Abstract
The implications of Lysine-Specific Demethylase-1 (LSD1) in tumorigenesis have urged scientists to develop diagnostic tools in order to explore the function of this enzyme. In this work, we present our efforts on the development of tranylcypromine (TCP)-based functionalized probes for activity-based protein profiling (ABPP) of LSD1 activity. Biotinylated forms of selected compounds enabled dose-dependent enzyme labeling of recombinant LSD1. However, treatment with LSD1 inhibitors did not clearly reduce the LSD1 labeling efficiency thus indicating that labelling using these probes in not activity dependent. This calls for alternative strategies to develop probes for ABPP of the enzyme LSD1.
Keywords: Lysine demethylation, Tumorigenesis, Tranylcypromine, Irreversible inhibition, Enzyme labeling
1. Introduction
Among post-translational modifications, histone lysine methylation has gained increasing attention as a major regulator of the transcriptional potential of eukaryotic cells. Lysine residues in histone tails (mainly of histones H3 and H4) can be mono-, di- or trimethylated on their ε-amino groups by lysine methyltransferases (KMTs) resulting in different functional outputs.1,2 Lysine demethylases (KDMs) act as erasers of these marks and, depending on the enzymatic mechanism, they are categorized to lysine-specific demethylases (LSDs) and JumonjiC (JmjC) demethylases.3 The first class are FAD (flavin adenine dinucleotide)-dependent amine oxidases that demethylate mono- or dimethylated lysine residues producing formaldehyde and hydrogen peroxide, whereas the second class utilizes Fe(II) and α-ketoglutarate as cofactors and can also act on trimethylated lysine side chains via its dioxygenase function.
Two mammalian flavin-containing demethylases have been identified: LSD1 (also known as KDM1A, AOF2, BHC110 or KIAA0601) and LSD2 (KDM1B or AOF1).4,5 The best studied is LSD1 which is now well documented to represent an important player in developmental processes, embryogenesis and differentiation of numerous cellular types.6–8 According to its substrates and interactions with other proteins, LSD1 is known to mediate transcriptional activation or repression.9,10 Accumulating data suggest that any imbalance of the dynamic regulation of lysine methylation due to aberrant expression of LSD1 can cause dramatic alterations in gene transcription and, consequently, in the development and progression of various cancer types.11–14 Nevertheless, several studies demonstrate that, in coordination with other proteins, LSD1 affects the growth of breast cancer cells negatively,15–17 while promoting effects have been described for viral infections.18,19
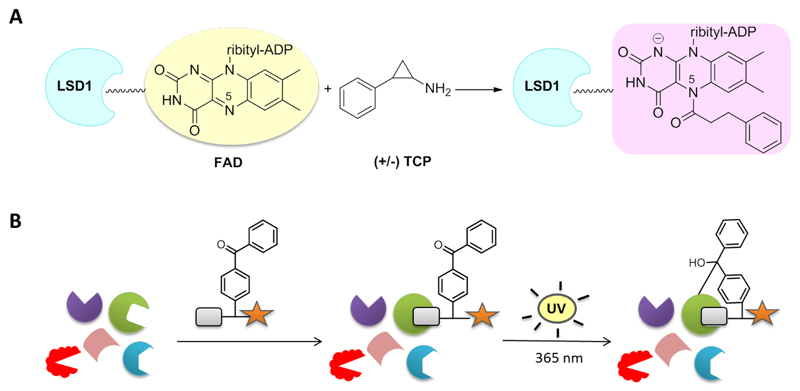
Due to its significant role in pathogenesis, LSD1 has been an emerging pharmacological target and thus, the development of potent inhibitors has attracted increasing research interest. Up to date, a wide variety of compounds has been reported to inactivate LSD1 in a reversible20–24 or irreversible way,25–27 which have been evaluated mainly for their antiproliferative effects. The majority of them was inspired by several anti-MAO (monoamine oxidase) agents found to inhibit LSD1 with tranylcypromine (trans-2-phenylcyclopropyl-1-amine, TCP) taking center stage. TCP is a non-selective irreversible MAO A/B inhibitor with a reported potency over LSD1 ranging from 20-400 µM.25,28 Structural and kinetic studies revealed a suicide-mechanism of time-dependent inhibition via opening of the cyclopropyl ring and formation of a stable covalent adduct with the reduced form of the cofactor FAD.25,28 Although different models have been proposed regarding the structure of this adduct,29 further structural analyses and crystallographic data from LSD1-TCP complex indicated the participation of the N(5) atom of the flavin ring (Fig. 1A).30,31 TCP has been employed by numerous research groups as the starting point for the development of more potent and selective derivatives with promising antitumor effects.29,32–38
Figure 1.
Irreversible enzyme binding through covalent linkage. (A) Proposed mechanism of LSD1 inactivation by racemic TCP.30 (B) Photocrosslinking with benzophenone-type activity-based probes.
Despite the tremendous progress on LSD1 inhibition, its controversial roles in gene expression and oncogenesis call for the discovery of novel diagnostic tools to gain a better insight on the biological function of this enzyme.39 Activity-based protein profiling (ABPP) has been proven to be a valuable approach to study intracellular enzyme activity.40–42 In this work, we report the design, synthesis and biological evaluation of activity-based functionalized probes for detection of human recombinant and endogenous LSD1.
2. Results and discussion
2.1. Molecular design
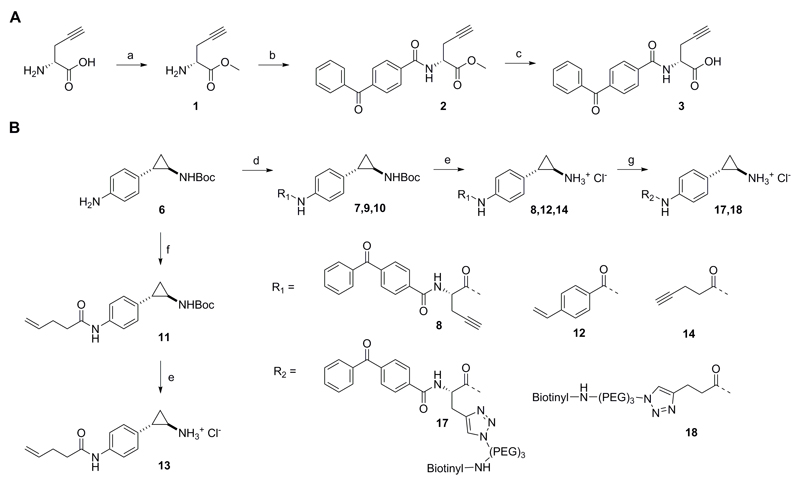
Two different approaches were followed in the molecular design of the probes. Activity-based probes are typically designed in a way to mimic the covalent binding of the substrate to the protein, modifying the latter in an irreversible manner. Protein visualization is then achieved directly after labeling, in case the probes contain a detection handle (i.e. fluorophore), or after bioorthogonal coupling to affinity tags. For instance, Breinbauer et al. developed functionalized probes based on the irreversible MAO inhibitors pargyline and deprenyl for the detection of MAO-A and B in tissues and live cells, while, later on, selective imaging and profiling of MAO-B in a variety of biological samples was achieved by Yao et al. employing a dual-purpose functionalized activity-based probe.43,44 For our purposes, all the developed compounds included a TCP moiety to ensure for covalent attachment to the cofactor FAD. However, unlike MAO and other flavoenzymes, the interactions between FAD and LSD1 are not covalent and thus we hypothesized that detection via the ABPP method would not be possible. Therefore, we firstly designed probe 8 where a benzophenone group was introduced aiming at photocrosslinking and covalent binding to the protein (Fig. 1B). Benzophenones are known to be activated upon ultraviolet irradiation at long wavelengths to generate a diradical that reacts irreversibly with neighboring C-H bonds, in particular those of methionine residues. They are generally characterized by fast activation and limited cross-reactivity.45–48 In addition, a polar linker was used to connect TCP with the photoactivatable part, whereas a protruding propargyl group was inserted to serve for subsequent linkage to a detection label via the ‘click reaction’ (Scheme 1).
Scheme 1.
Alternatively to probe 8, we developed three non-benzophenone-bearing compounds as controls to test the need for photocrosslinking. Recently Valente et al. developed derivatives of TCP, with a benzoylamino group at the para, meta or ortho position in respect to the phenyl ring.49 They observed that the para-substituted compounds exhibited the greatest potency at low nanomolar scale regarding MAO-A and LSD1 inhibition. Inspired by this, we employed a para-benzoylamino TCP analogue (Scheme. 2, compound 12) functionalized at position 4 (in respect to the benzoyl) with a terminal alkene and two alkoylamino derivatives (compounds 13 and 14) functionalized with terminal olefinic and a alkyl chains respectively. The alkyne-labeled compounds allow for detection via the ‘click’ reaction while the olefinic ones via the oxidative Heck reaction, recently introduced by our group as a successful bioorthogonal strategy for in vitro protein labeling in aqueous environment.50,51
2.2. Chemical synthesis
All compounds were synthesized from tert-butyl (trans-2-(4-aminophenyl)cyclopropyl)carbamate (6) after coupling to the corresponding carboxylic acids followed by acidic deprotection of the Boc group (Scheme 1). Regarding the synthesis of the photocrosslinking and linker part of probe 8, compound 3 was generated after coupling of D-propargylglycine to 4-benzoylbenzoic acid using HOBt/EDCI/triethylamine (TEA) in good yield (64%) (Scheme 1A). For the introduction of the TCP functional group, compound 6 was first synthesized according to previously reported methods,30 including p- nitration of trans-2-phenylcyclopropane-1-carboxylic acid (32%), Curtius rearrangement and Boc protection of the afforded cyclopropylamine (27%) and, finally, reduction of the nitro group to the corresporesponding aniline (27%) (see Experimental section). Afterwards, Boc-protected forms of probes 8, 12 and 14 were provided using EDC as a coupling reagent, while in the case of probe 13, the coupling was performed with the use of pent-4-enoyl chloride and TEA (Scheme 1B).
2.3. Inhibition studies
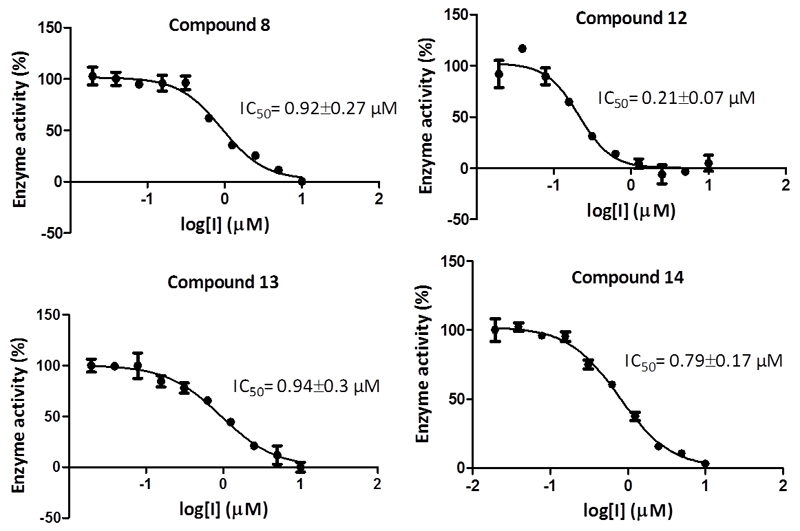
All synthesized compounds were screened for inhibition against human recombinant LSD1. The principle of the in vitro assay was based on the oxidative demethylation of the monomethylated histone peptide H3K4N(CH3) via a FAD/FADH2 mediated reduction of O2 to H2O2. The remaining activity of LSD1 was monitored via detection of the amount of H2O2 formed. This is done by Horseradish Peroxidase (HRP) which reduces H2O2 to H2O using 10-acetyl-3,7-dihydroxyphenoxazine (ADHP, Amplex Red, for synthesis see Experimental section) as an electron donor. The resulting product (resorufin) is highly fluorescent at 590 nm (Fig. S1). The inhibition assay was performed as described previously (see Experimental section).49 The compounds were pre-incubated at different concentrations with LSD1 for 15 min at room temperature in presence of HRP-Amplex Red. The substrate was then added and the fluorescence was measured for 30 min. The IC50 values were found to be in the high nanomolar range apart from compound 12 which exhibited a potency of 0.21 µM (Fig. 2).
Figure 2.
IC50 values of developed LSD1 probes and their respective standard deviations.
We then moved to the determination of the kinetic parameters Ki (equilibrium binding constant) and ki (inactivation rate constant) that characterize the irreversible type of inhibition through Kitz-Wilson analysis (see Experimental section). At start, irreversible inhibitors form a non-covalent complex with the enzyme (represented by Ki) which then reacts to generate the covalent ‘dead-end complex’ (represented by ki). For our purposes, the enzyme was incubated with different concentrations of each compound for different incubation times in presence of HRP-Amplex Red. Substrate was added and the enzyme activity was monitored as described above. In all cases, time-dependent inhibition was observed with Ki values ranging between 3-5 µM and the ki values between 0.3-0.4 min-1 indicating the rapid binding of the TCP derivatives to the FAD cofactor (reported ki for TCP 0.67 min-1).52
2.4. Labeling studies
2.4.1. Human recombinant LSD1
Having all the information required on the appropriate range of inhibitor concentration and incubation time necessary for binding, we set out to utilize them for labeling of human recombinant and cellular LSD1. Initial experiments included treatment of HeLa cell lysates with various concentrations of each compound and subsequent coupling to azide-PEG3-biotin via the ‘click’ reaction (compound 14)51 or 3-(biotinylamino)phenylboronic acid via the oxidative Heck reaction (compounds 12 and 13).51 Unfortunately, after imaging using the enhanced chemiluminescence assay, only endogenously biotinylated proteins were visualized, while the presence of LSD1 was confirmed with an anti-LSD1 antibody (data not shown). As supported from our previous findings, where incomplete conversion was observed for both bioorthogonal reactions at protein concentrations lower than 5 µM,51 we speculated that no labeling was due to the limiting performance of the reactions at the low protein concentrations in the case of cell lysates. Another reason may be that the reactive group was not properly positioned to react with the complementary coupling reagents.
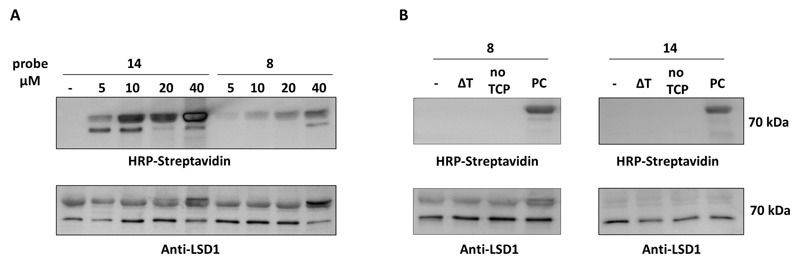
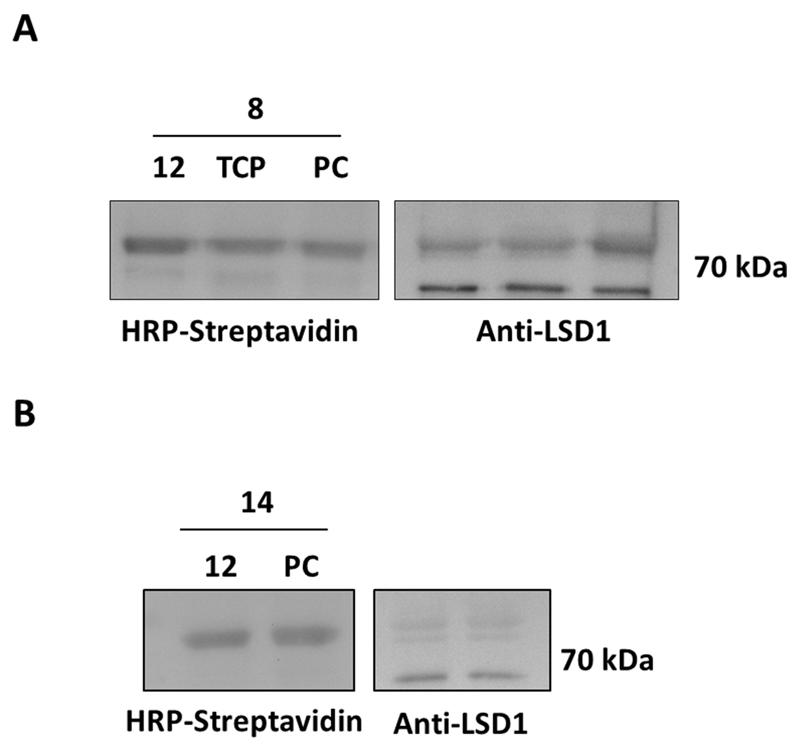
Therefore we continued our labeling efforts by employing a single-step process, assuring for the biotinylation step: benzophenone probe 8 and probe 14 were firstly subjected to ‘click’ reactions and coupling to azide-PEG3-biotin (see Experimental section) to generate the biotinylated compounds 17 and 18 accordingly. Human recombinant LSD1 was incubated with 0-40 µM of each compound for 10 min. In case of probe 17, the incubation was followed by 5 min UV irradiation at 365 nm. The enzyme was then visualized using anti-LSD1 antibody, proteins were stripped and subsequently detected via HRP-streptavidin. Surprisingly, a concentration-dependent luminescent signal was observed not only for probe 17 (Fig. 3A) but also for the non-benzophenone-carrying compound 18 (Fig. S3).. This finding is remarkable since previous studies where, in an attempt to prove the non-covalent binding of FAD to the enzyme, TCP-inactivated protein was denatured in presence of 0.3% SDS resulting in release of FAD and separation from the enzyme by HPLC.28 Our observation is difficult to explain at this point, nevertheless it indicates that benzophenone cross-linking is not key to labelling of LSD1 using the TCP-based probe applied in this study. We therefore decided to continue our studies using both probes 17 and 18 in order to explore which inhibitor-type is best for activity-based labeling.
Figure 3.
Detection of human recombinant LSD1. (A) Luminescence imaging after incubation with various concentrations of pre-clicked biotinylated probes 17 and 18. (B) Control reactions. ΔT; heat-denaturation control, PC; positive control.
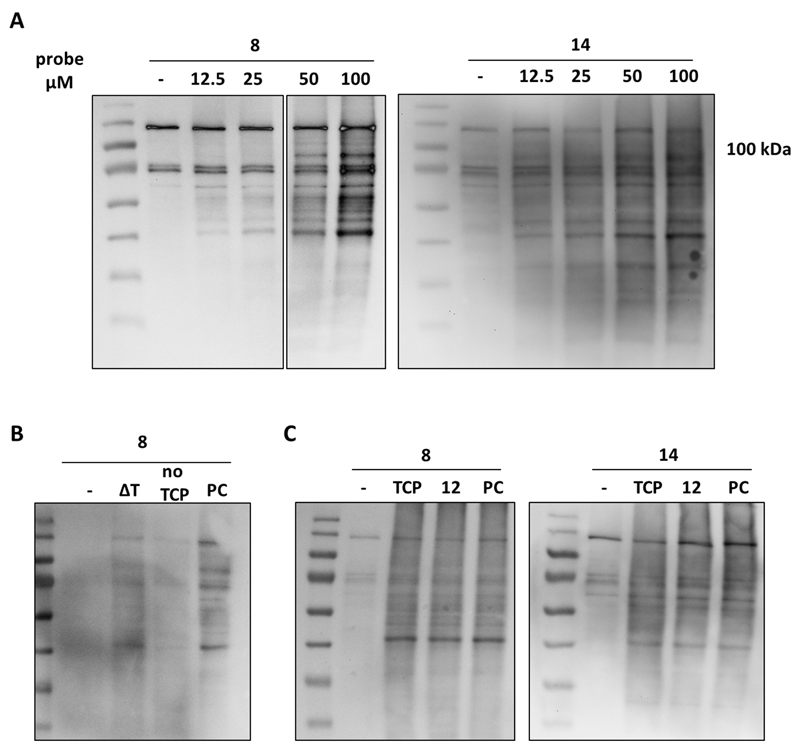
Control reactions were performed to ensure for activity-based labeling. For the negative control experiment, the enzyme was subjected to ‘click’ reaction conditions while, regarding the heat-denaturation control (ΔT), boiling for 15 min before labeling served for complete unfolding, protein aggregation and loss of activity as reported before.28,53 Compound 17 was added to the heat-denaturated and positive control samples at a final concentration of 40 µM and immediately all the samples were irradiated shortly (5 min). Furthermore, in order to confirm that the TCP-moiety is crucial for labelling, we did a control experiment in which the the TCP moiety was excluded by employing compound 2 that consists of a benzophenone and propargyl group but not the TCP (Scheme 1). Analogously to the rest of the controls, LSD1 was irradiated for 5 min in presence of 40 µM of pre-clicked compound 2 and detected in all cases as described above. Pleasingly, labeling occurred only in the positive control sample demonstrating that TCP is essential for enzyme labeling (Fig. 3B, Fig. S4). The same experiments were repeated for compound 18 (final concentration 10 µM) and pre-clicked compound 2 was used at a final concentration of 10 µM, where, again, luminescence was observed only in the case of the positive control sample.
Next, in order to further certify an activity-based labeling, we investigated the protein tagging after treatment with known LSD1 inhibitors (Fig. 4, Fig. S5). Compound 12 (1 mM) and TCP (1.5 mM) were pre-incubated with a low concentration of LSD1 for 30 min at room temperature. Compound 17 was added at a final concentration of 40 µM and the samples were immediately irradiated. The irradiation time was decreased from 5 to 2 min to induce the effect of the inhibitors. After detection via the usual procedure, no reduction in protein labeling was visible, compared to the positive control (Fig. 4A). This can be justified for TCP, known for its weak potency over LSD1 as mentioned above, but not for compound 12 whose equilibrium binding constant Ki was found to be 2.7 µM. The same experiment was repeated using probe 18 and only the potent compound 12 and, again, no clear difference was observed between the inhibitor-treated samples and the positive control (Fig. 4B). These results point to a lack of enzyme activity-dependence of the LSD1 labeling. However, as indicated by the control experiments, this can be attributed only to the TCP moiety and not the triazole-PEG3-biotin or the photocrosslinking one
Figure 4.
Detection of human recombinant LSD1 after treatment with known inhibitors. The enzyme was pre-incubated with compound 12 and TCP and subsequently treated with pre-clicked probes 17 (A) and 18 (B). PC; positive control.
2.4.2. HeLa nuclear extracts
Afterwards, we moved on to test the labeling efficiency of probes 17 and 18 on HeLa nuclear extracts, known as a source of endogenous LSD1.54,55 Various concentrations of each probe were incubated with protein extracts for 10 min followed by 5 min irradiation (for 17) or for 15 min at room temperature (for 18). Subsequent detection revealed a number of biotinylated proteins, potentially FAD-dependent (Fig. 5A, Fig. S6, S7). However, no visible labeling of LSD1 was observed, while its presence was confirmed via an anti-LSD1 antibody. This may be due to low LSD1 activity after nuclear extraction or to probe utilization by other enzymes, presumably amine oxidases. Additionally, it is possible that the probe does not bind to nuclear LSD1 as the enzyme exists in complex with other cofactors.54,56 Similar control experiments took place on nuclear extracts as described for the purified enzyme. Again, biotinylated proteins were visualized only in the case of the positive control sample, while heat-denaturated proteins hardly reacted and treatment with the non-TCP-bearing pre-clicked compound 2 resulted in no tagging (Fig. 5B, Fig. S8). This indicates that labeling occurs only in non-denatured enzymes and is due to the presence of TCP moiety.
Figure 5.
Labeling experiments on HeLa nuclear extracts. (A) Detection of endogenous proteins after treatment with various concentrations of pre-clicked probes 17 and 18. (B) Control reactions and (C) treatment with LSD1 inhibitors. Proteins were detected by using HRP-streptavidin-coupled enhanced chemiluminescence assay.
Finally, HeLa nuclear extracts were pre-treated with 1 mM of compound 12 or 1.5 mM of TCP and then labeled with the corresponding probe for 2 min. Nevertheless, no change in the level of biotinylation of endogenous proteins was shown upon incubation with the inhibitors (Fig. 5C). These findings are in line with the results obtained from the purified LSD1 and indicate a lack of activity-dependence in the observed labelling.
3. Conclusions
In conclusion, we have developed a series of alkene- and alkyne-labeled TCP-based probes for detection of LSD1 activity via activity-based protein profiling. Considering the reversible interactions between the enzyme and the cofactor FAD, we also designed a benzophenone-containing alkynic compound for covalent attachment to LSD1 after photocrosslinking with UV irradiation. Alkyne-bearing probes 8 and 14 were subjected to ‘click’ reactions for linkage to a biotin molecule and incubated with human recombinant LSD1. In both cases, a luminescence signal was observed while heat-inactivation or treatment with a non-TCP-containing probe resulted in no labeling. This demonstrates that tagging occurs only to non-denatured LSD1 and that the tranylcypromine group is crucial for enzyme inhibition. Unfortunately, we found no apparent reduction in labeling when LSD1 inhibitors were applied, a fact that implies that small molecule inhibition of the enzyme activity does not corroborate to the activity-based labeling. Further studies on HeLa nuclear extracts indicated that several proteins were effectively labeled by the probes, without a pronounced labeling of LSD1, while inhibition experiments, again, did not reveal a clear reduction in protein labeling. According to our control experiments, the developed molecules can be used as activity-based probes based on non-denatured enzymes but are disfavored in small molecule inhibition studies. Our data together demonstrate that photocrosslinking is not needed for enzyme labeling. This work sets the stage for further elucidation of the mechanism of inhibition of TCP derivatives and should be taken into account when designing novel analogues aiming at detecting LSD1 activity.
4. Experimental section
4.1. Chemistry
4.1.1. General
Chemicals were obtained from commercial suppliers (Sigma Aldrich, Acros Organics, Axon Medchem) and used without further purification, unless stated otherwise. Aluminum sheets of Silica Gel 60 F254 were used for Thin layer chromatography (TLC). Spots were visualized under ultraviolet light or stained with KMnO4 solution. MP Ecochrom Silica Gel 32-63 60 Å was used for flash column chromatography. NMR spectra were recorded on a Bruker Avance 500 spectrometer (1H NMR; 500 MHz), 13C NMR; 125 MHz)). Chemical shift values are reported in ppm (δ) relative to tetramethylsilane (TMS). Coupling constants (J) are reported in Hz with the following splitting abbreviations: s = singlet, d = doublet, t = triplet, q = quartet and m = multiplet. Abbreviations are defined as follows: dichloromethane (DCM), methanol (MeOH), thionyl chloride (SOCl2), dimethylformamide (DMF), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide (EDCI), 1-hydroxybenzotriazole hydrate (HOBt), trimethylamine (TEA), ethyl acetate (EtOAc), diphenylphosphoryl azide (DPPA) and magnesium sulphate (MgSO4).
4.1.2. (R)-methyl 2-aminopent-4-ynoate (1)
To an ice-cooled solution of D-propargylglycine (0.30 g, 2.7 mmol, 1.0 eq) in MeOH (5.0 mL), thionyl chloride (0.40 mL, 40 mmol, 15 eq) was added dropwise. The resulting mixture was heated at 60 °C for 4h. After completion of the reaction, the solvent was evaporated and the crude mixture was washed with DCM three times to obtain the pure compound 1 as a light brown oil (0.30 g, 89%). 1H NMR (500 MHz, CDCl3): δ = 8.73 (bs, 2H), 4.47 (s, 1H), 3.86 (s, 3H), 3.73 (m, 1H), 3.20-3.15 (m, 1H), 3.06-3.01 (m, 1H) ppm. 13C NMR (125 MHz, CDCl3): δ = 168.5, 76.4, 74.4, 53.7, 52.0, 20.6 ppm.
4.1.3. (R)-ethyl 2-(4-benzoylbenzamido)pent-4-ynoate (2)
To a solution of 4-benzoylbenzoic acid (0.30 g, 1.3 mmol, 1.0 eq) in DMF (3.5 mL) were added EDCI (0.33 g, 1.7 mmol, 1.3 eq), HOBt (0.27 g, 1.7 mmol, 1.3 eq) and TEA (0.65 mL, 4.6 mmol, 3.5 eq). After stirring for 30 min at room temperature, 1 (0.17 g, 1.3 mmol, 1.0 eq) was added and the mixture was stirred at room temperature for additional 17h. Then, the reaction was quenched with NaHCO3 (20 mL) and extracted with EtOAc (3 x 20mL). The organic layer was washed with brine, dried over MgSO4 and concentrated. The crude mixture was purified by flash chromatography on silica gel (EtOAc:petroleum ether 1:2) affording compound 2 as a white solid (0.29 g, 64%). Rf = 0.45 (EtOAc:petroleum ether 1:1). 1H NMR (500 MHz, CDCl3): δ = 7.93 (d, J = 8.2 Hz, 2H), 7.88 (d, J = 8.2 Hz, 2H), 7.80 (d, J = 7.8 Hz, 2H), 7.62 (t, J = 7.4 Hz, 1H), 7.52-7.49 (m, 2H), 7.05 (d, J = 7.3 Hz, 1H), 4.99-4.95 (m, 1H), 3.85 (s, 3H), 2.99-2.88 (m, 2H), 2.09-2.08 (m, 1H) ppm. 13C NMR (125 MHz, CDCl3): δ = 196.0, 170.9, 166.3, 140.7, 137.1, 137.0, 133.1, 130.3 (x2), 130.2 (x2), 128.6 (x2), 127.3 (x2), 78.4, 72.1, 53.2, 51.2, 22.7 ppm.
4.1.4. 2-(4-Benzoylbenzamido)pent-4-ynoic acid (3)
Compound 2 (0.29 g, 0.89 mmol, 1.0 eq) was dissolved in THF:MeOH (2.0 mL:1.0 mL) and a solution of LiOH (31 mg, 1.3 mmol, 1.5 eq) in H2O (1.0 mL) was added at 0 °C. After 3h stirring at room temperature, LiOH (31 mg, 1.3 mmol, 1.5 eq) was added. After further stirring for 5h, the mixture was acidified to pH 1.0 with a solution of HCl 1N and extracted with EtOAc (3 x 10 mL). The organic phase was then dried and evaporated to afford compound 3 as a white solid (0.27 g, 97%). 1H NMR (500 MHz, CD3OD): δ = 7.99 (d, J = 8.1 Hz, 2H), 7.86 (d, J = 8.1 Hz, 2H), 7.79 (d, J = 7.4 Hz, 2H), 7.67 (t, J = 7.4 Hz, 1H), 7.56-7.53 (m, 2H), 4.78-4.76 (m, 1H), 3.31-3.30 (m, 1H), 2.94-2.81 (m, 2H), 2.39-2.38 (m, 1H) ppm. 13H NMR (125 MHz, CD3OD): δ = 193.8, 175.8, 174.2, 140.5, 139.3, 136.8, 134.2, 131.1 (x2), 131.0 (x2), 129.7 (x2), 128.6 (x2), 80.4, 72.1, 53.4, 22.2 ppm.
4.1.5. trans-2-(4-Nitrophenyl)cyclopropane-1-carboxylic acid (4)
In a 50 mL round bottom flask equipped with a stirring bar, trans-2-phenylcyclopropane-1-carboxylic acid (2.5 g, 15 mmol, 1.0 eq) was added. A solution of HNO3 (70% in H2O) (15 mL, 0.23 mol, 15 eq) was added dropwise at 0 °C. After 3h stirring at room temperature, additional HNO3 (2.0 mL) was added and, after 3h, the mixture was filtrated and washed over the filter with H2O. The crude mixture contained para:ortho nitro substituted product in a ratio 4:1. It was triturated in toluene and then filtrated. The solid over the filter was washed with petroleum ether to afford compound 4 as a white solid (1.0 g, 32%). 1H NMR (500 MHz, CD3OD): δ = 8.15 (d, J = 7.6 Hz, 2H), 7.37 (d, J = 7.6 Hz, 2H), 2.58-2.57 (m, 1H), 2.00-1.98 (m, 1H), 1.66-1.62 (m, 1H), 1.49-1.45 (m, 1H) ppm. 13H NMR (125 MHz, CD3OD): δ = 176.1, 150.0, 147.9, 128 (x2), 124.7 (x2), 26.6, 26.1, 18.3 ppm.
4.1.6. tert-Butyl (trans-2-(4-nitrophenyl)cyclopropyl) carbamate (5)
To a suspension of compound 4 (0.50 g, 2.4 mmol, 1.0 eq) in dry toluene, TEA (0.40 mL, 2.9 mmol, 1.2 eq) and DPPA (0.60 mL, 2.8 mmol, 1.2 eq) were added dropwise at 0 °C. Then tert-butanol (3.5 mL, 36 mmol, 15 eq) was added at room temperature and the mixture was heated at 105 °C. After 16h, di-tert-butyl-dicarbonate (Boc2O, 0.53 g, 2.4 mmol, 1.0 eq) was added at room temperature and the mixture was stirred for another 2h at 105 °C. The reaction was quenched with H2O (5.0 mL) and extracted with EtOAc (6 x 20 mL). The organic layers were combined, dried over MgSO4 and evaporated under reduced pressure. The crude mixture was purified by flash chromatography on silica gel (EtOAc:petroleum ether 1:6) affording compound 5 as a yellowish solid (0.18 g, 27%). Rf = 0.28 (EtOAc:petroleum ether 1:2). 1H NMR (500 MHz, CDCl3): δ = 8.12 (d, J = 8.4 Hz, 2H), 7.25 (d, J = 8.4 Hz, 2H), 4.92 (bs, 1H), 2.78 (m, 1H), 2.13 (m, 1H), 1.44 (s, 9H), 1.32-1.24 (m, 2H) ppm. 13C NMR (125 MHz, CDCl3): δ = 149.1, 146.4, 127.1, 123.8 (x4), 71.7, 33.7, 28.5 (x3), 25.6, 17.3 ppm.
4.1.7. tert-Butyl (trans-2-(4-aminophenyl)cyclopropyl) carbamate (6)
To a solution of compound 5 (0.14 g, 0.66 mmol, 1.0 eq) in THF: H2O 1:1 (1.8 mL) were added K2CO3 (64 mg, 0.46 mmol, 0.7 eq) and Pd/C (10%, 3.7 mg). A solution of Na2HPO2 (0.27 g, 2.5 mmol, 3.8 eq) in H2O (0.50 mL) was added to the mixture under H2 atmosphere. The mixture was stirred at 60 °C for 1.5h and, after completion, the reaction was quenched with H2O (20 mL) and extracted with Et2O (3 x 20 mL). The organic phases were combined, dried over MgSO4, and evaporated under reduced pressure. The residue was purified by column chromatography (EtOAc:petroleum ether 1:2) to afford 6 as a red oil (45 mg, 27%). Rf = 0.28 (EtOAc:petroleum ether 1:2). 1H NMR (400 MHz, CDCl3): δ = 6.93 (d, J = 8.4 Hz, 2H), 6.60 (d, J = 8.4 Hz, 2H), 4.89 (bs, 1H), 3.59 (bs, 2H), 2.61 (m, 1H), 1.93 (m, 1H), 1.45 (s, 9H), 1.36 (m, 1H), 1.26 (m, 1H) ppm. 13C NMR (125 MHz, CDCl3): δ = 156.5, 144.6, 130.8, 127.8 (x2), 115.3 (x2), 79.6, 32.1, 29.8, 28.5 (x3), 15.8 ppm.
4.1.8. tert-Butyl (trans-2-(4-(2-(4-benzoylbenzamido)pent-4-ynamido)phenyl)cyclopropyl) carbamate (7)
In a 10 mL round bottom flask equipped with a stirring bar, compound 3 (68 mg, 0.2 mmol, 1.0 eq) and 6 (58 mg, 0.2 mmol, 1.1 eq) were dissolved in DCM (3.0 mL). EDC (49 mg, 0.3 mmol, 1.2 eq) was added at 0 °C and the mixture was left stirring at room temperature overnight. The solvent was then evaporated and the residue was dissolved in EtOAc (5.0 mL), washed with 5% citric acid (3 x 5 mL), NaHCO3 (3 x 5 mL), brine (1 x 5 mL). The organic layer was evaporated, dried over MgSO4 and evaporated under reduced pressure. The residue was further purified by flash column chromatography (EtOAc:petroleum ether 1:3) to afford 7 as a red oil (14 mg, 12%). Rf = 0.44 (EtOAc:petroleum ether 1:2). 1H NMR (500 MHz, CDCl3): δ = 8.54 (bs, 1H), 7.94 (d, J = 8.1 Hz, 2H), 7.87 (d, J = 8.1 Hz, 2H), 7.80 (d, J = 7.4 Hz, 2H), 7.62 (t, J = 7.4 Hz, 1H), 7.51-7.48 (m, 2H), 7.41 (d, J = 8.4 Hz, 2H), 7.40 (d, J = 7.4 Hz, 1H), 7.09 (d, J = 8.4 Hz, 2H), 4.96-4.90 (m, 1H), 4.88 (bs, 1H), 3.03-2.99 (m, 1H), 2.86-2.81 (m, 2H), 2.67 (m, 1H), 2.18-2.17 (m, 1H), 1.50 (m, 1H), 1.44 (s, 9H), 1.42 (m, 1H) ppm.
4.1.9. N-(1-((4-trans-(2-aminocyclopropyl)phenyl)amino)-1-oxopent-4-yn-2-yl)-4-benzoylbenzamide hydrochloride (8)
To a solution of 7 (14 mg, 0.03 mmol) in THF (1.0 mL) was added HCl 4N in dioxane (0.80 mL) at 0 °C and the mixture was left stirring at room temperature overnight. The solvent was then evaporated and the residue was triturated with THF (2 x 1 mL) and diethyl ether (1 x 1 mL) and then lyophilized to afford 8 as a brown solid (3.6 mg, 30%).1H NMR (400 MHz, CD3OD): δ = 8.04 (d, J = 8.1 Hz, 2H), 7.86 (d, J = 8.1 Hz, 2H), 7.80 (d, J = 7.4 Hz, 2H), 7.67 (t, J = 7.4 Hz, 1H), 7.59-7.53 (m, 2H), 7.33-7.32 (m, 2H), 7.14 (d, J = 8.4 Hz, 2H), 4.75-4.74 (m, 1H), 2.91-2.87 (m, 2H), 2.85-2.80 (m, 1H), 2.45-2.42 (m, 1H), 2.36-2.35 (m, 1H), 1.50-1.45 (m, 1H), 1.38-1.34 (m, 1H) ppm. 13C NMR (100 MHz, CD3OD): δ = 196.4, 175.9, 174.7, 140.7, 138.6, 135.3, 134.2, 131.1, 130.9, 129.7 (x2), 129.2 (x2), 128.7 (x2), 127.8 (x2), 123.9 (x2), 121.8 (x2), 80.2, 72.5, 55.1, 31.9, 22.2, 22.1, 13.7. HRMS: C28H26O3N3 expected mass [M+H]+ 452.19687, mass found 452.19653.
4.1.10. tert-Butyl (trans-2-(4-(4-vinylbenzamido)phenyl) cyclopropyl)carbamate (9), tert-butyl (trans-2-(4-(pent-4-ynamido)phenyl)cyclopropyl)carbamate (10)
Example: synthesis of 10. 4-Ethynylbenzoic acid (74 mg, 0.44 mmol, 1.2 eq), EDCI (0.10 g, 0.53 mmol, 1.4 eq), HOBt (82 mg, 0.53 mmol, 1.4 eq) and TEA (0.20 mL, 1.4 mmol, 3.8 eq) were added sequentially to a solution of 6 (94.2 mg, 0.38 mmol, 1.0 eq) in dry DMF (2.8 mL). The resulting mixture was then stirred for 25h at room temperature and, after completion of the reaction, quenched with NaHCO3 saturated solution (25 mL). The aqueous solution was extracted with EtOAc (4 x 20 mL), washed with KHSO4 solution 0.1N (2 x 10 mL), NaHCO3 saturated solution (1 x 10 mL) and brine (1 x 10 mL), dried over anhydrous Na2SO4 and finally concentrated under vacuum. The crude was then purified by column chromatography on silica gel eluting with a mixture EtOAc:hexane 22:78 to afford 10 as a pink solid (80 mg, 75%). Rf = 0.15 (EtOAc:n-hexane 1:2). 1H NMR (400 MHz, CDCl3): δ = 7.83-7.85 (d, J = 7.6 Hz, 2H), 7.59 (bs, 1H), 7.61-7.63 (d, J = 7.2 Hz, 2H), 7.53-7.55 (d, J = 7.2 Hz, 2H), 7.16-7.18 (d, J = 7.2 Hz, 2H), 4.80-4.89 (bs, 1H), 3.41 (s, 1H), 2.71-2.72 (m, 1H), 2.05-2.06 (m, 1H), 1.48 (s, 9H), 1.15-1.19 (m, 2H) ppm.
4.1.11. tert-Butyl (trans-2-(4-(pent-4-enamido)phenyl) cyclopropyl)carbamate (11)
Pent-4-enoyl chloride (0.047 mL, 0.43 mmol, 1.1 eq) was slowly added at 0 °C to a solution of 6 (90 mg, 0.39 mmol, 1.0 eq) and TEA (0.07 mL, 0.50 mmol, 1.3 eq.) in dry DCM (16 mL). The resulting mixture was stirred at room temperature for 1.5h. Afterwards, the reaction mixture was quenched with 10 mL of NaHCO3 saturated solution and extracted with EtOAc (4 x 10 mL). The organic layer was washed with KHSO4 solution 0.1M (2 x 5 mL), and brine (2 x 5 mL), dried over anhydrous Na2SO4 and concentrated in vacuum. The residue was purified by column chromatography on silica gel eluting with a mixture EtOAc:hexane 1:3 to afford 11 as a pink solid (56 mg, 76%). Rf = 0.15 (EtOAc:n-hexane 1:2). 1H NMR (400 MHz, CDCl3): δ = 7.40-7.42 (d, J = 8.4 Hz, 2H), 7.10-7.13 (m, 3H), 5.86-5.94 (m, 1H), 5.06-5.16 (m, 2H), 4.81-4.88 (bs, 1H), 2.67-2.69 (m, 1H), 2.41-2.43 (m, 4H), 2.02-2.04 (m, 1H), 1.47 (s, 9H), 1.12-1.14 (m, 2H) ppm.
4.1.12. N-(4-(trans-2-aminocyclopropyl)phenyl)-4-vinyl benzamide hydrochloride (12), N-(4-(trans-2-amino cyclopropyl)phenyl)pent-4-enamide (13), N-(4-(trans-2-aminocyclopropyl)phenyl)pent-4-ynamide hydrochloride (14)
The same deprotection procedure was followed as for the synthesis of compound 8. 12: 1H NMR (400 MHz, (CD3)2SO): δ = 10.23 (bs, 1H), 8.43 (s, 3H), 7.94-7.96 (d, J = 7.6 Hz, 2H), 7.71-7.73 (d, J = 7.6 Hz, 2H), 7.62-7.64 (d, J = 7.6 Hz, 2H), 7.13-7.16 (d, J = 8.4 Hz, 2H), 6.79-6.86 (m, Jtrans = 17.6 Hz, Jcis = 12 Hz, 1H), 5.97-6.02 (d, Jtrans = 17.2 Hz, 1H), 5.39-5.42 (d, Jcis = 11.2 Hz, 1H), 2.77-2.79 (m, 1H), 2.31-2.33 (m, 1H), 1.33-1.38 (m, 1H), 1.16-1.24 (m, 1H) ppm. 13C NMR (100 MHz, (CD3)2SO): δ = 164.9, 140.1, 137.5, 135.9, 134.4, 133.9, 128.1(x2), 126.4(x2), 126.0(x2), 120.4(x2), 116.5, 30.5, 20.4, 13.2 ppm. HRMS: C18H19ON2 expected mass [M+H]+ 279.14919, mass found 279.14923.
13: 1H NMR (400 MHz, (CD3)2SO): δ = 9.92 (bs, 1H), 8.13-8.41 (m, 3H), 7.50-7.52 (d, J = 8.4 Hz, 2H), 7.06-7.08 (d, J = 8.4 Hz, 2H), 5.80-5.90 (m, 1H), 5.04-5.08 (d, Jtrans = 16 Hz, 1H), 4.97-4.99 (d, Jcis = 10 Hz, 1H), 2.71-2.75 (m, 1H), 2.37-2.41 (m, 2H), 2.30-2.35 (m, 2H), 2.23-2.25 (m, 1H), 1.29-1.33 (m, 1H), 1.12-1.18 (m, 1H) ppm. 13C NMR (100 MHz, (CD3)2SO): δ = 170.4, 137.7, 137.6, 133.6, 126.5(x2), 119.0(x2), 115.2, 35.4, 30.4, 29.1, 20.3, 13.1 ppm. HRMS: C14H19ON2 expected mass [M+H]+ 231.14919, mass found 231.14922.
14: 1H NMR (400 MHz, (CD3)2SO): δ = 10.00 (bs, 1H), 8.41 (s, 3H), 7.52-7.54 (d, J = 8.4 Hz, 2H), 7.07-7.10 (d, J = 8.8 Hz, 2H), 2.80 (s, 1H), 2.73-2.77 (m, J = 5.8 Hz, 1H), 2.43-2.46 (m, 2H), 2.25-2.30 (m, 1H), 1.32-1.37 (m, J = 7.5 Hz, 1H), 1.13-1.18 (m, J = 7.0 Hz, 1H) ppm. 13C NMR (100 MHz, (CD3)2SO): δ = 169.2, 137.5, 133.8, 126.5(x2), 119.0(x2), 83.7, 71.5, 35.1, 30.4, 20.3, 14.1, 13.1 ppm. HRMS: C14H17ON2 expected mass [M+H]+ 229.13354, mass found 229.13354.
4.1.13. 10-Acetyl-10H-phenoxazine-3,7-diyl diacetate (15)
The synthesis of Amplex Red reagent was performed as described in literature.57 Yield 69%. 1H NMR (500 MHz, CDCl3): δ = 7.46 (m, 2H), 6.91-6.88 (m, 4H), 2.33 (s, 3H), 2.30 (s, 6H) ppm. 13C NMR (125 MHz, CDCl3): δ = 169.2 (x2), 151.2, 149.0 (x2), 127.0, 125.5, 116.9, 111.0, 23.1, 21.2 (x2) ppm.
4.1.14. 10-Acetyl-3,7-dihydroxyphenoxazine (ADHP) (16)
The synthetic procedure afforded a white solid (64%). 1H NMR (500 MHz, (CD3)2SO): δ = 9.7 (bs, 2H), 7.33 (m, 2H), 6.53 (m, 4H), 2.16 (s, 3H) ppm. 13C NMR (125 MHz, (CD3)2SO): δ = 169.0, 156.1, 151.0, 125.8, 121.0, 22.6 ppm.
4.2. LSD1 inhibition studies
4.2.1. General
LSD1 (KDM1A) (lysine (K)-specific demethylase 1A) or AOF2 was purchased from BPS Bioscience (Catalog No 50097). Monomethylated histone peptide H3K4N(CH3) was purchased from Pepscan and Horseradish Peroxidase (HRP) from Pierce (Catalog No 31490). The reagents for buffer preparation were purchased from Merck, Netherlands. Reactions were conducted in black 96-well flat bottom microplates (Corning® Costar®, Corning Incorporated, NY). The fluorescence measurements were carried out in a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek, USA) and the gain setting of the instrument was adjusted to 70. GraphPad Prism 5.0, GraphPad Software, Inc. GraphPad was used for the determination of kinetic values and of the half maximal inhibitory concentration (IC50). Non-linear regression was used for data fitting.
4.2.2. LSD1 assay
The assay buffer consisted of Tris-HCl 50 mM, pH 8.0 containing 0.05 mg/mL Bovine Serum Albumine (BSA). Shortly before use, LSD1 was diluted 500 times with LSD1 assay buffer. 35 μL of the diluted enzyme were used per well in the assay. Monomethylated histone peptide H3K4N(CH3) was dissolved in the assay buffer to afford 2 mM stock solution. After proper dilutions of this stock, 5 µL of the substrate were used per well in the assay. Horseradish Peroxidase (HRP, 300 U/mg) was diluted to 10 U/mL using the LSD1 assay buffer. A stock solution of 10 mM Amplex Red in DMSO was prepared, protected from light and used quickly. A solution of HRP-Amplex Red was prepared shortly before use, stored on ice and protected from light. 30 µL of the stock solution of Amplex Red and 180 µL of the solution of HRP were added in 2.790 mL of LSD1 buffer. The addition of 50 μL of this solution to the reaction wells is enough to use the H2O2 formed during the demethylation reaction for the oxidation of Amplex Red.
The assay was based on a procedure described previously.58 The experiment was performed in triplicate. The inhibitors’ stock solutions were prepared using the LSD1 assay buffer. 10 μL of each concentration of the inhibitor were added in the inhibitor wells. 10 μL of the solvent used to dissolve the inhibitor were added in the background and positive control wells. 50 μL of HRP-Amplex Red solution were added in all wells. 35 μL of the diluted LSD1 were added in the positive and inhibitor wells and the contents were mixed thoroughly. The plate was covered and incubated at room temperature for 15 min. 5 μL of a 30 µM solution of the substrate were added in all wells. The plate was shaken and the fluorescence was monitored for 30 minutes at λem= 460 nm and λex= 390 nm. The slope of the linear part was calculated and the value of the background wells was subtracted from itself and the values of the positive control and inhibitor wells. Non-linear regression was used to fit the data to the log(inhibitor) vs. response curve.
4.2.3. Testing for developer interference
10 μL of of appropriate stock solutions of the inhibitor were added in the inhibitor wells to afford final concentrations of 3-50 µM. 10 μL of the solvent of the inhibitor were added in the negative and positive control wells. 50 μL of HRP-Amplex Red Solution were added in all wells. 35 μL of LSD1 buffer were added in all wells and the contents were mixed thoroughly. The plate was covered and incubated at room temperature for 15 min. 5 μL of buffer were added in the negative control and 5 μL of 80 µM solution of H2O2 were added in the inhibitor and positive control wells. The plate was shaken and the fluorescence was measured at λem= 460 nm and λex= 390 nm. No interference was observed.
4.2.4. Kinetic studies-Kitz-Wilson analysis
LSD1 (41 ng) was incubated with four different concentrations of each compound for 0, 5, 10 and 15 min in presence of HRP-Amplex Red solution. Positive and negative control wells were also set up. The substrate was then added at a final concentration of 1.5 µM and the enzyme activity was monitored as described above. The experiments were performed in triplicate. The slope of the linear part was calculated and the value of the background wells was subtracted from itself and the values of the positive control and inhibitor wells. A time-dependent inhibition mechanism was observed in all cases. The natural logarithm (ln) values of the remaining enzyme activity for each inhibitor concentration were plotted versus the respective incubation time. The slope of the resulting linear graphs corresponded to the reaction rates. Finally, the 1/rate values were plotted against 1/[inhibitor] to generate linear graphs, fitting to the following equation: 1/rate=slope.(1/[inhibitor])+ki (ki=inactivation rate constant). The affinity rate constant was calculated from the following equation: Ki= slope.ki. For all the compounds the Ki values ranged between 3-5 µM and the ki between 0.3-0.4 min-1 indicating the very fast binding of the TCP-containing molecules.
4.3. Labeling studies
4.3.1. General
Human cervical cancer cells were used as a source of LSD1 activity (HeLa S3, ATCC® CCL-2.2™, USA). Dulbecco’s Modified Eagle Medium (DMEM) and Fetal Bovine Serum (FBS) were purchased from Life Technologies. The antibodies used against LSD1 were purchased from ThermoFischer scientific (Catalog No PA5-17361 for the identification of intracellular LSD1 and Catalog No PA5-23307 for the commercially available human recombinant LSD1). Fermentas PageRuler™ Prestained Protein Ladder was used as a ladder during sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The loading buffer (4x) consists of 20% of 0.2 M Tris-HCl pH 6.8, 8.9% of SDS, 40% of glycerol, 10% of 0.05 M EDTA, 0.09% of bromophenol blue, 21% of deionized H2O. Gels were stained with the coomassie based gel stain InstantBlue™ (Expedeon Ltd, Harston, Cambridgeshire, UK). Coomassie Protein Assay Reagent (950 mL) from Thermo Scientific was used for the Bradford assay and the absorbance was measured on a SPECTROstar Omega–UV/Vis absorbance spectrophotometer microplate reader from BMG Labtech. Chemiluminescence imaging was performed in G:BOX from Syngene under no light and no filter.
4.3.2. ‘Click’ reactions
10 µL of 10 mM solution of probe 8 in DMSO, 20 µL of azide-PEG3-biotin (5 mM in water), 40 µL of premixed catalyst solution (20 µL TABTA 20 mM in H2O, 20 µL CuSO4 5 mM in water), 20 µL of sodium acorbate (0.1 M in H2O) and 10 µL of DMSO were incubated at 32 °C for 5h. A blank reaction using 10 µL of DMSO was also set up. LC-MS revealed 50% conversion to the desired clicked-probe 17. C46H57N9O8S expected mass [M+H]+ 896.41, mass found 896.41.
The same conditions were applied for the ‘click’ reaction using compound 2 (90% conversion, HRMS: C38H49N7O9S expected mass [M+H]+ 780.33852, mass found 780.33706) and 14 (78% conversion, HRMS: C32H48N8O6S [M+H]+ 673.34948, pre-clicked probe 18).
4.3.3. Labeling of human recombinant LSD1
1.25 µg of LSD1 (0.25 mg/mL, 5 µL) were incubated with 5, 10, 20 and 40 µM (final concentration) of pre-clicked probes 17 and 18 for 10 min (50 µL reaction in assay buffer). A blank reaction with no probe was also set up using the equivalent amount of blank ‘click’ reaction conditions (see above). In case of probe 17 the samples were then irradiated at 365 nm for 5 min. For protein detection methods via the anti-LSD1 antibody or HRP-conjugated Streptavidin, see Supplementary Information.
4.3.4. Labeling of endogenous LSD1 in Hela nuclear extracts
HeLa cells were grown in DMEM supplemented with 10% FBS and 100 U/mL penicillin/streptomycin, and maintained at 37 °C and 5% CO2 to afford approximately 68x105 cells. Hela nuclear extraction was performed as described previously.59 12.5, 25, 50 and 100 µM of probe 17 were incubated with the protein extracts (3.0 mg/mL in 100 µL assay buffer) for 10 min at room temperature followed by 5 min irradiation at 365 nm. A blank reaction with no probe was also set up using the equivalent amount of blank ‘click’ reaction conditions. In samples with 0, 12.5 and 25 µM, small molecules were removed by centrifuge in Viva spin columns (Sartorius, 5 kDa membrane cut) at 4000 g for 12 min (x3). Samples were recollected by centrifuge at 3000 g for 2 min. Samples containing 50 and 100 µM of the probe were treated with DMF (400 µL) and the proteins were left precipitating overnight at 4 °C. Next morning, samples were centrifuged at 5200 rcf for 30 min at 4 °C (x3, pellets resuspended with ice-cold MeOH). The final protein pellets were resuspended in 10% SDS (150 µL). Further protein detection was performed as described in the Supplementary Information. Same experimental conditions were applied for labeling of nuclear extracts with probe 18, where 0, 12.5, 25, 50 and 100 µM were incubated with 3 mg/mL of protein extracts for 15 min at room temperature. Samples were then treated with DMF (400 µL) and the proteins were left precipitating overnight at 4 °C. Detection was performed as described for probe 17.
4.3.5. Control reactions
Regarding human recombinant LSD1, 1.25 µg of purified enzyme (0.25 mg/mL, 5 µL) in 41 µL of Tris 50 mM pH 8 were incubated with probe 17 (final concentration 40 µM) for 5 min under UV irradiation at 365 nm (positive control). The equivalent amount of blank ‘click’ reaction conditions (see above) were used as negative control. For heat-denaturation control (ΔT) experiment, the same amount of LSD1 was pre-heated at 96 °C for 15 min and subsequently incubated with the probe as described for positive control. To check that the probe targets the FAD, the same amount of LSD1 was incubated with pre-clicked compound 2 (final concentration 40 µM) for 5 min under UV irradiation at 365 nm. The same experiments were repeated for probe 18 (final concentration 10 µM, no UV irradiation) and pre-clicked compound 2 was used at a final concentration of 10 µM. Detection was performed as described above.
Nuclear extracts (3 mg/mL) were incubated with 100 µM of pre-clicked compound 2 or probe 17. The control reactions were performed as described above for human recombinant LSD1. Proteins were precipitated with DMF (400 µL) overnight. Further treatment and detection were performed as mentioned above for labeling of nuclear extracts.
4.3.6. Treatment with LSD1 inhibitors
Regarding human recombinant LSD1, 0.6 µg of purified enzyme (0.25 mg/mL, 2.5 µL) in 38.5 µL of Tris 50 mM pH 8 were incubated with 12 (10 mM, 5 µL), TCP (15 mM, 5 µL) or DMSO (5 µL, positive control) for 30 min at room temperature. Pobe 17 (final concentration 40 µM) was then added in all samples followed by UV irradiation at 365 nm for 2 min. The same experiment was repeated using pre-clicked probe 18 (final concentration 10 µM) incubated for 2 min after treatment with 10 mM of compound 12. Detection was performed as described above using longer exposure times for luminescence detection (1h).
HeLa nuclear extracts (3 mg/mL in LSD1 buffer) were incubated with 1.5 mM of TCP/1 mM of 12/ equivalent amount of DMSO for 30 min at room temperature followed by treatment with 100 µM of probe 17 and 2 min UV irradiation or 50 µM probe 18 and 2 min incubation at room temperature. Ablank reaction was also set up. Further treatment and detection were performed as mentioned above for labeling of nuclear extracts.
Supplementary Material
Acknowledgments
This work was financially supported by a VIDI grant (723.012.005) from the Netherlands Organization for Scientific Research and an ERC starting grant (309782) from the European Union to F. J. D. Funds from PRIN 2016 (prot. 20152TE5PK) and IIT-Sapienza Project grants are also acknowledged by A. M. PRIN 2012 (prot. 2012CTAYSY) and AIRC-TRIDEO 2015 (Id.17515) are also acknowledged by D. R. We acknowledge the COST actions ‘biomimetic radical chemistry’ CM1201 and ‘epigenetic chemical biology’ CM-1406.
References and notes
- 1.Bhaumik SR, Smith E, Shilatifard A. Nat Struct Mol Biol. 2007;14:1008. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 2.Sims RJ, 3rd, Nishioka K, Reinberg D. Trends Genet. 2003;19:629. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 3.Anand R, Marmorstein R. J Biol Chem. 2007;282:35425. doi: 10.1074/jbc.R700027200. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Cell. 2004;119:941. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. J Biol Chem. 2009;284:17775. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Y. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 7.Amente S, Lania L, Majello B. Biochim Biophys Acta. 2013;1829:981. doi: 10.1016/j.bbagrm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Burg JM, Link JE, Morgan BS, Heller FJ, Hargrove AE, McCafferty DG. Biopolymers. 2015;104:213. doi: 10.1002/bip.22643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballas N, Battaglioli E, Atouf F, Andres ME, Chenoweth J, Anderson ME, Burger C, Moniwa M, Davie JR, Bowers WJ, Federoff HJ, et al. Neuron. 2001;31:353. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- 10.Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. Nature. 2005;437:436. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 11.Kooistra SM, Helin K. Nat Rev Mol Cell Biol. 2012;13:297. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 12.Rotili D, Mai A. Genes Cancer. 2011;2:663. doi: 10.1177/1947601911417976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng Z, Yao Y, Zhou C, Chen F, Wu F, Wei L, Liu W, Dong S, Redell M, Mo Q, Song Y. J Hematol Oncol. 2016;9:24. doi: 10.1186/s13045-016-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BA, Nakamura Y, et al. Int J Cancer. 2011;128:574. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 15.Vasilatos SN, Katz TA, Oesterreich S, Wan Y, Davidson NE, Huang Y. Carcinogenesis. 2013;34:1196. doi: 10.1093/carcin/bgt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortez V, Mann M, Tekmal S, Suzuki T, Miyata N, Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK, Vadlamudi RK. Breast Cancer Res. 2012;14:R108. doi: 10.1186/bcr3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Breast Cancer Res Treat. 2012;131:777. doi: 10.1007/s10549-011-1480-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrisani OM. Viruses. 2013;5:858. doi: 10.3390/v5030858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill JM, Quenelle DC, Cardin RD, Vogel JL, Clement C, Bravo FJ, Foster TP, Bosch-Marce M, Raja P, Lee JS, Bernstein DI, et al. Sci Transl Med. 2014;6:265ra169. doi: 10.1126/scitranslmed.3010643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perkins B, Jones RJ, Woster PM, Casero RA., Jr Clin Cancer Res. 2009;15:721. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willmann D, Lim S, Wetzel S, Metzger E, Jandausch A, Wilk W, Jung M, Forne I, Imhof A, Janzer A, Kirfel J, et al. Int J Cancer. 2012;131:2704. doi: 10.1002/ijc.27555. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T, Zhang H. Cancer Res. 2011;71:7238. doi: 10.1158/0008-5472.CAN-11-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorna V, Theisen ER, Stephens B, Warner SL, Bearss DJ, Vankayalapati H, Sharma S. J Med Chem. 2013;56:9496. doi: 10.1021/jm400870h. [DOI] [PubMed] [Google Scholar]
- 24.Mould DP, McGonagle AE, Wiseman DH, Williams EL, Jordan AM. Med Res Rev. 2015;35:586. doi: 10.1002/med.21334. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt DM, McCafferty DG. Biochemistry. 2007;46:4408. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 26.Vianello P, Botrugno OA, Cappa A, Ciossani G, Dessanti P, Mai A, Mattevi A, Meroni G, Minucci S, Thaler F, Tortorici M, et al. Eur J Med Chem. 2014;86:352. doi: 10.1016/j.ejmech.2014.08.068. [DOI] [PubMed] [Google Scholar]
- 27.Zheng YC, Yu B, Jiang GZ, Feng XJ, He PX, Chu XY, Zhao W, Liu HM. Curr Top Med Chem. 2016;16:2179. doi: 10.2174/1568026616666160216154042. [DOI] [PubMed] [Google Scholar]
- 28.Yang M, Culhane JC, Szewczuk LM, Jalili P, Ball HL, Machius M, Cole PA, Yu H. Biochemistry. 2007;46:8058. doi: 10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 29.Khan MN, Suzuki T, Miyata N. Med Res Rev. 2013;33:873. doi: 10.1002/med.21269. [DOI] [PubMed] [Google Scholar]
- 30.Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, Edmondson DE, et al. J Am Chem Soc. 2010;132:6827. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- 31.Mimasu S, Sengoku T, Fukuzawa S, Umehara T, Yokoyama S. Biochem Biophys Res Commun. 2008;366:15. doi: 10.1016/j.bbrc.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 32.Ueda R, Suzuki T, Mino K, Tsumoto H, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N. J Am Chem Soc. 2009;131:17536. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- 33.Ogasawara D, Itoh Y, Tsumoto H, Kakizawa T, Mino K, Fukuhara K, Nakagawa H, Hasegawa M, Sasaki R, Mizukami T, Miyata N, et al. Angew Chem Int Ed Engl. 2013;52:8620. doi: 10.1002/anie.201303999. [DOI] [PubMed] [Google Scholar]
- 34.Valente S, Rodriguez V, Mercurio C, Vianello P, Saponara B, Cirilli R, Ciossani G, Labella D, Marrocco B, Ruoppolo G, Botrugno OA, et al. ACS Med Chem Lett. 2014;6:173. doi: 10.1021/ml500424z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vianello P, Botrugno OA, Cappa A, Dal Zuffo R, Dessanti P, Mai A, Marrocco B, Mattevi A, Meroni G, Minucci S, Stazi G, et al. J Med Chem. 2016;59:1501. doi: 10.1021/acs.jmedchem.5b01209. [DOI] [PubMed] [Google Scholar]
- 36.Mimasu S, Umezawa N, Sato S, Higuchi T, Umehara T, Yokoyama S. Biochemistry. 2010;49:6494. doi: 10.1021/bi100299r. [DOI] [PubMed] [Google Scholar]
- 37.Khan MN, Tsumoto H, Itoh Y, Ota Y, Suzuki M, Ogasawara D, Nakagawa H, Mizukami T, Miyataa N, Suzuki T. MedChemComm. 2015;6:407. [Google Scholar]
- 38.Feng Z, Yao Y, Zhou C, Chen F, Wu F, Wei L, Liu W, Dong S, Redell M, Mo Q, Song Y. J Hematol Oncol. 2016;9:24. doi: 10.1186/s13045-016-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch JT, Harris WJ, Somervaille TC. Expert Opin Ther Targets. 2012;16:1239. doi: 10.1517/14728222.2012.722206. [DOI] [PubMed] [Google Scholar]
- 40.Cravatt BF, Wright AT, Kozarich JW. Annu Rev Biochem. 2008;77:383. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 41.Speers AE, Adam GC, Cravatt BF. J Am Chem Soc. 2003;125:4686. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 42.Fonović M, Bogyo M. Expert Rev Proteomics. 2008;5:721. doi: 10.1586/14789450.5.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krysiak JM, Kreuzer J, Macheroux P, Hermetter A, Sieber SA, Breinbauer R. Angew Chem Int Ed Engl. 2012;51:7035. doi: 10.1002/anie.201201955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Zhang CW, Ge J, Qian L, Chai BH, Zhu Q, Lee JS, Lim KL, Yao SQ. Angew Chem Int Ed Engl. 2015;54:10821. doi: 10.1002/anie.201504441. [DOI] [PubMed] [Google Scholar]
- 45.Dormán G, Prestwich GD. Biochemistry. 1994;33:5661. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka Y, Bond MR, Kohler JJ. Mol Biosys. 2008;4:473. doi: 10.1039/b803218a. [DOI] [PubMed] [Google Scholar]
- 47.Martell J, Weerapana E. Molecules. 2014;19:1378. doi: 10.3390/molecules19021378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HS, Dimla RD, Schultz PG. Bioorg Med Chem Lett. 2009;19:5222. doi: 10.1016/j.bmcl.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valente S, Rodriguez V, Mercurio C, Vianello P, Saponara B, Cirilli R, Ciossani G, Labella D, Marrocco B, Monaldi D, Ruoppolo G, et al. Eur J Med Chem. 2015;94:163. doi: 10.1016/j.ejmech.2015.02.060. [DOI] [PubMed] [Google Scholar]
- 50.Ourailidou ME, van der Meer J-Y, Baas B-J, Jeronimus-Stratingh M, Gottumukkala AL, Poelarends GJ, Minnaard AJ, Dekker FJ. ChemBioChem. 2014;15:209. doi: 10.1002/cbic.201300714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ourailidou ME, Dockerty P, Witte M, Poelarends GJ, Dekker FJ. Org Biomol Chem. 2015;13:3648. doi: 10.1039/c4ob02502d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Mol Cell. 2006;23:377. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. FEBS Lett. 2005;579:2203. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 54.Luka Z, Moss F, Loukachevitch LV, Bornhop DJ, Wagner C. Biochemistry. 2011;50:4750. doi: 10.1021/bi200247b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsukada Y, Zhang Y. Methods. 2006;40:318. doi: 10.1016/j.ymeth.2006.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Mol Cell. 2005;19:857. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 57.Von der Eltz H, Guder H-J, Muhlegger K, inventors. US4900822 A. 1990
- 58.Valente S, Rodriguez V, Mercurio C, Vianello P, Saponara B, Cirilli R, Ciossani G, Labella D, Marrocco B, Monaldi D, Ruoppolo G, et al. Eur J Med Chem. 2015;94:163. doi: 10.1016/j.ejmech.2015.02.060. [DOI] [PubMed] [Google Scholar]
- 59.Szymanski W, Ourailidou ME, Velema WA, Dekker FJ, Feringa BL. Chem - A Eur J. 2015;21:16517. doi: 10.1002/chem.201502809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.