Abstract
Connexins or innexins form gap junctions, while claudins and occludins form tight junctions. In this study, statistical data, derived using novel software, indicate that these four junctional protein families and eleven other families of channel and channel auxiliary proteins are related by common descent and comprise the Tetraspan (4 TMS) Junctional Complex (4JC) Superfamily. These proteins all share similar 4 transmembrane α-helical (TMS) topologies. Evidence is presented that they arose via an intragenic duplication event, whereby a 2 TMS-encoding genetic element duplicated tandemly to give 4 TMS proteins. In cases where high resolution structural data were available, the conclusion of homology was supported by conducting structural comparisons. Phylogenetic trees reveal the probable relationships of these 15 families to each other. Long homologues containing fusions to other recognizable domains as well as internally duplicated or fused domains are reported. Large “fusion” proteins containing 4JC domains proved to fall predominantly into family-specific patterns as follows: (1) the 4JC domain was N-terminal; (2) the 4JC domain was C-terminal; (3) the 4JC domain was duplicated or occasionally triplicated and (4) mixed fusion types were present. Our observations provide insight into the evolutionary origins and subfunctions of these proteins as well as guides concerning their structural and functional relationships.
Keywords: gap junctions, tight junctions, connexins, occludins, Ca2+ channels, superfamily
Graphical abstract
1. INTRODUCTION
Connexins and innexins are the principal core proteins of gap junctions, while claudins and occludins are tight junctional core proteins [1]. All have the same topology with four α-helical transmembrane segments (TMSs), and all exhibit well-conserved extracytoplasmic cysteines that either are known to, or potentially can, form extracytoplasmic disulfide bridges [2, 3].
In metazoan tissues, adjacent cells are often connected by connexin- or innexin-containing gap junctional channels [4] as well as claudin- and occludin-containing tight junctions [2, 5–7]. All of these junctional proteins span the two plasma membranes. In the former cases, docking of the two half channels in the plasma membranes of two adjacent cells creates hexameric tori of junctional proteins enclosing an aqueous pore [8]. These densely packed gap junctional channels allow cells to exchange ions and small messenger molecules such as Ca2+ and cyclic nucleotides as well as oligonucleotides. They also coordinate electrical activities in excitable tissues [9].
In 2003, our laboratory published sequence, topological and phylogenetic analyses of the proteins that comprise the connexin, innexin, claudin and occludin families [1]. A multiple alignment of the sequences of each family was used to derive average hydropathy and similarity plots as well as a phylogenetic tree. Analyses led to the following conclusions: (1) In all four families, the most conserved regions of the proteins are the four TMSs, although the extracytoplasmic loops between TMSs 1 and 2, and TMSs 3 and 4 are usually well conserved [4]. (2) The phylogenetic trees revealed sets of orthologues except for the innexins where phylogeny primarily reflected the organismal source, probably due to a lack of close organismal sequence data [5]. (3) The two halves of the connexins exhibited similarities suggesting that they were derived from a common origin by an internal gene duplication event, but this possibility could not be demonstrated [6]. (4) Conserved cysteyl residues in the connexins and innexins pointed to a similar extracellular structure involved in hemichannel docking to create intercellular communication channels. Similar roles in homomeric interactions for conserved extracellular residues in the claudins and occludins were suggested. The apparent lack of obvious sequence and motif similarities between the four different families indicated that, if they did evolve from a common ancestral gene, they had diverged substantially to fulfill different functions.
In this work, statistical and other methods provide strong evidence that these four junctional protein families, as well as eleven additional families of ion (most frequently Ca2+) channel and channel-affiliated proteins have, in fact, arisen from a common origin. The fifteen families that comprise the 4JC superfamily are listed with their characteristics in Table 1.
Table 1.
Families included in the 4JC Superfamily.a
| Family Name | Family Abb’n | TC# | Phyla | Avg Seq. Length (aas) ±SD | #s of members | # of potential fusion proteins | Pfam designations | References |
|---|---|---|---|---|---|---|---|---|
| Connexin | Connexin | 1.A.24 | Animals | 323 ± 107 | 1690 | 12 | Connexin (PF00029), Connexin43 (PF03508), Connexin50 (PF03509) | [10] |
| Innexin | Innexin | 1.A.25 | Animals + dsDNA viruses (3%) | 389 ± 138 | 7971 | 16 | Innexin (PF00876), Pannexin_like (PF12534), LRR_8 (PF13855) | [11] |
| Intracellular Chloride Channel | ICC | 1.A.36 | Animals + dsDNA viruses (1%) | 494 ± 120 | 260 | 1 | MCLC (PF05394) | [12] |
| Plasmolipin | Plasmolipin | 1.A.64 | Animals | 169 ± 34 | 2413 | 1 | MARVEL (PF01284) | [13] |
| The Low Affinity Ca2+ Channel | LACC | 1.A.81 | Fungi | 277 ± 31 | 263 | 0 | Fig 1 (PF12351) | [14] |
| Hair Cell Mechanotransduction Channel | HCMC | 1.A.82 | Animals | 221 ± 34 | 511 | 0 | L_HMGIC_fpl (PF10242) | [15] |
| Calcium Homeostasis Modulator Ca2+ Channel | CALHM-C | 1.A.84 | Animals | 351 ± 56 | 537 | 4 | Ca_hom_mod (PF14798) | [16] |
| Claudin Tight Junction | Claudin | 1.H.1 | Animals | 225 ± 35 | 4770 | 1 | PMP22_Claudin (PF00822), SUR7 (PF06687) | [17] |
| Invertebrate PMP22- Claudin | Claudin2 | 1.H.2 | Animals | 243 ± 75 | 537 | 2 | Clc-like (PF07062), Claudin_2 (PF13903) | [18] |
| Ca2+ Channel Auxiliary Subunit γ1–γ8 | CCAγ | 8.A.16 | Animals | 231 ± 60 | 3839 | 3 | PMP22_Claudin (PF00822), GSG-1 (PF07803), Claudin_2 (PF13903), TMEM37 (PF15108) | [19] |
| Non-Classical Protein Exporter | NCPE | 9.A.27 | Fungi | 170 ± 13 | 584 | 1 | MARVEL (PF01284), NCE101 (PF11654) | [20] |
| Clarin | CLRN | 9.A.46 | Animals | 218 ± 42 | 407 | 1 | None | [21] |
| Occludin | Occludin | 9.B.41 | Animals | 531 ± 181 | 335 | 4 | MARVEL (PF01284), Occludin_ELL (PF07303) | [22] |
| Tetraspan Vesicle Membrane Protein | TVP | 9.B.130 | Animals | 258 ± 116 | 812 | 3 | MARVEL (PF01284) | [23] |
| MscS/DUF475 | DUF475 | 9.B.179 | Actinobacteria | 298 ± 159 | 361 | 0 |
Family names and abbreviations are provided in columns 1 and 2, respectively, while family numbers in the Transporter Classification Database (TCDB; www.tcdb.org) [24–26] are provided in column 3. Class 1 indicates a channel function. Class 8 indicates a transporter auxiliary function while class 9 indicates that insufficient information is available to establish the mechanisms of action of these proteins. Phylum representation for each protein family of the 4JC superfamily is provided in column 4. Average sizes of the proteins in each family ± standard deviations (SD) are provided in column 5, while estimates of family sizes, expressed in numbers of proteins retrieved with the Psi-BLAST program with two iterations and a cutoff of 90% to eliminate redundancies and very similar (>90%) sequences can b found in column 6. Potential fusion proteins (column 7) are those that are at least 2x larger than the familial average. Pfam designation(s) for memebers of a given family, when available, are provided in column 8, and a representative reference is given in column 9. Additional references for each family can be found in TCDB.
Evidence is presented that members of these families arose following a pathway involving duplication of a primordial 2 TMS element to give rise to the current 4 TMS proteins. The gap junctional innexins and connexins proved to be more closely related to each other although the tight junctional occludins and claudins do not appear to be closely related. We suggest that the innexins, present primarily in invertebrates, were the precursors of connexins in vertebrates. Vertebrate pannexins, members of the innexin family [4], may have been obtained by vertebrates from invertebrates via horizontal transfer after vertebrates diverged from invertebrates, giving rise to the current families of connexins and innexins [1].
2. METHODS
Representative members (the first member of each TC subfamily within the fifteen families included in the 4JC superfamily (Table 1) were obtained from the Transporter Classification Database (TCDB; www.tcdb.org) and expanded using a PSI-BLAST search tool within the Protocol1 program with an e-value cut-off of 0.005 and two iterations [27]. Redundant sequences were then removed using the CD-HIT component of Protocol1 with a 0.8 (80%) identity cutoff [28]. Using this approach, all sequences retained for analysis differ from each other by at least 20%.
Comparison scores, expressed in standard deviations (SD), were determined using the GSAT program [28]. GSAT performs a pairwise alignment using the Needleman-Wunsch algorithm, followed by 200 additional alignments using a shuffled sequence in each round. A standard score (z-score) is calculated and returned by the program. High scoring pairs (HSPs) were selected between families using the Protocol2 program [27]. Protocol2 performs a Smith-Waterman search between two FASTA files and selects the highest scoring pairs (HSPs) with overlapping TMSs. The HSPs are then analyzed with GSAT using 200 shuffles, and a standard score is determined for each. The greatest HSPs for each family comparison are then selected and again run through GSAT using 2,000 shuffles to confirm scores and gain greater accuracy.
The Web-based Hydropathy, Amphipathicity and Topology (WHAT) program was used to determine and plot the hydropathy, amphipathicity, secondary structure and predicted transmembrane topology of individual protein sequences [29]. All TMS predictions for individual proteins were performed using the WHAT program, which predicts integral membrane protein topology using a Hidden Markov Model approach [30, 31].
Multiple alignments were created using the ClustalX program [29]. Relative conservation was estimated using the AveHAS program [30], which generates average hydropathy, amphipathicity and similarity plots based on ClustalX multiple alignments, and also predicts topology with greater accuracy than is possible using the WHAT program. The plots obtained using this program are based on Clustal X multiple alignments (plots presented in Fig. 1 and 2).
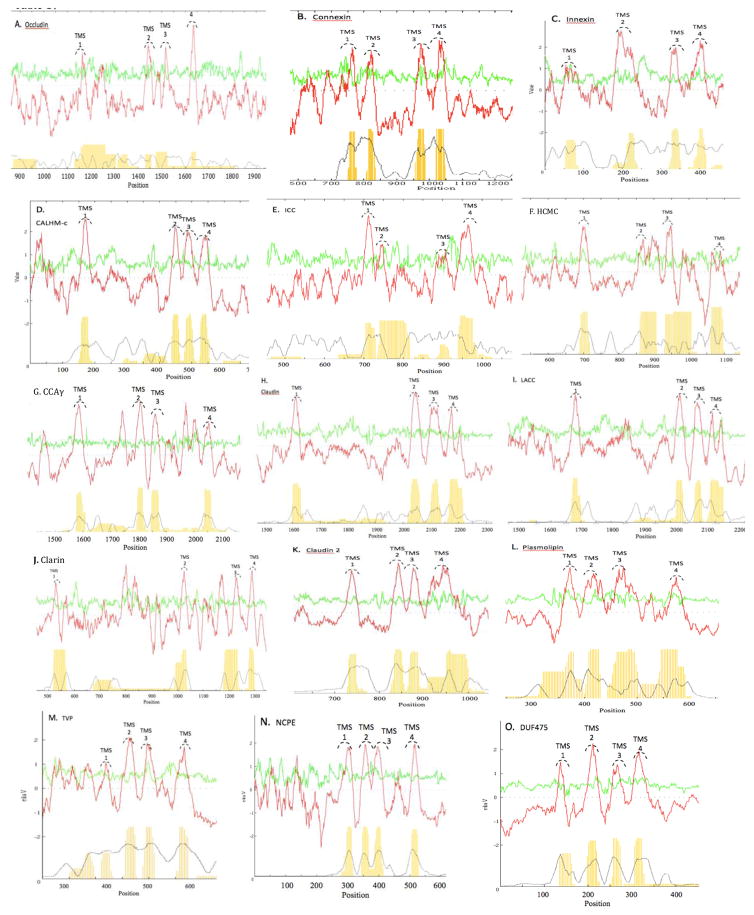
Figure 1.
AveHAS plots for all protein families in the 4JC superfamily. The families are indicated by their familial abbreviations (see Table 1). See Methods and the legend to Figure 2 for explanation of format.
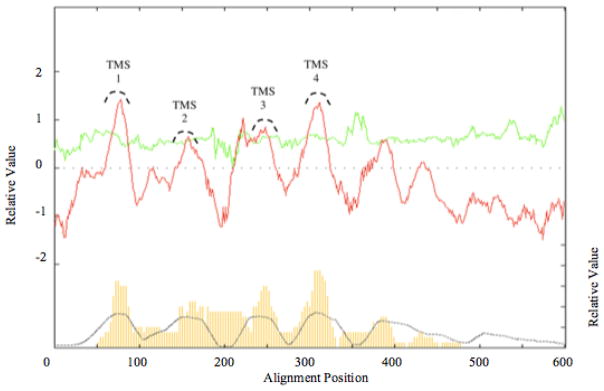
Fig 2. AveHAS plots for the entire 4JC Superfamily with sequences aligned using the Clustal X program.
See Methods section for procedures and references. The dark red line in the top plot represents average hydropathy, while the light green line represents average amphipathicity. The dotted black line in the lower plot shows the degree of conservation among the proteins at a particular location, while the thin vertical yellow lines indicate probable TMSs using a distinct program.
Phylogenetic superfamily trees were created using the SuperFamilyTree (SFT) programs [32–34]. SFT works by creating 100 distance matrices using tens of thousands of Blast bit scores. The matrices are then built using the Fitch program. The trees are averaged using the Consense program to produce a superfamily tree [32–34]. SFT1 creates a tree showing the individual proteins while SFT2 collapses this tree to show the relationships of the families to each other [32–34].
The Ancient Rep [27], REPRO [35] and HHRepID [36] programs were used to recognize distant transmembrane repeats within a single protein sequence. The former two programs use a variation of the Smith-Waterman local alignment strategy to find non-overlapping top-scoring alignments, but AncientRep also allows screening of multiple homologues for repeats after construction of ClustalX-generated multiple alignments, allowing comparison both within single proteins (horizontal comparisons) and between multiple homologues (vertical comparisons) [27]. TMS repeat units were located using these programs, and their common origin was established using the GSAT program as outlined above.
The phylum composition and average protein size ± S.D. for each of the 15 protein families of the 4JC superfamily were determined by using the Phylum-Size/Topology (PhyST) program [37]. The phyla of origin of the proteins in a family were automatically tabulated and used to quantitatively determine the phylum distribution for each family. The PhyST program was also used to determine the number of family members using a cut off of 90%, with the CD hit program and to identify large potential fusion proteins for each of the 15 families in the 4JC superfamily. This program was used precisely as describes previously [37].
Three-dimensional structures for members of TCDB families 1.A.24, 1.H.1 and 8.A.16 were obtained from RCSB PDB [38] through sequence and structural similarity searches. First, for family members without structures, we ran the online PDB sequence similarity tool (E-value ≤ 10−3) to find homologs. Second, for family members with structures, we ran the online PDB structural similarity tool and retrieved structures showing RMSD values of ≤ 4.0. Finally, we ran HMMTOP [39, 40] on all hits obtained and rejected structures with less than 4 predicted transmembrane segments (TMSs). Representative structural alignments based on pairwise combinations of structures between families were then computed using the Collaborative Computational Project 4 (CCP4) implementation of the Secondary Structure Matching (SSM) algorithm, which superposes structures with an emphasis on matching secondary structural elements as the name suggests, selecting for minimal RMSD values. [41–43].
3. RESULTS
3.1. Topological Predictions
To predict the common and distinctive topological features of each family found to belong to the 4JC superfamily, average hydropathy, amphipathicity and similarity (AveHAS) plots were generated using homologues obtained using Protocol 1 with a query protein from each family in TCDB, the first member of each subfamily (of the 15 families of the 4JC Superfamily) listed in TCDB (Table 1 and Figure 1) [32–34]. The red lines in the top plots represent hydropathy, while the green lines represent amphipathicity. The dotted black lines below show the degrees of conservation among the proteins at any one location in the alignment while the vertical yellow lines show an independent prediction of TMSs. The AveHAS plots for the fifteen families: Occludins (Figure 1A), Connexins (Figure 1B), Innexins (Figure 1C), CALHM-C (Figure 1D), ICC (Figure 1E), HCMC (Figure 1F), CCAγ (Figure 1G), Claudin (Figure 1H), LACC (Figure 1I), Clarin (Figure 1J), Claudin 2 (Figure 1K), Plasmolipin (Figure 1L), TVP (Figure 1M), NCPE (Figure 1N), and DUF475 (Figure 1O), all demonstrated a conserved 4 TMS topology. All 15 families showed comparable degrees of similarities for the four TMSs with slight differences being observed for a few families. For example, the connexins (Figure 1B) had TMSs 1 and 2 better conserved than TMSs 3 and 4 while the CALHM-C proteins (Figure 1D) showed the opposite behavior with TMSs 3 and 4 better conserved than TMSs 1 and 2.
3.2. Topological Correspondence among All Fifteen Families within the 4JC Superfamily
In addition to the AveHAS plots for the individual families, the AveHAS plot for the entire 4JC superfamily was generated as shown in Figure 2. Four clear peaks of hydrophobicity corresponding to four peaks of similarity can be visualized. All four peaks show similar degrees of conservation, but TMSs 3 and 4 may be somewhat better conserved than TMSs 1 and 2. The best conserved TMS appears to be TMS 4. In general, the peaks of similarity are broader than the peaks of hydropathy with similarity preceding peaks 1 and 3 but following peaks 2 and 4. This suggests that the cytoplasmic regions adjacent to the TMSs are better conserved than the remaining parts of the cytoplasmic loops or the corresponding extracellular regions. All four peaks exhibit moderate amphipathicity.
All members of each of the fifteen families of the 4JC superfamily are homologous throughout most of their lengths, although insertions, deletions and fusions, primarily in their hydrophilic regions, have occurred in various protein members during their evolutionary divergence. Proteins used for the initial PSI-BLAST searches were the first member of each subfamily within the 15 families of the 4JC Superfamily listed in the Transporter Classification Database (TCDB; www.tcdb.org) [24–26] under their respective families as indicated by abbreviation as summarized in Table 1. The values reported using this expanded dataset yielded scores that suggested homology between all fifteen families (Table 2). The criteria used for establishing homology were comparison scores of 14 standard deviations (SD) or greater, with an alignment of at least 60 amino acyl residues (aas) including corresponding TMSs [26, 44].
Table 2.
Comparison scores expressed in standard deviations (SD) for the fifteen families in the 4JC Superfamily.a
| Families Compared | Proteins Compared (UniProt # or GI #) | Comparison Score (SD) | ||||||
|---|---|---|---|---|---|---|---|---|
| Protein-1 (A) | Protein-2 (B) | Protein-3 (C) | Protein-4 (D) | A v. B | B v. C | C v. D | A v. D | |
| Connexin v. Innexin | Q8NFK1 | Q4SJR0 | K8LRA8 | Q8IWT6 | 62.6 | 17.9 | 32.3 | 0.8 |
| ICC v. Innexin | Q96S66 | J9JZG4 | K1QMI8 | Q96QZ0 | 26.5 | 14.9 | 15.3 | −0.2 |
| Occludin v. CCAγ | Q16625 | H2L4X0 | C3ZY32 | P54825 | 154.4 | 14.2 | 16.9 | 2.2 |
| HCMC v. CCAγ | Q8TAF8 | 291242472 | F7BS52 | Q06432 | 22.8 | 15.0 | 19.0 | 0.7 |
| CCAγ v. Claudin | Q9D563 | 701422218 | 657540378 | P56857 | 18.6 | 19.3 | 20.5 | 12.5 |
| LACC v. Claudin | I3VPY1 | S8ALD9 | G8C1J8 | P54003 | 16.3 | 16.0 | 95.4 | 9.5 |
| Clarin v. CCAγ | A7SGP9 | C1BSD7 | R7TFA9 | R7TFA9 | 21.8 | 14.3 | 126.5 | 9.5 |
| Claudin2 v. Claudin | Q9NGJ7 | U1MC19 | R4GBS8 | P56857 | 22.4 | 16.7 | 35.7 | 0.8 |
| CCAγ v. Claudin2 | Q9NY35 | 488549030 | 194750239 | F5HJC0 | 178.5 | 17.3 | 125.0 | 8.1 |
| TVP v. Connexin | P08247 | B3RIL2 | H3AJZ7 | P08050 | 47.6 | 15.6 | 282.5 | −1.0 |
| Occludin v. Plasmolipin | Q16625 | M7BW70 | C3ZW40 | P47897 | 18.5 | 15.5 | 23.0 | 4.2 |
| Occludin v. NCPE | Q16625 | F1QIE2 | Q6FKV0 | Q8NJ01 | 24.1 | 14.5 | 21.7 | 2.4 |
| Occludin v. TVP | Q16625 | 432884588 | 527260494 | P08247 | 152.9 | 15.7 | 63.3 | 1.7 |
| Plasmolipin v. NCPE | P47897 | 602664033 | K3VQ09 | Q8NJ01 | 59.0 | 14.5 | 37.7 | 6.3 |
| TVP v. DUF475 | B3RX02 | A0A077ZHE5 | 655407529 | Q9KXK6 | 14.5 | 14.4 | 65.6 | 3.7 |
| TVP v. Plasmolipin | P08247 | E9CJG0 | C3ZW39 | P47897 | 22.4 | 14.3 | 29.4 | −1.2 |
| TVP v. NCPE | P08247 | 641792620 | J7SAL5 | A5E332 | 119.4 | 14.4 | 14.2 | −1.1 |
| CALHM-c v. Claudin | Q8IU99 | 821384408 | E5R4H8 | Q06991 | 46.5 | 14.0 | 22.1 | −0.4 |
The Superfamily Principle, which states that if A is related to B, and B is related to C, then A must be related to C (The Transitivity Rule), was used to establish homology. Column 1 gives the family abbreviations (see Table 1). Accession numbers of the four proteins compared (based on Protocol1 and Protocol2 results) are provided in columns 2–5, and the comparison scores for the 3 comparisons (A vs. B, B vs. C, and C vs. D) are given in columns 6–8. Column 9 gives the value obtained when A was directly compared with D. Accession numbers provided are UniProt numbers when available or gi numbers when UniProt numbers were not available.
The phylum representation of each protein family within the 4JC superfamily is provided in Table 1. A majority of the protein families of the 4JC superfamily are from Metazoa. Two families, LACC and NCPE, have proteins derived from Fungi, while the DUF475 family includes proteins only from Actinobacteria. Table 1 also presents the average sizes of the proteins comprising the fifteen 4JC families (column 5) and the relative family sizes (in numbers of proteins recovered as described in the Methods section (column 6). The numbers of large proteins that could be fusion proteins as determined with the PhyST program were also tabulated. As illustrated in Figures 1 and 2, they all have at least four conserved TMSs, unifying characteristic of the 4JC Superfamily. If the 4TMS 4JC domain is fully or partially, duplicated, triplicated or fused to another transmembrane domain, there will be more of TMSs, but this occurs rarely (see Table 3). Table 1 also lists Pfam designations for members of the various TC families when available (column 8) and also provides a reference (column 9). Additional references can be found in TCDB for all of the families listed.
Table 3.
Fusion proteins containing 4TC Superfamily domains.a
| Gi # | Sizes (aas) | Domain/Order |
|---|---|---|
|
| ||
|
1.A.25 Connexins | ||
|
| ||
| 431890982 | 2729 | Connexin - PDZ (protein protein interaction domain) - Myosin-XVIIIa (MYSc = myosin motor) - Tropomyosin - Kinetochore (microtubule binding domain) - Opi1 (phosphorylated Tx factor) |
|
| ||
| 537123619 | 1178 | Connexin - Pkc-like cyclin-dependent Ser/Thr kinase |
|
| ||
| 530667615 | 994 | Connexin - SPRY/TRIM domain (regulator of immune system) - olfactory receptor |
|
| ||
| 521027364 | 760 | Connexin - SPRY/TRIM tripartite motif containing domain |
|
| ||
| 190358616 | 709 | Connexin - uncharacterized hydrophilic domain |
| 597731320 | 755 | |
| 736164105 | 700 | |
| 593734441 | 675 | |
|
| ||
| 528770327 | 839 | Connexin - DUF3735 - ABA-GPCR (golgi pH regulator) |
|
| ||
| 465977614 | 840 | 4 + 5 + 4 TMS topology; Connexin DUF3735-ABA-GPCR (abscisio acid receptor, G protein) |
|
| ||
| 444706902 | 930 | Connexin - Gtr1_RagA (P-loop NTPase) |
|
| ||
| 47222966 | 948 | LRR-RI-LRR-RI, Leucine-Rich Repeats (11 full repeats; protein-protein interaction domain) - Ribonuclease Inhibitor - Connexin |
|
| ||
|
1.A.25 Innexins | ||
|
| ||
| 669308587 | 795 | Two complete adjacent duplicated innexin domains. |
| 669225467 | 810 | |
| 339248393 | 813 | |
| 541046776 | 834 | |
| 568268171 | 797 | |
| 684378264 | 969 | |
|
| ||
| 669329541 | 1230 | DUF2045 - TAF7 - Innexin |
|
| ||
| 669313956 | 818 | Ndr-Innexin (Ndr may be an α, β-hydrolase (Pfam00561)). |
|
| ||
| 405960508 | 840 | AAT - I (aspartate amino transferease) - Innexin |
| 684379759 | 844 | Innexin fragments - Innexin (The innexin fragments precede the full length innexin domain) |
| 674266122 | 717 | |
| 734560734 | 836 | |
| 684386324 | 780 | |
| 684367491 | 884 | |
| 353231599 | 1023 | |
|
| ||
| 674595321 | 1006 | Innexin - pyruvate kinase |
|
| ||
|
1.A.36 ICC | ||
|
| ||
| 528765010 | 1089 | LisH (microtubule regulation) - W040 (signal transduction) - MCLC (ICC) Cl− channel |
|
| ||
|
1.A.64 Plasmolipin | ||
|
| ||
| 719732991 | 339 | N-terminal hydrophilic domain C-terminal MARVEL (plasmolipin domain) |
|
| ||
|
1.A.81 LACC |
No large proteins | |
|
| ||
|
1.A.82 HCMC |
No large proteins | |
|
| ||
|
1.A.84 CALHM-C | ||
|
| ||
| 465989358 | 1203 | Dermatansulfate epimerase - CALHM-C |
|
| ||
| 594679692 | 659 | Duplicated CALHM-C domains. C-terminal hydrophilic domain; The second CALHM-C domain is better conserved. |
| 537213670 | 668 | |
| 676278280 | 916 | |
|
| ||
|
1.H.1 Claudin | ||
|
| ||
| 521024295 | 908 | N-terminal 4 TMS Claudin domain - ARM repeat units (at least 5) (armadillo/β-cateinin repeats; protein protein interaction domains). |
|
| ||
|
1.H.2 Claudin 2 | ||
|
| ||
| 576700697 | 995 | N-terminal 4 TMSs Claudin 2 domain - C-terminal DM10 (3OUF1128) domain; function unknown |
|
| ||
| 322796000 | 627 | 1 + 4 + 4 + 3 TMSs. Three (triplicated) Claudin 2 domains |
|
| ||
|
8.A.16 CCAγ | ||
|
| ||
| 641736570 | 649 | Duplicated 4 TMS CCAγ domains. The N-terminal domain resembles 8.A.16.2, while the C-terminal domain more closely resembles 8.A.16.1 |
| 351715945 | 630 | |
| 555949535 | 487 | |
|
| ||
|
9.A.27 NCPE |
No large proteins | |
|
| ||
|
9.A.46 Clarin | ||
|
| ||
| 521024245 | 525 | Clarin - EEP (Endonuclease domain) - DUF4205 |
|
| ||
|
9.B.41 Fusions to Occludin | ||
|
| ||
| 528761243 | 1094 | Two fused Occludin domains. The N-terminal domain is like subfamily 9.B.41.2. The C-terminal domain is more like subfamily 9.B.41.1. |
| 465983309 | 1280 | |
|
| ||
| 528761243 | 1094 | Two full 4 TMS Occludin (MARVEL - Occludin) repeats |
| 465983309 | 1280 | |
|
| ||
|
9.B.130 TVP Family (MARVEL) | ||
|
| ||
| 528765018 | 1135 | MARVEL - SCA7 Zn2+ binding domain - Cytb561 |
| 431896453 | 1088 | |
|
| ||
| 432110159 | 980 | MARVEL - Prickle-like protein 3 (PET_Prickle - LIM2-LIM3 Zn2+ binding) |
The family and its TC# as well as the genbank ID number (gi#) are provided in Column 1. The protein size in number of amino acyl residues (aas) can be found in column 2, and the recognized domains, in order from N- to C-terminus, are presented in column 3.
3.3. Establishing Homology between Members of Different Families
The top comparison scores expressed in SD for each interfamilial comparison were obtained using the GSAT program with 2,000 random shuffles. Proteins in TCDB were checked for homology as shown in Table 2 with scores supporting the conclusion of homology. Global sequence alignments for several interfamilial comparisons are presented in Figure 3.
Fig 3. Global sequence alignments of various families within the 4JC superfamily demonstrating that corresponding TMSs align.
Accession numbers for the proteins compared are provided in Table 2(B vs. C). Residue numbers are provided at the beginning and end of each line. Shaded regions indicate the predicted TMSs which are numbered (1–4). CS, comparison score expressed in standard deviations (SD); %I = percent identity; %S = percent similarity. Vertical lines, identities; colons, similarities. The eight figures (A–H) shows eight binary comparisons for the families indicated at the top of each alignments.
Three patterns were observed when conducting binary alignments. The first demonstrated all or most of the four TMSs in a subject sequence aligning with their respective counterparts in the target sequence. Alignments of this type can be seen in Figures 3A, B and D, where (A) a CCAγ homologue is compared with a Claudin homologue, (B) an HCMC homologue is compared with a CCAγ homologue, and (D) a Claudin homologue is compared with a Claudin2 homologue. These three comparisons gave comparison scores of 19.3 SD, 15.0 SD, and 16.7 SD, respectively. The second pattern of binary alignments shown in Figure 3 always involved comparisons of corresponding TMSs (1 with 1; 2 with 2; 3 with 3; and 4 with 4, respectively) but with only two (Figures 3G and H) or three (Figure 3C, E and F) TMSs aligning. 10 SD has been reported to correspond to a probability of 10−24 that the observed degree of similarity has arisen by chance [45], but Gaussian skew can substantially increase this probability. Controls showed that 14 SD was 2 SD above the highest value that could be obtained when non-homologous sequences of similar topology were compared at the time these studies were conducted.
The remainder of the comparisons exhibited similar patterns. For example, the best comparison score obtained for the connexins and the innexins was 21 SD. A portion of this alignment is shown in Figure 3H. The full alignment had 28% identity (I) and 48% similarity (S) for a stretch of 539 residue positions.
The best comparison score for the ICC family compared to the Innexin family (not shown) was 14.9 SD with 24% I and 49% S, spanning 183 residue positions. For the comparison between the HCMC and CCAγ families, a score of 15.0 SD was achieved with 26% I and 46% S, spanning 187 residue positions (Figure 3B). The comparison of the clarin and CCAγ families gave a score of 14.3 SD with 25% I and 45% S, spanning 193 residue positions (not shown). The comparison between the claudin and claudin2 families (Figure 3D) gave a score of 16.7 SD with 23% I and 46% S, spanning 192 residue positions. The highest scores for the claudin and LACC families (Figure 3E) was 16.0 SD, with 24% I and 45% S, spanning 150 residues. All other comparisons are listed in Table 2.
3.4. Evidence for 2 TMS Repeat Units in Members of the 4JC Superfamily
The third pattern of aligned sequences showed TMSs 1 and 2 of a single 4JC superfamily member aligning with TMSs 3 and 4 of the same protein (or a homologous protein), as shown in Figure 4. Four alignments for the first and second halves of single proteins are presented, a connexin (A), an innnexin (B), an HCMC family member (C), and an occludin (D). Additionally, heterologous comparisons (i.e., the first half of protein A aligned with the second half of Protein B) gave convincing comparison scores. For example, an occludin homologue and a CCAγ homologue gave a comparison score of 14 SD with 25% I and 54% S for a stretch of 67 residue positions. The value of 14 SD was sufficient to establish homology when these studies were conducted [32, 37]. The AlignMe program was used to provide further evidence for similarity between the two halves of several of these proteins (See Figure S1).
Fig 4. Alignments of the two halves of several members of the 4JC superfamily.
The Repro program was used to identify potential repeats with default settings for gap penalties: 10 for open, 1 for extension, and 50 for N local alignments. A, a connexin, B, an innexin, C, an HMCM protein, D, an occludin. The UniProt accession and TC numbers of the proteins studied are provided above each alignment. TMSs are shaded and numbered.
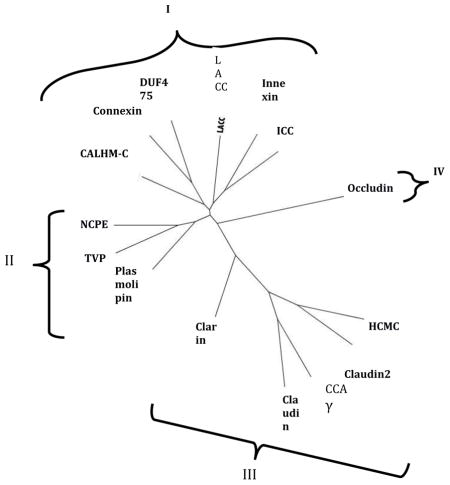
3.5. SFT-based Phylogenetic Trees
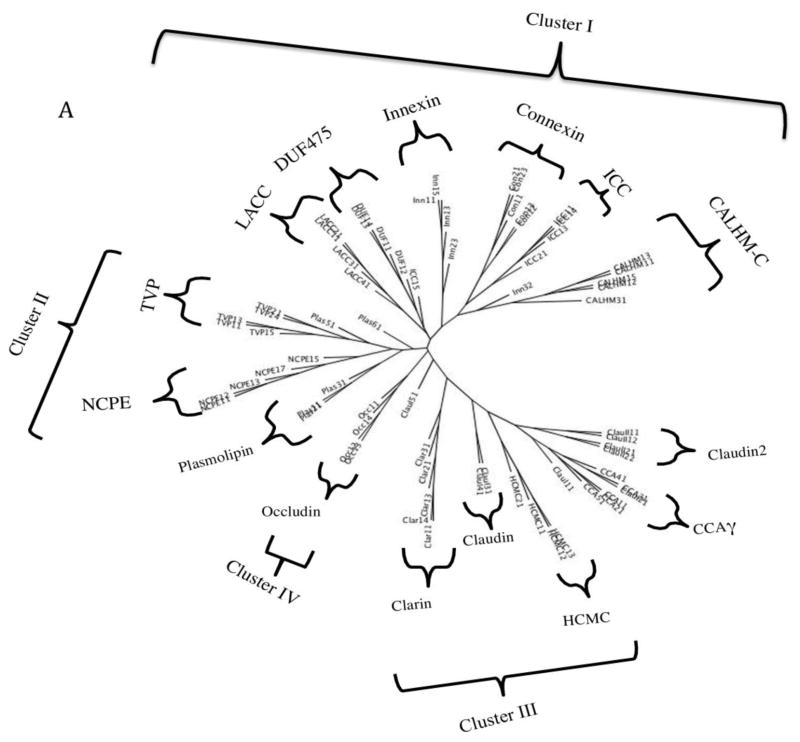
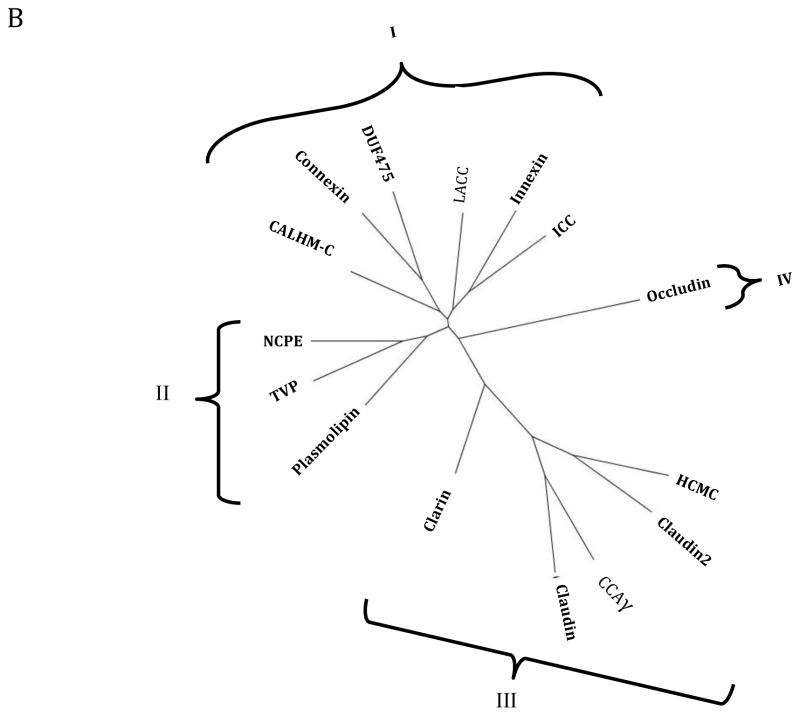
Phylogenetic trees for representative members of all 15 families in the 4JC Superfamily are shown in Figures 5A and B, where A shows the relationships of representative proteins while B shows the integrated family relationships [32–34]. Because the abbreviations for the individual proteins in Figure 5A may be too small to read, these are reproduced in clockwise order in supplementary Table S1. It will be noted that a very few proteins lie outside of the principle cluster that represents a particular family. The two trees show good agreement. In both trees, the families cluster into four major groups. Nine families cluster loosely into two groups. Thus, in the first group (Cluster I; top), six families cluster loosely together. These are (from left to right in B): CALHM-C, Connexin, DUF475, LACC, Innexin and ICC. In the second cluster (Cluster II, center left), the Plasmolipin, TVP and NCPE families cluster together. In the lower right hand cluster (Cluster III), the Claudin, Claudin2, CCAγ and HCMC families cluster together with the Clarins at the base of this cluster. Finally, the Occludins (Cluster IV) branch together by themselves from a point near the center of the tree (lwer left in A, right in B). These results provide evidence concerning the phylogenetic relationships of the fifteen families within the 4JC superfamily to each other.
Fig 5. Phylogenetic trees of representative proteins (A) and families (B) within the 4JC Superfamily.
The SuperfamilyTree programs (SFT1 and STF2) were used to generate the two trees, respectively (see 2. Methods). In A, the specific proteins examined in each of the 15 families have their protein abbreviations listed in Table S1 in supplementary materials, all listed in clockwise order in the tree. Those proteins that fall outside of their familial cluster are indicated by asterisks. Outside of these branches, indicating the positions of the individual proteins in the tree, the family abbreviation are provided. Finally, the four clusters (I–IV) are shown. B. The integrated tree in which each branch bears a single family. The same four clusters (I–IV) are indicated. The family abbreviation used with their full names are presented in Table 1.
3.6. 3D structural comparisons
Of the families believed to be members of the 4JC superfamily, high-resolution 3D structures are available for members of just three of these families. These families are the connexins (TC#1.A.24 [46]), type I claudins (TC#1.H.1, [47, 48]), and Ca2+ channel γ auxiliary subunits (CCAγ; TC#8.A.16 [49]). The results of comparisons, selecting for minimal RMSD values, are presented in Figure 6A–C. In each case, the front, back, and top views are shown. The superpositions were sufficient to provide strong evidence of homology. Here, comparison of Connexin-26 with Claudin-19 (A) gave an RMSD value of 2.67, that for Connexin-26 with CCAγ (B) gave an RMSD value of 3.35, and that for Claudin-15 and CCAγ (C) gave an RMSD value of 1.90. Other comparisons not shown were as follows: (1) Claudin-19 versus CCAγ, 2.78 over 126 aas (3X29.C versus 3JBR.E), (2) Connexin-26 versus Claudin-15, 2.87 over 114 aas (2ZW3 versus 4P79A), (3) Claudin-19 versus Claudin 15, 1.36 over 151 aas (3X29.A versus 4P79.A). These values are highly suggestive of homology, confirming the conclusions based on primary sequence analyses.
Fig 6. Representative alignments of available 4JC structures.
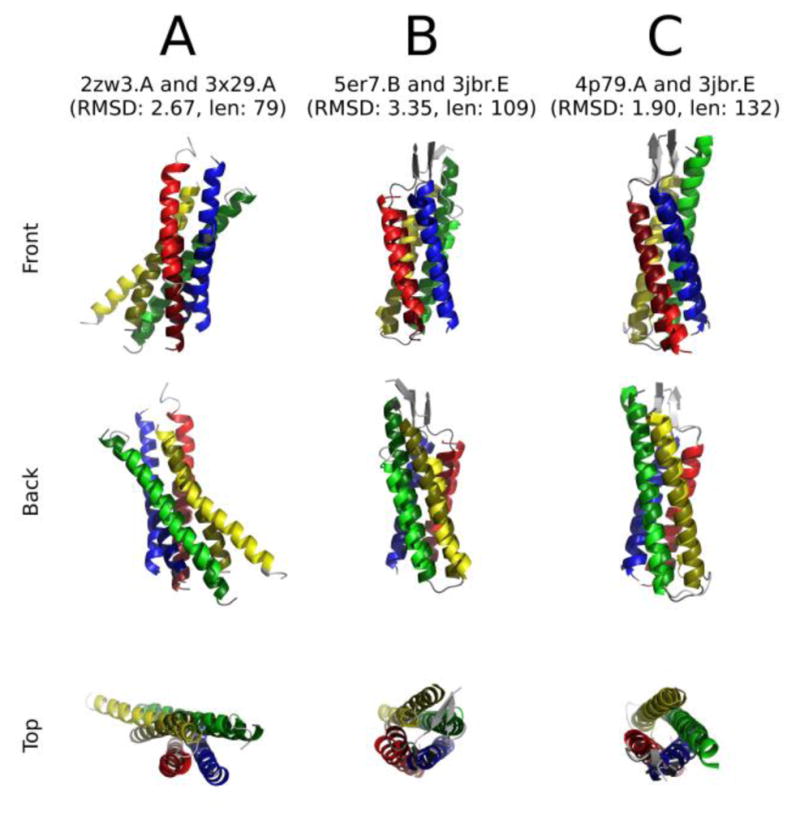
Left to right: A. Connexin-26 (TCDB: 1.A.24.1.3) and claudin-19 (TCDB: 1.H.1.1.5); B. connexin-26 (TCDB: 1.A.24.1.3) and CCAγ (TCDB: 8.A.16.1.1); C. claudin-15 (TCDB: 1.H.1.1.9) and CCAγ (TCDB: 8.A.16.1.1). Hydrophilic domains and unaligned loops have been excluded for clarity.
The color-coding is as follows:
| Color | TMS | A | B | C |
|---|---|---|---|---|
| Light red | 1 | 2ZW3.A | 5ER7.B | 4P79 |
| Light yellow | 2 | |||
| Light green | 3 | |||
| Light blue | 4 | |||
| Dark red | 1 | 3X29.A | 3JBR.E | 3JBR.E |
| Dark yellow | 2 | |||
| Dark green | 3 | |||
| Dark blue | 4 |
3.7. POTENTIAL FUSION PROTEINS INCLUDING 4JC DOMAINS
No large homologues were identified for the LACC (1.A.81), HCMC (1.A.82), NCPE (9.A.27) and DUF475 (9.B.179) families, but such proteins were found for all other 4JC families (Table 1).
3.7.1. Connexins (1.A.24)
Twelve connexin homologues from animals proved to be more than two-fold in size relative to the average size of these proteins, and all were examined (Table 3). All but one had full length connexin domains at their N-terminal ends; four of these had long C-terminal hydrophilic domains of unknown function. Two long homologues had an N-terminal connexin domain followed by a 7–9 TMS “Golgi pH Regulatory” domain (TC#1.A.38), which consists of a DUF3735 domain followed by an abscisic acid GPCR receptor (Aba_GPCR) domain. One protein had a C-terminal P-loop NTPase (Gtr1_RagA) domain. Two proteins had an SPRY/TRIM immune system regulatory domain C-terminal to the connexin domain, and one of these also had a C-terminal 7 TMS olfactory receptor domain (TC#9.A.14.8). The largest of these connexins had a size of 2729 aas. Following the connexin domain in this protein was (1) a PDZ protein-protein interaction domain, (2) a myosin-XVIIIa (MYSc myosin motor) domain, (3) a tropomyosin domain, (4) a microtubule binding kinetochore domain, and (5) an Opi1 (phosphorylated transcription factor) domain in this order.
Only one large homologue had the connexin domain at its C-terminus. This protein of 948 aas had an N-terminal multiply repeated leucine-rich domain. The NCBI database also contained shorter 2 TMS proteins corresponding to much of the N-terminal 2 TMS domain or the C-terminal 2 TMS domain, each of 200–400 aas in length. The potential existence of such proteins, although functionally uncharacterized, supports the conclusion presented above, that 4JC proteins arose by intragenic duplication of a 2 TMS-encoding element.
3.7.2. Innexins (1.A.25)
Sixteen large Innexin homologues were identified. Six proteins, each from a different invertebrate species, proved to have internal duplications, containing two complete adjacent innexin domains, each with 4 TMSs (Table 3). One such protein was entered into TCDB with TC#1.A.25.1.11. Several other large proteins appeared to contain complete C-terminal innexin domains with N-terminal fragmentary innexin domains of variable sizes. These frequently included partial innexin fragments or sequences that were too distantly related to known proteins to allow their identification, even though some of them represented conserved domains. Recognized N-terminal domains included (1) a DUF2047-TAF7 region and (2) an Ndr domain, thought to be involved in cell differentiation, possibly an α, β-hydrolase (Pfam 00561). One protein had an N-terminal innexin domain with a C-terminal pyruvate kinase domain.
It is interesting to note that while most connexin fusion proteins have their extra domains fused C-terminal to the connexin domain, the innexin fusion proteins have their extra domains fused N-terminal to the innexin domain. Additionally, while no protein was identified with two full length connexin domains, six homologues with two full length innexin domains were detected, and six more had full length C-terminal innexin domains with what appeared to be N-terminal innexin fragments.
3.7.3. ICC (1.A.36)
Only one large (>2x average) protein was identified for the ICC family. This protein, of 1089 aas, had an N-terminal LisH microtubule regulation domain, a central WD40 signal transduction domain and a C-terminal MCLC chloride channel domain (TC#1.A.36; Table 3).
3.7.4. Plasmolipin (1.A.64)
One protein of 339 aas was substantially larger than its plasmolipin homologues. This protein had an N-terminal hydrophilic domain, not associated with a conserved domains in CDD or Pfam, followed by a single C-terminal plasmolipin domain.
3.7.5. CALHM-C (1.A.84)
Four large CALHM-C homologues were identified (Table 3). The first had a C-terminal CALHM-C domain fused to a large hydrophilic dermatan sulfate epimerase. The other three proteins, each from a different animal species, possessed two internally duplicated, full length, 4 TMS CALHM-C domains. In these cases, the C-terminal domains were better conserved (90–95% identical to its best TC hit) with the N-terminal domain exhibiting only about 30% identity with the same TC CALHM-C homologue. One of these proteins, with gi# 676278280, has a duplicated domain with 5 rather than 4 putative TMSs (peaks of hydrophobicity). Possibly, the domain duplication events occurred late during the evolution of these proteins.
3.7.6. Claudins (1.H.1 and 1.H.2)
Only three large claudin proteins were identified, one for the Claudin (TC#1.H.1) family and two for the Claudin 2 (TC#1.H.2) Family. The large claudin homologue (Table 3) was of 908 aas and had an N-terminal claudin domain of 4 TMSs followed by at least 5 ARM (Armadillo/β-cateinin) repeat units. The ARM domains are probably protein-protein interaction domains. The first of the large Claudin2 homologues, of 995 aas, had at least 3 DM10 (DUF1128) domains of unknown function, C-terminal to the Claudin2 domain. The second large Claudin 2 protein, of 627 aas, had a Claudin2 triplication in a 1+ 4 + 4 + 3 TMS arrangement. Repeat #3 showed 52% identity, repeat #1 showed 32% identity and repeat #2 showed 29% identity with TC#1.H.2.1.1.
3.7.7. CCAγ (8.A.16)
Three large homologues in the CCAγ family were identified, and all shared the same domain patterns. They exhibited duplicated 4 TMS Claudin2-like domains, the first belonging to subfamily 8.A.16.2, and the second belonging to subfamily 8.A.16.1. This fact suggests that they arose by gene fusion rather than by intragenic duplication. Finding multiple homologues with the same domain order increases confidence that these proteins did not result from artifacts of sequencing or exon identification.
3.7.8. Clarins (9.A.46)
Only one large homologue of the Clarins was found. This protein, of 525 aas, had three domains in the order: Clarin - EEP - DUF4205, where the EEP domain is found in endonucleases while the DUF4205 domain has not been characterized.
3.7.9. Occludins (9.B.41)
Two of four large occludin homologues, of 1094–1280 aas, displayed two full length occludin repeats. These proteins appeared to be fusions of two occludins rather than duplications because their N-terminal occludin domains resembled subfamily 9.B.41.2 proteins while the C-terminal domains most closely resembled subfamily 9.B.41.1 occludins. Two such proteins are listed in Table 3. This situation is similar to that observed for the CCAγ family. Two additional proteins also had two full length 4 TMS occludin domains, but their origins could not be ascertained.
3.7.10. Tetraspan Vesicle Membrane (9.B.130)
The 4 TMS 4JC domain of this family is referred to as MARVEL in CDD. Two proteins had their N-terminal MARVEL domains fused to a long hydrophilic domain with a Zn2+ binding SCA7 domain, and one of them had a C-terminal Cytochrome b561 domain. One other homologue had the N-terminal MARVEL domain fused to a hydrophilic Prickle-like protein 3 (PET_Prickle-LIM1-LIM2-LIM3) series of domains.
4. DISCUSSION
The statistical analyses presented in this article provide the first evidence that four families of junctional proteins, the innexins, connexins, claudins and occludins, as well as eleven channel, transport auxiliary protein and uncharacterized families (see Tables 1 and 2) all arose from a common ancestor via the same pathway. In all cases, a 2 TMS hairpin structure with its N- and C-termini inside, probably duplicated to give the 4 TMS proteins. Interestingly, additional duplication or fusion events giving 8 TMS proteins with two 4JC domains and even 12 TMS proteins with three 4JC domains (with some variations) were identified (see section entitled “Potential Fusion Proteins” and Table 3). The 4 TMS topology is therefore the basic characteristic of all members of the 4JC superfamily, although in several cases, particularly putative “fusion” proteins, more TMSs were observed (see below). The relatively high frequencies of duplicated or fused 4JC domains, especially among the junctional connexins, innexins, claudins and occludins, is consistent with the conclusion that the proteins of each family form (hetero)oligomeric structures in the intact cell [50–58].
The evidence for homology between the fifteen families of the 4JC Superfamily was substantial (all scores at or above the 14 SD cutoff; see Fig. 2 and Table 2), and in the few cases where 3-d structures were available (connexins, claudins, and CCAγ), structural comparisons confirmed this conclusion. Analysis of the phylogenetic trees for the proteins and families of the 4JC Superfamily revealed some interesting details (Figures 5A and B). First, all but two of the known channel proteins (T.C. Class 1.A) occur in cluster I at the tops of the two phylogenetic trees. The two exceptions are the Plasmolipin Family which can be found in Cluster III (middle left), and the HCMC Family, present in Cluster II (bottom). The DUF475 Family, with proteins derived from Actinobacteria, of unknown function, is found in Cluster I, suggesting that these proteins may be ion channels. Second, the plasmolipins cluster with the occludins and two poorly defined families, the TVP and NCPE families. None of the proteins in these families are mechanistically defined. Third, cluster III includes both Claudin families (Claudin and Claudin2) as well as the CCAγ and HCMC families with the Clarin family branching from a position much closer to the center of this radial tree. Fourth, the Occludin family branches from the center of the tree by itself (Branch IV). While Claudins and Occludins are known to be constituents of tight junctions, HCMC family members are mechanosensitive ion channels, while CCAγ proteins are believed to be auxiliary subunits of Ca2+ channels. Clarin 1 is a component of the USH complex involved in mechanotransduction [59], responsible, when defective, for deaf-blindness [60]. It is the causative protein which when mutated gives rise to the human Usher syndrome type 3A [61].
Of the 15 families in the 4JC Superfamily, a search for proteins at least 2-fold larger than the average size of all members of the family revealed a few proteins that proved to have more than a single domain. Four of these families had no such recognizable fusion proteins, nine families had between 1 and 4 such members, and two, the connexins and innexins, had more, 12 and 15, respectively. These numbers indicate that almost all members of the 4JC superfamily consist of single domain proteins; there are only a few exceptions. Some of these large proteins may have resulted from errors in sequencing, incorrect intron/exon assignment or misinterpretation of the sequence data.
The most common occurrence among large putative fusion proteins were internal duplications, having two, or in a few cases, three, 4JC domains. These proteins often have family-specific characteristics. For example, no internally duplicated connexin was identified, although several putative connexin proteins were fused to other domains, almost always with the connexin domain N-terminal. By contrast, six innexin homologues were internally duplicated, and six more displayed C terminal innexin domains with N-terminal fragments of innexins. In contrast to the connexins, where N-terminal 4JC domains were fused to other domains, the innexin domains were almost always C-terminal.
In each of the ICC and Plasmolipin Families, only a single fusion protein was identified, and in both cases, the 4JC domain was C-terminal. A single CALHM-C protein had a C-terminal 4JC domain with an N-terminal dermatan sulfate epimerase domain, but three other proteins had duplicated 4JC domains, where the C-terminal domains were better conserved. By contrast, Claudins, when fused to other domains, had the 4JC domain N-terminal. A single Claudin2 homologue seemed to have three (triplicated) 4JC domains. Three large CCAγ homologues had the 4JC domains C-terminal with the other domains being N-terminal. The single Clarin fusion protein identified also had its 4JC domain C-terminal.
The large occludins and TVP proteins contained 4JC domains recognized by CDD as MARVEL/occludin domains. Of the occludins, two had two C-terminal 4JC domains with these 4JC domains fused to hydrophilic N-terminal domains. Of the three large TVP homologues, all had the 4JC domain N-terminal to the other domains.
It is apparent, that the occurrence of internally duplicated (or fused) or partially duplicated 4JC domains represented a fairly high percentage of the large proteins and that several of these appeared to have arisen by fusion of two dissimilar 4JC domains, rather than duplication of a single such domain.
Whether these fusions occur with the 4JC domain C-terminal or N-terminal depended on the family to which these proteins belong. One primary function of the fusions could be to anchor a soluble enzyme or structural protein to the membrane with formation of a multiprotein complex. However, it is also possible that these fusions provide cooperative metabolic regulatory functions. Systematic identification of these fusion proteins provides food for thought and future research prospects concerning their evolution and functions.
The observations reported in this communication suggest that the 4 TMS topology, conserved in all members of the 4JC superfamily, is important for a structure and/or function common to all of its members. The two patterns of topological alignment, one showing the same TMSs (1–4) aligning with the corresponding TMSs in members of another family, and the other showing TMSs 1 and 2 aligning with TMSs 3 and 4, substantiate the conclusion of homology and also provide evidence for the conclusion that the proteins of this superfamily arose via intragenic duplication. These suggestions were substantiated using rigorous statistical criteria, and by the identification of 2 TMS elements in some homologues.
In general, we observed that the 4 TMSs in any one family of the 4JC superfamily were conserved to similar degrees. This suggests that these 4 TMSs are of comparable importance, both structurally and functionally, in all 15 families. In some families, differing degrees of conservation were observed, but these were not large differences. In these instances, however, parts of the proteins may serve extra functions not assumed by the others. We suggest that family-specific functions could include subunit:subunit interactions, channel formation, and hemichannel docking. Superfamily-generalized functions could include overall 3-dimensional structural features, subunit stability, and proper biogenesis.
Supplementary Material
Highlights.
We define the Tetraspan Junctional Complex (4JC) Superfamily.
The superfamily includes 15 previously recognized families.
The basic unit always consists of 4 transmembrane α-helices.
Large duplicated or fused proteins have been identified.
Acknowledgments
This work was supported by NIH grant GM077402. We thank Joshua Asiaban, Anne Chu and Sabrina Phan for assistance with manuscript preparation.
Footnotes
CONFLICT OF INTEREST
This work was supported by NIH grant GM077402. The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hua VB, Chang AB, Tchieu JH, Kumar NM, Nielsen PA, Saier MH., Jr Sequence and phylogenetic analyses of 4 TMS junctional proteins of animals: connexins, innexins, claudins and occludins. J Membr Biol. 2003;194:59–76. doi: 10.1007/s00232-003-2026-8. [DOI] [PubMed] [Google Scholar]
- 2.Morrow CM, Mruk D, Cheng CY, Hess RA. Claudin and occludin expression and function in the seminiferous epithelium. Philos Trans R Soc Lond B Biol Sci. 2010;365:1679–1696. doi: 10.1098/rstb.2010.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suga M, Maeda S, Nakagawa S, Yamashita E, Tsukihara T. A description of the structural determination procedures of a gap junction channel at 3.5 A resolution. Acta Crystallogr D Biol Crystallogr. 2009;65:758–766. doi: 10.1107/S0907444909014711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yen MR, Saier MH., Jr Gap junctional proteins of animals: the innexin/pannexin superfamily. Prog Biophys Mol Biol. 2007;94:5–14. doi: 10.1016/j.pbiomolbio.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummins PM. Occludin: one protein, many forms. Mol Cell Biol. 2012;32:242–250. doi: 10.1128/MCB.06029-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Findley MK, Koval M. Regulation and roles for claudin-family tight junction proteins. IUBMB Life. 2009;61:431–437. doi: 10.1002/iub.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overgaard CE, Daugherty BL, Mitchell LA, Koval M. Claudins: control of barrier function and regulation in response to oxidant stress. Antioxid Redox Signal. 2011;15:1179–1193. doi: 10.1089/ars.2011.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oshima A, Tani K, Hiroaki Y, Fujiyoshi Y, Sosinsky GE. Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proc Natl Acad Sci U S A. 2007;104:10034–10039. doi: 10.1073/pnas.0703704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herve JC, Phelan P, Bruzzone R, White TW. Connexins, innexins and pannexins: bridging the communication gap. Biochim Biophys Acta. 2005;1719:3–5. doi: 10.1016/j.bbamem.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Moore KB, O’Brien J. Connexins in neurons and glia: targets for intervention in disease and injury. Neural Regen Res. 2015;10:1013–1017. doi: 10.4103/1673-5374.160092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahl G, Muller KJ. Innexin and pannexin channels and their signaling. FEBS Lett. 2014;588:1396–1402. doi: 10.1016/j.febslet.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Al Khamici H, Brown LJ, Hossain KR, Hudson AL, Sinclair-Burton AA, Ng JP, Daniel EL, Hare JE, Cornell BA, Curmi PM, Davey MW, Valenzuela SM. Members of the chloride intracellular ion channel protein family demonstrate glutaredoxin-like enzymatic activity. PLoS One. 2015;10:e115699. doi: 10.1371/journal.pone.0115699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yaffe Y, Hugger I, Yassaf IN, Shepshelovitch J, Sklan EH, Elkabetz Y, Yeheskel A, Pasmanik-Chor M, Benzing C, Macmillan A, Gaus K, Eshed-Eisenbach Y, Peles E, Hirschberg K. The myelin proteolipid plasmolipin forms oligomers and induces liquid-ordered membranes in the Golgi complex. J Cell Sci. 2015;128:2293–2302. doi: 10.1242/jcs.166249. [DOI] [PubMed] [Google Scholar]
- 14.Cavinder B, Trail F. Role of Fig1, a component of the low-affinity calcium uptake system, in growth and sexual development of filamentous fungi. Eukaryot Cell. 2012;11:978–988. doi: 10.1128/EC.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao B, Wu Z, Grillet N, Yan L, Xiong W, Harkins-Perry S, Muller U. TMIE is an essential component of the mechanotransduction machinery of cochlear hair cells. Neuron. 2014;84:954–967. doi: 10.1016/j.neuron.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Z, Siebert AP, Cheung KH, Lee RJ, Johnson B, Cohen AS, Vingtdeux V, Marambaud P, Foskett JK. Calcium homeostasis modulator 1 (CALHM1) is the pore-forming subunit of an ion channel that mediates extracellular Ca2+ regulation of neuronal excitability. Proc Natl Acad Sci U S A. 2012;109:E1963–1971. doi: 10.1073/pnas.1204023109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capaldo CT, Nusrat A. Claudin switching: Physiological plasticity of the Tight Junction. Semin Cell Dev Biol. 2015;42:22–29. doi: 10.1016/j.semcdb.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Jaspers MH, Nolde K, Behr M, Joo SH, Plessmann U, Nikolov M, Urlaub H, Schuh R. The claudin Megatrachea protein complex. J Biol Chem. 2012;287:36756–36765. doi: 10.1074/jbc.M112.399410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacLean DM, Ramaswamy SS, Du M, Howe JR, Jayaraman V. Stargazin promotes closure of the AMPA receptor ligand-binding domain. J Gen Physiol. 2014;144:503–512. doi: 10.1085/jgp.201411287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tovaranonte PP, Beasley SW, Maoate K, Blakelock R, Skinner A. Trends in the use of minimally invasive surgery in children. N Z Med J. 2010;123:15–22. [PubMed] [Google Scholar]
- 21.Gopal SR, Chen DH, Chou SW, Zang J, Neuhauss SC, Stepanyan R, McDermott BM, Jr, Alagramam KN. Zebrafish Models for the Mechanosensory Hair Cell Dysfunction in Usher Syndrome 3 Reveal That Clarin-1 Is an Essential Hair Bundle Protein. J Neurosci. 2015;35:10188–10201. doi: 10.1523/JNEUROSCI.1096-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krug SM, Schulzke JD, Fromm M. Tight junction, selective permeability, and related diseases. Semin Cell Dev Biol. 2014;36:166–176. doi: 10.1016/j.semcdb.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Arthur CP, Stowell MH. Structure of synaptophysin: a hexameric MARVEL-domain channel protein. Structure. 2007;15:707–714. doi: 10.1016/j.str.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saier MH, Jr, Reddy VS, Tamang DG, Vastermark A. The transporter classification database. Nucleic Acids Res. 2014;42:D251–258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saier MH, Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res. 2016;44:D372–379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy VS, Saier MH., Jr BioV Suite--a collection of programs for the study of transport protein evolution. FEBS J. 2012;279:2036–2046. doi: 10.1111/j.1742-4658.2012.08590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai Y, Saier MH., Jr A web-based program for the prediction of average hydropathy, average amphipathicity and average similarity of multiply aligned homologous proteins. J Mol Microbiol Biotechnol. 2001;3:285–286. [PubMed] [Google Scholar]
- 31.Zhai Y, Saier MH., Jr A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotechnol. 2001;3:501–502. [PubMed] [Google Scholar]
- 32.Chen JS, Reddy V, Chen JH, Shlykov MA, Zheng WH, Cho J, Yen MR, Saier MH., Jr Phylogenetic characterization of transport protein superfamilies: superiority of SuperfamilyTree programs over those based on multiple alignments. J Mol Microbiol Biotechnol. 2011;21:83–96. doi: 10.1159/000334611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yen MR, Chen JS, Marquez JL, Sun EI, Saier MH. Multidrug resistance: phylogenetic characterization of superfamilies of secondary carriers that include drug exporters. Methods Mol Biol. 2010;637:47–64. doi: 10.1007/978-1-60761-700-6_3. [DOI] [PubMed] [Google Scholar]
- 34.Yen MR, Choi J, Saier MH., Jr Bioinformatic analyses of transmembrane transport: novel software for deducing protein phylogeny, topology, and evolution. J Mol Microbiol Biotechnol. 2009;17:163–176. doi: 10.1159/000239667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.George RA, Heringa J. The REPRO server: finding protein internal sequence repeats through the Web. Trends Biochem Sci. 2000;25:515–517. doi: 10.1016/s0968-0004(00)01643-1. [DOI] [PubMed] [Google Scholar]
- 36.Biegert A, Soding J. De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics. 2008;24:807–814. doi: 10.1093/bioinformatics/btn039. [DOI] [PubMed] [Google Scholar]
- 37.Kuppusamykrishnan H, Chau LM, Moreno-Hagelsieb G, Saier MH., Jr Analysis of 58 Families of Holins Using a Novel Program, PhyST. J Mol Microbiol Biotechnol. 2016;26:381–388. doi: 10.1159/000448040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rose PW, Prlic A, Bi C, Bluhm WF, Christie CH, Dutta S, Green RK, Goodsell DS, Westbrook JD, Woo J, Young J, Zardecki C, Berman HM, Bourne PE, Burley SK. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikeda M, Arai M, Lao DM, Shimizu T. Transmembrane topology prediction methods: a reassessment and improvement by a consensus method using a dataset of experimentally-characterized transmembrane topologies. In Silico Biol. 2002;2:19–33. [PubMed] [Google Scholar]
- 40.Reddy A, Cho J, Ling S, Reddy V, Shlykov M, Saier MH. Reliability of nine programs of topological predictions and their application to integral membrane channel and carrier proteins. J Mol Microbiol Biotechnol. 2014;24:161–190. doi: 10.1159/000363506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 42.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aasum E, Lathrop DA, Henden T, Sundset R, Larsen TS. The role of glycolysis in myocardial calcium control. J Mol Cell Cardiol. 1998;30:1703–1712. doi: 10.1006/jmcc.1998.0732. [DOI] [PubMed] [Google Scholar]
- 44.Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filipovic L, Hlavka M. Our experiences and results in the surgical treatment of metacarpal bone and had phalangeal fractures. Acta Chir Iugosl. 1977;24:485–489. [PubMed] [Google Scholar]
- 46.Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- 47.Krause G, Protze J, Piontek J. Assembly and function of claudins: Structure-function relationships based on homology models and crystal structures. Semin Cell Dev Biol. 2015;42:3–12. doi: 10.1016/j.semcdb.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y. Crystal structure of a claudin provides insight into the architecture of tight junctions. Science. 2014;344:304–307. doi: 10.1126/science.1248571. [DOI] [PubMed] [Google Scholar]
- 49.Wu J, Yan Z, Li Z, Yan C, Lu S, Dong M, Yan N. Structure of the voltage-gated calcium channel Cav1.1 complex. Science. 2015;350:aad2395. doi: 10.1126/science.aad2395. [DOI] [PubMed] [Google Scholar]
- 50.Ayad WA, Locke D, Koreen IV, Harris AL. Heteromeric, but not homomeric, connexin channels are selectively permeable to inositol phosphates. J Biol Chem. 2006;281:16727–16739. doi: 10.1074/jbc.M600136200. [DOI] [PubMed] [Google Scholar]
- 51.Bonander N, Jamshad M, Oberthur D, Clare M, Barwell J, Hu K, Farquhar MJ, Stamataki Z, Harris HJ, Dierks K, Dafforn TR, Betzel C, McKeating JA, Bill RM. Production, purification and characterization of recombinant, full-length human claudin-1. PLoS One. 2013;8:e64517. doi: 10.1371/journal.pone.0064517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falk MM. Cell-free synthesis for analyzing the membrane integration, oligomerization, and assembly characteristics of gap junction connexins. Methods. 2000;20:165–179. doi: 10.1006/meth.1999.0934. [DOI] [PubMed] [Google Scholar]
- 53.Hou J, Goodenough DA. Claudin-16 and claudin-19 function in the thick ascending limb. Curr Opin Nephrol Hypertens. 2010;19:483–488. doi: 10.1097/MNH.0b013e32833b7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCaffrey G, Staatz WD, Quigley CA, Nametz N, Seelbach MJ, Campos CR, Brooks TA, Egleton RD, Davis TP. Tight junctions contain oligomeric protein assembly critical for maintaining blood-brain barrier integrity in vivo. J Neurochem. 2007;103:2540–2555. doi: 10.1111/j.1471-4159.2007.04943.x. [DOI] [PubMed] [Google Scholar]
- 55.McCaffrey G, Willis CL, Staatz WD, Nametz N, Quigley CA, Hom S, Lochhead JJ, Davis TP. Occludin oligomeric assemblies at tight junctions of the blood-brain barrier are altered by hypoxia and reoxygenation stress. J Neurochem. 2009;110:58–71. doi: 10.1111/j.1471-4159.2009.06113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milatz S, Piontek J, Schulzke JD, Blasig IE, Fromm M, Gunzel D. Probing the cis-arrangement of prototype tight junction proteins claudin-1 and claudin-3. Biochem J. 2015;468:449–458. doi: 10.1042/BJ20150148. [DOI] [PubMed] [Google Scholar]
- 57.Oshima A, Matsuzawa T, Nishikawa K, Fujiyoshi Y. Oligomeric structure and functional characterization of Caenorhabditis elegans Innexin-6 gap junction protein. J Biol Chem. 2013;288:10513–10521. doi: 10.1074/jbc.M112.428383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stebbings LA, Todman MG, Phelan P, Bacon JP, Davies JA. Two Drosophila innexins are expressed in overlapping domains and cooperate to form gap-junction channels. Mol Biol Cell. 2000;11:2459–2470. doi: 10.1091/mbc.11.7.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogun O, Zallocchi M. Clarin-1 acts as a modulator of mechanotransduction activity and presynaptic ribbon assembly. J Cell Biol. 2014;207:375–391. doi: 10.1083/jcb.201404016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reiners J, Nagel-Wolfrum K, Jurgens K, Marker T, Wolfrum U. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 61.Phillips JB, Vastinsalo H, Wegner J, Clement A, Sankila EM, Westerfield M. The cone-dominant retina and the inner ear of zebrafish express the ortholog of CLRN1, the causative gene of human Usher syndrome type 3A. Gene Expr Patterns. 2013;13:473–481. doi: 10.1016/j.gep.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.