Abstract
The metabolic syndrome (MetS) is a constellation of metabolic disorders that increase the risk of developing several diseases including type 2 diabetes and cardiovascular diseases. Although genome-wide association studies (GWAS) have successfully identified variants associated with individual traits comprising MetS, the genetic basis and pathophysiological mechanisms underlying the clustering of these traits remain unclear. We conducted GWAS of MetS in 1,427 Africans from Ghana and Nigeria followed by replication testing and meta-analysis in another continental African sample from Kenya. Further replication testing was performed in an African American sample from the Atherosclerosis Risk in Communities (ARIC) study. We found two African-ancestry specific variants that were significantly associated with MetS: SNP rs73989312[A] near CA10 that conferred increased risk (P=3.86x10−8, OR=6.80) and SNP rs77244975[C] in CTNNA3 that conferred protection against MetS (P=1.63x10−8, OR=0.15). Given the exclusive expression of CA10 in the brain, our CA10 finding strengthens previously reported link between brain function and MetS. We also identified two variants that are not African specific: rs76822696[A] near RALYL associated with increased MetS risk (P=7.37x10−9, OR=1.59) and rs7964157[T] near KSR2 associated with reduced MetS risk (P=4.52x10−8, Pmeta=7.82x10−9, OR=0.53). The KSR2 locus displayed pleiotropic associations with triglyceride and measures of blood pressure. Rare KSR2 mutations have been reported to be associated with early onset obesity and insulin resistance. Finally, we replicated the LPL and CETP loci previously found to be associated with MetS in Europeans. These findings provide novel insights into the genetics of MetS in Africans and demonstrate the utility of conducting trans-ethnic disease gene mapping studies for testing the cosmopolitan significance of GWAS signals of cardio-metabolic traits.
Keywords: Metabolic syndrome, Pleiotropy, Genome-wide association study, African ancestry
1. Introduction
The metabolic syndrome (MetS) manifests as a clustering of risk factors including abdominal obesity, atherogenic dyslipidemia, elevated blood pressure, insulin resistance, and prothrombotic and proinflammatory states [1, 2]. Individuals with MetS have at least a fivefold increased risk of developing type 2 diabetes (T2D), a twofold increased risk of cardiovascular diseases [3], and increased susceptibility to several other disorders including fatty liver disease [4], sleep apnea [5], and some forms of cancer [6].
Recent studies suggest that complex networks of metabolic pathways modulated by interacting genetic and environmental factors underlie MetS [7, 8]. Early evidence for the potential contribution of genetics to MetS was provided in a seminal study of twin pairs with 31.6% of monozygotic twin pairs compared to 6.3% of dizygotic pairs displaying concordance for clustering of three MetS components (hypertension, diabetes, and obesity) [9]. Heritability estimates for MetS vary, with estimates as low as 13% in the Dutch [10], 24% in Caribbean-Hispanic families [10, 11], and as high as 48% in Omani families [12]. Earlier genome-wide linkage studies reported links between MetS and several chromosomal regions including 1p34.1, 10p11.2 and 19q13.4 [13], 1q21–q25 [14], and 1q23–q31 [15]. Candidate gene and whole exome sequencing studies have also identified mutations in patients with rare forms of MetS [16, 17]. Meta-analyses of genome-wide association studies (GWAS) in Europeans and Asians have reported associations between MetS and variants in or near several loci including ZPR1, BUD13, APOA5, LPL, and CETP genes [18–20].
In contrast to the growing success in the identification of variants associated with the individual components of MetS, little progress has been made in the identification of variants underlying the syndromic clustering of the component traits of MetS and variants with pleiotropic effect that may shed light on dysregulated pathways in MetS [21, 22]. Furthermore, the prevalence of MetS shows ethnic disparity in individuals of African descent. For example, analysis of the US National Health and Nutrition Survey (NHANES) serial data from 1999–2000 to 2009–2010 revealed modest decline in prevalence of MetS in Caucasians (25.6% to 21.8%) but a slight increase in African-Americans (22.0% to 22.7%) [23, 24]. Paradoxically, the high prevalence of hypertension and diabetes in African-Americans contrasts with the observed low prevalence of high triglyceride levels [25]. Low prevalence of high triglyceride levels is also observed in west Africans, the ancestral populations of African Americans despite dietary and other differences between the two groups. These observed ethnic disparities in the burden of MetS [25] and other cardiometabolic traits [26] persists even after adjustment for modifiable risk factors, implying the role of background genetic predisposition.
In this study, we performed a GWAS of MetS in continental Africans enrolled from Ghana and Nigeria (AF1), and replication testing and meta-analysis with another continental African sample from Kenya (AF2) using ~15 million directly genotyped and imputed single nucleotide polymorphisms (SNPs). Further replication was tested in an African American sample from the Atherosclerosis Risk in Communities (ARIC) study. We also performed a GWAS of MetS in a subset of the samples in the tails of the continuous metabolic syndrome risk scores (cMetS) derived from the sum of standardized residuals of MetS component traits.
2. Materials and methods
2.1. Study samples
Individuals included in this study were participants enrolled in the Africa America Diabetes Mellitus (AADM) study with centers in Ghana and Nigeria (AF1) and Kenya (AF2) [27]. Although the AADM study has been ongoing for over a decade, the majority of participants included in the present analysis was recruited in the year 2008. The study populations, data collection procedures, and ethical processes have been described in detail elsewhere [27, 28].
For developing continuous metabolic syndrome scores and testing their predictive accuracy, we analyzed all 4,820 individuals in the cohorts with non-missing phenotype values (4,023 AF1 and 797 AF2). The discovery genome-wide association analysis was done in 1,427 AF1. Independent replication of genome-wide significant loci was tested in 174 AF2 and 2,475 African Americans enrolled in the ARIC study.
2.2. MetS phenotypes
Based on the definition of the National Cholesterol Education Program (NCEP) improved threshold, an individual was considered to have MetS if they have the following measures for three or more of the five component traits [1]: waist circumference ≥ 102 cm for men or ≥ 88 cm for women; fasting plasma glucose ≥ 100 mg/dL; plasma triglyceride levels ≥ 150 mg/dL; HDL cholesterol < 40 mg/dL for men or < 50 mg/dL for women; systolic BP ≥ 130 mmHg or diastolic BP ≥ 85 mmHg. In our analysis, cases were individuals with MetS and controls were individuals without MetS. We also developed a continuous metabolic syndrome risk scores (cMetS). Previous studies used different approaches including Z-scores, principal components, and percentile rankings to derive cMetS; the scores obtained by using these methods displayed strong correlation with one another [29]. We developed cMetS using the sum of the standardized scores from the components of MetS. Prior to deriving cMetS, normality of each trait was assessed using descriptive statistics and distribution plots (histograms and scatter plots). Plasma glucose and triglyceride values were log10-transformed because their distributions did not conform to normality. Next, standardized residuals were obtained by regressing each trait on age, sex, and broad ethnicity. The sign of the standardized residuals for HDL was reversed so that higher values indicate greater risk of metabolic syndrome. Finally, all standardized residuals of MetS component traits were summed to form cMetS. The higher the cMetS, the higher the tendency for the components to cluster indicating a higher MetS risk. To evaluate the accuracy of cMetS, we developed another continuous metabolic syndrome score based on principal components analysis of MetS component traits. Previous studies have shown that the power of detecting pleiotropic effects of genetic variants on correlated traits can be enhanced by reducing the dimension of the original traits into a single trait using principal components analysis and subsequent analysis of the top principal component in place of the original traits[30, 31]. We used the loadings of the first principal component (PC1) that explain most of the total trait variance as PC-based continuous metabolic syndrome score (pc1MetS).
2.3. Genotyping, quality control, and imputation
The discovery GWAS was done using 1,637 unrelated AF1 in the AADM study genotyped on the Affymetrix Axiom® PANAFR SNP array. The array was chosen because it offers ≥90% genetic coverage of variants with minor allele frequency (MAF) > 2% of the Yoruba (West African) genome and >85% coverage of the Luhya and Maasai (East African) genome. DNA sample preparation and quality control were done in our laboratory at the National Institutes of Health (NIH). We excluded 210 samples (one duplicated, three sex discordant, and 206 with one or more missing MetS component trait values), leaving 1,427 AF1 for the discovery GWAS. Replication of top signals and meta-analysis were done using 185 unrelated AF2 genotyped on the Affymetrix Axiom® PANAFR SNP array. We excluded 11 samples (10 sex discordant, and one with one or more missing MetS component trait values), leaving 174 AF2 for independent replication and meta-analysis. Additional replication was done using 2,612 African Americans in the ARIC study with ~24 million imputed SNP dosage data accessed through the Database of Genotypes and Phenotypes. We excluded 137 samples with one or more missing MetS component trait values, leaving 2,475 ARIC study individuals for replication.
Of the 2,217,748 SNPs in the raw genotype data of the AADM cohort, we excluded 46,562 SNPs that had a minor allele frequency (MAF) of <1%, 22,509 that were missing in more than 10% of individuals, 10,282 that had a Hardy-Weinberg p-value <1x10−6, and 61,087 non-autosomal SNPs. The remaining 2,077,552 SNPs that passed quality filers were used as the basis for imputation. Imputation was performed using the 1000 Genomes Consortium phase 1, version 3 cosmopolitan reference using the programs MaCH [32]/MaCH-ADMIX [33]. The resulting imputed dosage data were filtered for imputed allelic dosage frequency < 0.01 and r2 < 0.3, yielding ~15 million SNPs for analysis.
We generated principal components (PCs) from multi-dimensional scaling analysis of a pruned set of uncorrelated genome-wide SNPs (i.e., r2 threshold of 0.2, and a sliding window of 50 SNPs by skipping 5 SNPs between consecutive windows) as implemented in the program PLINK. A plot of the first three PCs is shown in Fig. S1. To minimize the effect of population structure in the GWAS, all analyses were adjusted for the first three PCs in AF1 and AF2 and the first two PCs in AA. Quantile-quantile plot of the distribution of the test statistic was consistent with the expected distribution under the null hypothesis (Fig. S2), and the genomic control inflation was 1.014 (AF1) and 1.017 (AF2), indicating a lack of population structure. The power of the study at different scenarios is shown in Fig. S3.
2.4. Statistical analyses
Statistical significance of differences between groups was tested using student’s t-test for continuous variables and the chi-square test for discrete variables. The level of significance was set at 0.05.
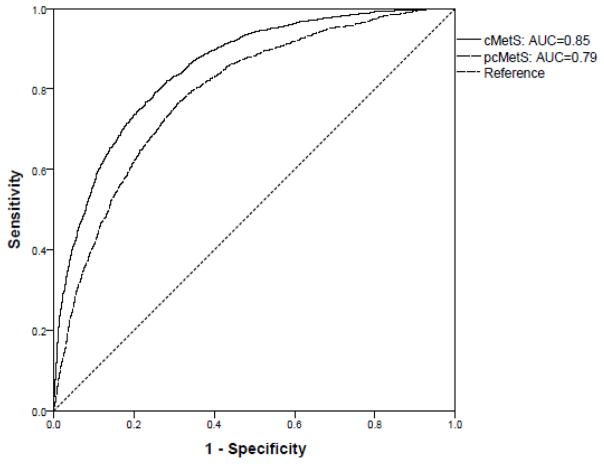
The ability of the cMetS variable to be discriminative between individuals with and without MetS was evaluated using a receiver operating characteristic (ROC) curve. The area under the curve (AUC) of the ROC curve was estimated using a nonparametric approach [34] to determine the probability that cMetS was higher in individuals with MetS than those without MetS.
The imputed SNP dosage data were analyzed using a logistic regression model with adjustment for age, sex, and the first three PCs as implemented in mach2dat. For association analyses of MetS component traits, in addition to the above covariates, adjustment was done for BMI and the analysis was performed using linear regression models under the additive genetic model as implemented in mach2qtl. Genome-wide significance was set as P-value < 5x10−8.
Meta-analysis of the discovery and replication datasets was done using the METAL software program [35]. We combined the evidence of association in two sided p-values using a fixed-effects model. Sample size of each SNP was used as a weight, and the sign of the beta value was used as the direction of association of the effect allele. Evidence for the between-study heterogeneity was tested using Cochran’s Q statistic and I2, and the P-value of the chi-square was used to declare significance [36].
3. Results
3.1. Characteristics of the study participants
Characteristics of the AF1 subjects included in the discovery GWAS and the AF2 subjects included in the replication testing is presented in Tables S1 & S2. Of the 1,427 AF1, 602 were cases (i.e., had MetS) and 825 were controls (i.e., did not have MetS). Of the 174 AF2, 118 were cases and 56 were controls. On average, subjects with MetS were ~3 years older than those without MetS (Tables S1 & S2); as a result, age was adjusted for in all analyses models. The prevalence of MetS was 35.6% in the overall sample, and was greater in women than men (43.2% vs. 25.0%; P<0.001) (Tables 1 & S3).
Table 1.
Characteristics of subjects included in developing and testing accuracy of the continuous metabolic syndrome score
| All (n=4,820) | Sample AF1 (Ghana and Nigeria) (n=4,023) | Sample AF2 (Kenya) (n=797) | P-valuea | |
|---|---|---|---|---|
| Mean ± SD | ||||
| Age, year | 47.33±15.64 | 45.73±16.02 | 55.43±10.33 | <0.001 |
| Waist circumference, cm | 88.86±13.07 | 88.53±12.94 | 90.55±13.61 | <0.001 |
| Systolic BP, mmHg | 134.13±23.65 | 133.56±23.76 | 137.00±22.87 | <0.001 |
| Diastolic BP, mmHg | 80.05±13.98 | 79.39±14.27 | 83.37±11.88 | <0.001 |
| Plasma glucose, mg/dL | 127.22±78.62 | 125.69±75.35 | 134.97±93.05 | 0.008 |
| HDL cholesterol, mg/dL | 46.50±16.92 | 46.24±16.89 | 47.83±17.04 | 0.016 |
| Triglyceride, mg/dL | 107.43±73.13 | 94.24±51.18 | 173.97±117.59 | <0.001 |
| cMetSb | −0.028±3.20 | −0.06±3.14 | 0.14±3.46 | 0.095 |
| pcMetSc | −0.016±1.00 | −0.04±1.00 | 0.09±1.02 | 0.001 |
| n (%) | ||||
| MetSd | 1716 (35.6) | 1318 (32.8) | 398 (49.9) | <0.001 |
| Women | 2811 (58.3) | 2389 (59.4) | 422 (52.9) | <0.001 |
Based on t-test for differences of means and Pearson’s chi-square test statistic for differences of proportions between AF1 (sample from Ghana and Nigeria) and AF2 (sample from Kenya).
cMetS: continuous metabolic syndrome score based on sum of standardized components of the metabolic syndrome.
pcMetS: continuous metabolic syndrome score based on the first principal component loadings of the metabolic syndrome components in principal components analysis.
MetS: the metabolic syndrome (present/absent) based on definition of the National Cholesterol Education Program (NCEP).
Characteristics of the study participants included in the development of cMetS and testing of its predictive accuracy are presented in Tables 1, S3 & S4. Our assessment of cMetS showed that it is a valid predictor of MetS as follows: (i) cMetS for individuals with MetS was significantly higher than for those without MetS (mean difference 3.86, P<0.001) (Table S5); (ii) cMetS was strongly correlated with another continuous metabolic syndrome score metric derived from the first principal component loadings of MetS component traits (Pearson correlation (r) =0.92, Fig. S4); (iii) the AUC of cMetS corresponding to MetS was 0.85 (95% CI 0.84–0.87) (Figs. 1 & S5); and (iv) cMetS was moderately correlated with individual component traits of MetS ranging from a correlation coefficient (r) of 0.40 for fasting plasma glucose to 0.62 for diastolic blood pressure (Table S6).
Fig. 1.
Receiver operating characteristics (ROC) curves of continuous metabolic risk scores corresponding to the metabolic syndrome.
3.2. Genome-wide significant associations
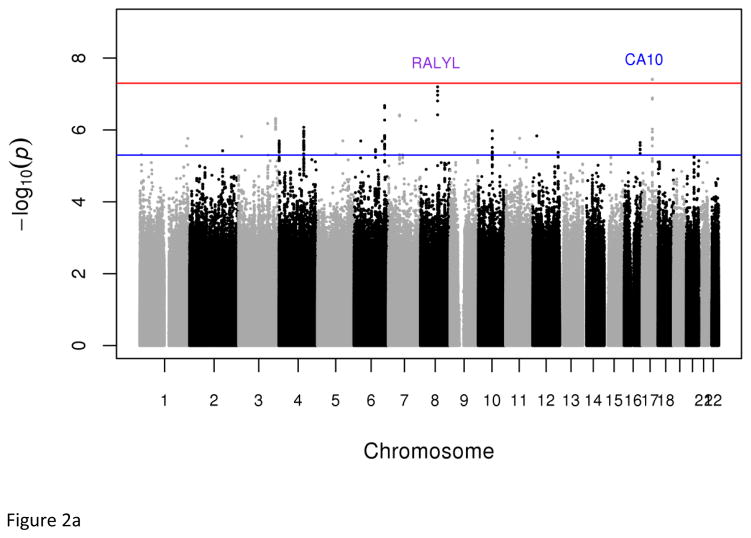
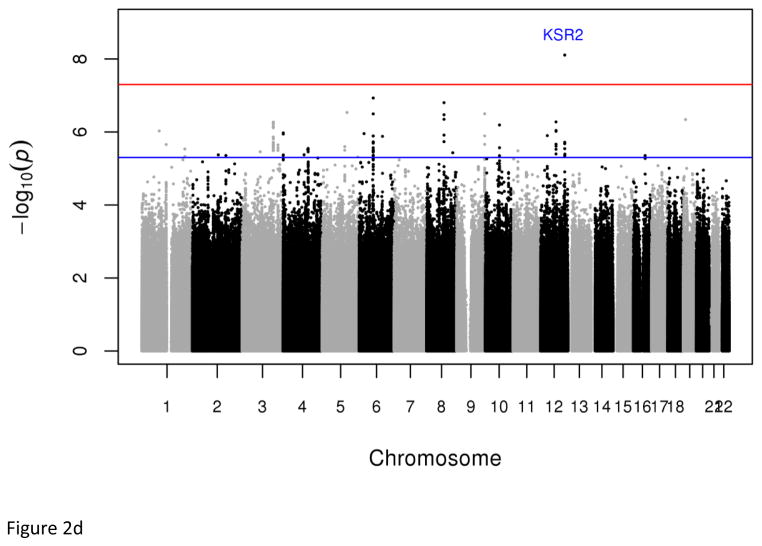
In the discovery sample, two strongly correlated low-frequency variants [rs73989312 (1.64%) and rs73989319 (1.63%)] near CA10 conferred a more than six-fold increased risk of developing MetS (P=3.86x10−8, OR=6.80; and P=3.97x10−8; OR=6.85 respectively) (Figs. 2 & S6, Tables 1 & S7). These variants had frequencies of 1.70% and 1.64% respectively in the 1000 Genomes west African ancestry samples (YRI- Yoruba in Ibadan, Nigeria and ASW- African ancestry in Southwest USA), but were absent in all non-African and east African (LWK - Luhya in Webuye, Kenya) samples. In general, we observed that variant frequencies were similar between our samples and those of west African ancestry samples in the 1000 Genomes dataset, validating the genotype accuracy in our datasets (Table S8).
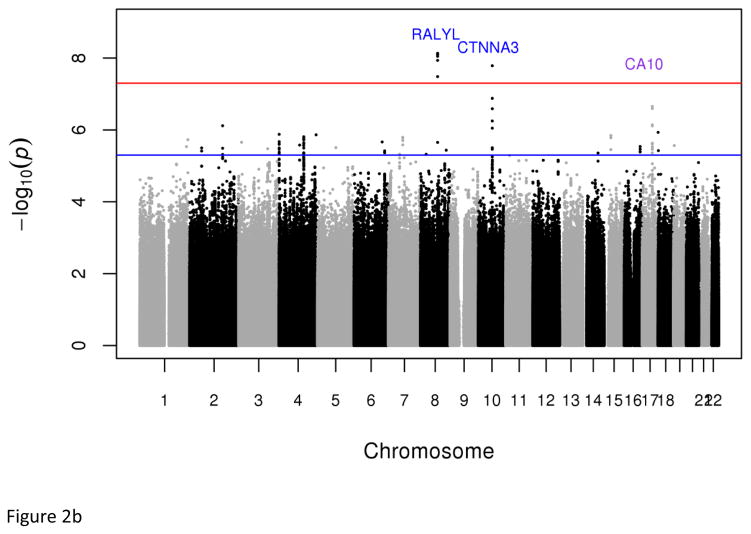
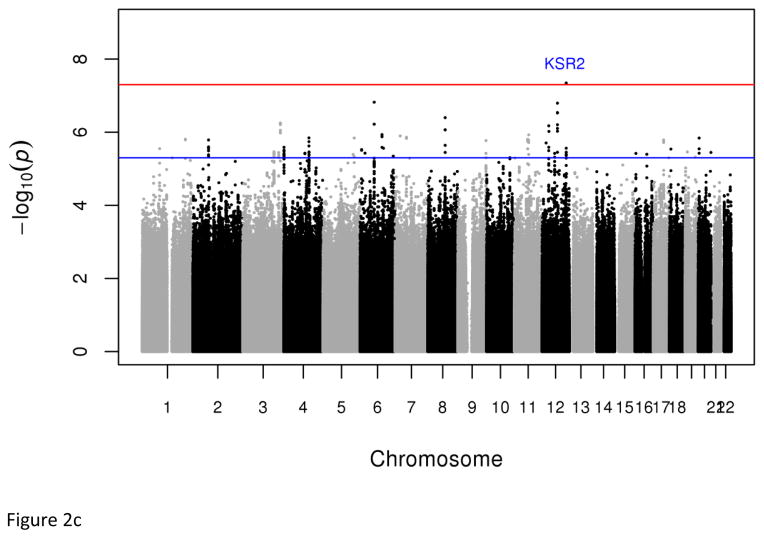
Fig. 2.
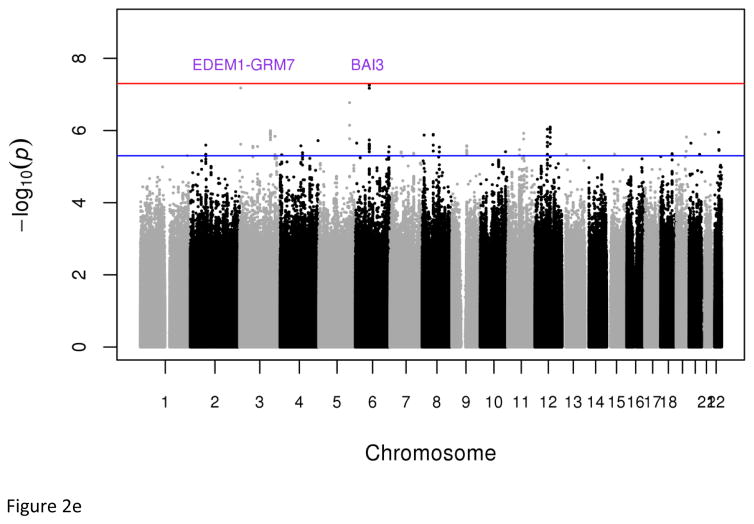
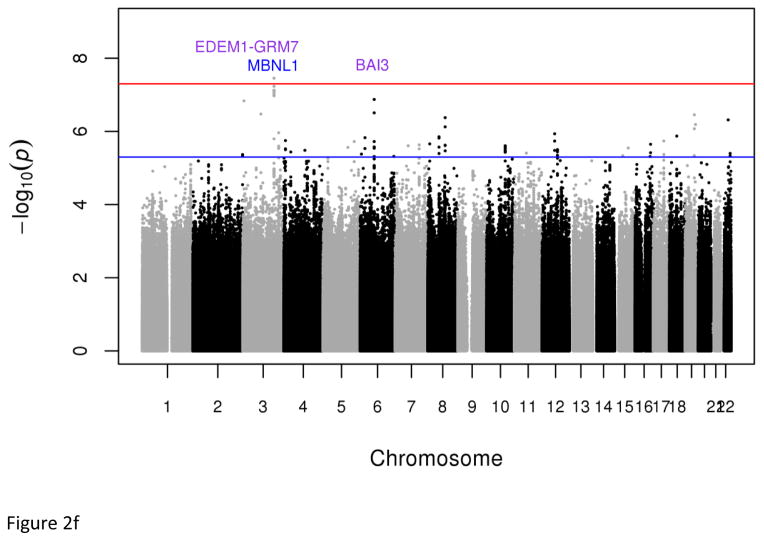
Plots of genome-wide association of single nucleotide polymorphisms with the metabolic syndrome. Panels A and B represent plots from analyses of the full data set from the discovery GWAS and meta-analysis, respectively. Panels C and D represent plots from analyses of the two 33% tails of the data set from the discovery GWAS and meta-analysis, respectively. Panels C and D represent plots from analyses of the two 25% tails of the data set from the discovery GWAS and meta-analysis, respectively. Each single nucleotide polymorphism (SNP) is represented by a point, with higher points (higher negative log10 P values), showing more significant association. Points above the blue horizontal line represent SNPs with a P value of less than 10−5 and points above the red horizontal line indicate SNPs with a P value of less than 5×10−8.
The meta-analysis of AF1 and AF2 identified six variants with genome-wide significant associations with MetS: rs77244975 in the intron of CTNNA3 (P=1.63x10−8, OR=0.15, C allele frequency=1.71%) and five strongly correlated (r2>=0.8) common variants (frequencies from 53.61% to 58.54%) near RALYL (lead SNP=rs76822696, P=7.37x10−9, OR=1.59) (Tables 1 & S9). The CTNNA3 rs77244975[C] variant was present at low frequency (<3%) in the 1000 Genomes African ancestry samples (YRI, ASW, and LWK) but was absent in all non-African samples (Table S8). The rs77244975[C] in CTNNA3 (allele frequency 1.82%) was successfully replicated in African Americans (ARIC study) with directionally consistent significant association with MetS (P=0.011, OR=0.52).
Dichotomous analyses of the extreme tails (⅓ −33.3% and ¼ – 25%) of the empirical distribution of cMetS identified two genome-wide significant associations; the first is an intronic SNP (rs7964157) in KSR2 (Pdiscovery =4.52x10−8, Pmeta=7.82x10−9, OR=0.53, frequency=29.73%) and the second is a SNP (rs146816516) near MBNL1 (Pmeta=3.51x10−8, OR=0.22, G allele frequency=3.95%) (Tables 1, S7 & S9). The rs146816516[G] variant was absent in the 1000 Genome’s non-African samples (Table S8). We also identified variants with suggestive genome-wide significant associations (5x10−8 < P < 9x10−8) including rs9354671 (P=5.56x10−8) and rs2061117 (P=6.74x1−8) near BAI3 and rs149307971 (P=6.66x10−8) near EDEM1-GRM7 (Table S7).
3.3. MetS-associated variants displaying pleiotropic effects
The GWAS significant MetS variants and neighboring SNPs within ± 100kb were tested for association with the individual traits used to define the MetS. We found several SNPs near or within two genes displaying Bonferroni corrected significant pleiotropic associations: 1) KSR2 SNPs were associated with both triglycerides and measures of blood pressure; 2) MBNL1 SNPs were associated with BMI and measures of blood pressure.
3.4. Replication of European MetS meta-analyses GWAS loci in African ancestry individuals
We used the local replication strategy to test whether three loci reported to be associated with MetS in Europeans were also associated with MetS in our African ancestry samples. The index European associated SNPs along with neighboring (± 100kb) variants were evaluated for replication [18, 19]. Correlated variants (r2≥0.4 in the 1000 Genomes’ CEU samples) in two of the three loci were replicated by five SNPs: LPL (rs294, P=8.1x10−3; rs4523270, P=2.2x10−2; rs2165558, P=2.3x10−2; rs35237252, P=3.3x10−2) and CETP (rs4783961, P=1.3x10−2). The BUD13-ZPR1-APOA5 locus did not replicate, mainly because the variants showing significant associations in Europeans are very rare in our African-ancestry population dataset (e.g., rs10790162 and rs2266788 have minor allele frequencies of 0.0045 and 0.0028 in our dataset). Directionally consistent and stronger replication P-values were achieved with meta-analysis of the west Africans (AF1) and African Americans (ARIC) (LPL: rs294, P=3.5x10−3; CETP: rs4783961, P=3.0x10−4). Interestingly, we found neighboring SNPs uncorrelated with the index SNPs that had lower P-values: rs306 (LPL, P=8.16x10−4), rs734104 (APOC3, P=1.52x10−4), and rs149959798 (NLRC5, P=1.75x10−3) (Table S10). Notably, the rs306[A] allele associated with reduced risk of MetS was present in African ancestry populations at a frequency of about 10% but was absent in non-African 1000 Genomes’ samples. In the ENCODE and Roadmap projects, these variants have been shown to be enhancer histone marks: rs306 in left ventricle and adipose cells, and rs734104 in liver, adipose and pancreatic cells.
4. Discussion
This first GWAS of MetS in continental Africans identified novel common and low-frequency variants that conferred increased risk as well as protection against MetS. Two of these variants (CA10 and CTNNA3) are African ancestry specific. The identified variants are functionally plausible given their regulatory functional predictions and their locations in or near genes involved in glucose homeostasis (KSR2), adipocyte differentiation (RALYL), brain function (CA10, EDEM1-GRM7 locus in 3p26–25 & BAI3), and cardiac and skeletal muscle contractions (CTNNA3, MBNL1). KSR2 and MBNL1 variants displayed pleiotropic properties given their association with two or more cardio-metabolic traits. In addition to these novel findings, we replicated the LPL and CETP loci previously found to be associated with MetS in European ancestry individuals providing evidence for the trans-ethnic properties of these variants.
The rs76822696 A variant (RALYL) associated with increased risk of MetS is predicted to alter binding motifs of C/EBPβ and the glucocorticoid receptor that modulate adipocyte differentiation by regulating the transcriptional induction of PPARG [37, 38]. PPARG is a master regulator of adipogenesis and rare variants in PPARG that reduce adipocyte differentiation have been associated with increased risk of T2D, a major consequence of MetS [39]. Our finding parallels the hypothesis that reduced adipocyte differentiation leads to diabetes [40], and further suggests that reduced adipocyte differentiation attributed to a novel common variant in RALYL increases the risk of the metabolic syndrome.
We found a common variant (rs7964157) in KSR2 associated with a 47% reduced risk of developing MetS. KSR2 (Kinase suppressor of Ras 2) is a molecular scaffold of the RAS-MEK-ERK pathway that regulates glucose uptake and glycolysis [41, 42], and promotes activation of the AMPK enzyme that regulates cellular energy homeostasis [43]. Rare KSR2 loss-of-function mutations have been linked with early onset obesity and insulin resistance in humans [44]. These KSR2 mutations impair the RAS-MEK-ERK pathway similar to gain-of-function mutations in DYRK1B that was recently associated with a form of metabolic syndrome characterized by early onset coronary artery disease, central obesity, hypertension, and diabetes [17]. Individuals carrying the KSR2 loss-of-function mutations exhibited childhood hyperphagia, reduced basal metabolic rate, and severe insulin resistance [44]. Recapitulating these metabolic phenotypes observed in humans, mice that are deficient in Ksr2 develop profound obesity [43, 45], glucose intolerance, and insulin resistance in liver, muscle and adipose tissues [46, 47], possibly due to reduced AMPK activation leading to impairment of fatty acid oxidation and enhancement of triglyceride storage [43]. Identification of a novel variant associated with MetS in our study showed that, in addition to the previously identified rare KSR2 mutations [44], common variants in KSR2 contribute to metabolic disorders. Our KSR2 GWAS locus rs7964157 is predicted to alter binding motifs of Maf, a family of transcription factors involved in regulating pancreatic β cell function, and β-cell-specific insulin gene transcription (ENCODE/RoadMap project). MafA knockout mice displayed pancreatic islet abnormalities and glucose intolerance, and developed T2D [48].
An important pleiotropic role of the KSR2 locus is shown by our findings of statistically significant associations of KSR2 variants with MetS component traits (systolic, diastolic blood pressure, and triglycerides) (Table 3) as well as published associations with LDL cholesterol and waist circumference (Tables S13 & S14). Previously, we reported linkage signals at chromosome 12q24, the locus harboring KSR2, in a genome-wide linkage study of T2D in a cohort of 343 affected sibling pairs from west Africa [27]. Other family-based studies have also linked the 12q23–24 locus to obesity [49–51], T2D [52–54], percent body fat [55], and total cholesterol [56]. Genome-wide studies have found suggestive associations (that fell short of genome-wide significance threshold) of KSR2 SNPs with visceral adipose tissue [57], LDL cholesterol [58], and total cholesterol [58] (Table S11). A recent meta-analysis reported a pleiotropic effect of a nearby locus in 12q24.31 (rs12310367 in ZNF664) for lipid and inflammatory traits (triglycerides, HDL cholesterol, and adiponectin) [22]. Taken together, our finding and those of others provide strong evidence in support of the pleiotropic properties of the KSR2-dependent signaling.
Table 3.
Metabolic syndrome-associated loci showing Bonferroni-corrected significant association with individual components of the MetS and related cardiometabolic traitsa
| Gene | MetSb GWAS SNP (chr: position) | Replication trait | Replicated SNP | Position | Alleles | A1c freq | P-value | Data set |
|---|---|---|---|---|---|---|---|---|
| MBNL1 | rs146816516 (chr3:152262078) | Systolic BP | rs76204242 | 152291018 | A,C | 0.98 | 3.74E-05 | A |
| Body Mass Index | rs116815255 | 152192393 | T,A | 0.98 | 1.29E-05 | B | ||
| Body Mass Index | rs145202083 | 152193084 | C,A | 0.98 | 1.32E-05 | B | ||
| Body Mass Index | rs150947994 | 152195940 | T,A | 0.98 | 1.25E-05 | B | ||
| Body Mass Index | rs114991118 | 152199365 | T,A | 0.98 | 1.16E-05 | B | ||
| Body Mass Index | rs184693776 | 152201330 | C,T | 0.98 | 1.07E-05 | B | ||
| Body Mass Index | rs114284899 | 152203823 | G,T | 0.98 | 1.32E-05 | B | ||
| Body Mass Index | rs192244039 | 152209252 | C,T | 0.98 | 1.17E-05 | B | ||
| Body Mass Index | rs183566325 | 152221527 | G,T | 0.98 | 2.66E-05 | B | ||
| Body Mass Index | rs6804557 | 152334938 | C,A | 0.98 | 1.44E-05 | B | ||
| Body Mass Index | rs6773118 | 152339917 | C,T | 0.98 | 1.05E-05 | B | ||
| KSR2 | rs7964157 (chr12:117982220) | Systolic BP | rs78683574 | 117980191 | T,A | 0.89 | 1.39E-05 | A |
| Systolic BP | rs7964157 | 117982220 | C,T | 0.70 | 3.56E-05 | A | ||
| Diastolic BP | rs7964157 | 117982220 | C,T | 0.70 | 3.56E-05 | A | ||
| Systolic BP | rs1501635 | 117973298 | T,C | 0.60 | 2.70E-05 | B | ||
| Systolic BP | rs78683574 | 117980191 | T,A | 0.89 | 1.35E-05 | B | ||
| Triglycerides | rs11610896 | 117973915 | A,G | 0.64 | 1.70E-05 | B | ||
| Triglycerides | rs7964157 | 117982220 | C,T | 0.70 | 9.92E-06 | B | ||
| Triglycerides | rs11610896 | 117973915 | A,G | 0.64 | 1.14E-05 | C | ||
| Triglycerides | rs7964157 | 117982220 | C,T | 0.70 | 2.06E-05 | C | ||
| EDEM1-GRM7 | rs149307971 (chr3:5854876) | HDL cholesterol | rs1377212 | 5767686 | G,C | 0.27 | 7.20E-06 | A |
| HDL cholesterol | rs1377212 | 5767686 | G,C | 0.27 | 1.54E-05 | B | ||
| HDL cholesterol | rs1377212 | 5767686 | G,C | 0.27 | 1.71E-05 | C | ||
| HDL cholesterol | rs116357511 | 5769812 | G,T | 0.87 | 2.16E-05 | B | ||
| HDL cholesterol | rs116357511 | 5769812 | G,T | 0.87 | 2.11E-05 | C | ||
| CTNNA3 | rs77244975 (chr10:69207975) | Waist circumference | rs188455459 | 69273893 | C,T | 0.98 | 1.40E-05 | B |
SNPs within 100kb distance from the variants showing genome-wide significant association with MetS were tested for association with components of MetS in the discovery datasets. Here we reported those showing Bonferroni-corrected (0.05/number of SNPs tested in the respective locus) significant association.
MetS: the metabolic syndrome based on definition of the National Cholesterol Education Program (NCEP).
A1 freq: frequency of the first of the two alleles listed under the ‘Alleles’ column in the West African discovery dataset.
Position: base-pair position in NCBI Build 37 coordinates
A: Full dataset (602 cases from Ghana and Nigeria (AF1), 825 AF1 controls)
B: 33.3% tails of cMetS (402 AF1 cases, 550 AF1 controls)
C: 25% tails of cMetS (310 AF1 cases, 405 AF1 controls)
cMetS: continuous metabolic syndrome score based on the sum of standardized residuals of the metabolic syndrome component traits.
Two Africa-specific GWAS variants we found in the CTNNA3 and MBNL1 genes that modulate cardiac muscle contraction are likely relevant to the understanding of the patho-biology of heart diseases. The alpha-T-catenin protein encoded by CTNNA3 is involved in muscle contraction and is predominantly expressed in the heart [59]. Published GWAS reported suggestive associations between CTNNA3 variants and serum levels of the cardiac function markers alanine aminotransferase/aspartate aminotransferase [60] (Tables S11 & S12). Furthermore, the 10q21 locus harboring CTNNA3 is a common fragile site linked to dilated cardiomyopathy [61]. Similarly, MBNL1 is highly expressed in cardiac muscle [62]. MBNL1 inhibits pre-mRNA expression of cardiac troponin T (cTNT) that regulates contractile function of cardiac muscle [63, 64]. Overexpression of MBNL1 in muscle cells reduces inhibition of insulin receptor (IR) protein [63, 65] leading to enhanced glucose uptake in skeletal muscle [66]. According to the ENCODE project, our GWAS associated variant rs146816516 [G] in MBNL1 is predicted to alter binding motifs of Foxa and STAT transcription factors that regulate glucose homeostasis [67] and adipocyte metabolism [68]. SNP rs116264158 which is in strong LD (r2=0.87) with rs146816516 has been shown to be a promoter/enhancer histone mark in adipose and skeletal muscle cells, in a DNAse hypersensitivity site in pancreatic islets, and is predicted to alter binding motifs of the glucocorticoid receptor protein. SNP rs150505636, which is strongly correlated with rs146816516 (r2=0.87), is a weak enhancer in pancreatic islets (ENCODE and RoadMap).
Our finding that variants near CA10 (Carbonic anhydrase X) substantially increased the risk (OR=6.80) of developing MetS provides evidence in support of the reported pathophysiological link between brain function and metabolic syndrome. CA10 is exclusively expressed in the brain and is considered to have an essential, yet unknown, functional role because it is highly conserved across animal species [69]. The encoded CA10 protein is catalytically inactive due to absence of zinc-binding histidine residues. When the missing histidines are restored in the sequence, the mutated CA10 exhibits high catalytic activity [70]. Most carbonic anhydrases (CAs) are involved in important biosynthetic processes including lipogenesis and gluconeogenesis. Inhibition of mitochondrial CA isozymes using sulfonamide analogues such as the antiepileptic drugs topiramate (TPM) and zonisamide (ZNS) induces weight loss [71–75]. FDA approved chronic weight management drug Qsymia (phentermine and TPM extended-release) leads to weight loss, and improvements in weight-related comorbidities such as improved glycemia, decreased blood pressure, and improved cholesterol levels [76]. The rs73989312 [A] variant that we found to be associated with a substantially increased risk (OR=6.80) of MetS is predicted to alter binding motifs of transcription factor activator protein-2 (AP-2) (ENCODE). The AP-2 transcription factors are involved in the pathogenesis of metabolic diseases through dysregulation of adipocyte function [77] and regulates nervous system and other developmental processes [78]. The obesity-linked Drosophila homolog TfAP-2 is expressed in octopaminergic neurons that regulate satiation and the mouse homolog AP-2β is located in parts of the brain involved in modulating feeding behavior [79].
Our EDEM1-GRM7 and BAI3 findings provide additional evidence for the link between neural connectivity in the brain and MetS. GRM7 is the major excitatory neurotransmitter in the central nervous system, and BAI3 regulates excitatory synapses and is involved in brain angiogenesis. EDEM1 is directly involved in endoplasmic reticulum-associated degradation (ERAD) of misfolded proteins. Dysfunctional ERAD mediates the link between chronic ER stress and impairment of cellular functions that lead to metabolic dysregulations and neurodegenerative diseases [80–82]. The 3p26–25 locus that harbors the EDEM1-GRM7 region has been shown to have pleiotropic effects on obesity, lipids, and blood pressure traits [83]. These SNPs are located 3kb-17kb from the less frequent Africa-specific SNP (rs149307971) that showed association with MetS in our study. In all, our findings open novel research areas for understanding the genetic basis of the roles of the brain in regulating energy metabolism, body fat content, and glucose metabolism and the link between defects in brain function and metabolic disorders [84].
We acknowledge that our replication sample from Africa is small because of the paucity of well phenotyped genome-wide data from continental Africa. Despite this limitation, our study identified novel low frequency and common variants in genes with plausible pathophysiological role in metabolic and cardiovascular diseases. The functional diversity of the proteins encoded by the identified genes including glucose homeostasis, adipocyte differentiation, cardiac and muscle contractions, neuro-endocrine signaling, brain and pancreatic β cell function indicates that the underlying pathology in MetS is a complex network of dysregulated pathways. The identification of African ancestry specific variants influencing MetS is noteworthy for several reasons including providing strong supportive evidence for including multi-ethnic global populations in disease mapping strategies to facilitate discovery of novel variants influencing complex traits. Finally, we showed that a continuous measure of metabolic syndrome (cMetS) can be used to identify individuals at the extremes of metabolic disease risk profile in GWAS, and that cMetS is a valid predictor of MetS in continental Africans as previously observed in other ancestral populations [85–87].
Supplementary data to this article can be found online at Molecular Genetics and Metabolism.
Supplementary Material
Table 2.
SNPs showing genome-wide significant association with the metabolic syndrome
| Variant | Chr | Position | Frequency (%) | Nearest gene | P-value | ORa | SE |
|---|---|---|---|---|---|---|---|
|
Discovery
| |||||||
| Full datasetb | |||||||
| rs73989312[A]c | 17 | 50464800 | 1.64 | CA10 | 3.86E-08 | 6.80 | 0.398 |
| rs73989319[C]c | 17 | 50477498 | 1.63 | CA10 | 3.97E-08 | 6.85 | 0.401 |
| Extreme 33.3% tails of cMetSd | |||||||
| rs7964157[T] | 12 | 117982220 | 29.73 | KSR2 | 4.52E-08 | 0.53 | 0.121 |
|
| |||||||
|
Meta-analysis
| |||||||
| Full dataset | |||||||
| rs77244975[C] | 10 | 69207975 | 1.71 | CTNNA3 | 1.63E-08 | 0.15 | 0.430 |
| rs76822696[A] | 8 | 84929136 | 53.61 | RALYL | 7.37E-09 | 1.59 | 0.080 |
| rs16912410[T] | 8 | 84926391 | 55.42 | RALYL | 8.00E-09 | 1.56 | 0.077 |
| rs62526240[G] | 8 | 84927369 | 56.22 | RALYL | 8.94E-09 | 1.58 | 0.079 |
| rs55752635[A] | 8 | 84926274 | 56.46 | RALYL | 1.16E-08 | 1.55 | 0.077 |
| rs188622356[T] | 8 | 84931781 | 58.54 | RALYL | 3.28E-08 | 1.56 | 0.080 |
| Extreme 33.3% tails of cMetS | |||||||
| rs7964157[T] | 12 | 117982220 | 29.73 | KSR2 | 7.82E-09 | 0.52 | 0.116 |
| Extreme 25% tails of cMetSe | |||||||
| rs146816516[G] | 3 | 152262078 | 3.95 | MBNL1 | 3.51E-08 | 0.22 | 0.363 |
Chr: chromosome; Position: base-pair position in NCBI Build 37 coordinates; OR: odds ratio; SE: standard error
In the meta-analysis, odds ratio was calculated using the SCHEME STDERR command that weights effect size estimates using the inverse of the corresponding standard errors as implemented in METAL
Full dataset (602 cases from Ghana and Nigeria (AF1 sample), 825 AF1 controls)
This has not replicated because the variant allele is absent in east African populations.
Extreme 33.3% tails of cMetS (402 AF1 cases, 550 AF1 controls)
Extreme 25% tails of cMetS (310 AF1 and 54 cases from Kenya (AF2), 405 AF1 and 32 AF2 controls)
Highlights.
African-specific CA10 and CTNNA3 variants is associated with metabolic syndrome.
A common KSR2 variant is associated with a 47% reduced risk for metabolic syndrome.
KSR2 and MBNL1 loci display pleiotropic effects across two or more traits.
A continuous metabolic score identifies loci implicated in metabolic profiles.
Acknowledgments
Funding
This work was supported in part by the Intramural Research Program of the National Institutes of Health in the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is supported by the National Human Genome Research Institute, the National Institute of Diabetes and Digestive and Kidney Diseases, the Center for Information Technology, and the Office of the Director at the National Institutes of Health (1ZIAHG200362). Support for participant recruitment and initial genetic studies of the AADM study was provided by NIH grant No. 3T37TW00041-03S2 from the Office of Research on Minority Health. The project was also supported in part by the National Center for Research Resources. The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C). The authors thank the staff and participants of the ARIC study for their important contributions. Funding for ARIC Gene Environment Association Studies (GENEVA) was provided by National Human Genome Research Institute grant U01HG004402 (E. Boerwinkle).
Abbreviations
- cMetS
continuous metabolic syndrome score
- GWAS
genome-wide association study
- MetS
the metabolic syndrome as defined by the National Cholesterol Education Program
- NCEP
the National Cholesterol Education Program
- T2D
type 2 diabetes
Footnotes
Disclosures
The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Grundy SM, Brewer HB, Jr, Cleeman JI, Smith SC, Jr, Lenfant C. Definition of metabolic syndrome: report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Arterioscler Thromb Vasc Biol. 2004;24:e13–18. doi: 10.1161/01.ATV.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 2.National Cholesterol Education Program (NCEP) Expert Panel. Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults (Adult Treatment Panel III) final report. Circulation. 2002;106:3143–3421. [PubMed] [Google Scholar]
- 3.Eckel RH, Grundy SM, Zimmet PZ. The metabolic syndrome. Lancet. 2005;365:1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- 4.Kotronen A, Yki-Jarvinen H. Fatty liver: a novel component of the metabolic syndrome. Arterioscler Thromb Vasc Biol. 2008;28:27–38. doi: 10.1161/ATVBAHA.107.147538. [DOI] [PubMed] [Google Scholar]
- 5.Vgontzas AN, Bixler EO, Chrousos GP. Sleep apnea is a manifestation of the metabolic syndrome. Sleep Med Rev. 2005;9:211–224. doi: 10.1016/j.smrv.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Esposito K, Chiodini P, Colao A, Lenzi A, Giugliano D. Metabolic syndrome and risk of cancer: a systematic review and meta-analysis. Diabetes Care. 2012;35:2402–2411. doi: 10.2337/dc12-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joy T, Hegele RA. Genetics of metabolic syndrome: is there a role for phenomics? Curr Atheroscler Rep. 2008;10:201–208. doi: 10.1007/s11883-008-0032-0. [DOI] [PubMed] [Google Scholar]
- 8.Lusis AJ, Attie AD, Reue K. Metabolic syndrome: from epidemiology to systems biology. Nat Rev Genet. 2008;9:819–830. doi: 10.1038/nrg2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmelli D, Cardon LR, Fabsitz R. Clustering of hypertension, diabetes, and obesity in adult male twins: same genes or same environments? Am J Hum Genet. 1994;55:566–573. [PMC free article] [PubMed] [Google Scholar]
- 10.Henneman P, Aulchenko YS, Frants RR, van Dijk KW, Oostra BA, van Duijn CM. Prevalence and heritability of the metabolic syndrome and its individual components in a Dutch isolate: the Erasmus Rucphen Family study. J Med Genet. 2008;45:572–577. doi: 10.1136/jmg.2008.058388. [DOI] [PubMed] [Google Scholar]
- 11.Lin HF, Boden-Albala B, Juo SH, Park N, Rundek T, Sacco RL. Heritabilities of the metabolic syndrome and its components in the Northern Manhattan Family Study. Diabetologia. 2005;48:2006–2012. doi: 10.1007/s00125-005-1892-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Alvarenga JC, Solis-Herrera C, Kent JW, Jaju D, Albarwani S, Al Yahyahee S, Hassan MO, Bayoumi R, Comuzzie AG. Prevalence and heritability of clusters for diagnostic components of metabolic syndrome: the Oman family study. Metab Syndr Relat Disord. 2008;6:129–135. doi: 10.1089/met.2007.0039. [DOI] [PubMed] [Google Scholar]
- 13.Loos RJ, Katzmarzyk PT, Rao DC, Rice T, Leon AS, Skinner JS, Wilmore JH, Rankinen T, Bouchard C. Genome-wide linkage scan for the metabolic syndrome in the HERITAGE Family Study. J Clin Endocrinol Metab. 2003;88:5935–5943. doi: 10.1210/jc.2003-030553. [DOI] [PubMed] [Google Scholar]
- 14.Ng MC, So WY, Lam VK, Cockram CS, Bell GI, Cox NJ, Chan JC. Genome-wide scan for metabolic syndrome and related quantitative traits in Hong Kong Chinese and confirmation of a susceptibility locus on chromosome 1q21–q25. Diabetes. 2004;53:2676–2683. doi: 10.2337/diabetes.53.10.2676. [DOI] [PubMed] [Google Scholar]
- 15.Langefeld CD, Wagenknecht LE, Rotter JI, Williams AH, Hokanson JE, Saad MF, Bowden DW, Haffner S, Norris JM, Rich SS, Mitchell BD. Linkage of the metabolic syndrome to 1q23–q31 in Hispanic families: the Insulin Resistance Atherosclerosis Study Family Study. Diabetes. 2004;53:1170–1174. doi: 10.2337/diabetes.53.4.1170. [DOI] [PubMed] [Google Scholar]
- 16.Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum Mol Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- 17.Keramati AR, Fathzadeh M, Go GW, Singh R, Choi M, Faramarzi S, Mane S, Kasaei M, Sarajzadeh-Fard K, Hwa J, Kidd KK, Babaee Bigi MA, Malekzadeh R, Hosseinian A, Babaei M, Lifton RP, Mani A. A form of the metabolic syndrome associated with mutations in DYRK1B. N Engl J Med. 2014;370:1909–1919. doi: 10.1056/NEJMoa1301824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristiansson K, Perola M, Tikkanen E, Kettunen J, Surakka I, Havulinna AS, Stancakova A, Barnes C, Widen E, Kajantie E, Eriksson JG, Viikari J, Kahonen M, Lehtimaki T, Raitakari OT, Hartikainen AL, Ruokonen A, Pouta A, Jula A, Kangas AJ, Soininen P, Ala-Korpela M, Mannisto S, Jousilahti P, Bonnycastle LL, Jarvelin MR, Kuusisto J, Collins FS, Laakso M, Hurles ME, Palotie A, Peltonen L, Ripatti S, Salomaa V. Genome-wide screen for metabolic syndrome susceptibility Loci reveals strong lipid gene contribution but no evidence for common genetic basis for clustering of metabolic syndrome traits. Circ Cardiovasc Genet. 2012;5:242–249. doi: 10.1161/CIRCGENETICS.111.961482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraja AT, Vaidya D, Pankow JS, Goodarzi MO, Assimes TL, Kullo IJ, Sovio U, Mathias RA, Sun YV, Franceschini N, Absher D, Li G, Zhang Q, Feitosa MF, Glazer NL, Haritunians T, Hartikainen AL, Knowles JW, North KE, Iribarren C, Kral B, Yanek L, O’Reilly PF, McCarthy MI, Jaquish C, Couper DJ, Chakravarti A, Psaty BM, Becker LC, Province MA, Boerwinkle E, Quertermous T, Palotie L, Jarvelin MR, Becker DM, Kardia SL, Rotter JI, Chen YD, Borecki IB. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes. 2011;60:1329–1339. doi: 10.2337/db10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong SW, Chung M, Park SJ, Cho SB, Hong KW. Genome-wide association study of metabolic syndrome in koreans. Genomics Inform. 2014;12:187–194. doi: 10.5808/GI.2014.12.4.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kraja AT, Chasman DI, North KE, Reiner AP, Yanek LR, Kilpelainen TO, Smith JA, Dehghan A, Dupuis J, Johnson AD, Feitosa MF, Tekola-Ayele F, Chu AY, Nolte IM, Dastani Z, Morris A, Pendergrass SA, Sun YV, Ritchie MD, Vaez A, Lin H, Ligthart S, Marullo L, Rohde R, Shao Y, Ziegler MA, Im HK, Schnabel RB, Jorgensen T, Jorgensen ME, Hansen T, Pedersen O, Stolk RP, Snieder H, Hofman A, Uitterlinden AG, Franco OH, Ikram MA, Richards JB, Rotimi C, Wilson JG, Lange L, Ganesh SK, Nalls M, Rasmussen-Torvik LJ, Pankow JS, Coresh J, Tang W, Linda Kao WH, Boerwinkle E, Morrison AC, Ridker PM, Becker DM, Rotter JI, Kardia SL, Loos RJ, Larson MG, Hsu YH, Province MA, Tracy R, Voight BF, Vaidya D, O’Donnell CJ, Benjamin EJ, Alizadeh BZ, Prokopenko I, Meigs JB, Borecki IB. Pleiotropic genes for metabolic syndrome and inflammation. Mol Genet Metab. 2014 doi: 10.1016/j.ymgme.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falkner B, Cossrow ND. Prevalence of metabolic syndrome and obesity-associated hypertension in the racial ethnic minorities of the United States. Curr Hypertens Rep. 2014;16:449. doi: 10.1007/s11906-014-0449-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beltran-Sanchez H, Harhay MO, Harhay MM, McElligott S. Prevalence and trends of metabolic syndrome in the adult U.S. population, 1999–2010. J Am Coll Cardiol. 2013;62:697–703. doi: 10.1016/j.jacc.2013.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park YW, Zhu S, Palaniappan L, Heshka S, Carnethon MR, Heymsfield SB. The metabolic syndrome: prevalence and associated risk factor findings in the US population from the Third National Health and Nutrition Examination Survey, 1988–1994. Arch Intern Med. 2003;163:427–436. doi: 10.1001/archinte.163.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karter AJ, Ferrara A, Liu JY, Moffet HH, Ackerson LM, Selby JV. Ethnic disparities in diabetic complications in an insured population. Jama. 2002;287:2519–2527. doi: 10.1001/jama.287.19.2519. [DOI] [PubMed] [Google Scholar]
- 27.Rotimi CN, Chen G, Adeyemo AA, Furbert-Harris P, Parish-Gause D, Zhou J, Berg K, Adegoke O, Amoah A, Owusu S, Acheampong J, Agyenim-Boateng K, Eghan BA, Jr, Oli J, Okafor G, Ofoegbu E, Osotimehin B, Abbiyesuku F, Johnson T, Rufus T, Fasanmade O, Kittles R, Daniel H, Chen Y, Dunston G, Collins FS. A genome-wide search for type 2 diabetes susceptibility genes in West Africans: the Africa America Diabetes Mellitus (AADM) Study Diabetes. 2004;53:838–841. doi: 10.2337/diabetes.53.3.838. [DOI] [PubMed] [Google Scholar]
- 28.Rotimi CN, Dunston GM, Berg K, Akinsete O, Amoah A, Owusu S, Acheampong J, Boateng K, Oli J, Okafor G, Onyenekwe B, Osotimehin B, Abbiyesuku F, Johnson T, Fasanmade O, Furbert-Harris P, Kittles R, Vekich M, Adegoke O, Bonney G, Collins F. In search of susceptibility genes for type 2 diabetes in West Africa: the design and results of the first phase of the AADM study. Ann Epidemiol. 2001;11:51–58. doi: 10.1016/s1047-2797(00)00180-0. [DOI] [PubMed] [Google Scholar]
- 29.Okosun IS, Lyn R, Davis-Smith M, Eriksen M, Seale P. Validity of a continuous metabolic risk score as an index for modeling metabolic syndrome in adolescents. Ann Epidemiol. 2010;20:843–851. doi: 10.1016/j.annepidem.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Klei L, Luca D, Devlin B, Roeder K. Pleiotropy and principal components of heritability combine to increase power for association analysis. Genet Epidemiol. 2008;32:9–19. doi: 10.1002/gepi.20257. [DOI] [PubMed] [Google Scholar]
- 31.Aschard H, Vilhjalmsson BJ, Greliche N, Morange PE, Tregouet DA, Kraft P. Maximizing the power of principal-component analysis of correlated phenotypes in genome-wide association studies. Am J Hum Genet. 2014;94:662–676. doi: 10.1016/j.ajhg.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu EY, Li M, Wang W, Li Y. MaCH-admix: genotype imputation for admixed populations. Genet Epidemiol. 2013;37:25–37. doi: 10.1002/gepi.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 35.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamm JK, Park BH, Farmer SR. A role for C/EBPbeta in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J Biol Chem. 2001;276:18464–18471. doi: 10.1074/jbc.M100797200. [DOI] [PubMed] [Google Scholar]
- 38.Park SH, Choi HJ, Yang H, Do KH, Kim J, Lee DW, Moon Y. Endoplasmic reticulum stress-activated C/EBP homologous protein enhances nuclear factor-kappaB signals via repression of peroxisome proliferator-activated receptor gamma. J Biol Chem. 2010;285:35330–35339. doi: 10.1074/jbc.M110.136259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ou CY, Chen TC, Lee JV, Wang JC, Stallcup MR. Coregulator cell cycle and apoptosis regulator 1 (CCAR1) positively regulates adipocyte differentiation through the glucocorticoid signaling pathway. J Biol Chem. 2014;289:17078–17086. doi: 10.1074/jbc.M114.548081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Danforth E., Jr Failure of adipocyte differentiation causes type II diabetes mellitus? Nat Genet. 2000;26:13. doi: 10.1038/79111. [DOI] [PubMed] [Google Scholar]
- 41.Dougherty MK, Ritt DA, Zhou M, Specht SI, Monson DM, Veenstra TD, Morrison DK. KSR2 is a calcineurin substrate that promotes ERK cascade activation in response to calcium signals. Mol Cell. 2009;34:652–662. doi: 10.1016/j.molcel.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, Schmidt K, Willson JK, Markowitz S, Zhou S, Diaz LA, Jr, Velculescu VE, Lengauer C, Kinzler KW, Vogelstein B, Papadopoulos N. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costanzo-Garvey DL, Pfluger PT, Dougherty MK, Stock JL, Boehm M, Chaika O, Fernandez MR, Fisher K, Kortum RL, Hong EG, Jun JY, Ko HJ, Schreiner A, Volle DJ, Treece T, Swift AL, Winer M, Chen D, Wu M, Leon LR, Shaw AS, McNeish J, Kim JK, Morrison DK, Tschop MH, Lewis RE. KSR2 is an essential regulator of AMP kinase, energy expenditure, and insulin sensitivity. Cell Metab. 2009;10:366–378. doi: 10.1016/j.cmet.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pearce LR, Atanassova N, Banton MC, Bottomley B, van der Klaauw AA, Revelli JP, Hendricks A, Keogh JM, Henning E, Doree D, Jeter-Jones S, Garg S, Bochukova EG, Bounds R, Ashford S, Gayton E, Hindmarsh PC, Shield JP, Crowne E, Barford D, Wareham NJ, O’Rahilly S, Murphy MP, Powell DR, Barroso I, Farooqi IS. KSR2 mutations are associated with obesity, insulin resistance, and impaired cellular fuel oxidation. Cell. 2013;155:765–777. doi: 10.1016/j.cell.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brommage R, Desai U, Revelli JP, Donoviel DB, Fontenot GK, Dacosta CM, Smith DD, Kirkpatrick LL, Coker KJ, Donoviel MS, Eberhart DE, Holt KH, Kelly MR, Paradee WJ, Philips AV, Platt KA, Suwanichkul A, Hansen GM, Sands AT, Zambrowicz BP, Powell DR. High-throughput screening of mouse knockout lines identifies true lean and obese phenotypes. Obesity (Silver Spring) 2008;16:2362–2367. doi: 10.1038/oby.2008.361. [DOI] [PubMed] [Google Scholar]
- 46.Henry MD, Costanzo-Garvey DL, Klutho PJ, Lewis RE. Obesity-dependent dysregulation of glucose homeostasis in kinase suppressor of ras 2−/− mice. Physiol Rep. 2014;2 doi: 10.14814/phy2.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Revelli JP, Smith D, Allen J, Jeter-Jones S, Shadoan MK, Desai U, Schneider M, van Sligtenhorst I, Kirkpatrick L, Platt KA, Suwanichkul A, Savelieva K, Gerhardt B, Mitchell J, Syrewicz J, Zambrowicz B, Hamman BD, Vogel P, Powell DR. Profound obesity secondary to hyperphagia in mice lacking kinase suppressor of ras 2. Obesity (Silver Spring) 2011;19:1010–1018. doi: 10.1038/oby.2010.282. [DOI] [PubMed] [Google Scholar]
- 48.Kaneto H, Matsuoka TA, Kawashima S, Yamamoto K, Kato K, Miyatsuka T, Katakami N, Matsuhisa M. Role of MafA in pancreatic beta-cells. Adv Drug Deliv Rev. 2009;61:489–496. doi: 10.1016/j.addr.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 49.Li WD, Dong C, Li D, Zhao H, Price RA. An obesity-related locus in chromosome region 12q23–24. Diabetes. 2004;53:812–820. doi: 10.2337/diabetes.53.3.812. [DOI] [PubMed] [Google Scholar]
- 50.Chagnon YC, Merette C, Bouchard RH, Emond C, Roy MA, Maziade M. A genome wide linkage study of obesity as secondary effect of antipsychotics in multigenerational families of eastern Quebec affected by psychoses. Mol Psychiatry. 2004;9:1067–1074. doi: 10.1038/sj.mp.4001537. [DOI] [PubMed] [Google Scholar]
- 51.Wilson SG, Adam G, Langdown M, Reneland R, Braun A, Andrew T, Surdulescu GL, Norberg M, Dudbridge F, Reed PW, Sambrook PN, Kleyn PW, Spector TD. Linkage and potential association of obesity-related phenotypes with two genes on chromosome 12q24 in a female dizygous twin cohort. Eur J Hum Genet. 2006;14:340–348. doi: 10.1038/sj.ejhg.5201551. [DOI] [PubMed] [Google Scholar]
- 52.Bowden DW, Sale M, Howard TD, Qadri A, Spray BJ, Rothschild CB, Akots G, Rich SS, Freedman BI. Linkage of genetic markers on human chromosomes 20 and 12 to NIDDM in Caucasian sib pairs with a history of diabetic nephropathy. Diabetes. 1997;46:882–886. doi: 10.2337/diab.46.5.882. [DOI] [PubMed] [Google Scholar]
- 53.Shaw JT, Lovelock PK, Kesting JB, Cardinal J, Duffy D, Wainwright B, Cameron DP. Novel susceptibility gene for late-onset NIDDM is localized to human chromosome 12q. Diabetes. 1998;47:1793–1796. doi: 10.2337/diabetes.47.11.1793. [DOI] [PubMed] [Google Scholar]
- 54.Lindgren CM, Mahtani MM, Widen E, McCarthy MI, Daly MJ, Kirby A, Reeve MP, Kruglyak L, Parker A, Meyer J, Almgren P, Lehto M, Kanninen T, Tuomi T, Groop LC, Lander ES. Genomewide search for type 2 diabetes mellitus susceptibility loci in Finnish families: the Botnia study. Am J Hum Genet. 2002;70:509–516. doi: 10.1086/338629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai F, Keighley ED, Sun G, Indugula SR, Roberts ST, Aberg K, Smelser D, Tuitele J, Jin L, Deka R, Weeks DE, McGarvey ST. Genome-wide scan for adiposity-related phenotypes in adults from American Samoa. Int J Obes (Lond) 2007;31:1832–1842. doi: 10.1038/sj.ijo.0803675. [DOI] [PubMed] [Google Scholar]
- 56.Shmulewitz D, Heath SC, Blundell ML, Han Z, Sharma R, Salit J, Auerbach SB, Signorini S, Breslow JL, Stoffel M, Friedman JM. Linkage analysis of quantitative traits for obesity, diabetes, hypertension, and dyslipidemia on the island of Kosrae, Federated States of Micronesia. Proc Natl Acad Sci U S A. 2006;103:3502–3509. doi: 10.1073/pnas.0510156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fox CS, Liu Y, White CC, Feitosa M, Smith AV, Heard-Costa N, Lohman K, Johnson AD, Foster MC, Greenawalt DM, Griffin P, Ding J, Newman AB, Tylavsky F, Miljkovic I, Kritchevsky SB, Launer L, Garcia M, Eiriksdottir G, Carr JJ, Gudnason V, Harris TB, Cupples LA, Borecki IB. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zemunik T, Boban M, Lauc G, Jankovic S, Rotim K, Vatavuk Z, Bencic G, Dogas Z, Boraska V, Torlak V, Susac J, Zobic I, Rudan D, Pulanic D, Modun D, Mudnic I, Gunjaca G, Budimir D, Hayward C, Vitart V, Wright AF, Campbell H, Rudan I. Genome-wide association study of biochemical traits in Korcula Island, Croatia. Croat Med J. 2009;50:23–33. doi: 10.3325/cmj.2009.50.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janssens B, Goossens S, Staes K, Gilbert B, van Hengel J, Colpaert C, Bruyneel E, Mareel M, van Roy F. alphaT-catenin: a novel tissue-specific beta-catenin-binding protein mediating strong cell-cell adhesion. J Cell Sci. 2001;114:3177–3188. doi: 10.1242/jcs.114.17.3177. [DOI] [PubMed] [Google Scholar]
- 60.Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, Butte NF. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vanpoucke G, Goossens S, De Craene B, Gilbert B, van Roy F, Berx G. GATA-4 and MEF2C transcription factors control the tissue-specific expression of the alphaT-catenin gene CTNNA3. Nucleic Acids Res. 2004;32:4155–4165. doi: 10.1093/nar/gkh727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 63.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. Embo J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Omland T, de Lemos JA, Sabatine MS, Christophi CA, Rice MM, Jablonski KA, Tjora S, Domanski MJ, Gersh BJ, Rouleau JL, Pfeffer MA, Braunwald E. A sensitive cardiac troponin T assay in stable coronary artery disease. N Engl J Med. 2009;361:2538–2547. doi: 10.1056/NEJMoa0805299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paul S, Dansithong W, Kim D, Rossi J, Webster NJ, Comai L, Reddy S. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. Embo J. 2006;25:4271–4283. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryder JW, Gilbert M, Zierath JR. Skeletal muscle and insulin sensitivity: pathophysiological alterations. Front Biosci. 2001;6:D154–163. doi: 10.2741/ryder. [DOI] [PubMed] [Google Scholar]
- 67.Friedman JR, Kaestner KH. The Foxa family of transcription factors in development and metabolism. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Richard AJ, Stephens JM. The role of JAK-STAT signaling in adipose tissue function. Biochim Biophys Acta. 2014;1842:431–439. doi: 10.1016/j.bbadis.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aspatwar A, Tolvanen ME, Ortutay C, Parkkila S. Carbonic anhydrase related proteins: molecular biology and evolution. Subcell Biochem. 2014;75:135–156. doi: 10.1007/978-94-007-7359-2_8. [DOI] [PubMed] [Google Scholar]
- 70.Nishimori I, Vullo D, Minakuchi T, Scozzafava A, Capasso C, Supuran CT. Restoring catalytic activity to the human carbonic anhydrase (CA) related proteins VIII, X and XI affords isoforms with high catalytic efficiency and susceptibility to anion inhibition. Bioorg Med Chem Lett. 2013;23:256–260. doi: 10.1016/j.bmcl.2012.10.103. [DOI] [PubMed] [Google Scholar]
- 71.Supuran CT. Carbonic anhydrases as drug targets--an overview. Curr Top Med Chem. 2007;7:825–833. doi: 10.2174/156802607780636690. [DOI] [PubMed] [Google Scholar]
- 72.Arechederra RL, Waheed A, Sly WS, Supuran CT, Minteer SD. Effect of sulfonamides as carbonic anhydrase VA and VB inhibitors on mitochondrial metabolic energy conversion. Bioorg Med Chem. 2013;21:1544–1548. doi: 10.1016/j.bmc.2012.06.053. [DOI] [PubMed] [Google Scholar]
- 73.Supuran CT, Di Fiore A, De Simone G. Carbonic anhydrase inhibitors as emerging drugs for the treatment of obesity. Expert Opin Emerg Drugs. 2008;13:383–392. doi: 10.1517/14728214.13.2.383. [DOI] [PubMed] [Google Scholar]
- 74.Scozzafava A, Supuran CT, Carta F. Antiobesity carbonic anhydrase inhibitors: a literature and patent review. Expert Opin Ther Pat. 2013;23:725–735. doi: 10.1517/13543776.2013.790957. [DOI] [PubMed] [Google Scholar]
- 75.Kramer CK, Leitao CB, Pinto LC, Canani LH, Azevedo MJ, Gross JL. Efficacy and safety of topiramate on weight loss: a meta-analysis of randomized controlled trials. Obes Rev. 2011;12:e338–347. doi: 10.1111/j.1467-789X.2010.00846.x. [DOI] [PubMed] [Google Scholar]
- 76.Heal DJ, Gosden J, Smith SL. What is the prognosis for new centrally-acting anti-obesity drugs? Neuropharmacology. 2012;63:132–146. doi: 10.1016/j.neuropharm.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 77.Fuke T, Yoshizaki T, Kondo M, Morino K, Obata T, Ugi S, Nishio Y, Maeda S, Kashiwagi A, Maegawa H. Transcription factor AP-2beta inhibits expression and secretion of leptin, an insulin-sensitizing hormone, in 3T3-L1 adipocytes. Int J Obes (Lond) 2010;34:670–678. doi: 10.1038/ijo.2009.295. [DOI] [PubMed] [Google Scholar]
- 78.Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams MJ, Goergen P, Rajendran J, Zheleznyakova G, Hagglund MG, Perland E, Bagchi S, Kalogeropoulou A, Khan Z, Fredriksson R, Schioth HB. Obesity-linked homologues TfAP-2 and Twz establish meal frequency in Drosophila melanogaster. PLoS Genet. 2014;10:e1004499. doi: 10.1371/journal.pgen.1004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Otoda T, Takamura T, Misu H, Ota T, Murata S, Hayashi H, Takayama H, Kikuchi A, Kanamori T, Shima KR, Lan F, Takeda T, Kurita S, Ishikura K, Kita Y, Iwayama K, Kato K, Uno M, Takeshita Y, Yamamoto M, Tokuyama K, Iseki S, Tanaka K, Kaneko S. Proteasome dysfunction mediates obesity-induced endoplasmic reticulum stress and insulin resistance in the liver. Diabetes. 2013;62:811–824. doi: 10.2337/db11-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Costes S, Huang CJ, Gurlo T, Daval M, Matveyenko AV, Rizza RA, Butler AE, Butler PC. beta-cell dysfunctional ERAD/ubiquitin/proteasome system in type 2 diabetes mediated by islet amyloid polypeptide-induced UCH-L1 deficiency. Diabetes. 2011;60:227–238. doi: 10.2337/db10-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Muriach M, Flores-Bellver M, Romero FJ, Barcia JM. Diabetes and the brain: oxidative stress, inflammation, and autophagy. Oxid Med Cell Longev. 2014;2014:102158. doi: 10.1155/2014/102158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kraja AT, Lawson HA, Arnett DK, Borecki IB, Broeckel U, de las Fuentes L, Hunt SC, Province MA, Cheverud J, Rao DC. Obesity-insulin targeted genes in the 3p26–25 region in human studies and LG/J and SM/J mice. Metabolism. 2012;61:1129–1141. doi: 10.1016/j.metabol.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwartz MW, Porte D., Jr Diabetes, obesity, and the brain. Science. 2005;307:375–379. doi: 10.1126/science.1104344. [DOI] [PubMed] [Google Scholar]
- 85.Wijndaele K, Beunen G, Duvigneaud N, Matton L, Duquet W, Thomis M, Lefevre J, Philippaerts RM. A continuous metabolic syndrome risk score: utility for epidemiological analyses. Diabetes Care. 2006;29:2329. doi: 10.2337/dc06-1341. [DOI] [PubMed] [Google Scholar]
- 86.Klein BE, Klein R, Lee KE. Components of the metabolic syndrome and risk of cardiovascular disease and diabetes in Beaver Dam. Diabetes Care. 2002;25:1790–1794. doi: 10.2337/diacare.25.10.1790. [DOI] [PubMed] [Google Scholar]
- 87.Agarwal S, Jacobs DR, Jr, Vaidya D, Sibley CT, Jorgensen NW, Rotter JI, Chen YD, Liu Y, Andrews JS, Kritchevsky S, Goodpaster B, Kanaya A, Newman AB, Simonsick EM, Herrington DM. Metabolic Syndrome Derived from Principal Component Analysis and Incident Cardiovascular Events: The Multi Ethnic Study of Atherosclerosis (MESA) and Health, Aging, and Body Composition (Health ABC) Cardiol Res Pract. 2012;2012:919425. doi: 10.1155/2012/919425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.