Abstract
Macrophages are ubiquitous innate immune cells that play a central role in health and disease by functionally “polarizing” to distinct phenotypes, which are broadly divided into classical inflammatory responses (M1) and alternative responses (M2) that promote immune suppression and wound healing. Although macrophages are attractive therapeutic targets, incomplete understanding of polarization limits clinical manipulation. While individual stimuli, pathways, and genes involved in polarization have been identified, how macrophages evaluate complex in vivo milieus comprising multiple divergent stimuli remains poorly understood. Here, we used combinations of “incoherent” stimuli – those that individually promote distinct macrophage phenotypes – to elucidate how the immunosuppressive, IL-10-driven macrophage phenotype is induced, maintained, and modulated under such combinatorial stimuli. The IL-10-induced immunosuppressive phenotype was largely dominant but required sustained IL-10 signaling to maintain this phenotype. Our data also implicate the intracellular protein, BCL3, as a key mediator of the IL-10-driven phenotype. IL-12 did not directly impact polarization of IL-10-treated macrophages, but IFNγ disrupted a positive feedback loop that may reinforce the IL-10-driven phenotype in vivo. This novel combinatorial perturbation approach thus generated new insights into macrophage decision making and local immune network function.
Keywords: interleukin-10 (IL-10), macrophages, macrophage polarization, tumor-associated macrophages (TAM)
Introduction
Macrophages are ubiquitous cells of the innate immune system that play central roles in both host defense and disease. A central feature of macrophage biology is the capacity to functionally “polarize” to various phenotypes in response to diverse environmental stimuli.1–5 Although these phenotypes span a continuum of states, macrophage heterogeneity may be broadly divided into classic inflammatory responses (M1), such as those mounted to combat microbial infections, and alternative responses (M2), with the latter type mediating killing of parasites, activation of Th2 helper T cells, and immuno-suppression and tissue remodeling during wound healing.6 In many chronic diseases, macrophage-mediated responses become dysfunctional and promote disease, as is perhaps best illustrated in cancer.
Many types of cancer involve dysfunctional macrophage responses, which drive tissue remodeling, tumor proliferation, invasion, and metastasis.3, 7–9 However, macrophages may also limit cancer growth and promote clearance of established tumors. Tumor infiltrating macrophages (TIM), tumor-associated macrophages (TAM, which are also present at sites away from the tumor bed, such as the peritoneum), and related myeloid-derived suppressor cells (MDSC) help to establish and propagate the dysfunctional immunological microenvironment at tumor sites and proximal sites of peripheral immune function including lymph nodes10–13, where other immune cell types such as T cells and dendritic cells are converted to phenotypes that promote tumor survival. Dysfunctional local immune networks are generally considered to be one of the most significant barriers to immunotherapies for cancer, including both adoptive T cell-based therapies (such as those in which T cells are engineered to express chimeric antigen receptors)14, 15 and induced immunity through cancer vaccines16. For these reasons, macrophages are attractive targets for therapeutic intervention.17–19 The promise of such a strategy was highlighted by a recent report that shifting macrophage phenotype was sufficient to provide benefits in a pre-clinical model of cancer treatment.20 However, despite the importance of macrophage polarization to a broad range of diseases, a mechanistic and systems-level understanding of these phenomena is generally lacking.4, 21, 22
To date, investigations into macrophage functional polarization generally involve stimulating cells with one or more microenvironmental stimuli known to promote a particular functional phenotype.1, 3, 23–27 Such cues are predominantly cytokines or chemokines but may also be metabolites or physical conditions such as hypoxia.3 An important limitation of these approaches is that nearly all natural microenvironments in vivo are characterized by multiple stimuli that, taken individually, would promote polarization into distinct macrophage phenotypes. Therefore, in this study, we investigated how macrophages resolve physiologically and therapeutically relevant combinations of “incoherent” stimuli – those that individually promote divergent phenotypes – at both phenomenological and mechanistic levels. In particular, we investigated how the IL-10-driven macrophage phenotype is modulated by co-exposure to stimuli that may be considered incoherent with IL-10. IL-10 directs macrophage polarization to an immunosuppressive phenotype that has been likened to that observed in TAM.1, 21, 23, 28, 29 Moreover, IL-10 is prevalent in both tumors and other immunosuppressive environments established by MDSC and regulatory T cells (Treg).30, 31 Although stimuli such as IL-4 or IL-13 drive polarization to a distinct M2 phenotype1, here we use “M2” as a shorthand refer to the IL-10-induced macrophage phenotype that is the focus of this study. We identified key molecular mechanisms by which the IL-10-driven macrophage phenotype is induced, maintained, and modulated under incoherent stimuli. Together, these findings provide new insights into macrophage decision making and interactions within local immune networks.
Materials and methods
Cell culture
RAW264.7 cells (RAW cells) were a gift from David Segal, Experimental Immunology Branch, NIH (Bethesda, MD). RAW cells were cultured in complete DMEM medium (DMEM + 10% heat-inactivated FBS + 4 mM L-glutamine) on tissue culture treated dishes. Cells were passaged via incubating in PBS-EDTA (0.05 M EDTA) for 5 min followed by gentle cell scraping. M-CSF-containing L929-conditioned medium was made by culturing L929 cells (ATCC) in complete RPMI (RPMI + 10% heat-inactivated FBS + 4 mM L-glutamine) for 7 days. Bone marrow-derived macrophage (BMDM) differentiation medium comprised 10% L929-conditioned medium added to complete RPMI. To generate BMDM, 4–8 week old C57BL/6 male mice (Jackson Labs) were sacrificed and bone marrow cells in the femur were harvested and cultured in differentiation medium.32, 33 Medium was replaced with fresh differentiation medium every 3 days of culture. At day 7, BMDM were harvested and characterized by flow cytometry to confirm differentiation using antibodies against surface antigens F4/80 (BD bioscience) and CD11b (BD bioscience) (Fig. S1). Where indicated, cells were activated with 100 ng/mL E. Coli 055:B5 LPS (Sigma-Aldrich). Cytokine treatments (IL-10, IL-12p70, IFNγ) were conducting using recombinant cytokines purchased from R&D Systems.
Real-time quantitative PCR
RNA was isolated from cells (Omega Bio-Tek) and reverse transcribed into cDNA (Quanta Bioscience). Real-time quantitative PCR (qPCR) was conducted using SYBR green super mix (Bio-Rad). Approximately 10 ng of mRNA template were loaded per run. Primer concentrations were 200 nM for both forward and reverse primers, and primer sequences are presented in Table S1. Cycling conditions were as follows: 95 °C 5 min, 40 cycles (95 °C for 30 s, 55 °C for 30 s). qPCR data were analyzed using normalized ΔΔCt analysis34, for which GAPDH was selected as an internal reference “housekeeping” gene. Experiments were conducted in biological triplicate and, unless otherwise stated, were normalized to samples from cells treated with LPS only. Error bars represent one standard deviation. Statistical analyses were conducted using two-tailed Student’s t-tests.
Western blots
Western blots were performed on whole cell lysates. RIPA buffer was used for protein extraction followed by centrifugation at 4 °C for 20 min to pellet cell debris. Protein lysate was boiled at 95 °C for 5 min in Laemmli buffer. Protein concentration was quantified by Bradford assay (Thermo Scientific), and 30 μg per sample was loaded and run in pre-cast gels (Bio-Rad). Samples were transferred to PVDF membranes overnight at 50 V. Antibodies used for Western blot analyses were: anti-SOCS1 (Abcam), anti-SOCS3 (Abcam), anti-pSTAT3 (Abcam), anti-pSTAT4 (Y693) (Abcam) anti-STAT3 (Cell Signaling), anti-STAT4 (Cell Signaling), anti-GAPDH (Abcam), anti-rabbit HRP-conjugated secondary antibody (Invitrogen). Transferred membranes were blocked using 5 % BSA in TBST for 2 h followed by primary antibody labeling in 5% BSA in TBST overnight. Membranes that were incubated with primary antibodies overnight were washed with TBST three times followed by secondary antibody staining in 5% BSA in TBST for 1 h. Membranes were treated with ECL solution (Bio-Rad) for 5 min and then exposed on films (GE).
Flow cytometry
Surface staining: cells were incubated in PBS-EDTA followed by gentle scraping to suspend, pelleted, and re-suspended in flow buffer (0.1 % BSA + PBS). Cells were re-pelleted, suspended in 50 ul of flow buffer and incubated in fixing buffer (1 % paraformaldehyde in PBS) for 30 min at 4 °C before washing 2 times in flow buffer. Normal mouse serum (Thermo Scientific) was used to block non-specific binding. Primary antibodies were added to the mouse serum-containing cell samples and incubated for 1 h at 4 °C. Cells were washed with flow buffer three times and stained with secondary antibodies for 30 min at 4 °C before washing 2 times with flow buffer and collecting for flow analysis. Positive signal were determined using isotype control samples to determine negative gating. Intracellular staining: Cells were harvested, washed and fixed as for surface staining. Cells were then suspended in permeabilization wash buffer (0.5 % saponin and 0.2 % BSA in PBS). Normal mouse serum was used to block both surface and intracellular non-specific binding. Primary conjugated antibodies were added to mouse serum-containing cell samples and incubated for 1 h at 4 °C before washing 2 times with flow buffer and collecting for flow analysis. Antibodies used for flow cytometry analyses were: surface antigens F4/80 (BD bioscience) and CD11b (BD bioscience) and intracellular antigen TNFα (BD bioscience). Samples were analyzed on an LSR II (BD), and mean fluorescent intensity (MFI) was calculated using FlowJo software (Treestar) and used to quantify relative protein expression levels. Statistical analyses were conducted using two-tailed Student’s t-tests.
Results
Resolution of incoherent stimulation by IL-10 and IL-12
To begin investigating how IL-10 drives macrophage phenotype in the presence of incoherent stimuli, we first examined a “competition” between the cytokines IL-10 and IL-12. IL-12 is one of the few cues that has been reported to drive phenotypic conversion of TAM, TIM, and MDSC in vivo, promoting expression of M1-associated genes and suppressing M2-associated genes.35–37 Some of these effects may be driven indirectly, since IL-12 induces IFNγ production in multiple cell types that may appear at the tumor site or in proximal lymph nodes, including NK cells, type I T helper (Th1) cells and dendritic cells38, 39, and IFNγ promotes expression of M1-associated genes1, 21, 40. Nonetheless, IL-12 may also act upon macrophages directly and has been reported to drive phenotypic conversion of TAM and TIM in vitro (Fig. S2).35, 41 However, the specific mechanisms by which IL-12 might induce phenotypic conversion, especially in the presence of immunosuppressive signals such as IL-10, are not known.
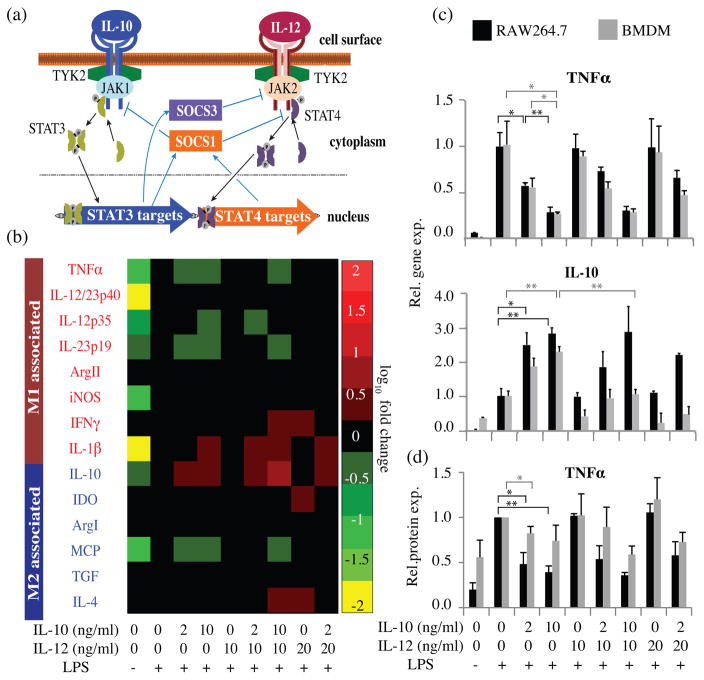
The downstream pathways through which the IL-10 and IL-12 receptors signal exhibit several potential modalities for cross-talk and competition, via the suppressors of cytokine signaling proteins, SOCS1 and SOCS3 (Fig. 1a). IL-10 induces signaling via the transcription factor Signal Transducer and Activator of Transcription 3 (STAT3), which drives the production of SOCS1 and SOCS3.42, 43 IL-12 induces signaling via STAT4, which promotes the production of SOCS1.44 SOCS1 inhibits signaling downstream of both IL-10 and IL-12 receptors45, while SOCS3 inhibits signaling downstream from IL-12 but not IL-1043. Therefore, SOCS1 and SOCS3 may mediate asymmetric feedback regulation of signaling pathways that regulate macrophage phenotype determination. This network has also been reported to exhibit positive feedback in response to IL-10 or IL-12, via increased IL-10 secretion46 and increased expression of the IL-12 receptor47, respectively. Notably, this qualitative conceptual model (Fig. 1a) cannot predict the outcome of a competition between incoherent stimuli IL-10 and IL-12.
Figure 1. Dose dependent responses to incoherent stimuli IL-10 and IL-12.
(a) Conceptual model of potential motifs of cross-talk and competition in the IL-10/IL-12 signaling network in macrophages. (b) RAW cells were treated with combinations of IL-10 and IL-12 for 12 h, activated with LPS for 3 h, and profiled by qPCR. Red text: genes typically annotated as M1, blue text: genes typically annotated as M2. Heat map colors indicate up-regulation or down-regulation vs. the LPS-only control as per the scale bar. All data, including non-responsive genes not shown here, are shown with error bars in Figure S3. (c) Comparative gene expression responses of RAW cells and BMDM to IL-10/IL-12 competition. Error bars represent +/−1 standard deviation, from biological triplicates. (d) Comparative intracellular TNFα protein expression by RAW cells and BMDM following IL-10/IL-12 competition, quantified by flow cytometry. Error bars represent +/−1 standard deviation, from three independent experiments. For additional statistical analysis of panels C–D, see Figure S4. (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001)
To experimentally investigate how macrophages resolve an IL-10/IL-12 competition, murine RAW264.7 (RAW) macrophages were pre-treated with IL-10, IL-12, or both at biologically-relevant doses35, 39, 41 for 12 h, and then cells were “activated” with lipopolysaccharide (LPS). LPS-induced Toll-like Receptor 4 (TLR4) signaling generally promotes macrophage M1 responses and can also be induced via endogenous signals, for example in necrotic tissue.48, 49 However, in the presence of IL-10, TLR4 signaling enhances expression of M2-associated genes that are not induced by LPS or IL-10 alone.50 In this experiment, gene expression was profiled 3 h post-LPS activation to focus on early phase responses (Figs. 1b, S3). In general, pre-treatment with IL-10 suppressed some M1-associated gene expression and enhanced M2-associated expression of IL-10. Inclusion of IL-12 in the pre-treatment shifted expression patterns somewhat, and several genes (e.g., IL-4, IDO, and IFNγ) were moderately up-regulated by IL-12 pretreatment. However, in general, IL-12 neither promoted M1 genes nor suppressed M2 genes. To investigate whether this dominance was an artifact of the RAW cell line, we compared responses of RAW cells to those of primary bone marrow derived macrophages (BMDM) following IL-10/IL-12 pre-treatment, focusing on the most dynamically-regulated genes (TNF α and IL-10). Both cell types exhibited similar patterns of TNFα gene and protein expression in response to IL-10 dose, which was irrespective of IL-12 co-treatment (Figs. 1c–d, S4). In BMDM, IL-10 expression was moderately suppressed by IL-12 pre-treatment, although this effect was not observed in RAW cells. Because TNFα expression is regulated post-transcriptionally by mRNA stabilization51, 52, we also investigated whether TNF α protein levels correlated with mRNA levels. TNF α protein expression was assessed by intracellular flow cytometry in order to capture patterns evident at this early time point (3 h post-LPS activation), at which time secreted protein levels are expected to be low53, 54. Intracellular TNFα protein levels largely mirrored the pattern observed at the mRNA level. Controls in which cells were treated with IL-10/IL-12 without LPS activation exhibited similar patterns (Fig. S3b). To investigate whether this apparent dominance of IL-10 was limited to the early response, an additional IL-10/IL-12 competition was conducted in which gene expression was profiled at 3, 6, and 12 h after LPS activation (Fig. S5). Overall, macrophages exposed to IL-10 and IL-12 simultaneously again exhibited gene expression patterns similar to those induced by IL-10 alone.
Having characterized the IL-10/IL-12 competition at a phenomenological level, we next investigated potential molecular mechanisms underlying the dominant role of IL-10 in determining macrophage phenotype in this system. We hypothesized that IL-10 may exert its dominant effect via either “upstream” or “downstream” mechanisms, or both. An “upstream” mechanism would block the effects of IL-12 by blocking signaling via its receptor (e.g., induction of SOCS3 expression) (Fig. 1a). A “downstream” mechanism would comprise IL-10 signaling-induced expression of a regulator of transcription or translation that suppresses the expression of M1 genes, such that expression of this regulator is not sensitive to IL-12-induced signaling.
To first investigate upstream mechanisms, we examined the involvement of SOCS proteins, given their potential roles discussed above (Fig. 1a). SOCS1, which blocks both STAT3 and STAT4 phosphorylation18, 55, was expressed at a basal level that did not vary substantially (at either protein or mRNA levels) following treatment with various combinations of IL-10, IL-12, and LPS (Fig. 2a, b). In contrast, SOCS3 mRNA and protein expression increased proportionally with IL-10 treatment (Fig. 2a, b). Elevated SOCS3 expression did not require LPS activation, suggesting that this mechanism could confer blockade of IL-12-mediated STAT4 phosphorylation prior to activation with LPS. To investigate this possibility, STAT4 phosphorylation and SOCS3 expression were monitored over time during an IL-10/IL-12 competition in the absence of LPS (Fig. 2c–e). At both low and high doses of IL-10 (2, 10 ng/mL, respectively), IL-12-mediated STAT4 phosphorylation was impaired for times greater than ~30 minutes, which correlated with the elevated SOCS3 gene and protein expression observed beginning 30–60 minutes post-cytokine treatment (Fig. 2d–f) and is consistent with known mechanisms of SOCS343. However, high IL-10 doses impaired STAT4 phosphorylation at time points preceding elevated SOCS3 expression (e.g., 5 minutes post-cytokine treatment), and this effect was not observed for low IL-10 doses. Together, these results indicate that an “upstream” mechanism mediated by IL-10-induced SOCS3 expression may explain long-term blockade of IL-12-mediated signaling, and additional upstream mechanisms may exist (see Discussion).
Figure 2. Potential regulation of IL-10-dominated polarization.
(a) SOCS1 and SOCS3 gene expression in response to 12 h of IL-10 and IL-12 treatment (+/− subsequent activation by LPS for 3 h). Experiments in panel (a) were conducted in biological triplicate in RAW cells; the left panel was normalized to the LPS-only control, and the right panel was normalized to the “no treatment” control. (b) SOCS1 and SOCS3 protein expression in response to IL-10/IL-12 treatment followed by 3 h activation with LPS (c) pSTAT4 protein expression post IL-12 exposure (d, e) pSTAT4 and SOCS3 protein expression after IL-10/IL-12 treatment (f) SOCS3 gene expression following IL-10/IL-12 treatment. Experiments in panel (f) were conducted in biological triplicate in RAW cells and normalized to the “no treatment” control. (g) RAW cells were treated with IL-10 and IL-12 for 15 h (+/− activation with LPS for the final 3 h) and harvested for qPCR analysis. Each data series was normalized to its own “no cytokine” control. For additional statistical analysis, see Figure S6. (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001)
Since these upstream mechanisms would not explain why IL-10 induced a dose-dependent suppression of M1 genes and induction of M2-associated IL-10 (Fig. 1), we next investigated whether potential “downstream” regulators could explain these observations. Note that such a proposed IL-10-induced regulator need not necessarily promote the expression of IL-10, since IL-10-induced STAT3 phosphorylation already drives expression of IL-10 and other M2 genes56. One candidate regulator is cyclic AMP-responsive element-binding protein (CREB), which indirectly induces M2 genes and suppresses M1 genes. CREB induces the expression CEBPβ, which drives expression of M2 genes (e.g., IL-10). CREB also drives the expression of a negative regulator of M1 gene expression termed dual specificity protein phosphatase 1 (DUSP1).4, 57, 58 An additional candidate regulator is BCL3, since expression of this protein is induced by STAT3 phosphorylation, and BCL3 blocks NF-κB (p50/p65)-mediated induction of TNFα gene expression.59, 60 A final candidate examined is interferon regulatory factor (IRF5), which is a transcriptional activator for multiple inflammatory cytokines including TNFα, IL-12 and IFNγ, and IRF5 also suppresses expression of anti-inflammatory cytokines including IL-10.4, 61
We investigated whether the expression of these candidate genes fit the pattern predicted for a downstream regulator: dose-dependence on IL-10 and independence of IL-12 treatment. Of all candidates examined, only BCL3 and DUSP1 expression fit this pattern (Figs. 2g, S6). However, DUSP1 induction was dependent on activation of cells with LPS, while BCL3 induction did not require LPS activation, suggesting that only BCL3 might mediate the dominant effects of IL-10 prior to activation by LPS. Note that BCL3 gene expression was enhanced about 4-fold by LPS activation, but the pattern of dose-responsiveness to IL-10 was unaltered by LPS activation (Figs. 2g, S6). Thus, BCL3 may play an important role in the induction of the IL-10-driven macrophage phenotype both before and after these cells become activated.
Phenotypic reversion upon IL-10 removal
We next investigated how the IL-10-driven macrophage phenotype may be sustained or altered following initial functional commitment. In prior work, when TIM or TAM were removed from tumor-bearing mice and cultured in vitro, addition of IL-12 to the in vitro culture suppressed M2-associated expression (e.g., IL-10 secretion by TAM) and enhanced M1-associated expression (e.g., TNF α secretion by both TIM and TAM).35 In such experiments, removal of macrophages from an animal may reduce the concentration of cytokines to which these cells are exposed, potentially rendering them more directly responsive to IL-12. By analogy, we next investigated whether removing extracellular IL-10 from cultures of macrophages previously driven to M2 in our distinct in vitro model would alter phenotype and responsiveness to IL-12. RAW cells and BMDM were exposed to IL-10 for 12 h, then exogenous IL-10 was removed from the system by replacement with fresh medium (+/− IL-12), and 12 h later, cells were activated with LPS (Figs. 3a, S7). Notably, removal of IL-10 was generally sufficient to drive reversion of both BMDM and RAW cells to a more M1-like phenotype; addition of IL-12 induced no significant changes in RAW cell phenotype, and with the exception of IL-12/IL-23p40, IL-1β and IL-23p19, BMDM gene expression was also largely unaltered by IL-12 addition. Interestingly, IL-12 treatment resulted in a modest decrease in IL-23p19 gene expression in BMDM, via unknown mechanisms. Thus, sustaining the IL-10-induced M2 phenotype required sustained IL-10-induced signaling, reversion to a phenotype that could be driven to M1 upon LPS activation was spontaneous upon IL-10 removal, and neither of these processes was directly impacted by IL-12.
Figure 3. Maintenance of macrophage phenotype via sustained IL-10 exposure.
(a) RAW cells (black) and BMDM (grey) were exposed to the various sequential treatment conditions described in the schematic at left. Cells were harvested 3 h after LPS activation for qPCR analysis. Error bars represent +/−1 standard deviation, from biological triplicates. (MC: Media change, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001) (b) BCL3 gene expression was measured at various time points after IL-10 removal. Data were normalized to a no treatment control, shown by a grey bar spanning +/−1 standard deviation, from biological triplicates. (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001)
We next investigated whether this spontaneous phenotypic reversion was reflected in the expression of the key regulator BCL3. We hypothesized that if BCL3 is a central regulator of IL-10-induced polarization, then since removal of IL-10 was sufficient to restore M1-like responses to LPS (Fig. 3a), then BCL3 expression would be expected to decrease following removal of IL-10 from the environment. As predicted, when macrophages were pre-treated with IL-10 for 12 h, and then IL-10 was removed from the culture medium, BCL3 gene expression dropped to baseline within 3 h (Fig. 3b). This pattern is thus consistent with our proposed model in which BCL3 plays a central role in mediating the dose-dependent and reversible effects of IL-10-induced suppression of M1-associated gene expression.
Resolution of incoherent stimulation by IL-10 and IFNγ
Since IL-12 did not directly modulate IL-10-induced macrophage polarization in our model, we next investigated a competition between IL-10 and the potent pro-M1 stimulus IFNγ. As noted above, IL-12-induced production of IFNγ by neighboring immune cells may mediate indirect effects of IL-12 on macrophages in vivo.38, 39 Previously, the primary insight into this interaction was that when macrophages were pre-polarized to M1 via exposure to IFNγ during differentiation from primary monocytes, subsequent exposure to IL-10 + LPS did not result in induction of M2 genes associated with immunosuppression.62 Furthermore, this IFN γ-induced pre-polarization prevented IL-10-mediated inhibition of the expression of M1-associated genes and proteins. How macrophages would resolve simultaneous exposure to this set of incoherent stimuli, however, was not clear. To investigate, RAW cells and BMDM were treated with combinations of IL-10 and IFNγ, followed by activation with LPS (Figs. 4, S8, S9). Interestingly, co-stimulation with IFNγ did not block IL-10-mediated suppression of TNFα gene expression, although IFNγ did suppress IL-10-induced expression of IL-10. These effects were similar in RAW cells and BMDM (Fig. 4b, S9). In addition, IFNγ induced the expression of iNOS and IL-12/IL-23p40 even in the presence of IL-10 (Figs. 4a, S8), indicating that IFNγ promoted some M1-associated gene expression even in IL-10-rich environments.
Figure 4. Macrophage polarization in response to incoherent stimuli IL-10 and IFNγ.
(a) RAW cells were treated with combinations of IL-10 and IFNγ for 12 h, activated with LPS for 3 h, and profiled by qPCR. Heat map is formatted as in Figure 1, and all data, including non-responsive genes not shown here, are reported with error bars in Figure S8. (b) Comparative gene expression for RAW cells and BMDM treated as in (a). Error bars represent +/−1 standard deviation, from biological triplicates. For additional statistical analyses, see Figure S9. (*p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001)
We next investigated potential mechanisms by which macrophages may resolve incoherent stimulation by IL-10 and IFNγ. Because IFNγ failed to reverse IL-10-mediated suppression of TNFα expression following activation with LPS, we hypothesized the absence of any “upstream” mechanism (e.g., induction of SOCS1 expression) that would block signaling downstream of the IL-10 receptor. To investigate, we examined SOCS1 expression and STAT3 phosphorylation following treatment with IL-10 and/or IFNγ (Fig. 5). Although IFNγ treatment enhanced expression of SOCS1 mRNA after LPS-mediated activation, IFNγ treatment alone did not drive increased production of SOCS1 protein, nor did IFNγ treatment substantially impair IL-10-mediated phosphorylation of STAT3. These observations support the lack of an upstream mechanism in the absence of LPS, and thus downstream regulator(s) may mediate the resolution of incoherent stimulation by IL-10 and IFNγ (see Discussion).
Figure 5. Evaluating upstream regulation of the competition between incoherent stimuli IL-10 and IFNγ.
(a) SOCS1 gene expression after 12 hours treatment with IL-10 and IFNγ followed by 3 hours of activation with LPS. Experiments were conduced in biological triplicate in RAW cells and normalized to the LPS only control. (b) SOCS1 protein expression and STAT3 phosphorylation following treatment with IL-10 and IFNγ without LPS activation.
Discussion
This investigation revealed several important features of macrophage responses to complex stimuli. First, direct competition between IL-12 and IL-10 yielded a general dominance of the IL-10-driven M2 phenotype (Figs. 1, S2, S3, S4). Moreover, removal of IL-10 was sufficient to drive reversion of cells to a phenotype resembling M1 after activation with LPS (Fig. 3a). While IL-10-induced expression of SOCS3 may confer long-term non-responsiveness to IL-12, IL-10-mediated reduction in IL-12-induced STAT4 phosphorylation was sometimes apparent as early as 5 minutes post cytokine treatment (Fig. 4). It is possible that the IL-10 and IL-12 receptors compete for downstream signaling molecules, such as Tyrosine kinase 2 (TyK2), which is involved in both pathways.63–65 We also present evidence that BCL3 is a central “downstream” mediator of IL-10-induced suppression of the M1 phenotype59, and BCL3 expression increased upon exposure to IL-10 and decreased upon IL-10 removal (Figs. 2g, 3b, S6). Together, these results suggest that BCL3 may be an attractive therapeutic target for disrupting at least some features of the dominant IL-10-induced immunosuppressive phenotype. These results also suggest that comparing our observations with a direct IL-10/IL-12 competition in TIM and TAM, which are directly responsive to IL-12 in vitro35, 41, may elucidate whether IL-10 exerts a similarly dominant influence in these macrophages.
In a distinct pairing of incoherent stimuli, IFNγ suppressed IL-10-induced expression of IL-10 (Fig. 4, S8, S9), which could disrupt an important positive feedback loop within immunosuppressive networks46. Since IFNγ production is induced by IL-12 in vivo38, 39, it may thereby mediate a key indirect effect of IL-12 on macrophage polarization, even in the presence of IL-10 (Fig. 6a). Although IFNγ also drove the expression of M1-associated genes iNOS and IL-12/IL-23p40 in the presence of IL-10, some M1 genes (including TNFα) were suppressed (Figs. 4, S8), suggesting that co-exposure to this pairing of incoherent stimuli partially mutes both IFNγ and IL-10-induced macrophage phenotypes. Our data did not support the existence of an upstream mechanism by which IFNγ induces expression of SOCS1 to inhibit IL-10-mediated signaling (Fig. 5), and thus downstream regulator(s) may mediate resolution of co-stimulation by IL-10 and IFNγ. One candidate regulator is glycogen synthase kinase 3 (GSK3), which is activated by dephosphorylation in response to IFNγ treatment.66, 67 GSK3 blocks LPS-induced expression of IL-10 by impairing the MAPK-mediated phosphorylation of CREB downstream from TLR4 (Fig. S10).58, 67, 68 Since GSK3 does not block TLR4-mediated activation of NF-κB, this mechanism would also explain why IFNγ does not render cells non-responsive to LPS40. Ultimately, such models of core regulatory networks can also motivate the investigation of clinically relevant strategies. For example, if we speculate (or determine) that the core IFNγ/IL-10 regulatory network proposed here (Fig. 6b) applies to TAM and TIM in vivo, it would be informative to investigate whether IL-10 receptor blockade can boost the efficacy of macrophage-targeted drugs. For example, in a model of glioblastoma multiforme (GBM), BLZ945 (a colony stimulating factor-1 receptor inhibitor) drove suppression of M2-associated genes but not expression of M1-associated genes, despite elevated IFNγ at the tumor69. Our model suggests that sustained IL-10 signaling could explain this effect, and perhaps blockade of this pathway could promote enhanced M1-associated phenotypes.
Figure 6. Resolution of incoherent stimuli by multiscale networks.
(a) Proposed multicellular network model by which IL-12 may indirectly promote an M1-like phenotype. (b) Proposed core intracellular regulatory network mediating resolution of IL-10/IFN γ-induced signaling. Dotted lines indicate potential mechanisms requiring further investigation (see Fig. S10 for additional detail).
This investigation employed a novel approach to studying macrophage decision-making by applying “incoherent” stimuli to illuminate dominant features of cellular regulation. This strategy has previously proven informative for elucidating networks regulating processes such as apoptosis/survival70, stem cell differentiation71, T cell lineage commitment72, and B cell fate outcome73. An important feature of these investigations, including ours, is that qualitative descriptions of cellular networks are insufficient to predict the outcomes of such inherently quantitative experimental investigations. Indeed many related cells types including macrophages of different types, MDSC, monocytes and dendritic cells likely share similar intracellular signaling topologies. Thus, experimental interrogation by incoherent stimuli could reveal whether similar patterns of dominance are present in related cells, perhaps most importantly in TIM, TAM, and the distinct IL-4/IL-13 induced M2 phenotypes1. In general, the degree to which conclusions reached using pairwise combinatorial stimuli hold as environmental complexity increases must also be evaluated. Ultimately, such an approach may identify additional experimentally testable hypotheses and help to bridge the divide between in vitro experiments and complex environments in vivo.
Supplementary Material
Acknowledgments
RAW264.7 cells were a generous gift from David Segal (NIH). This work was supported by the Northwestern University Physical Sciences-Oncology Center (http://www.psoc.northwestern.edu) through National Institutes of Health award U54 CA143869-05 (http://www.nih.gov) and startup funds from Northwestern University (JNL). This work was supported by the Northwestern University Flow Cytometry Facility and a Cancer Center Support Grant (NCI CA060553). Traditional sequencing services were performed at the Northwestern University Genomics Core Facility. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–86. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Porta C, Rimoldi M, Raes G, et al. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc Natl Acad Sci USA. 2009;106:14978–83. doi: 10.1073/pnas.0809784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sica A, Larghi P, Mancino A, et al. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18:349–55. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–61. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 5.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray PJ, Allen JE, Biswas SK, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gajewski TF. Failure at the effector phase: immune barriers at the level of the melanoma tumor microenvironment. Clin Cancer Res. 2007;13:5256–61. doi: 10.1158/1078-0432.CCR-07-0892. [DOI] [PubMed] [Google Scholar]
- 8.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–12. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–12. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 10.Mantovani A, Sica A, Allavena P, Garlanda C, Locati M. Tumor-associated macrophages and the related myeloid-derived suppressor cells as a paradigm of the diversity of macrophage activation. Hum Immunol. 2009;70:325–30. doi: 10.1016/j.humimm.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–22. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin RA, Liao W, Sarkar A, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–5. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Cao X. The origin and function of tumor-associated macrophages. Cellular & molecular immunology. 2015;12:1–4. doi: 10.1038/cmi.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gattinoni L, Powell DJ, Jr, Rosenberg SA, Restifo NP. Adoptive immunotherapy for cancer: building on success. Nat Rev Immunol. 2006;6:383–93. doi: 10.1038/nri1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudek RM, Chuang Y, Leonard JN. Engineered cell-based therapies: a vanguard of design-based medicine. In: Corey SJ, Kimmel M, Leonard JN, editors. A Systems Biology Approach to Blood. Heidelberg: Springer; in press. [Google Scholar]
- 16.Goldman B, DeFrancesco L. The cancer vaccine roller coaster. Nat Biotechnol. 2009;27:129–39. doi: 10.1038/nbt0209-129. [DOI] [PubMed] [Google Scholar]
- 17.Hill HC, Conway TF, Jr, Sabel MS, et al. Cancer immunotherapy with interleukin 12 and granulocyte-macrophage colony-stimulating factor-encapsulated microspheres: coinduction of innate and adaptive antitumor immunity and cure of disseminated disease. Cancer Res. 2002;62:7254–63. [PubMed] [Google Scholar]
- 18.Luo Y, Zhou H, Krueger J, et al. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J Clin Invest. 2006;116:2132–41. doi: 10.1172/JCI27648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemann T, Lawrence T, McNeish I, et al. “Re-educating” tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205:1261–8. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garris C, Pittet MJ. Therapeutically reeducating macrophages to treat GBM. Nat Med. 2013;19:1207–8. doi: 10.1038/nm.3355. [DOI] [PubMed] [Google Scholar]
- 21.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–96. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 22.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sica A, Bronte V. Altered macrophage differentiation and immune dysfunction in tumor development. J Clin Invest. 2007;117:1155–66. doi: 10.1172/JCI31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sica A, Saccani A, Bottazzi B, et al. Autocrine production of IL-10 mediates defective IL-12 production and NF-kappa B activation in tumor-associated macrophages. J Immunol. 2000;164:762–7. doi: 10.4049/jimmunol.164.2.762. [DOI] [PubMed] [Google Scholar]
- 25.Mantovani A, Sica A, Locati M. Macrophage polarization comes of age. Immunity. 2005;23:344–6. doi: 10.1016/j.immuni.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–61. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 27.Geissmann F, Gordon S, Hume DA, Mowat AM, Randolph GJ. Unravelling mononuclear phagocyte heterogeneity. Nat Rev Immunol. 2010;10:453–60. doi: 10.1038/nri2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guiducci C, Vicari AP, Sangaletti S, Trinchieri G, Colombo MP. Redirecting in vivo elicited tumor infiltrating macrophages and dendritic cells towards tumor rejection. Cancer Res. 2005;65:3437–46. doi: 10.1158/0008-5472.CAN-04-4262. [DOI] [PubMed] [Google Scholar]
- 29.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–22. [PubMed] [Google Scholar]
- 30.Garcia-Hernandez ML, Hernandez-Pando R, Gariglio P, Berumen J. Interleukin-10 promotes B16-melanoma growth by inhibition of macrophage functions and induction of tumour and vascular cell proliferation. Immunology. 2002;105:231–43. doi: 10.1046/j.1365-2567.2002.01363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–81. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 32.Weischenfeldt J, Porse B. Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. CSH Protoc. 2008:2008. doi: 10.1101/pdb.prot5080. pdb prot5080. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14(Unit 14):1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Watkins SK, Egilmez NK, Suttles J, Stout RD. IL-12 rapidly alters the functional profile of tumor-associated and tumor-infiltrating macrophages in vitro and in vivo. J Immunol. 2007;178:1357–62. doi: 10.4049/jimmunol.178.3.1357. [DOI] [PubMed] [Google Scholar]
- 36.Stout RD, Watkins SK, Suttles J. Functional plasticity of macrophages: in situ reprogramming of tumor-associated macrophages. J Leukoc Biol. 2009;86:1105–9. doi: 10.1189/jlb.0209073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerkar SP, Goldszmid RS, Muranski P, et al. IL-12 triggers a programmatic change in dysfunctional myeloid-derived cells within mouse tumors. J Clin Invest. 2011;121:4746–57. doi: 10.1172/JCI58814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nastala CL, Edington HD, McKinney TG, et al. Recombinant IL-12 administration induces tumor regression in association with IFN-gamma production. J Immunol. 1994;153:1697–706. [PubMed] [Google Scholar]
- 39.Macatonia SE, Hsieh CS, Murphy KM, O’Garra A. Dendritic cells and macrophages are required for Th1 development of CD4+ T cells from alpha beta TCR transgenic mice: IL-12 substitution for macrophages to stimulate IFN-gamma production is IFN-gamma-dependent. Int Immunol. 1993;5:1119–28. doi: 10.1093/intimm/5.9.1119. [DOI] [PubMed] [Google Scholar]
- 40.Bosisio D, Polentarutti N, Sironi M, et al. Stimulation of toll-like receptor 4 expression in human mononuclear phagocytes by interferon-gamma: a molecular basis for priming and synergism with bacterial lipopolysaccharide. Blood. 2002;99:3427–31. doi: 10.1182/blood.v99.9.3427. [DOI] [PubMed] [Google Scholar]
- 41.Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175:342–9. doi: 10.4049/jimmunol.175.1.342. [DOI] [PubMed] [Google Scholar]
- 42.Ding Y, Chen D, Tarcsafalvi A, Su R, Qin L, Bromberg JS. Suppressor of cytokine signaling 1 inhibits IL-10-mediated immune responses. J Immunol. 2003;170:1383–91. doi: 10.4049/jimmunol.170.3.1383. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto K, Yamaguchi M, Miyasaka N, Miura O. SOCS-3 inhibits IL-12-induced STAT4 activation by binding through its SH2 domain to the STAT4 docking site in the IL-12 receptor beta2 subunit. Biochem Biophys Res Commun. 2003;310:1188–93. doi: 10.1016/j.bbrc.2003.09.140. [DOI] [PubMed] [Google Scholar]
- 44.Athie-Morales V, Smits HH, Cantrell DA, Hilkens CM. Sustained IL-12 signaling is required for Th1 development. J Immunol. 2004;172:61–9. doi: 10.4049/jimmunol.172.1.61. [DOI] [PubMed] [Google Scholar]
- 45.Evel-Kabler K, Song XT, Aldrich M, Huang XF, Chen SY. SOCS1 restricts dendritic cells’ ability to break self tolerance and induce antitumor immunity by regulating IL-12 production and signaling. J Clin Invest. 2006;116:90–100. doi: 10.1172/JCI26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staples KJ, Smallie T, Williams LM, et al. IL-10 induces IL-10 in primary human monocyte-derived macrophages via the transcription factor Stat3. J Immunol. 2007;178:4779–85. doi: 10.4049/jimmunol.178.8.4779. [DOI] [PubMed] [Google Scholar]
- 47.Becskei A, Grusby MJ. Contribution of IL-12R mediated feedback loop to Th1 cell differentiation. FEBS Lett. 2007;581:5199–206. doi: 10.1016/j.febslet.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campana L, Bosurgi L, Rovere-Querini P. HMGB1: a two-headed signal regulating tumor progression and immunity. Curr Opin Immunol. 2008;20:518–23. doi: 10.1016/j.coi.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 49.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–88. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 50.Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. Journal of immunology. 2002;169:2253–63. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- 51.Han J, Brown T, Beutler B. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J Exp Med. 1990;171:465–75. doi: 10.1084/jem.171.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol Cell Biol. 2001;21:721–30. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rouzer CA, Jacobs AT, Nirodi CS, Kingsley PJ, Morrow JD, Marnett LJ. RAW264.7 cells lack prostaglandin-dependent autoregulation of tumor necrosis factor-alpha secretion. J Lipid Res. 2005;46:1027–37. doi: 10.1194/jlr.M500006-JLR200. [DOI] [PubMed] [Google Scholar]
- 54.Yuk JM, Shin DM, Lee HM, et al. The orphan nuclear receptor SHP acts as a negative regulator in inflammatory signaling triggered by Toll-like receptors. Nat Immunol. 2011;12:742–51. doi: 10.1038/ni.2064. [DOI] [PubMed] [Google Scholar]
- 55.Eyles JL, Metcalf D, Grusby MJ, Hilton DJ, Starr R. Negative regulation of interleukin-12 signaling by suppressor of cytokine signaling-1. J Biol Chem. 2002;277:43735–40. doi: 10.1074/jbc.M208586200. [DOI] [PubMed] [Google Scholar]
- 56.Williams L, Bradley L, Smith A, Foxwell B. Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J Immunol. 2004;172:567–76. doi: 10.4049/jimmunol.172.1.567. [DOI] [PubMed] [Google Scholar]
- 57.Ruffell D, Mourkioti F, Gambardella A, et al. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A. 2009;106:17475–80. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. J Immunol. 2010;185:6413–9. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuwata H, Watanabe Y, Miyoshi H, et al. IL-10-inducible Bcl-3 negatively regulates LPS-induced TNF-alpha production in macrophages. Blood. 2003;102:4123–9. doi: 10.1182/blood-2003-04-1228. [DOI] [PubMed] [Google Scholar]
- 60.Palmer S, Chen YH. Bcl-3, a multifaceted modulator of NF-kappaB-mediated gene transcription. Immunol Res. 2008;42:210–8. doi: 10.1007/s12026-008-8075-4. [DOI] [PubMed] [Google Scholar]
- 61.Krausgruber T, Blazek K, Smallie T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–8. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 62.Herrero C, Hu X, Li WP, et al. Reprogramming of IL-10 activity and signaling by IFN-gamma. J Immunol. 2003;171:5034–41. doi: 10.4049/jimmunol.171.10.5034. [DOI] [PubMed] [Google Scholar]
- 63.Ragimbeau J, Dondi E, Vasserot A, Romero P, Uze G, Pellegrini S. The receptor interaction region of Tyk2 contains a motif required for its nuclear localization. J Biol Chem. 2001;276:30812–8. doi: 10.1074/jbc.M103559200. [DOI] [PubMed] [Google Scholar]
- 64.Shaw MH, Freeman GJ, Scott MF, et al. Tyk2 negatively regulates adaptive Th1 immunity by mediating IL-10 signaling and promoting IFN-gamma-dependent IL-10 reactivation. J Immunol. 2006;176:7263–71. doi: 10.4049/jimmunol.176.12.7263. [DOI] [PubMed] [Google Scholar]
- 65.Tokumasa N, Suto A, Kagami S, et al. Expression of Tyk2 in dendritic cells is required for IL-12, IL-23, and IFN-gamma production and the induction of Th1 cell differentiation. Blood. 2007;110:553–60. doi: 10.1182/blood-2006-11-059246. [DOI] [PubMed] [Google Scholar]
- 66.Beurel E, Michalek SM, Jope RS. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3) Trends Immunol. 2010;31:24–31. doi: 10.1016/j.it.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu X, Paik PK, Chen J, et al. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006;24:563–74. doi: 10.1016/j.immuni.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 68.Martin M, Rehani K, Jope RS, Michalek SM. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat Immunol. 2005;6:777–84. doi: 10.1038/ni1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pyonteck SM, Akkari L, Schuhmacher AJ, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19:1264–72. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–53. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 71.Kattman SJ, Witty AD, Gagliardi M, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell stem cell. 2011;8:228–40. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 72.Antebi YE, Reich-Zeliger S, Hart Y, et al. Mapping differentiation under mixed culture conditions reveals a tunable continuum of T cell fates. PLoS Biol. 2013;11:e1001616. doi: 10.1371/journal.pbio.1001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sciammas R, Li Y, Warmflash A, Song Y, Dinner AR, Singh H. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol Syst Biol. 2011;7:495. doi: 10.1038/msb.2011.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.