Abstract
The objective of this study was to evaluate molecular and epidemiologic factors associated with Escherichia coli sequence type 131 (ST131) among long-term care facility (LTCF) residents who acquired gastrointestinal tract colonization with fluoroquinolone-resistant E. coli (FQREC). Colonizing isolates from 37 residents who newly developed FQREC colonization at three LTCFs from 2006–2008 were evaluated. Twenty-nine (78%) of 37 total FQREC colonizing isolates were ST131. Most ST131 isolates had a distinctive combination of gyrA and parC replacement mutations. The ST131 and non-ST131 isolates differed significantly for the prevalence of many individual virulence factors but not for the proportion that qualified molecularly as extraintestinal pathogenic E. coli (ExPEC) or aggregate virulence factor scores. E. coli ST131 was highly prevalent among LTCF residents with FQREC colonization. Future studies should determine the risk factors for infection among ST131-colonized residents, and assess the potential for increased transmissibility of ST131 in the long-term care setting.
Keywords: Escherichia coli, ST131, Long-term care
INTRODUCTION
The increasing prevalence of multidrug-resistant Escherichia coli has been driven largely by the widespread emergence of a single clonal group, sequence type 131 (ST131) [1–3]. E. coli ST131 is characterized by multidrug resistance, including resistance to fluoroquinolones (FQs) and extended-spectrum cephalosporins, the latter usually mediated by CTX-M-15 extended-spectrum beta-lactamase (ESBL)-production [4–7]. In addition, ST131 has been associated with expression of multiple virulence factors, including various toxins, adhesins, and siderophores [1, 7, 8]. More recently, E. coli ST131 has been shown to be associated with older age and long-term care facility (LTCF) residence [5].
The recent rapid increase in the elderly population has led to a corresponding rise in LTCF utilization in the United States [9]. Due to certain host and healthcare factors (e.g., immune senescence, comorbidities, and antibiotic use), LTCF residents are at significant risk for colonization and/or infection with antibiotic-resistant organisms [10–12]. In particular, fluoroquinolone-resistant Escherichia coli (FQREC) has dramatically increased in prevalence in LTCFs over the past decade, representing a major cause of infections in this vulnerable population [13–15].
FQREC infections are most likely preceded by gastrointestinal tract colonization with the causative strain [16, 17]. E. coli isolates within phylogenetic group B2, including specifically E. coli ST131, may have a greater capacity for successful and persistent colonization of the gastrointestinal tract [18, 19]. Although FQ resistance mechanisms and virulence factors associated with ST131 status have been described in clinical isolates [6], very little is known about these traits in colonizing isolates. Furthermore, despite the increasing burden of FQREC in LTCFs, the molecular and clinical epidemiology of E. coli ST131 colonization has only recently been characterized in the long-term care setting in the United States [20].
To improve understanding of E. coli ST131 in the long-term care setting, we assessed the molecular and clinical epidemiology of ST131 and non-ST131 E. coli colonizing isolates from residents who acquired FQREC colonization during LTCF stay. In addition, within individual residents we evaluated clonal relatedness between newly acquired FQREC isolates and preceding FQ-susceptible E. coli isolates to assess for possible in situ development of resistance (i.e., conversion of a FQ-susceptible clone to a FQ-resistant clone). We also compared the present LTCF isolates with a large library of clinical ST131 isolates.
SUBJECTS AND METHODS
Study design and setting
This study was conducted from 2006 to 2008 at three LTCFs within the University of Pennsylvania: 1) LTCF #1, a 124-bed facility; 2) LTCF #2, a 240-bed facility; and 3) LTCF #3, a 200-bed facility. As previously described [21], LTCF residents who were colonized initially with FQ-susceptible E. coli (FQSEC) were followed longitudinally for up to 12 months. Serial fecal samples were obtained approximately every 14 days until recovery of FQREC, discharge, or death. The study was approved by the institutional review board of the University of Pennsylvania.
Study population
For the present study, we selected for further characterization the initial FQREC E. coli isolate from each of the 37 LTCF residents in the parent study who were documented previously to have developed incident FQREC colonization while under surveillance, i.e., who were colonized initially only with FQSEC and were found subsequently to be colonized with FQREC, +/− FQSEC (21). Additionally, to assess whether the subject’s first detected FQREC isolate arose within the patient from a preceding colonizing FQ-susceptible isolate (i.e., conversion of a clone from FQSEC to FQREC), for each subject the most recent FQ-susceptible E. coli isolate prior to the index FQREC isolate was included for pulsed-field gel electrophoresis (PFGE) analysis if the sampling date was ≤ 30 days prior to the first fecal swab with FQREC.
Microbiologic methods
Given the multi-step nature of the development of FQ resistance (e.g., accumulation of mutations in the quinolone resistance-determining regions [QRDR] of the gyrA and parC genes), isolates with elevated FQ minimum inhibitory concentrations (MICs) may exhibit mutations in FQ target genes [22–25]. Therefore, isolates with elevated FQ MICs are important in evaluating development of FQ resistance. Accordingly, as described previously [21], FQ resistance was defined operationally as a levofloxacin MIC ≥ 0.25 µg/mL. FQ susceptibility was therefore defined as a levofloxacin MIC < 0.25 µg/mL.
Furthermore, LTCF residents have previously been shown to have high rates of gastrointestinal colonization with multiple distinct strains of E. coli [26]. To identify, isolate, and characterize multiple E. coli colonies with diverse FQ susceptibility profiles from a given patient, stool samples were inoculated and replica-plated, as follows: a perirectal swab with freshly (≤ 24 hours) collected stool sample was inoculated by triple-streaking onto a MacConkey agar plate and incubated at 37°C overnight. Single colonies suspected of being E. coli (up to 25 per plate) were replica-plated onto 4 MacConkey agar plates, each supplemented with a different concentration of levofloxacin (8 µg/mL, 1 µg/mL, 0.25µg/mL, no antibiotic). Each colony underwent PYR and indole testing. Indole-positive and PYR-negative colonies were preliminarily considered E. coli and were saved in 30% glycerol stock at −80°C for further analyses.
From each specimen, up to six each presumed FQ-susceptible (levofloxacin MIC <0.25 µg/mL) and FQ-resistant (levofloxacin MIC ≥0.25 µg/mL) E. coli isolates, as available, were analyzed further. Isolates were identified definitively as E. coli and underwent standardized susceptibility testing using the semi-automated Vitek 2 identification and susceptibility system (bioMérieux, Durham, NC). A patient was considered to be colonized with FQREC if at least one stool isolate was confirmed as FQREC.
Evaluation for specific mechanisms of FQ resistance was performed as described previously [27–29]. Overexpression of AcrAB was measured using the organic solvent tolerance (OST) assay [30, 31]. The genetic relatedness of E. coli isolates was determined by PFGE [27], with all profiles analyzed using the Bionumerics v6.6 (Applied Maths, Austin, TX) and interpreted according to established criteria [32]. Strains were defined as those isolates sharing a pulsotype. Pulsotypes were assigned based on > 94% similarity to reference strains within an established PFGE library [33]. Susceptibility testing was performed using the semiautomated Vitek 2 system and interpreted according to Clinical and Laboratory Standards Institute criteria.
Major E. coli phylogenetic group (A, B1, B2, D) was determined by triplex PCR [34]. ST131 status was determined by detection of ST131-specific single-nucleotide polymorphisms (SNPs) in mdh and gyrB [35]. ST131 isolates were screened by PCR for presence of the O25b rfb variant [8] and for membership in the H30 ST131 subclone [6, 36]. The prevalence of the (CTX-M-15-associated) H30Rx subclone [37] was assessed by PCR-based detection of a subclone-specific SNP (G723) within the allantoin protein-encoding gene, ybbW [38].
Fifty extraintestinal virulence-associated genes were detected by multiplex PCR [35, 39]. Virulence scores were calculated as the total number of virulence genes detected, adjusted for multiple detection of the pap (P fimbriae), sfa/foc (S and F1C fimbriae), kpsM II (group 2 capsule), and clb (colibactin) operons. Isolates were regarded as extraintestinal pathogenic E. coli (ExPEC) if positive for ≥ 2 of the following genes: papA and/or papC (P fimbriae); afa/draBC (Dr-family adhesins); sfa/focDE; iutA (aerobactin receptor); and kpsM II (group 2 capsule synthesis) [36].
Data collection
Baseline demographic and clinical characteristics were extracted from the LTCF medical record using a standardized data abstraction form [21]. Clinical variables included comorbidities, fecal incontinence, bed-bound status, and presence of a urinary or central venous catheter at the time of study enrollment. All antimicrobial therapy received within 30 days of initial detection of FQREC colonization, was documented, as was receipt of immunosuppressive agents.
Statistical analysis
Among FQREC isolates, associations were explored between E. coli ST131 and 1) mechanisms of FQ resistance, 2) antibiotic susceptibility profiles, and 3) virulence factors. Clinical characteristics were compared between subjects with and without an ST131 FQREC colonizing isolate. Categorical variables were compared using the Fisher exact test, and continuous variables were compared using the Wilcoxon rank-sum test. Odds ratios (OR) and 95% confidence intervals (CI) were calculated to evaluate the strength of associations between ST131 status and clinical variables. All statistical calculations were performed using commercially available software (STATA v13.0; StataCorp LP, College Station, Texas).
RESULTS
Prevalence of ST131
E. coli isolates were analyzed from all 37 LTCF residents who were identified as having incident FQREC colonization (levofloxacin MIC ≥0.25 µg/mL) during the study period. Most (n = 24; 65%) subjects had multiple strains of E. coli detected. The median number of distinct E. coli strains identified on testing from each initial FQREC-containing fecal sample for these 24 residents was 2 (interquartile range [IQR], 2, 3), most commonly one FQ-susceptible and one FQ-resistant strain.
The first-identified FQREC isolate was characterized for each of the 37 residents; of these, 29 (78%) were identified as ST131 by dual-SNP PCR (Table 1). The only other ST131 isolate identified among the study subjects was a FQSEC isolate (levofloxacin MIC of 0.047 µg/mL) from a multiply-colonized patient who had one non-ST131 FQ-resistant strain and two FQ-susceptible strains, including this ST131 isolate. Thus, 29 (97%) of the study’s 30 total ST131 isolates were FQREC.
Table 1.
Microbiologic characteristics of ST131 and non-ST131 E. coli fecal isolates from long-term care facility residents colonized with fluoroquinolone-resistant Escherichia coli.
| Variable | ST131 (n = 29) |
Non-ST131 (n = 8) |
P value |
|---|---|---|---|
| Median levofloxacin MIC (IQR) | 24 (16, 32) | 20 (8, 32) | 0.58 |
| Median no. of gyrA mutations (IQR)a | 2 (2, 2) | 2 (1, 2) | 0.01 |
| Median no. of parC mutations (IQR)b | 2 (2, 2) | 1 (0, 1) | < 0.001 |
| OST-positive status, no. (%) | 6 (21) | 4 (50) | 0.17 |
NOTE. IQR, interquartile range; OST, organic solvent tolerance.
One ST131 isolate could not be characterized in regard to gyrA mutations.
One ST131 isolate could not be characterized in regard to parC mutations.
All 29 ST131 FQREC isolates represented the H30 subclone within ST131, and 9 (31%) belonged specifically to the H30Rx subgroup within the H30 subclone. In contrast, the single ST131 FQSEC isolate did not belong to the H30 ST131 subclone.
Fluoroquinolone resistance mechanisms
The 29 ST131 and 8 non-ST131 FQREC isolates did not differ significantly for levofloxacin MIC (median value, 24 µg/mL versus 20 µg/mL, respectively, P = 0.58) (Table 1). ST131 FQREC isolates had a greater number of replacement gyrA mutations compared to non-ST131 FQREC isolates, with a median of 2 (interquartile range [IQR], 2, 2), versus 2 (IQR, 1, 2) (P = 0.01). All isolates with two gyrA mutations exhibited Ser83Leu and Asp87Asn, regardless of ST131 status. Similarly, ST131 isolates had a greater number of replacement parC mutations compared to non-ST131 isolates, with a median of 2 (IQR, 2, 2), versus 1 (IQR, 0, 1) (P < 0.001). The most common combination of replacement parC mutations among ST131 FQREC isolates, present in all but two of the characterized isolates, was Ser80Ile and Glu84Val.
Virulence genes
Differences in virulence gene profiles between ST131 and non-ST131 FQREC isolates are shown in Table 2. Virulence genes significantly associated with ST131 included fimH, sat, usp, ompT, and malX. A similar proportion of ST131 and non-ST131 isolates qualified as ExPEC (45% and 50%, respectively; P > 0.99). There was no significant difference in virulence gene scores between ST131 isolates and non-ST131 isolates, with median scores of 9 (IQR, 8, 10) and 8 (IQR, 4, 12), respectively (P = 0.64). The virulence gene profile of the one FQ-susceptible ST131 E. coli isolate included adhesins (iha, fimH, afa/draBC), toxins (hylD, sat, tsh), siderophore receptors (iroN, fyuA, iutA), and other factors (kpsM II, K5, iss, ompT, ibeA, usp, traT, rfc, cvaC), qualifying this isolate molecularly as ExPEC.
Table 2.
Distribution of virulence-associated traits among ST131 and non-ST131 E. coli fecal isolates from long-term care facility residents colonized with fluoroquinolone-resistant Escherichia coli.
| Trait | Prevalence of trait, no. (%) | P valuea,b | ||
|---|---|---|---|---|
| ST131 (n = 29) |
Non-ST131 (n = 8) |
|||
| Adhesins | papAHc | 0 (0) | 1 (13) | 0.22 |
| iha | 21 (72) | 3 (38) | 0.10 | |
| fimH | 29 (100) | 6 (75) | 0.04 | |
| Toxins | sat | 23 (79) | 3 (38) | 0.04 |
| vat | 0 (0) | 5 (63) | < 0.001 | |
| hlyFd | 0 (0) | 1 (13) | 0.22 | |
| Siderophores | iroN | 0 (0) | 1 (13) | 0.22 |
| fyuA | 29 (100) | 7 (88) | 0.22 | |
| Miscellaneous | usp | 27 (93) | 4 (50) | 0.01 |
| traT | 27 (93) | 5 (63) | 0.06 | |
| cvaC | 9 (0) | 1 (13) | 0.22 | |
| K1 | 29 (100) | 1 (13) | 0.22 | |
| ompT | 29 (100) | 5 (63) | 0.007 | |
| iss | 0 (0.0) | 1 (13) | 0.22 | |
| H7 fliC | 0 (0) | 2 (25) | 0.04 | |
| malX | 49 (100) | 6 (75) | 0.04 | |
| clbB/N | 0 (0) | 1 (13) | 0.22 | |
Only traits that yielded a P value < 0.30 (by Fisher exact test) are shown. papAH (P fimbria structural subunit), iha (adhesin-siderophore), fimH (type 1 fimbria adhesin), sat (secreted autotransporter toxin), vat (vacuolating toxin), hlyF (hemolysin F), iroN (catecholate siderophore receptor), fyuA (yersiniabactin receptor), usp (uropathogenic-specific protein), traT (serum resistance outer membrane protein), cvaC (colicin V precursor), K1 (group 2 capsule variant), ompT (outer membrane protease), iss (increased serum survival protein), H7 (flagellar variant), malX (pathogenicity island marker), and clbB/N (colibactin synthesis system).
The presence of the following traits were evaluated but not detected in any isolate: papG allele I (P adhesin variant), papG allele III (P adhesin variant), sfa/focDE (S and F1C fimbriae), sfaS (S fimbriae), focG (F1C fimbriae), afa/draBC (Dr-binding adhesins), afaE8 (variant afimbrial adhesin), bmaE (M-agglutinin subunit), gafD (G fimbriae), F17 (fimbrial adhesin), clpG (CS31A surface antigen), hlyD (hemolysin D), cnf1 (cytotoxic-necrotizing factor 1), cdtB (cytolethal distending toxin), tsh (temperature sensitive hemagglutinin), ireA (siderophore receptor), iutA (aerobactin receptor), kpsM II (group 2 capsule), kpsM III (group 3 capsule), K5 (group 2 capsule variant), K15 (group 2 capsule variant), K2/K100 (group 2 capsule variants), rfc (O4 antigen polymerase), and ibeA (invasion of brain endothelium).
papC (P fimbria assembly), papEG (P fimbria tip pilins), papG allele II (P adhesin variant), and hra (heat-resistant agglutinin) demonstrated similar results to those for papAH.
pic (protein associated with intestinal colonization) and astA (enteroaggregative E. coli toxin) demonstrated similar results to those for hlyF.
Antibiotic susceptibility
Overall, ST131 and non-ST131 FQREC isolates (as defined by a levofloxacin MIC ≥0.25 µg/mL) exhibited no significant differences in antibiotic susceptibility profiles (Table 3). Eight (28%) ST131 isolates were ceftazidime-resistant; four (50%) of these belonged to the H30Rx subgroup within the H30 subclone. The prevalence of ceftazidime resistance was numerically but not statistically significantly higher among H30Rx isolates compared to the other ST131 isolates (44% versus 20%; P = 0.20).
Table 3.
Antibiotic resistance by ST131 status among fluoroquinolone-resistant fecal Escherichia coli isolates from long-term care facility residents.
| Antibiotica | Prevalence of resistance, no. (%) | P value | ||
|---|---|---|---|---|
| All isolates (n = 37) |
ST131 (n = 29) |
Non-ST131 (n = 8) |
||
| Ampicillin-sulbactam | 31 (84) | 25 (86) | 6 (75) | 0.59 |
| Cefazolin | 13 (35) | 10 (35) | 3 (38) | > 0.99 |
| Ceftazidime | 10 (27) | 8 (28) | 2 (25) | > 0.99 |
| Ceftriaxone | 9 (24) | 7 (24) | 2 (25) | > 0.99 |
| Gentamicin | 13 (35) | 8 (28) | 5 (63) | 0.10 |
| Tobramycin | 16 (43) | 11 (38) | 5 (63) | 0.25 |
| Piperacillin-tazobactam | 4 (11) | 2 (7) | 2 (25) | 0.20 |
| Trimethoprim-sulfamethoxazole | 17 (46) | 14 (48) | 3 (38) | 0.70 |
Antibiotics listed are those to which ≥ 1 isolate was resistant. No isolate was resistant to imipenem.
PFGE analysis
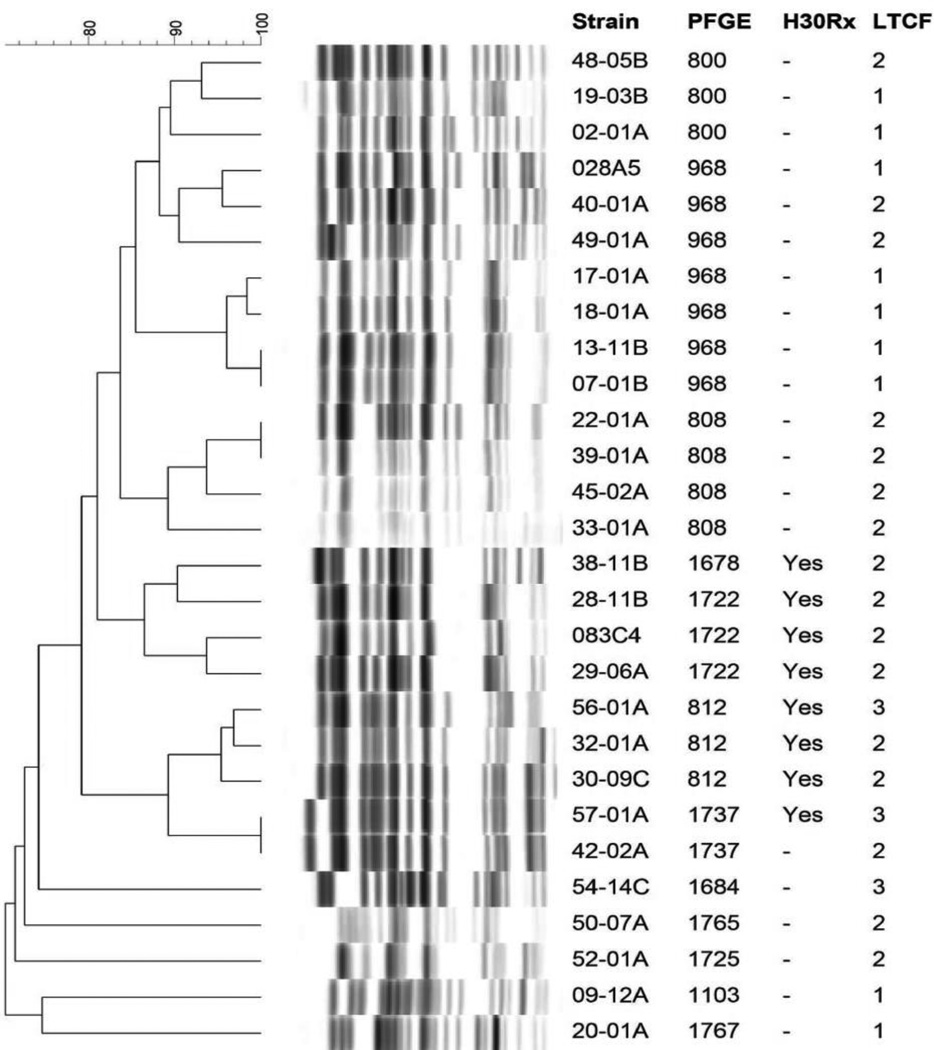
PFGE was performed for 28 of the 29 FQREC ST131 isolates, and pulsotypes were compared to those of previously characterized clinical ST131 isolates from a large, established library [33]. Among the present ST131 colonizing isolates the predominant pulsotypes (defined based on ≥ 94% similarity to an index isolate) were 968 and 808, with 7 and 4 isolates respectively (Figure 1), which also are the leading pulsotypes among the diverse-source clinical ST131 isolates in the reference library [33].
Figure 1. Pulsed field gel electrophoresis patterns of 28 fluoroquinolone-resistant ST131 fecal Escherichia coli isolates from long-term care facility residents.
Data columns to the right of the PFGE profiles are the strain number, pulsotype, H30Rx subclone status, and long-term care facility of origin (1, 2, or 3). Pulsotypes were assigned by comparison to an existing large private PFGE profile reference library (33). All isolates represented the H30 subclone within ST131.
In assessing longitudinal development of resistance, 26 (70%) of 37 subjects had a fecal swab that yielded FQSEC (levofloxacin MIC <0.25 µg/mL) within 30 days prior to the first fecal swab that yielded FQREC (levofloxacin MIC ≥0.25 µg/mL). On PFGE analysis, none of these FQSEC isolates exhibited ≥ 76% profile similarity to the corresponding patient’s subsequent FQREC isolate. Furthermore, none of the FQSEC isolates were ST131.
Risk factors for ST131
Among the 37 subjects with incident FQREC colonization, clinical characteristics were compared between the 29 subjects whose new FQREC strain was ST131 and the 8 for whom it was not (Table 4). These two subgroups did not differ significantly for LTCF of residence, presence of various comorbidities, device use, or receipt of various antibiotic agents or classes (P > 0.10 for all comparisons). There was a trend approaching statistical significance toward an association between female gender and having an ST131 incident colonizing FQREC strain (OR, 0.12; 95% CI, 0.01–1.03; P = 0.05).
Table 4.
Bivariable analyses of risk factors for ST131 Escherichia coli among long-term care facility residents colonized with fluoroquinolone-resistant E. coli.
| Variable | ST131 (n = 29)a |
Non-ST131 (n = 8)a |
OR (95% CI) | P value |
|---|---|---|---|---|
| Year of culture | ||||
| 2006 | 11 (38) | 4 (50) | ||
| 2007 | 8 (28) | 3 (38) | 0.61 | |
| 2008 | 10 (34) | 1 (13) | ||
| Mean age (SD) | 75 (12) | 70 (15) | -------- | 0.28 |
| Female sex | 3 (10) | 4 (50) | 0.12 (0.01–1.03) | 0.05 |
| Non-white race | 19 (66) | 4 (50) | 1.90 (0.28–12.5) | 0.45 |
| Site | ||||
| LTCF #1 | 9 (31) | 4 (50) | ||
| LTCF #2 | 16 (55) | 4 (50) | 0.62 | |
| LTCF #3 | 4 (14) | 0 (0) | ||
| Hospitalization in previous year | 22 (76) | 5 (63) | 1.89 (0.23–12.8) | 0.67 |
| Comorbidities and medicationsb | ||||
| Chronic pulmonary disease | 11 (38) | 1 (13) | 4.28 (0.43–210) | 0.23 |
| Malignancy | 12 (41) | 1 (13) | 4.94 (0.50–241) | 0.22 |
| Any antibioticc | 11 (38) | 1 (13) | 4.28 (0.43–210) | 0.23 |
OR, odds ratio; CI, confidence interval; SD, standard deviation; LTCF, long-term care facility.
Data are presented as numbers (percentages) except for age.
Only variables with P < 0.30 are shown.
≤ 30 days prior to recovery of FQREC.
DISCUSSION
During a 3-year period of screening for colonization with FQREC among residents of 3 LTCFs in Pennsylvania (2006–2008) we evaluated the molecular and clinical epidemiology of ST131 and non-ST131 E. coli colonizing isolates from residents who acquired FQREC colonization during their LTCF stay. We found a strikingly high prevalence of E. coli ST131 (nearly 80%) among the incident FQREC isolates (. Although ST131 and non-ST131 isolates differed significantly for the prevalence of many individual virulence genes, they did not differ for the proportion of isolates that qualified molecularly as ExPEC or for aggregate VF scores. In addition, there were no significant differences in clinical risk factors, including recent antibiotic use, for recovery of ST131 versus non-ST131 FQREC. Lastly, the PFGE pulsotypes for the ST131 colonizing isolates from residents in this study corresponded to those that were common among previously characterized clinical ST131 isolates.
The prevalence of antibiotic-resistant organisms in LTCFs is high, likely due in part to high rates of antibiotic use, elderly hosts, and significant opportunities for person-to-person transmission [40]. In a recent study, LTCF residence and older age were predictors of infection with E. coli ST131 [5]. Similarly, the prevalence of E. coli ST131 in our study cohort of LTCF residents was notably high, with nearly 80% of residents who developed new colonization with FQREC during longitudinal sampling acquiring E. coli ST131. Clinical infection with FQREC is most likely preceded by gastrointestinal tract colonization [16, 17]. Therefore, this finding is particularly concerning given the elderly, vulnerable population, and suggests that LTCFs may serve as an important reservoir of E. coli ST131.
The high prevalence of E. coli ST131 among incident colonizing FQREC isolates in our study may have been due in part to a greater propensity for colonization of the gastrointestinal tract by E. coli isolates within phylogenetic group B2 [18, 19] and ST131 specifically, in combination with other risk factors present in the LTCF setting (e.g., antibiotic selection pressure). Notably, a significant proportion (~40%) of LTCF residents who acquired ST131 FQREC colonization received at least one antibiotic prior to ST131 E. coli recovery. Lastly, increased colonization pressure with E. coli ST131 may have increased the risk of subsequent acquisition of ST131 FQREC by LTCF residents.
As shown previously for clinical ST131 FQREC isolates [6], the majority of fecal ST131 FQREC isolates in our study demonstrated a distinct gyrA/parC allele combination. In addition, common ST131-associated pulsotypes from a large reference library were the predominant pulsotypes (968 and 808) represented in our LTCF cohort. These findings support the likely clinical relevance of the present colonization isolates.
Notably, among the 26 study subjects who had an available FQ-susceptible fecal E. coli isolate from within 30 days prior the subject’s incident FQREC isolate, in no instance was the antecedent FQSEC isolate genetically related to the subject’s later incident FQREC isolate. These findings establish that person-to-person or environmental transmission of already-resistant strains was a more important determinant of FQREC acquisition in these LTCFs than in situ development of de novo FQ resistance (e.g., by stepwise acquisition of gyrA and parC mutations in a given strain during sustained colonization). In this regard, ST131 strains, or the particular ST131 lineages identified here, may be characterized by increased transmissibility, compared with other FQREC. This potentially greater risk of dissemination should be further evaluated in future studies, focusing particularly on the LTCF setting, which is notable for increased opportunity for person-to-person contact.
A recent study demonstrated that, among clinical FQREC isolates, a significantly greater proportion of ST131 compared to non-ST131 isolates were classified as ExPEC [36]. In contrast, among the present fecal FQREC isolates, although the ST131 and non-ST131 isolates differed significantly for the distribution of specific virulence genes, they did not differ for the proportion that qualified molecularly as ExPEC. This finding may be explained by differences in virulence potential between colonizing isolates versus those causing clinical infections. Furthermore, it is possible that the rapid dissemination of ST131 may be a result more of ST131’s ability for successful and persistent intestinal colonization than for greater virulence. Nevertheless, data are needed from ST131-colonized LTCF residents regarding the incidence of and clinical and molecular characteristics associated with progression to clinical infection.
Finally, both ST131 and non-ST131 FQREC isolates exhibited resistance to multiple non-fluoroquinolone antibiotics, including aminoglycosides, ampicillin-sulbactam, and trimethoprim-sulfamethoxazole. The multidrug resistance seen in the present ST131 isolates confirms high rates of colonization with multidrug-resistant gram-negative pathogens seen in the long-term care population [14, 40], and has important implications for selection of empiric therapy for infections in LTCF residents.
Potential limitations of our study must be considered. First, the small sample size, especially for the non-ST131 group, limited statistical power for identification of risk factors for ST131 colonization. Second, since the present study was conducted in LTCFs that were part of a single healthcare system, the results may not be generalizable to other LTCFs with differing characteristics. Third, since our study assessed only incident FQREC isolates, it is uninformative the distribution of ST131 vs. non-ST131 E. coli in prevalent FQREC colonization, which might favor ST131 even more disproportionately than seen here if ST131 strains tend to out-persist other FQREC in the gut.
In conclusion, we found a high prevalence of ST131 among FQREC strains newly acquired by LTCF residents during their LTCF stay. Given the predominantly elderly, vulnerable population in the long-term care setting, future studies are needed to evaluate potentially modifiable risk factors for such acquisition and for subsequent infection in ST131 colonized residents. Furthermore, elucidating the potential for increased transmissibility of ST131 isolates, particularly in long-term care settings characterized by frequent patient-to-patient contact, will be particularly important for development of effective infection control strategies to limit the spread of this emerging multidrug-resistant pathogen.
A prevalence of E. coli ST131 of nearly 80% was found among incident FQREC isolates in long-term care facility residents.
ST131 and non-ST131 isolates differed significantly in the prevalence of virulence factors.
ST131 and non-ST131 isolates did not differ in the proportion of isolates that qualified molecularly as ExPEC.
PFGE pulsotypes for colonizing ST131 isolates were common among previously characterized clinical ST131 isolates.
Acknowledgments
Funding. This material is based upon work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, grant # 1 I01 CX000192 01 (to J.R.J.). This work was also supported by the National Institutes of Health: K01-AI103028 (to J.H.H.) and R01-AG023792 and K24-AI080942 (to E.L.). This study was also supported, in part, by the Centers for Disease Control and Prevention Epicenters Program (U54-CK000163 to E.L.).
Role of the funding agency. The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript.
Potential conflicts of interest. J.R.J has had research grants or contracts with Actavis, Crucell/Jannsen, ICET, Merck, Syntiron, and Tetraphase, and has patent applications for diagnostic tests to detect ST131 and other E. coli clonal groups.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis. 2010;51:286–294. doi: 10.1086/653932. [DOI] [PubMed] [Google Scholar]
- 2.Peirano G, Pitout JD. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents. 2010;35:316–321. doi: 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Lautenbach E. Editorial commentary: flying under the radar: the stealth pandemic of Escherichia coli sequence type 131. Clin Infect Dis. 2013;57:1266–1269. doi: 10.1093/cid/cit505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rogers BA, Sidjabat HE, Paterson DL. Escherichia coli O25b–ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother. 2011;66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 5.Banerjee R, Johnston B, Lohse C, Porter SB, Clabots C, Johnson JR. Escherichia coli sequence type 131 is a dominant, antimicrobial-resistant clonal group associated with healthcare and elderly hosts. Infect Control Hosp Epidemiol. 2013;34:361–369. doi: 10.1086/669865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson J, Tchesnokova V, Johnston B, et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis. 2013;207:919–928. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson JR, Urban C, Weissman SJ, et al. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4), blaCTX-M-15 among extended-spectrum-beta-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother. 2012;56:2364–2370. doi: 10.1128/AAC.05824-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont O, Lavollay M, Vimont S, et al. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother. 2008;61:1024–1028. doi: 10.1093/jac/dkn084. [DOI] [PubMed] [Google Scholar]
- 9.Jones AL, Dwyer LL, Bercovitz AR, Strahan GW. The National Nursing Home Survey: 2004 overview. Vital Health Stat. 2009;13:1–155. [PubMed] [Google Scholar]
- 10.Pakyz AL, Dwyer LL. Prevalence of antimicrobial use among United States nursing home residents: results from a national survey. Infect Control Hosp Epidemiol. 2010;31:661–662. doi: 10.1086/653072. [DOI] [PubMed] [Google Scholar]
- 11.Strausbaugh LJ, Joseph CL. The burden of infection in long-term care. Infect Control Hosp Epidemiol. 2000;21:674–679. doi: 10.1086/501712. [DOI] [PubMed] [Google Scholar]
- 12.Tsan L, Davis C, Langberg R, et al. Prevalence of nursing home-associated infections in the Department of Veterans Affairs nursing home care units. Am J Infect Control. 2008;36:173–179. doi: 10.1016/j.ajic.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Viray M, Linkin D, Maslow JN, et al. Longitudinal trends in antimicrobial susceptibilities across long-term-care facilities: emergence of fluoroquinolone resistance. Infect Control Hosp Epidemiol. 2005;26:56–62. doi: 10.1086/502487. [DOI] [PubMed] [Google Scholar]
- 14.O’Fallon E, Pop-Vicas A, D’Agata E. The emerging threat of multidrug-resistant gram-negative organisms in long-term care facilities. J Gerontol A Biol Sci Med Sci. 2009;64:138–141. doi: 10.1093/gerona/gln020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lautenbach E, Marsicano R, Tolomeo P, Heard M, Serrano S, Stieritz DD. Epidemiology of antimicrobial resistance among gram-negative organisms recovered from patients in a multistate network of long-term care facilities. Infect Control Hosp Epidemiol. 2009;30:790–793. doi: 10.1086/599070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donskey CJ. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin Infect Dis. 2006;(43 Suppl 2):S62–S69. doi: 10.1086/504481. [DOI] [PubMed] [Google Scholar]
- 17.Richard P, Delangle MH, Raffi F, Espaze E, Richet H. Impact of fluoroquinolone administration on the emergence of fluoroquinolone-resistant gram-negative bacilli from gastrointestinal flora. Clin Infect Dis. 2001;32:162–166. doi: 10.1086/317551. [DOI] [PubMed] [Google Scholar]
- 18.Nowrouzian FL, Adlerberth I, Wold AE. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 2006;8:834–840. doi: 10.1016/j.micinf.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 19.Vimont S, Boyd A, Bleibtreu A, et al. The CTX-M-15-producing Escherichia coli clone O25b: H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One. 2012;7:e46547. doi: 10.1371/journal.pone.0046547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess MJ, Johnson JR, Porter SB, et al. Long-term care facilities are reservoirs for antimicrobial-resistant sequence type 131 Escherichia coli. Open Forum Infect Dis. 2015;2:ofv011. doi: 10.1093/ofid/ofv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han JH, Maslow J, Han X, et al. Risk factors for the development of gastrointestinal colonization with fluoroquinolone-resistant Escherichia coli in residents of long-term care facilities. J Infect Dis. 2014;209:420–425. doi: 10.1093/infdis/jit471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang TM, Lu PL, Li HH, Chang CY, Chen TC, Chang LL. Characterization of fluoroquinolone resistance mechanisms and their correlation with the degree of resistance to clinically used fluoroquinolones among Escherichia coli isolates. J Chemother. 2007;19:488–494. doi: 10.1179/joc.2007.19.5.488. [DOI] [PubMed] [Google Scholar]
- 23.Singh R, Swick MC, Ledesma KR, et al. Temporal interplay between efflux pumps and target mutations in development of antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 2012;56:1680–1685. doi: 10.1128/AAC.05693-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kern WV, Oethinger M, Jellen-Ritter AS, Levy SB. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2000;44:814–820. doi: 10.1128/aac.44.4.814-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gales AC, Gordon KA, Wilke WW, Pfaller MA, Jones RN. Occurrence of single-point gyrA mutations among ciprofloxacin-susceptible Escherichia coli isolates causing urinary tract infections in Latin America. Diagn Microbiol Infect Dis. 2000;36:61–64. doi: 10.1016/s0732-8893(99)00121-2. [DOI] [PubMed] [Google Scholar]
- 26.Lautenbach E, Tolomeo P, Black N, Maslow JN. Risk factors for fecal colonization with multiple distinct strains of Escherichia coli among long-term care facility residents. Infect Control Hosp Epidemiol. 2009;30:491–493. doi: 10.1086/597234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lautenbach E, Fishman NO, Metlay JP, et al. Phenotypic and genotypic characterization of fecal Escherichia coli isolates with decreased susceptibility to fluoroquinolones: results from a large hospital-based surveillance initiative. J Infect Dis. 2006;194:79–85. doi: 10.1086/503046. [DOI] [PubMed] [Google Scholar]
- 28.Lautenbach E, Metlay JP, Weiner MG, et al. Gastrointestinal tract colonization with fluoroquinolone-resistant Escherichia coli in hospitalized patients: changes over time in risk factors for resistance. Infect Control Hosp Epidemiol. 2009;30:18–24. doi: 10.1086/592703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han JH, Nachamkin I, Tolomeo P, Mao X, Bilker WB, Lautenbach E. Risk factors for efflux pump overexpression in fluoroquinolone-resistant Escherichia coli. J Infect Dis. 2012;206:1597–1603. doi: 10.1093/infdis/jis567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6126. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001;45:1515–1521. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goering RV, Tenover FC. Epidemiological interpretation of chromosomal macro-restriction fragment patterns analyzed by pulsed-field gel electrophoresis. J Clin Microbiol. 1997;35:2432–2433. doi: 10.1128/jcm.35.9.2432-2433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson JR, Nicolas-Chanoine MH, DebRoy C, et al. Comparison of Escherichia coli ST131 pulsotypes, by epidemiologic traits, 1967–2009. Emerging infectious diseases. 2012;18:598–607. doi: 10.3201/eid1804.111627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JR, Menard M, Johnston B, Kuskowski MA, Nichol K, Zhanel GG. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother. 2009;53:2733–2739. doi: 10.1128/AAC.00297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colpan A, Johnston B, Porter S, et al. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis. 2013;57:1256–1265. doi: 10.1093/cid/cit503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price LB, Johnson JR, Aziz M, et al. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio. 2013;4(6) doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bannerjee R, Robicsek A, Kuskowski MA, et al. Molecular epidemiology of Escherichia coli sequence type ST131 and its H30 H30-Rx subclones among extended-spectrum beta-lactamase-positive and -negative E. coli clinical isolates from the Chicago region (2007–2010) Antimicrob Agents Chemother. 2013;57:6385–6388. doi: 10.1128/AAC.01604-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson JR, Johnston B, Kuskowski MA, Nougayrede JP, Oswald E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J Clin Microbiol. 2008;46:3906–3911. doi: 10.1128/JCM.00949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Fallon E, Kandel R, Schreiber R, D’Agata EM. Acquisition of multidrug-resistant gram-negative bacteria: incidence and risk factors within a long-term care population. Infect Control Hosp Epidemiol. 2010;31:1148–1153. doi: 10.1086/656590. [DOI] [PubMed] [Google Scholar]