Abstract
Melatonin, a small organic molecule synthesized by the pineal gland and the retina, has a variety of physiologic functions such as circadian clock pacemaker and antioxidant. Retinal melatonin is down-regulated by light and is barely detectable during the day. The absence of melatonin in the retina during prolonged light exposure may contribute to light-induced retinal degeneration. We sought to investigate the impact of melatonin in the light-exposed retina using proteomic approaches. We exposed mice to either light (250–300 lux) for 12 h followed by 12 h of darkness or the same intensity of continuous light for 7 days. In half of the animals exposed to continuous light, melatonin was injected each night. Proteomic analysis of the retina from these three groups of animals showed that five proteins prominently up-regulated by constant light were down-regulated by melatonin treatment. These five proteins were identified as vimentin, serine/threonine-protein phosphatase 2A, Rab GDP dissociation inhibitor alpha, guanine nucleotide-binding protein Go alpha, and retinaldehyde-binding protein.
These five proteins are known to be involved in several cellular processes that may contribute to light-induced retinal degeneration. Identification of melatonin target proteins in our study provides a basis for future studies on melatonin’s potential in preventing or treating light-induced retinal degeneration.
Keywords: Melatonin, Light, Proteomics, Retina, Mass spectrometry
1. Introduction
Light isomerizes cis-retinyl imine chromophore to trans isomer of cone opsin and rhodopsin, which is essential for visual perception. However, light can also accelerate photoreceptor cell death by apoptosis. Even though light has been proposed as a risk factor for many eye diseases, including age-related macular degeneration (AMD) and retinitis pigmentosa (RP), the signaling mechanisms that lead to degeneration are not understood at the molecular level.
Melatonin (N-acetyl-5-methoxytryptamine) is a paracoid and autocoid molecule derived from tryptophan, nocturnally synthesized in the pineal gland and the retina of mammals [1,2]. Melatonin interacts with G-protein coupled melatonin receptors in amacrine, horizontal, ganglion, and photoreceptor cells in the retina [3,4]. Melatonin also interacts with nuclear receptors and intracellular proteins such as calmodulin, tubulin, and calreticulin [5]. Daily production and secretion of melatonin shows striking daily organization with peaks during each night and very low levels during the day. This pattern is regulated by both the endogenous circadian clock and environmental tuning by light [6]. As a circadian pacemaker, melatonin modulates many rhythmic processes in the eye such as retinomotor movements, neurotransmitter release, and retinal pigment epithelium (RPE) phagocytosis [7,8]. Furthermore, ocular melatonin is an effective anti-oxidative molecule that crosses cell membranes and the blood–retina-barrier to react with many reactive oxygen and nitrogen species, including hydroxyl radicals, superoxide anions, and nitric oxide [9,10]. Since many eye diseases are caused by oxidative stress, the retinal melatonin cycle is a prime candidate for normal maintenance of retinal health and a potential therapy for oxidative retinal diseases.
In this work, we investigate the effects of continuous light and melatonin repletion upon protein targets of melatonin in the retina. Proteomic strategies are used to compare protein expression patterns among mice maintained under three different conditions: 12 h light/12 h dark cyclic conditions; and constant light conditions with or without daily melatonin repletion. By comparing the constant light group to the light/dark group, we identified 22 proteins, the levels of which were markedly changed by constant light. We identified five of these proteins that showed a reversal of the constant light-induced expression changes after melatonin treatment. These five proteins, up-regulated by constant light and down-regulated by repletion of melatonin, are involved in phototransduction, rhodopsin trafficking, and 11-cis retinal regeneration. Identification of melatonin targets in our study provides a foundation for future studies to determine melatonin’s potential in the treatment of certain retinal degenerations.
2. Materials and methods
Animals were handled in compliance with the Association for Research in Vision and Ophthalmology (ARVO) statement on the Use of Animals in Ophthalmic and Vision Research. 12-Week-old female C3HeB/FeJ mice were purchased from Jackson Laboratory (Bar Harbor, ME) and housed under a 12 h light/12 h dark cyclic lighting condition (250–300 lux of full spectra fluorescent room light) for 2 weeks. The time is referred to as HALO (Hour After Light On), with light on at HALO 0, and off at HALO 12. Animals were divided into three groups (12 mice/group). The first group of mice was housed in the 12 h light/12 h dark condition and treated with melatonin vehicle solution (0.3% ethanol–saline) at HALO 14 for 7 days. The second group was housed in constant room light (250–300 lux) for 7 days and also treated with vehicle solution (0.3% ethanol–saline) at HALO 14. The third group was also kept in constant room light as above but treated with melatonin (Sigma–Aldrich) via intraperitoneal injection at a dose of 50 μg/kg body wt of mouse at HALO 14.
2.1. Sample preparation, electrophoresis and western blot
After treatment all mice were euthanized at HALO 18. After euthanasia, the eyes were rapidly removed from animals and retinas were isolated by microscopic dissection. Retinas were washed with a solution (250 mM sucrose, 10 mM Tris–HCl, pH 7.0) to remove contaminants before addition of lysis buffer (30 mM TrisCl, 2 M thiourea, 7 M urea, 4% CHAPS, and protease inhibitors). Samples were then sonicated in pulses for 30 seconds until cells were completely lysed. The crude lysate was centrifuged at 20,817 × g for 30 min at 4 °C. Proteomic procedures including mass spectrometry analysis were performed as previously described [11–13]. Blot membranes were incubated overnight at 4 °C with a goat polyclonal antibody against vimentin (1:1000 dilution, Santa Cruz) in blocking buffer. Protein concentrations of bands on the blot were analyzed semi-quantitatively by densitometer, PDQuest, and ImageJ software, and compared with positive controls including actin and tubulin (Neomarkers).
2.2. Statistical analysis
Values are presented as the mean ± SEM in three independent experiments. Statistical significance of differences among the analyzed groups was determined by performing post hoc comparison and one-way ANOVA. p < 0.05 was considered statistically significant.
3. Results
3.1. Constant light alters proteomic profile in the retina
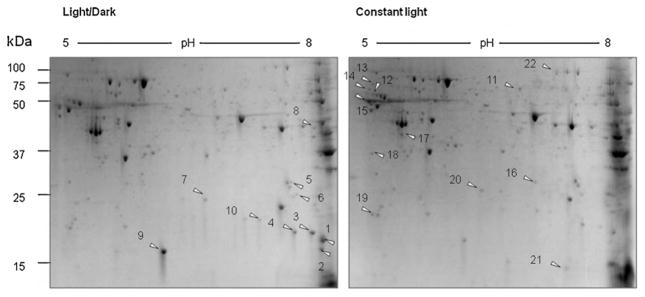
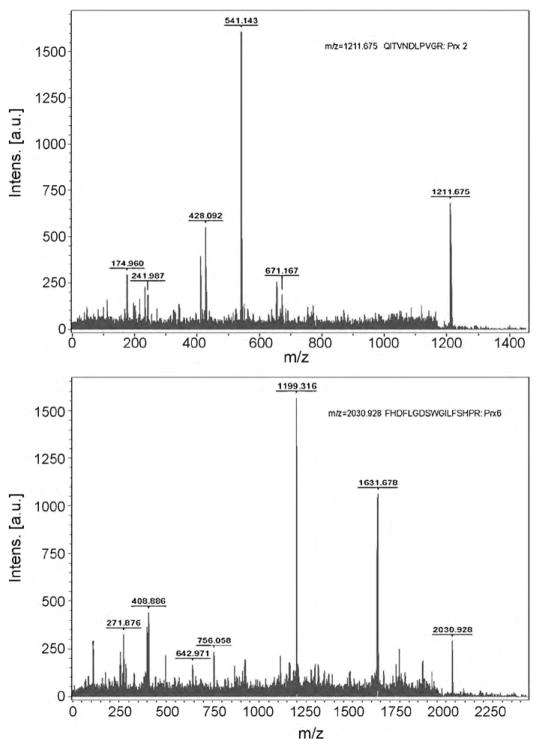
We compared retinal proteome between mice that were kept in the 12 h light/12 h dark cyclic condition and in continuous light. Retinas from three mice (n = 3) were isolated from each group of animals and retinal proteins were separated by two-dimensional electrophoresis (Fig. 1). Constant light exposure markedly decreased the expression of 10 proteins and increased the expression of 12 proteins, compared to the light/dark control group. Each of the differentially expressed protein spots were excised from the gels and analyzed by MALDI-TOF mass spectrometry for identification. Twenty-two proteins that were down- or up-regulated by constant light are listed in Tables 1 and 2. Tandem time-of-flight mass spectrometry (TOF–TOF) identified three proteins that were up-regulated by constant light as peroxiredoxin (Prx) 2, Prx 6, and retinaldehyde-binding protein (CRALBP). Representative spectra are shown in Fig. 2A and B. The peptide with m/z = 1211.675 was identified as an amino acid sequence of QITVNDLPVGR in Prx 2, and the peptide with m/z = 2030.928 corresponds to FHDFLGDSWGILFSHPR in Prx 6.
Fig. 1.
Two-dimensional electrophoretic analysis of retinal proteins from the light/dark group and the constant light group. Retinas were isolated from mice that were exposed to 12 h light/12 h dark cyclic condition or constant room light condition. Two hundred microgram proteins were separated by using 11 cm isoelectric focusing gradient strip (pH 5–8) and 8–12% precast gradient gel. Proteins were visualized by Coomassie blue staining. Arrows are showing spots with differential intensities. (A) Retinal proteome of mice exposed to cyclic light/dark condition; (B) retinal proteome of mice exposed to constant light condition.
Table 1.
Down-regulated proteins by constant light compared to light/dark group.
| Spot number | Protein identified | Scorea | Coverageb | Mass | pI |
|---|---|---|---|---|---|
| N1 | Gamma crystallin B | 89 | 44 | 21,581 | 7.55 |
| N2 | Gamma-crystallin F | 98 | 47 | 21,577 | 6.78 |
| N3 | Alpha-crystallin B chain | 106 | 49 | 20,056 | 6.76 |
| N4 | Beta-crystallin S | 72 | 39 | 21,236 | 6.89 |
| N5 | Beta-crystallin B1 | 107 | 44 | 28,213 | 6.84 |
| N6 | Beta-crystallin B3 | 112 | 54 | 24,447 | 6.71 |
| N7 | Beta-crystallin A1 | 89 | 56 | 23,843 | 5.96 |
| N8 | Aspartate aminotransferase | 113 | 31 | 46,488 | 6.68 |
| N9 | Alpha-crystallin A chain | 98 | 45 | 22,532 | 6.35 |
| N10 | Triosephosphate isomerase | 78 | 27 | 27,038 | 6.90 |
Probability-based MASCOT MOWSE score is −10 Log(P), where P is the probability that the observed match is a random event. Protein scores greater than 55 are significant (p < 0.05).
Amino acids sequence % identified.
Table 2.
Up-regulated proteins by constant light compared to light/dark group.
| Spot number | Protein identified | Score | Coverage | Mass | pI |
|---|---|---|---|---|---|
| N11 | Dihydropyrimidinase-related protein 2 | 123 | 28 | 62,638 | 5.95 |
| N12 | Vimentin | 125 | 30 | 53,712 | 5.06 |
| N13 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | 94 | 28 | 66,079 | 5.00 |
| N14 | Rab GDP dissociation inhibitor alpha | 92 | 26 | 51,059 | 4.96 |
| N15 | Tubulin beta-5 chain | 101 | 25 | 50,095 | 4.78 |
| N16 | Phosphoglycerate mutase 1 | 100 | 48 | 28,928 | 6.67 |
| N17 | Guanine nucleotide-binding protein G (o) subunit alpha | 70 | 30 | 40,629 | 5.34 |
| N18 | Retinaldehyde-binding protein 1 | 79 | 29 | 36,670 | 4.98 |
| N19 | Peroxiredoxin-2 | 94 | 51 | 21,936 | 5.20 |
| N20 | Peroxiredoxin-6 | 90 | 34 | 24,969 | 5.71 |
| N21 | Nucleoside diphosphate kinase A | 74 | 41 | 17,311 | 6.84 |
| N22 | Vesicle-fusing ATPase | 105 | 15 | 83,131 | 6.52 |
Fig. 2.
Peptide sequencing using TOF–TOF mass spectrometry. Protein spots that were identified by MALDI-TOF as Prx 2 and Prx 6 were further analyzed by TOF–TOF mass spectrometry. Selected peptides were sequenced as Prx 2, and Prx 6, respectively. Representative mass spectra for Prx 2 and Prx 6 are shown. X-Axis represents mass to charge ratio (m/z) and Y-axis shows relative intensity.
3.2. Melatonin treatment reprogrammed proteomic profile in light-exposed retina
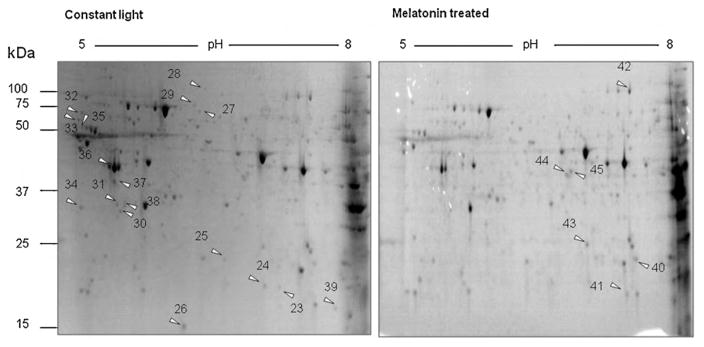
We then compared proteomic profiles between mice that were kept in continuous light ± melatonin treatment. Retinas were isolated from each group of animals (n = 3) and retinal proteins were separated by 2D-PAGE (Fig. 3). Melatonin treatment lowered the expression of 17 proteins and raised levels of 6 proteins, when compared to mice in constant light without melatonin. Twenty-three proteins that were down- or up-regulated by melatonin treatment are shown in Tables 3 and 4.
Fig. 3.
2D-SDS-PAGE analysis of retinal proteins from constant light group and melatonin treated group. Retinas were isolated from mice that were housed in constant light conditions treated with or without melatonin. Arrows are showing spots of up- or down-regulation. (A) Retinal proteome of mice exposed to constant light and treated with control vehicle; (B) retinal proteome of mice exposed to constant light and treated with melatonin.
Table 3.
Down-regulated proteins by melatonin/constant light treatment compared to constant light group.
| Spot number | Protein identified | Score | Coverage | Mass | pI |
|---|---|---|---|---|---|
| N23 | Alpha-crystallin A chain | 55 | 38 | 22,532 | 6.35 |
| N24 | Beta-crystallin A2 | 86 | 44 | 22,622 | 6.30 |
| N25 | Beta-crystallin A1 | 101 | 56 | 23,843 | 5.96 |
| N26 | Alpha-crystallin A chain | 98 | 45 | 22,532 | 6.35 |
| N27 | Dihydropyrimidinase-related protein 2 | 63 | 21 | 62,638 | 5.95 |
| N28 | Mitochondrial inner membrane protein | 88 | 18 | 84,247 | 6.18 |
| N29 | Serum albumin precursor | 62 | 17 | 70,700 | 5.75 |
| N30 | Beta-soluble NSF attachment protein | 117 | 50 | 33,878 | 5.32 |
| N31 | Inorganic pyrophosphatase | 60 | 28 | 33,102 | 5.37 |
| N32 | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | 94 | 28 | 66,079 | 5.00 |
| N33 | Rab GDP dissociation inhibitor alpha | 92 | 26 | 51,059 | 4.96 |
| N34 | Retinaldehyde-binding protein 1 | 79 | 29 | 36,670 | 4.98 |
| N35 | vimentin | 125 | 30 | 53,712 | 5.06 |
| N36 | Actin | 141 | 54 | 42,052 | 5.29 |
| N37 | Guanine nucleotide-binding protein G(o) alpha | 70 | 30 | 40,629 | 5.34 |
| N38 | Guanine nucelotide-binding protein G(I)/G(S)/G(T) subunit beta-3 | 79 | 33 | 38,185 | 5.41 |
| N39 | Alpha-crystallin B chain | 106 | 49 | 20,056 | 6.76 |
Table 4.
Up-regulated proteins by melatonin/constant light treatment compared to constant light group.
| Spot number | Protein identified | Score | Coverage | Mass | pI |
|---|---|---|---|---|---|
| N40 | Triosephosphate isomerase | 78 | 27 | 27,038 | 6.90 |
| N41 | Ribosomal protein S6 kinase alpha-3 | 62 | 23 | 83,983 | 6.41 |
| N42 | Serotransferrin precursor | 143 | 28 | 78,841 | 6.94 |
| N43 | Phosphoglycerate mutase 1 | 100 | 48 | 28,928 | 6.67 |
| N44 | Fas apoptotic inhibitory molecule 3 precursor | 57 | 26 | 47,898 | 9.57 |
| N45 | Septin-5 | 68 | 29 | 43,177 | 6.21 |
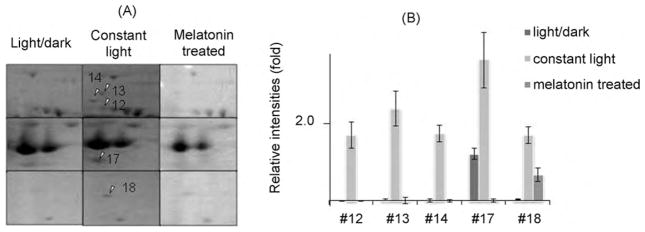
3.3. Five light-elevated proteins in the retina were down-regulated by melatonin treatment
Five of the proteins up-regulated by continuous light were either completely or partially down-regulated by melatonin treatment. Magnified 2D-PAGE gel images of these five proteins from all three groups of mice are shown in Fig. 4A, and spot densities are analyzed semi-quantitatively using the total intensity of all spots from the corresponding gel for normalization. The relative intensities of each spot are shown in Fig. 4B. These five proteins were identified as vimentin, serine/threonine-protein phosphatase 2A, Rab GDP dissociation inhibitor alpha (GDI), guanine nucleotide-binding protein Go subunit alpha and retinaldehyde-binding protein (CRALBP). The identity of one of these five proteins, CRALBP, was further confirmed by TOF–TOF mass spectrometry. The peptide with m/z = 2425.301 was identified as ELQELVQAQAASGEELALAVAER in CRALBP1.
Fig. 4.
2D-SDS-PAGE and quantitative analysis of the target proteins. (A) Magnified gel images with arrows showing five proteins, up-regulated by constant light and down-regulated by melatonin. #12, vimentin; #13, serine/threonine-protein phosphatase 2A; #14, Rab GDP dissociation inhibitor alpha; #17, guanine nucleotide-binding protein; #18, retinaldehyde-binding protein. Left panel: cyclic light/dark; middle panel: constant light; right panel: constant light and melatonin. (B) Semi-quantitative analysis of five proteins. Number indicates the relative intensity of each spot. Values are shown in mean ± SD (n = 3).
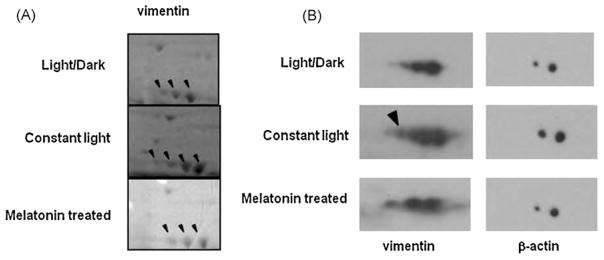
3.4. Melatonin treatment eliminates post-translational modifications of vimentin induced by constant light
Coomassie blue stained 2D gels showed a series of spots with a similar molecular weight to vimentin. Mass spectrometry identified all of these spots, shown by arrows in Fig. 5A, as vimentin. Typically, the most basic spot of this charge train is the unmodified form, and the additional spots are assumed to be modified post-translationally. By comparing the Coomassie blue stained 2D gel images among the three groups of mice, we found an additional spot at the acidic end that was increased in constant light and disappeared after melatonin repletion (Fig. 5A). Subsequently, 2D-western blotting using an anti-vimentin antibody confirmed our observation (Fig. 5B). Hence, our result suggests that vimentin undergoes additional post-translational modification when exposed to continuous light, and melatonin reverts the vimentin modification to the original form.
Fig. 5.
2D-SDS-PAGE and western blot analysis. (A) Magnified Coomassie blue stained 2D gel image. Arrows represent vimentin. (B) 100 μg retinal proteins were separated by using 7 cm IPG strips and 10% SDS-PAGE gel and visualized by western blot with anti-vimentin polyclonal antibody. Arrow shows new spots of vimentin induced by constant light. Upper panel: cyclic light/dark condition; middle panel: constant light; bottom panel: constant light and melatonin.
4. Discussion
We have identified five major melatonin target proteins. These proteins are up-regulated by constant light and diminished by supplementation of melatonin during the subjective night. These results indicate that melatonin acts as a downstream regulator of light and confers its beneficial effects by blocking constant light effects on target protein expression. Since these five proteins are known to participate in several light-induced cellular processes that may trigger retinal degeneration, melatonin may protect the retina from light-induced damage by modulating these five proteins after usual daily light exposure.
Exposure to continuous light at intensities ordinarily encountered during daytime causes retinal degeneration [14]. The equivalent light hypothesis suggests that a subset of inherited retinal degenerations, featured with a variety of photoreceptor molecular defects that produce electrical signals equivalent to those produced by real light, is triggered by the same mechanism [15]. How constant light induces retinal degeneration remains unknown, although many mechanisms have been proposed, including defects in rhodopsin regeneration, accumulation of free radicals, and a continuous low level of Ca2+ resulting from an excessively active phototransduction cascade [16]. Considering the versatile functions of melatonin in the retina, it is possible that deprivation of retinal melatonin contributes substantially to light-induced retinal degeneration. In our study, we identified 22 proteins differentially expressed in the retina during constant light exposure. Most of the down-regulated proteins belong to the crystallin family. Alpha A and B crystallins have chaperone functions to prevent heat or oxidative stress-induced protein aggregation [17]. Alpha crystallin also inhibits apoptosis and enhances the resistance of cells to stress. Decreased levels of alpha A and B crystallins indicate a general loss of chaperone function and a disruption of homeostasis in retinas that have been continuously exposed to light. Prx 2 and Prx 6, antioxidant enzymes using cysteine residues to decompose peroxides, are up-regulated by constant light, which indicates increased oxidative stress in retinas exposed to constant light [18].
Melatonin interferes with various apoptotic pathways through an anti-apoptotic signaling mechanism [19]. The daily administration of melatonin to rds/rds mouse, an animal model for retinitis pigmentosa, reduces the rate of photoreceptor cell loss [20]. A case control study on 100 age-related macular degeneration patients showed that melatonin, taken each night at bedtime for 3 months at a dosage of 3 mg/day, delays macular degeneration in most patients [21].
Our results show that melatonin depletion, a major effect of constant light, may cause changes of the key protein expressions in the retina. The melatonin target proteins, up-regulated by constant light, may be key mediators of light-induced retinal degeneration. By down-regulation of the five target proteins, melatonin may slow or block the process of light-induced retinal degeneration.
Reverse regulation of light-induced retinaldehyde-binding protein (CRALBP) by melatonin might imply a key protection mechanism against light-induced retinal degeneration. CRALBP is a key retinoid chaperone involved in the visual cycle, to regenerate 11-cis-retinol, the precursor of the chromophore at a high rate [22]. Regeneration of the chromophore 11-cis retinal is a key pathway in the process of light-induced retinal degeneration [16]. Excess absorption of photons by rhodopsin triggers light-induced retinal damage. Mice that lack rhodopsin or mice that were treated with the visual cycle inhibitors are protected from light-induced retinal damage [23]. Light-induced CRALBP may accelerate the retinal damage through the higher rate of the visual cycle.
Rab proteins, up-regulated by melatonin, are members of the Ras superfamily of GTPases and are involved in rhodopsin trafficking, a process of rhodopsin translocalization to the site of outer segment disk formation [24]. After completion of rhodopsin trafficking and GTP hydrolysis, Rab is retrieved by Rab GDP dissociation inhibitor (GDI), and is delivered to its initial compartment. We observed up-regulation of GDI in constant light which suggests a potential role for a higher turnover of Rab. This process may lead to a higher trafficking rate of rhodopsin and triggering of retinal degeneration. Strikingly, melatonin attenuated the light-induced GDI up-regulation and therefore expected to protect the retina from light-induced pathological pathways.
Up-regulation of Go by constant light may lead to continuous activation of transduction pathway in ipRGC and disrupt the circadian clock in retinal recovery during the nocturnal dark phase for outer segment disk shedding and phagocytosis. Disruption of this circadian organization may induce an imbalance of daily maintenance of rod outer segment and eventually lead to apoptotic photoreceptor cell death. Melatonin-induced decrease of Go expression may resynchronize these nocturnal protective mechanics in the retina during constant light exposure.
Modifications of vimentin are essential reactions of filament dynamics. Changes of vimentin phosphorylation are directed to reorganization of the intermediate filament network and altered function of Müller cells. Hyperphosphorylation of vimentin can disassemble intermediate filament into soluble monomeric form. Müller cells are the major glial cells in the retina and play crucial roles in maintaining neuroretinal architecture and support neuronal survival. Our results suggest that vimentin and Müller cells might be critically involved in the process of light-induced damage in the retina. By reducing the light-induced post-translational modification of vimentin, melatonin may assist to maintain the proper filament network in Müller cells, which subsequently supports neuronal survival and architecture in the retina.
Several kinases and phosphatases are involved in regulating the phosphorylation state of vimentin. Serine/threonine protein phosphatase 2A (PP2A) directly interacts with vimentin and is involved in regulating the dynamics of intermediate filament. PP2A interacts with Bcl-2, a potent anti-apoptotic protein activated by phosphorylation on Ser70. PP2A dephosphorylates and inactivates Bcl-2, and induces apoptosis. PP2A also negatively regulates the Erk signaling pathway. In our study, PP2A was markedly up-regulated in constant light, possibly due to post-translationally modified vimentin. Melatonin-mediated down-regulation of PP2A may regulate the formation of more stabilized vimentin post-translationally and negatively regulate apoptosis.
In this study, we noticed that there were melatonin-induced up-regulated proteins, but not affected initially by constant light. The proteomic changes caused by constant light during the subjective night at 18 HALO may be different at the other circadian time point. In the absence of round-the-clock sampling in 12 h light/dark, dark/dark and constant light with and without melatonin, we cannot conclude whether the protein changes seen result from circadian proteomic phase shifting. The study was, however, involved a carefully selected and controlled circadian time, indicating that the changes observed are real at 18 HALO, when melatonin or placebo is given daily at 14 HALO 4 h before sampling time. These experimental discoveries of proteomic changes caused by light at night and reversed by melatonin, despite constant light, are provocative. They provide the potential mechanisms responsible for light-induced retinal changes by melatonin. Profound protein changes occur with constant light at night, and many of them are modulated or reversed by melatonin. Regardless of the mechanisms, this study clearly raises therapeutic possibilities for melatonin in the treatment of retinal degeneration, especially initiated by prolonged exposure of light.
Taken together, our results clearly demonstrate that supplementation of melatonin to mice with constant light-exposed retina in vivo partially reverses light-induced protein changes significantly. Constant light eventually results in retinal dysfunction, at least in our rodent model. This study provides a stimulus to understand constant light-induced retinal pathologies as well as whether melatonin provided at night may prevent or alleviate degenerative pathway.
Acknowledgments
The authors thank Dr. Ping Chen for assistance in isolating mouse retina, and Dr. Xiaoming Yang for valuable suggestions. Dr. Hilal Arnouk, Hyunju Lee, and Folami Lamoke are acknowledged for their technical assistance. The authors thank Dr. Elizabeth Hager and Dr. Steven Bailey for their critical reading. This work was supported by a start-up package, the Century II Equipment Fund, and the Research Excellent Fund from Michigan Technological University, and start-up funds, Centenary Plan from the University of South Carolina, and the New Investigator Award from the International Foundation.
Footnotes
Competing interest
None to declare.
References
- 1.Bubenik GA, Purtill RA, Brown GM, Grota LJ. Exp Eye Res. 1978;27:323–333. doi: 10.1016/0014-4835(78)90166-5. [DOI] [PubMed] [Google Scholar]
- 2.Vivien-Roels B, Pévet P, Dubois MP, Arendt J, Brown GM. Cell Tissue Res. 1981;217:105–115. doi: 10.1007/BF00233830. [DOI] [PubMed] [Google Scholar]
- 3.Reppert SM, Weaver DR, Ebisawa T. Neuron. 1994;13:1177–1185. doi: 10.1016/0896-6273(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 4.Fujieda H, Hamadanizadeh SA, Wankiewicz E, Pang SF, Brown GM. Neuroscience. 1999;93:793–799. doi: 10.1016/s0306-4522(99)00111-6. [DOI] [PubMed] [Google Scholar]
- 5.Carrillo-Vico A, García-Pergañeda A, Naji L, Calvo JR, Romero MP. Cell Mol Life Sci. 2003;60:2272–2278. doi: 10.1007/s00018-003-3207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein DC, Weller JL. Science. 1972;177:532–533. doi: 10.1126/science.177.4048.532. [DOI] [PubMed] [Google Scholar]
- 7.White MP, Fisher LJ. Vision Res. 1989;29:167–179. doi: 10.1016/0042-6989(89)90122-3. [DOI] [PubMed] [Google Scholar]
- 8.Lundmark PO, Pandi-Perumal SR, Srinivasan V, Cardinali DP. Visual Neurosci. 2006;23:853–862. doi: 10.1017/S0952523806230189. [DOI] [PubMed] [Google Scholar]
- 9.Tan DX, Manchester LC, Terron MP, Flores LJ, Reiter RJ. J Pineal Res. 2007;42:28–42. doi: 10.1111/j.1600-079X.2006.00407.x. [DOI] [PubMed] [Google Scholar]
- 10.Hardeland R. Endocrine. 2005;27:119–130. doi: 10.1385/endo:27:2:119. [DOI] [PubMed] [Google Scholar]
- 11.Jahng WJ, David D, Nesnas N, Nakanishi K, Rando RR. Biochemistry. 2003;42:6159–6168. doi: 10.1021/bi034002i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, Golan DE, Marto JA, Coen dm. PLoS Pathog. 2009;5:e1000275. doi: 10.1371/journal.ppat.1000275. (Epub 2009 January 23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung H, Lee H, Lamoke F, Hrushesky WJ, Wood PA, Jahng WJ. J Neurosci Res. 2009;87:2365–2374. doi: 10.1002/jnr.22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noell WK, Walke VS, Kang BS, Berman S. Invest Ophthalmol Vis Sci. 1966;5:450–473. [PubMed] [Google Scholar]
- 15.Lisman J, Fain G. Nat Med. 1995;1:1254–1255. doi: 10.1038/nm1295-1254. [DOI] [PubMed] [Google Scholar]
- 16.Grimm C, Wenzel A, Hafezi F, Yu S, Redmond TM, Reme CE. Nat Genet. 2000;25:63–66. doi: 10.1038/75614. [DOI] [PubMed] [Google Scholar]
- 17.Klemenz R, Fröhli E, Steiger RH, Schäfer R, Aoyama A. Proc Natl Acad Sci USA. 1991;88:3652–3656. doi: 10.1073/pnas.88.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhee SG, Chae HZ, Kim K. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 19.Osborne NN, Nash MS, Wood JP. Invest Ophthalmol Vis Sci. 1998;39:2374–2383. [PubMed] [Google Scholar]
- 20.Liang FQ, Aleman TS, Yang Z, Cideciyan AV, Jacobson SG, Bennett J. Neuroreport. 2001;12:1011–1014. doi: 10.1097/00001756-200104170-00029. [DOI] [PubMed] [Google Scholar]
- 21.Yi C, Pan X, Yan H, Guo M, Pierpaoli W. Ann NY Acad Sci. 2005;1057:384–392. doi: 10.1196/annals.1356.029. [DOI] [PubMed] [Google Scholar]
- 22.Saari JC, Bredberg DL, Noy N. Biochemistry. 1994;33:3106–3112. doi: 10.1021/bi00176a045. [DOI] [PubMed] [Google Scholar]
- 23.Maeda A, Maeda T, Golczak M, Imanishi Y, Leahy P. Mol Pharmacol. 2006;70:1220–1229. doi: 10.1124/mol.106.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deretic D. Electrophoresis. 1997;18:2537–2541. doi: 10.1002/elps.1150181408. [DOI] [PubMed] [Google Scholar]