relationships between specific antibody types and high antibody function were apparent. For example, hierarchical clustering of subjects demonstrated the ability of this unsupervised approach to capture IgG subclass and magnitude differences that were associated with divergent ADCC activity.
In order to gain insights into the properties of antibodies that support recruitment of effective functional responses, another study developed and applied an ML framework to identify and model associations among properties of antibodies and corresponding functional responses from antibody subclass-specificity data collected from RV144 vaccine recipients (32). This study demonstrated that models trained to encapsulate antibody feature-function relationships were able to robustly predict the quality of the polyclonal functional response using information about the specific antibody subtypes that were present. Additionally, this study explicitly compared different modeling strategies, including methods ranging from penalized linear regression to regularized random forest and support vector machines, among others for classification and regression. Prediction accuracy and predictive features were generally conserved across methods, and identified IgG3 as making an outsized contribution to antibody function, and IgG4 as a marker of decreased activity. These predictions were experimentally tested and validated (23), suggesting the ability of learned models to identify biologically meaningful rather than simply computationally useful relationships.
A complementary study evaluated a panel of monoclonal Abs that recognized different epitopes and exhibited differential glycosylation, and observed that glycoprofiles were more highly correlated to various antibody functions than epitope (31). Studies such as this represent small data sets, but given the complexity of IgG glycosylation, are nonetheless well suited to ML approaches. Other efforts have evaluated longitudinal changes in IgG subclass and specificity, neutralization and effector activity and identified how response patterns changed over time, strengthening observations relating specific features of the polyclonal response to various activities as infection passed from acute into chronic stages (40).
In sum, the integration of antibody feature and function data within an ML framework provides a new, objective approach to discovering and assessing multivariate immune correlates and hallmarks of highly functional humoral responses (Figure 1). The ability to build predictive models of relationships in these and other studies may distill humoral correlates in diverse data sets and ultimately lead to the development of vaccines and antibody therapies that will better steer the immune system to produce antibodies with beneficial activities.
Profiling vaccine-induced immunity
One recent application of this method compared the humoral immune responses induced by recent human HIV vaccine trials including the RV144 vaccine trial that showed partial effectiveness (ALVAC/AIDSVAX B/E (46)), two trials the did not demonstrate efficacy in phase 2b trials (VAX003; AIDSVAX B/E (60); and HVTN204; DNA/rAD5 (61)), and a phase 1 study designed to evaluate the safety and immunogenicity of the Ad26 vector (IPCAVD001; Ad26.ENVA.01 (34)). In this study, a collection of modeling techniques was applied to a rich dataset comprising both Ab features and Ab effector functions assessed in vitro with a suite of cell-based assays (22). Importantly, this analysis identified unexpected shared features among vaccine trials utilizing similar immunogens and among non-protective vaccine trials that differentiated them from the protective RV144 vaccine trial. Moreover, the Systems Serology approach applied there begins to point to unique humoral response architectures induced by each vaccine regimen and highlight complex interactions and relationships between individual Ab parameters that may underlie “protective” correlates in RV144.
Specifically, Chung et al. identified response clusters defined by adenovirus versus protein immunogens in both unsupervised and supervised approaches. VAX003 and RV144 recipients were well- differentiated by disparate env-specific IgG3 versus IgG4 subclass responses in a supervised analysis that used LASSO (least absolute feature shrinkage and selection operator, (62)) for feature selection followed by PLSDA (partial least-squares discriminant analysis, (63)) to define the degree to which subjects from each trial could be differentiated and the relative contributions of humoral features responsible for this discrimination. A similar approach was used to discriminate among all four trials, and identified features related to vaccine antigen(s) as key to discriminating between adenovirus and protein vaccines. Intriguingly, discrimination between recipients of the Ad26 vaccine, whose efficacy is unknown in humans, and the protective RV144 vaccine, from subjects receiving either ineffective vaccine was attributed to differences in the Env-specific IgG3 response. A similar approach was used to differentiate between RV144 recipients with high or low V1V2 responses, and high or low expected degrees of risk based on original correlates, and to use them, their correlates, and the network of structure and topology of humoral response feature correlations to provide a more nuanced picture of vaccine-induced immunity that may be of critical comparative value in systematically revealing features of vaccine-induced immunity.
Thus, this analytical approach provided initial insights into the extent and identities of differences in the overall features and function of polyclonal immune responses elicited by candidate vaccines to help guide the evaluation, down-selection, and rational improvement of future vaccine concepts against HIV and other pathogens for which immune correlates of protection have yet to be elucidated. For example, this type of analytical approach begins to round out the concept of the “immune space”, a rubric developed to identify whether and how unique candidate vaccines are relative to those evaluated previously (64).
Learning from vaccine studies in the NHP model
A number of studies have identified correlations between binding antibodies and protection (56, 65, 66). A recent example of application of systems serology to one such NHP study involves the evaluation of the Ad26 prime, Env protein boost HIV vaccine candidate, which is currently in phase 2a clinical trials and is under consideration for advancement into phase 2b/3 efficacy trials. The SIV version of this vaccine provided 50% protection against acquisition of infection against repetitive, heterologous, intrarectal SIVmac251 challenges (33). Protection correlated with both binding and functional antibodies as measured by systems serology, and functionality was substantially increased by the addition of the Env protein boost. In fact, protection correlated significantly with a composite measure of antibody functionality, which included ADCC, ADCP, ADCD, and antibody dependent NK cell activation. Promisingly, ADCD and ADCP were correlated with protection in a previous study (12). Thus, repeated protection studies have collectively pointed to convergent signatures associated with antibody-mediated complement activation and phagocytic activity, potentially highlighting the mode of action of protective antibodies raised by this particular Ad26 prime/protein boost vaccine strategy.
Similarly, to gain insights into qualitative differences in the humoral immune response induced in the context of an NHP immunization regimen modeled on RV144. A systems serology approach helped to define fundamental humoral signature differences in the quality of humoral immune response induced by different adjuvants that may alter protective immunity. A combination of systemic and mucosal SIV-specific antibody data were collected, representing a suite of Fc-functional assays (ADCC, phagocytosis, complement deposition, cytokine and chemokine secretion), IgG glycosylation, and Fc-biophysical assays to assess both human and rhesus Fc-receptor and complement component binding to antigen-specific antibodies. Multivariate differences were assessed using ML approaches and indicated that the two regimens induced fundamentally different antibody Fc-profiles, and logistic regression analyses comparing Fc profiles of macaques that were infected in 3 or fewer challenges (≤3) or after 4 or more challenges (≥4) identified composite humoral profiles associated with protection. Collectively, these data highlighted divergence in the mucosal Fc-profile response as a key distinguishing profile separating protected from unprotected animals within each arm, potentially associated with the distinct mucosal plasma cell programming induced by these different adjuvants.
Further, in this study, transcriptional profiles could be related to specific humoral characteristics. In this case, this effort identified a RAS-associated genetic signature. In doing so, it points toward a future in which such models are also built using genetic, cell type-specific transcriptional profiles, Ig sequencing, and other data types, resulting in a more complete understanding of mechanisms of vaccination. Because other NHP studies have identified multiple (66), sex-specific (67), tissue-specific (68), conditional (if a but not b) (69), and insights may be gained from profiling non-protective regimens, there is a rich landscape of studies with data regarding responses and outcomes that can be mined. Overall, more sophisticated vaccine development strategies offer benefits as well as pose challenges. Most critically, they offer a path forward where traditional methods have failed. As the analytical tools to interrogate the humoral immune response grow- these ML tools will evolve in concert to provide more and more info to help rationally prioritize future vaccines or designing better immunotherapeutics. In the meantime, the convergence of human studies in identifying functionally coordinate IgG3 and IgG1 antibodies suggests that continued evaluation of human and NHP studies may likewise identify conserved features of effective responses across studies and vaccine strategies, pointing to robust mechanisms of vaccine-mediated protection from HIV infection.
Systems Serology 2.0
Like cellular transcriptional profiling analyses that have provided a deeper understanding of mechanisms of protective vaccine-induced immunity, the systematic evaluation of serological immune responses, through investment in high-throughput tools that comprehensively enable the dissection of humoral immune responses at unprecedented depth and that broadly capture the enormous biodiversity that exists within the humoral immune response, have begun to identify novel correlates of protection and highly functional antibody responses. Using these high-throughput biophysical and functional profiling, “systems serology” approaches, we have begun to define protective humoral immune response signatures among vaccinees and naturally infected cohorts (12, 22–35). In parallel, diverse vaccine regimens with varying levels of protective efficacy have been tested in non-human primates (33, 34, 66, 69, 70). Application of these systems serology approaches has begun to identify novel correlates of vaccine-mediated protection (33, 34) and provides a strong rationale for their continued use and further development in support of future clinical vaccine development. Critically, we posit that these systematic antibody profiling efforts will continue to expand our understanding of the diversity of antibody responses observed in the context of different vaccination strategies and lead to the development of enabling understanding of the antibody features that may be mechanistically associated with protection. However, we may only be at the inflection point in truly interrogating the complexity of the humoral immune responses. Thus over the next decade, major advances in mass-spectrometry, single cell profiling, and other technologies, will combine with today’s advanced methods to enable the interrogation of the full spectrum of differences that exist within the humoral immune system, offering a deeper understanding of the immune correlates of protection against HIV and beyond.
Acknowledgments
We acknowledge support from the NIH (AI060354, AI078526, AI080289, AI084794, AI095985, AI096040, AI102660, AI102691, OD011170, and HHSN261200800001E), the Bill and Melinda Gates Foundation (OPP1032817 and OPP1114729), and the Ragon Institute of MGH, MIT, and Harvard. The authors gratefully acknowledge Dr. Karen Dowell for figure preparation.
Footnotes
Conflict of Interest
The authors declare no competing financial interests. D.H.B. is a named co-inventor on vector, antigen, and protein patents (PCT/EP2007/052463, PCT/US2009/060494, PCT/US2009/064999).
References
- 1.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17(7):1055–65. doi: 10.1128/CVI.00131-10. Epub 2010/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, et al. A Blueprint for HIV Vaccine Discovery. Cell host & microbe. 2012;12(4):396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rappuoli R. Reverse vaccinology. Current opinion in microbiology. 2000;3(5):445–50. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- 4.Kulp DW, Schief WR. Advances in structure-based vaccine design. Current opinion in virology. 2013;3(3):322–31. doi: 10.1016/j.coviro.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rappuoli R, Bottomley MJ, D’Oro U, Finco O, De Gregorio E. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. The Journal of experimental medicine. 2016;213(4):469–81. doi: 10.1084/jem.20151960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33(4):516–29. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nature immunology. 2009;10(1):116–25. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin L, Finak G, Ushey K, Seshadri C, Hawn TR, Frahm N, et al. COMPASS identifies T-cell subsets correlated with clinical outcomes. Nature biotechnology. 2015;33(6):610–6. doi: 10.1038/nbt.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plotkin SA. Vaccines: correlates of vaccine-induced immunity. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2008;47(3):401–9. doi: 10.1086/589862. [DOI] [PubMed] [Google Scholar]
- 10.Plotkin SA. Correlates of protection induced by vaccination. Clinical and vaccine immunology: CVI. 2010;17(7):1055–65. doi: 10.1128/CVI.00131-10. Epub 2010/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449(7158):101–4. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 12.Barouch DH, Stephenson KE, Borducchi EN, Smith K, Stanley K, McNally AG, et al. Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell. 2013;155(3):531–9. doi: 10.1016/j.cell.2013.09.061. Epub 2013/11/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158(6):1243–53. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forthal DN, Landucci G, Daar ES. Antibody from patients with acute human immunodeficiency virus (HIV) infection inhibits primary strains of HIV type 1 in the presence of natural-killer effector cells. J Virol. 2001;75(15):6953–61. doi: 10.1128/JVI.75.15.6953-6961.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nature medicine. 2014;20(2):143–51. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol. 2004;172(9):5598–605. doi: 10.4049/jimmunol.172.9.5598. [DOI] [PubMed] [Google Scholar]
- 17.Kohl S, Loo LS, Pickering LK. Protection of neonatal mice against herpes simplex viral infection by human antibody and leukocytes from adult, but not neonatal humans. J Immunol. 1981;127(4):1273–5. Epub 1981/10/01. [PubMed] [Google Scholar]
- 18.Kohl S, Loo LS. Protection of neonatal mice against herpes simplex virus infection: probable in vivo antibody-dependent cellular cytotoxicity. J Immunol. 1982;129(1):370–6. Epub 1982/07/01. [PubMed] [Google Scholar]
- 19.Warfield KL, Swenson DL, Olinger GG, Kalina WV, Aman MJ, Bavari S. Ebola virus-like particle-based vaccine protects nonhuman primates against lethal Ebola virus challenge. The Journal of infectious diseases. 2007;196(Suppl 2):S430–7. doi: 10.1086/520583. [DOI] [PubMed] [Google Scholar]
- 20.Bouharoun-Tayoun H, Oeuvray C, Lunel F, Druilhe P. Mechanisms underlying the monocyte-mediated antibody-dependent killing of Plasmodium falciparum asexual blood stages. The Journal of experimental medicine. 1995;182(2):409–18. doi: 10.1084/jem.182.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi J, McIntosh RS, Pleass RJ. Antibody- and Fc-receptor-based therapeutics for malaria. Clinical science. 2006;110(1):11–9. doi: 10.1042/CS20050136. [DOI] [PubMed] [Google Scholar]
- 22.Chung AW, Kumar MP, Arnold KB, Yu WH, Schoen MK, Dunphy LJ, et al. Dissecting Polyclonal Vaccine-Induced Humoral Immunity against HIV Using Systems Serology. Cell. 2015;163(4):988–98. doi: 10.1016/j.cell.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung AW, Ghebremichael M, Robinson H, Brown E, Choi I, Lane S, et al. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Science translational medicine. 2014;6(228):228ra38. doi: 10.1126/scitranslmed.3007736. [DOI] [PubMed] [Google Scholar]
- 24.Ackerman ME, Crispin M, Yu X, Baruah K, Boesch AW, Harvey DJ, et al. Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. The Journal of clinical investigation. 2013;123(5):2183–92. doi: 10.1172/JCI65708. Epub 2013/04/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ackerman ME, Dugast AS, McAndrew EG, Tsoukas S, Licht AF, Irvine DJ, et al. Enhanced phagocytic activity of HIV-specific antibodies correlates with natural production of immunoglobulins with skewed affinity for FcgammaR2a and FcgammaR2b. J Virol. 2013;87(10):5468–76. doi: 10.1128/JVI.03403-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai JI, Licht AF, Dugast AS, Suscovich T, Choi I, Bailey-Kellogg C, et al. Divergent antibody subclass and specificity profiles but not protective HLA-B alleles are associated with variable antibody effector function among HIV-1 controllers. J Virol. 2014;88(5):2799–809. doi: 10.1128/JVI.03130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ackerman ME, Moldt B, Wyatt RT, Dugast AS, McAndrew E, Tsoukas S, et al. A robust, high-throughput assay to determine the phagocytic activity of clinical antibody samples. Journal of immunological methods. 2011;366(1–2):8–19. doi: 10.1016/j.jim.2010.12.016. Epub 2011/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brown EP, Licht AF, Dugast AS, Choi I, Bailey-Kellogg C, Alter G, et al. High-throughput, multiplexed IgG subclassing of antigen-specific antibodies from clinical samples. Journal of immunological methods. 2012;386(1–2):117–23. doi: 10.1016/j.jim.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sajadi MM, Farshidpour M, Brown EP, Ouyang X, Seaman MS, Pazgier M, et al. lambda Light Chain Bias Associated With Enhanced Binding and Function of Anti-HIV Env Glycoprotein Antibodies. The Journal of infectious diseases. 2015 doi: 10.1093/infdis/jiv448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Storcksdieck Genannt Bonsmann M, Niezold T, Temchura V, Pissani F, Ehrhardt K, Brown EP, et al. Enhancing the Quality of Antibodies to HIV-1 Envelope by GagPol-Specific Th Cells. J Immunol. 2015;195(10):4861–72. doi: 10.4049/jimmunol.1501377. [DOI] [PubMed] [Google Scholar]
- 31.Chung AW, Crispin M, Pritchard L, Robinson H, Gorny MK, Yu X, et al. Identification of antibody glycosylation structures that predict monoclonal antibody Fc-effector function. Aids. 2014;28(17):2523–30. doi: 10.1097/QAD.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi I, Chung AW, Suscovich TJ, Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Machine learning methods enable predictive modeling of antibody feature:function relationships in RV144 vaccinees. PLoS computational biology. 2015;11(4):e1004185. doi: 10.1371/journal.pcbi.1004185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, et al. HIV-1 vaccines. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science. 2015;349(6245):320–4. doi: 10.1126/science.aab3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barouch DH, Liu J, Peter L, Abbink P, Iampietro MJ, Cheung A, et al. Characterization of humoral and cellular immune responses elicited by a recombinant adenovirus serotype 26 HIV-1 Env vaccine in healthy adults (IPCAVD 001) The Journal of infectious diseases. 2013;207(2):248–56. doi: 10.1093/infdis/jis671. Epub 2012/11/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackerman ME, Mikhailova A, Brown EP, Dowell KG, Walker BD, Bailey-Kellogg C, et al. Polyfunctional HIV-Specific Antibody Responses Are Associated with Spontaneous HIV Control. PLoS pathogens. 2016;12(1):e1005315. doi: 10.1371/journal.ppat.1005315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340(6133):751–6. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- 37.Ackerman ME, Dugast AS, McAndrew EG, Tsoukas S, Licht AF, Irvine DJ, et al. Enhanced phagocytic activity of HIV-specific antibodies correlates with natural production of immunoglobulins with skewed affinity for FcgammaR2a and FcgammaR2b. Journal of virology. 2013 doi: 10.1128/JVI.03403-12. Epub 2013/03/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ackerman ME, Crispin M, Yu X, Baruah K, Boesch AW, Harvey DJ, et al. Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. The Journal of clinical investigation. 2013 doi: 10.1172/JCI65708. Epub 2013/04/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baum LL, Cassutt KJ, Knigge K, Khattri R, Margolick J, Rinaldo C, et al. HIV-1 gp120-specific antibody-dependent cell-mediated cytotoxicity correlates with rate of disease progression. J Immunol. 1996;157(5):2168–73. Epub 1996/09/01. [PubMed] [Google Scholar]
- 40.Dugast AS, Stamatatos L, Tonelli A, Suscovich TJ, Licht AF, Mikell I, et al. Independent evolution of Fc- and Fab-mediated HIV-1-specific antiviral antibody activity following acute infection. Eur J Immunol. 2014;44(10):2925–37. doi: 10.1002/eji.201344305. Epub 2014/07/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollara J, Bonsignori M, Moody MA, Liu P, Alam SM, Hwang KK, et al. HIV-1 vaccine-induced C1 and V2 Env-specific antibodies synergize for increased antiviral activities. J Virol. 2014;88(14):7715–26. doi: 10.1128/JVI.00156-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forthal DN, Landucci G, Ding H, Kappes JC, Wang A, Thung I, et al. IgG2 inhibits HIV-1 internalization by monocytes, and IgG subclass binding is affected by gp120 glycosylation. Aids. 2011;25(17):2099–104. doi: 10.1097/QAD.0b013e32834b64bd. Epub 2011/08/13. [DOI] [PubMed] [Google Scholar]
- 43.Chung AW, Ghebremichael M, Robinson H, Brown E, Choi I, Lane S, et al. Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci Transl Med. 2014;6(228):228ra38. doi: 10.1126/scitranslmed.3007736. Epub 2014/03/22. [DOI] [PubMed] [Google Scholar]
- 44.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. The New England journal of medicine. 2012;366(14):1275–86. doi: 10.1056/NEJMoa1113425. Epub 2012/04/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomaras GD, Ferrari G, Shen X, Alam SM, Liao HX, Pollara J, et al. Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(22):9019–24. doi: 10.1073/pnas.1301456110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. The New England journal of medicine. 2009;361(23):2209–20. doi: 10.1056/NEJMoa0908492. Epub 2009/10/22. [DOI] [PubMed] [Google Scholar]
- 47.Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490(7420):417–20. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li SS, Gilbert PB, Tomaras GD, Kijak G, Ferrari G, Thomas R, et al. FCGR2C polymorphisms associate with HIV-1 vaccine protection in RV144 trial. The Journal of clinical investigation. 2014;124(9):3879–90. doi: 10.1172/JCI75539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prentice HA, Tomaras GD, Geraghty DE, Apps R, Fong Y, Ehrenberg PK, et al. HLA class II genes modulate vaccine-induced antibody responses to affect HIV-1 acquisition. Science translational medicine. 2015;7(296):296ra112. doi: 10.1126/scitranslmed.aab4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yates NL, Liao HX, Fong Y, deCamp A, Vandergrift NA, Williams WT, et al. Vaccine-induced Env V1–V2 IgG3 correlates with lower HIV-1 infection risk and declines soon after vaccination. Science translational medicine. 2014;6(228):228ra39. doi: 10.1126/scitranslmed.3007730. Epub 2014/03/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zolla-Pazner S, deCamp A, Gilbert PB, Williams C, Yates NL, Williams WT, et al. Vaccine-induced IgG antibodies to V1V2 regions of multiple HIV-1 subtypes correlate with decreased risk of HIV-1 infection. PloS one. 2014;9(2):e87572. doi: 10.1371/journal.pone.0087572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tarca AL, Carey VJ, Chen XW, Romero R, Draghici S. Machine learning and its applications to biology. PLoS computational biology. 2007;3(6):e116. doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19(1):55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 54.Tarca AL, Cooke JE, Mackay J. A robust neural networks approach for spatial and intensity-dependent normalization of cDNA microarray data. Bioinformatics. 2005;21(11):2674–83. doi: 10.1093/bioinformatics/bti397. [DOI] [PubMed] [Google Scholar]
- 55.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 56.Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NP, Cameron M, et al. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nature medicine. 2016;22(7):762–70. doi: 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nature immunology. 2014;15(2):195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ovsyannikova IG, Oberg AL, Kennedy RB, Zimmermann MT, Haralambieva IH, Goergen KM, et al. Gene signatures related to HAI response following influenza A/H1N1 vaccine in older individuals. Heliyon. 2016;2(5):e00098. doi: 10.1016/j.heliyon.2016.e00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boesch AW, Brown EP, Ackerman ME. The role of Fc receptors in HIV prevention and therapy. Immunological reviews. 2015;268(1):296–310. doi: 10.1111/imr.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. The Journal of infectious diseases. 2006;194(12):1661–71. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 61.Churchyard GJ, Morgan C, Adams E, Hural J, Graham BS, Moodie Z, et al. A phase IIA randomized clinical trial of a multiclade HIV-1 DNA prime followed by a multiclade rAd5 HIV-1 vaccine boost in healthy adults (HVTN204) PloS one. 2011;6(8):e21225. doi: 10.1371/journal.pone.0021225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tibshirani R. The lasso method for variable selection in the Cox model. Statistics in medicine. 1997;16(4):385–95. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 63.Brereton R, Lloyd G. Partial least squares discriminant analysis: taking the magic away. J Chemometrics. 2014;28(4):213–25. [Google Scholar]
- 64.Manrique A, Adams E, Barouch DH, Fast P, Graham BS, Kim JH, et al. The immune space: a concept and template for rationalizing vaccine development. AIDS research and human retroviruses. 2014;30(11):1017–22. doi: 10.1089/AID.2014.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482(7383):89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roederer M, Keele BF, Schmidt SD, Mason RD, Welles HC, Fischer W, et al. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature. 2014;505(7484):502–8. doi: 10.1038/nature12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mohanram V, Demberg T, Musich T, Tuero I, Vargas-Inchaustegui DA, Miller-Novak L, et al. B Cell Responses Associated with Vaccine-Induced Delayed SIVmac251 Acquisition in Female Rhesus Macaques. J Immunol. 2016;197(6):2316–24. doi: 10.4049/jimmunol.1600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Q, Zeng M, Duan L, Voss JE, Smith AJ, Pambuccian S, et al. Live simian immunodeficiency virus vaccine correlate of protection: local antibody production and concentration on the path of virus entry. J Immunol. 2014;193(6):3113–25. doi: 10.4049/jimmunol.1400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fouts TR, Bagley K, Prado IJ, Bobb KL, Schwartz JA, Xu R, et al. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(9):E992–9. doi: 10.1073/pnas.1423669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fourati S, Vaccari M, Gordon SN, Schifanella L, Cameron M, Keele BF, et al. Modulation of RAS Pathways as a Biomarker of Protection against HIV and as a Means to Improve Vaccine Efficacy. AIDS research and human retroviruses. 2014;30:A99-A. [Google Scholar]