Abstract
We determined MICs of antibiotics against Anaplasma phagocytophilum, Ehrlichia chaffeensis, and Ehrlichia canis by real-time quantitative PCR. The doubling times of the organisms were established: 19 h for E. chaffeensis, 26 h for A. phagocytophilum, and 28 h for E. canis. In comparison to the reference method for determining sensitivities, which uses Diff-Quick staining, our PCR assay was very sensitive and specific. We confirmed that doxycycline and rifampin are highly active against these bacteria and found variable susceptibilities to fluoroquinolones; A. phagocytophilum was susceptible, but E. canis and E. chaffeensis were only partly susceptible. β-Lactam compounds, cotrimoxazole, macrolide compounds, and telithromycin showed no activity against any of the three organisms. Thiamphenicol was found to be more active than chloramphenicol. For the first time, we showed that these three species have numerous point mutations in their 23S RNA genes, with those at positions 754, 2057, 2058, 2059, and 2611 (Escherichia coli numbering) known to confer resistance to macrolide compounds in other bacteria. The role of each of these mutations in resistance to these drugs should be investigated in the future. Our study confirms previous reports that quantitative PCR is a reliable method for determining antibiotic susceptibility; therefore, it might be useful for screening new drugs.
Ehrlichioses are emerging infectious diseases caused by gram-negative, obligately intracellular bacteria belonging to the α subgroup of Proteobacteria (15). Following a recent reorganization, three genera are recognized in the family Anaplasmataceae. Neorickettsia species include Neorickettsia sennetsu, Neorickettsia risticii, and Neorickettsia helminthoeca; Ehrlichia species include Ehrlichia canis, Ehrlichia chaffeensis, Ehrlichia ruminantium, Ehrlichia muris, and Ehrlichia ewingii; and Anaplasma species include Anaplasma platys, Anaplasma marginale, and Anaplasma phagocytophilum (13, 15). These species are agents of ehrlichioses or anaplasmoses which are human and/or animal diseases; for example, E. chaffeensis is the agent of human monocytic ehrlichiosis (HME), A. phagocytophilum is the agent of human granulocytic anaplasmosis (HGA), and E. canis is the agent of canine ehrlichiosis.
Human ehrlichioses are severe, influenza-like febrile illnesses that frequently require hospitalization; 2 to 4% of reported cases are fatal (14). Recently, the first case of human anaplasmosis presenting as atypical pneumonitis was reported in France (37). HME was first reported in the United States in 1987, and more than 400 cases are now reported. HGA is reported in the United-States, and serological studies have shown its presence in Switzerland, the United Kingdom, Italy, Norway, and Slovenia (2). The HGA agent has also been cultivated from patients in Sweden and the Czech Republic (25, 27), and PCR evidence of human infection is now widespread in Europe (37).
The organisms are transmitted to humans or animals by ticks which also feed on wild animals. The latter act as reservoirs of infection (13). For example, A. phagocytophilum is transmitted by Ixodes ticks, E. chaffeensis is transmitted by Amblyomma americanum, and E. canis is transmitted by Rhipicephalus sanguineus. In their mammalian hosts, these small, nonmotile gram-negative bacteria live and grow in cytoplasmic vacuoles and form loose to condensed aggregates of bacteria termed morulae. The morulae of different species are found in different cells in their hosts; E. canis and E. chaffeensis morulae are found in monocytes and macrophages, while A. phagocytophilum infects neutrophils of humans and many animals (46).
Tetracyclines are highly active in vitro against E. canis, E. chaffeensis, and A. phagocytophilum, and doxycycline is currently the first-line antibiotic in the treatment of patients infected with these organisms (13, 45). Rifampin is active against Ehrlichia in vitro and may represent a safe alternative for patients who should not receive tetracycline, for example, pregnant women and children (9). In vitro susceptibility to fluoroquinolones differs by species; A. phagocytophilum is susceptible while E. chaffeensis is resistant (7, 28, 33). This difference in susceptibility to fluoroquinolones was recently related to a natural DNA gyrase-mediated resistance (32).
In vitro tests currently used to assess the antibiotic susceptibilities of the organisms are based on microscopic counting of morulae in cells in tissue cultures before and after exposure to serial dilutions of antibiotics. These tests, however, are not standardized, are not sensitive, are time-consuming, and are not adapted for the screening of new drugs or strains of organisms. Recently, PCR and real-time quantitative PCR have been used successfully to study the antibiotic susceptibilities of other intracellular bacteria such as Rickettsia species (41), Wolbachia pipientis (18), Coxiella burnetii (3, 5), and Tropheryma whipplei (4, 31). These methods are simple, rapid, sensitive, reproducible, and specific and provide the possibility of automation. In this report, we describe the antibiotic susceptibilities of three reference strains of Ehrlichia and Anaplasma determined by real-time quantitative PCR with primers of the 16S ribosomal DNA (rDNA). We compare our findings with susceptibilities obtained by using the classical method in which morulae are counted microscopically. Furthermore, we aligned and compared the 23S rDNA sequences of these bacteria in order to seek a potential genetic correlation with resistance to macrolide compounds, which would be the subject of future investigation.
MATERIALS AND METHODS
Strains.
Strains used in the study were E. canis strain Oklahoma, E. chaffeensis strain Arkansas, and A. phagocytophilum strain Webster (J. S. Dumler, The Johns Hopkins Medical Institutions, Baltimore, Md.). E. canis and E. chaffeensis were grown in a DH82 canine malignant histiocytic cell line (ATTC number CR5L-10389) (47). Percentages of infected cells were monitored twice a week by Diff-Quick staining as previously described (7).
A. phagocytophilum was grown in the human promyelocytic cell line HL60 at 37°C under a 5% CO2 atmosphere (20). Three times a week, the percentage of infected cells was determined by counting cells with morulae in cytocentrifuged preparations stained with Diff-Quick. Concentrations of uninfected and infected cells were maintained at 4 × 105 and 2 × 105/ml, respectively.
Source and preparation of antibiotic solutions.
Antibiotics and their concentration ranges used in this study were as follows: doxycycline, 0.03 to 4 μg/ml (Pfizer, Neuilly, France); levofloxacin, 0.5 to 2 μg/ml (Hoechst Marion Roussel, Romainville, France); ciprofloxacin, 0.5 to 2 μg/ml (Bayer, Pharma, Sebs, France); ofloxacin, 0.5 to 2 μg/ml (Diamant, Puteaux, France); erythromycin, 4 μg/ml (Abbott, Rungis, France); telithromycin, 1 μg/ml (Hoechst Marion Roussel); thiamphenicol, 0.5 to 4 μg/ml (Sanofi, Winthrop, Gentilly, France); rifampin, 0.03 to 1 μg/ml (Cassenne, Puteaux, France); cotrimoxazole (trimethoprim and sulfamethoxazole), 8 to 32 μg/ml (Roche, Paris, France); gentamicin, 2.5 to 20 μg/ml (Dakota Pharm, Creteil, France); amoxicillin, 10 to 100 μg/ml (Beecham-Sevigne, Paris, France); ceftriaxone, 10 to 100 μg/ml (Roche, Neuilly sur Seine, France). Stock solutions were prepared as described by the manufacturer and stored at −20°C. Final antibiotic solutions were made up by diluting concentrated stock solutions in a medium adapted to the bacteria.
Growth kinetics.
Microplates (24-well plates; final volume, 1 ml) were inoculated with cell cultures, prepared as above, in which about 5% of cells contained morulae. HL60 cells were maintained at 2 × 105/ml. Percentages of infected cells were monitored daily over 5 days by removing 100 μl from a well for cytocentrifugation, staining with Diff-Quick, and preserving the remaining 900 μl at −20°C for real-time PCR.
Antimicrobial susceptibility testing.
The growth curves were analyzed as described above to determine the optimal time for testing antibiotic susceptibilities of bacteria. This optimal time was defined as the number of days necessary to reach the plateau of the growth curve. Initially, antibiotics were tested at the MIC breakpoint for resistance as defined by the current NCCLS guidelines. If the bacteria were inhibited at this concentration, MICs were then determined by using twofold serial dilutions. The MIC was defined as the lowest antibiotic concentration required for inhibition of the growth of the bacteria compared to that of the control (without antibiotics) at day 0. An infected culture without antibiotics served as a positive growth control, and an uninfected cell culture served as a negative control. All experiments were performed in triplicate and repeated three times to confirm results. Wells were sampled each day, and samples were divided into aliquots and stored at −20°C until genomic DNA was extracted for PCR assays.
Antibiotic activity.
The activities of the antibiotics were verified at the end of each experiment against two reference strains, Staphyloccocus aureus CIP 103429 and Escherichia coli CIP 76.24, as previously described (31, 39).
LightCycler PCR assay.
Total genomic DNA of each harvested well was extracted from aliquots by using a MaGNA Pure instrument as described by the manufacturer (Roche Diagnostics, Courtaboeuf, France). DNA was extracted from 100 μl of cell culture, and a total of 100 μl of elution buffer was used to resuspend the DNA. PCR was performed using a LightCycler instrument (Roche Biochemicals, Mannheim, Germany) to amplify a 345-bp fragment of the 16S rRNA of Ehrlichia or Anaplasma (35). The PCR mixture had a final volume of 20 μl in each capillary tube and contained 2 μl of DNA Master Mix (DNA Master SYBR Green I kit [Roche Diagnostics] including SYBR Green, deoxynucleoside triphosphates, and Taq polymerase), 1.6 μl of MgCl2, 12.4 μl of sterile water, 1 μl of each primer (EHR16S D [5′ GGTACCYACAGAAGAAGTCC 3′] and EHR16S R [5′ TAGCACTCATCGTTTACAGC 3′] [Eurogentec, Seraing, Belgium]) (35), and 2 μl of extracted DNA. Each PCR mixture included sterile distilled water and uninfected cells as a negative control. Amplification conditions were as follows: an initial denaturation step at 95°C for 10 min, followed by 30 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 20 s, and extension for15 s at 72°C. Melting curve analysis was done from 45 to 95°C (ramping time, 20°C/s) with stepwise fluorescence acquisition by real-time measurement of the fluorescence directly in the clear glass capillary tubes.
Sequence-specific standard curves were generated by using 10-fold serial dilutions of an extracted DNA sample containing 100% infected cells of E. canis, E. chaffeensis, and A. phagocytophilum, respectively. The number of DNA copies of each sample transcript was then calculated from the standard curve with the LightCycler software. For each experiment, the mean number (± standard deviation) of DNA copies in cultures was determined at day 0 and compared to the mean number (± standard deviation) of DNA copies in antibiotic-treated cultures at day 4. Lack of bacterial inhibition was defined as a mean number of DNA copies at day 4 statistically higher than that at the start of the experiment (by the Student t test). The MIC was defined as the lowest antibiotic concentration allowing inhibition of bacterial growth, i.e., a number of DNA copies at day 4 not statistically different from that at day 0. The specificity of amplification was then confirmed by agarose gel electrophoresis of PCR products and DNA sequencing.
Study of 23S rRNA in A. phagocytophilum, E. chaffeensis, and Wolbachia pipientis.
Sequences of the 23S rRNA gene of A. phagocytophilum strain USG3 (GenBank accession number AF416766), E. chaffeensis strain Arkansas (GenBank accession number AF416765), and Wolbachia pipientis (GenBank accession number U23710) were compared and aligned by using the E. coli numbering system (GenBank accession number AF278710) to look at possible mutations known to be associated with macrolide resistance (44). The 23S rRNA sequences were aligned by using the CLUSTALW program supported by the Infobiogen website (www.infobiogen.fr).
RESULTS
Specificity, sensitivity, and reproducibility of the quantitative PCR assay.
Primers used in this study were specific for the 16S rDNA of Ehrlichia and Anaplasma (35). A 345-bp DNA fragment was consistently amplified when the PCR was carried out with DNA from all the experiments. PCRs were also negative for the negative controls (sterile water instead of DNA, uninfected host cells HL60 and DH82). We confirmed the specificity of PCR products by agarose gel electrophoresis, and DNA sequencing of PCR products always gave the expected sequence (data not shown).
Determination of growth kinetics of bacteria by real-time PCR and Diff-Quick staining.
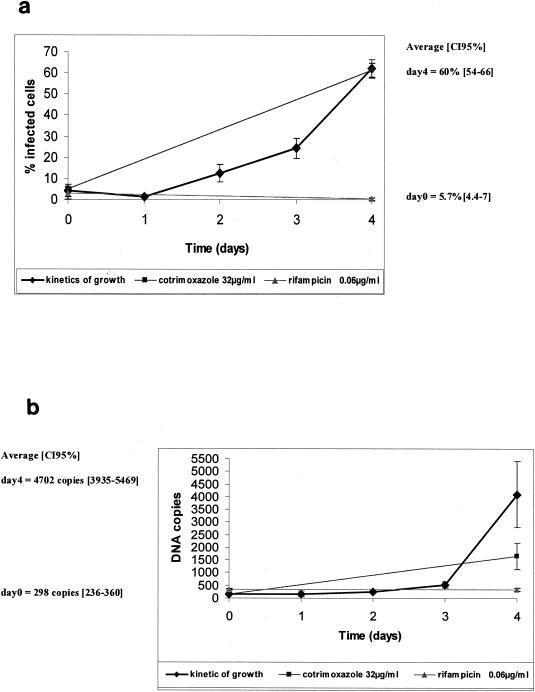
Our experiments enabled us to study and compare the growth kinetics of each bacterium by two methods. The growth curves of A. phagocytophilum as determined by Diff-Quick staining and real-time quantitative PCR are shown in Fig. 1a and b, respectively. In cultures infected with A. phagocytophilum but not treated with antibiotics, the mean percentages of infected cells were 5.7% (95% confidence interval [95% CI], 4.4 to 7%) at day 0 and 60.0% (95% CI, 54 to 66%) on day 4 (Fig. 1a). By the real-time PCR assay, the mean numbers of DNA copies found were 298 (95% CI, 236 to 360) on day 0 and 4,702 (95% CI, 3,935 to 5,469) on day 4 (Fig. 1b). Doubling times were estimated to be 23.5 ± 4.5 h for E. chaffeensis, 24.4 ± 1.4 h for A. phagocytophilum, and 26.0 ± 2.0 h for E. canis. Based on the data given above, we selected day 4 to be the most appropriate time to evaluate antibiotic susceptibilities, because at this time the growth curve had reached a plateau.
FIG. 1.
Susceptibilities of A. phagocytophilum to cotrimoxazole (32 μg/ml) and rifampin (0.06 μg/ml) as determined by enumeration of infected cells after Diff-Quick staining (a) or by quantification of DNA copies using the LightCycler instrument (b).
MICs.
In Fig. 1, we show examples of the growth kinetics of A. phagocytophilum in cultures without antibiotics, with rifampin at 0.06 μg/ml, and with cotrimoxazole at 32 μg/ml. In this experiment, cotrimoxazole at 32 μg/ml was not active: the geometric mean number of DNA copies at day 4 (1,754 copies) was significantly higher than that at day 0 (162 copies) (n = 9 experiments [95% CI, 8.4 to 14.2]). In contrast, rifampin was active against A. phagocytophilum at 0.06 μg/ml, with no growth of the organism detected by Diff-Quick staining or by real-time PCR between days 0 and 4 (no statistical difference between mean numbers of DNA copies on days 0 and 4).
The MICs we found for the 12 antibiotics we tested and those reported previously are shown in Tables 1 and 2. The results we obtained by the two methods were very similar and never differed by more than 2 dilutions. Doxycycline and rifampin were the most active antibiotics, with very low MICs (0.03 to 0.06 μg/ml). β-Lactam compounds, erythromycin, cotrimoxazole, and telithromycin were not active against the three bacteria, whereas MICs of thiamphenicol ranged from 1 to 4 μg/ml. Gentamicin was moderately active, with MICs from 2.5 to 10 μg/ml for E. chaffeensis and a MIC of 10 μg/ml for E. canis. In vitro susceptibility to fluoroquinolones differed among the organisms; A. phagocytophilum was susceptible, while E. canis and E. chaffeensis were less susceptible.
TABLE 1.
MICs of antibiotics against E. canis and E. chaffeensis determined in the present study and previous studies
| Antibiotic | MIC (μg/ml) determineda for the following strain in the indicated study:
|
||||||
|---|---|---|---|---|---|---|---|
|
E. canis Oklahoma isolate
|
E. chaffeensis Arkansas isolate
|
||||||
| This study
|
Brouqui and Raoult (8) | This study
|
Brouqui and Raoult (7) | Rolain et al. (40) | |||
| DQ | PCR | DQ | PCR | ||||
| Gentamicin | 10 | 10 | >100 | 10 | 2.5 | >32 | ND |
| Ceftriaxone | >100 | >100 | ND | >100 | >100 | ND | ND |
| Amoxicillin | >100 | >100 | 1,000 | >100 | >100 | ND | ND |
| Ofloxacin | >1 | >1 | ND | >1 | >1 | ND | ND |
| Ciprofloxacin | 1 | 1 | ND | >1 | >1 | >2 | ND |
| Pefloxacin | ND | ND | >2 | ND | ND | ND | ND |
| Levofloxacin | >1 | >1 | ND | >1 | >1 | ND | ND |
| Rifampin | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | 0.125 | ND |
| Doxycycline | 0.03 | 0.03 | 0.03 | 0.03 | 0.03 | <0.5 | ND |
| Cotrimoxazole | >32 | >32 | >4 | >32 | >32 | >4 | ND |
| Thiamphenicol | 2 | 1 | ND | 2 | 1 | ND | ND |
| Chloramphenicol | ND | ND | >4 | ND | ND | >4 | ND |
| Telithromycin | >1 | >1 | ND | >1 | >1 | ND | >1 |
| Erythromycin | >8 | >8 | >4 | >8 | >8 | >8 | ND |
DQ, Diff-Quick staining; PCR, real-time PCR assay; ND, not done.
TABLE 2.
MICs of antibiotics against A. phagocytophilum strains determined in the present study and previous studies
| Antibiotic | MIC (μg/ml) determined in the following study for the indicated A. phagocytophilum strain(s):
|
||||
|---|---|---|---|---|---|
| This study (Webster)a
|
Klein et al. (28) (3 strains)b | Horowitz et al. (23) (6 N.Y. strains)c | Maurin et al. (33) (Webster) | ||
| DQ | PCR | ||||
| Gentamicin | >10 | >10 | 50 | NDd | ND |
| Amikacin | ND | ND | ND | >16 | >64 |
| Ceftriaxone | >100 | >100 | >64 | >64 | >128 |
| Amoxicillin | >100 | >100 | >32 | >32 | >128 |
| Ofloxacin | 1 | 2 | 2 | <2 | ND |
| Ciprofloxacin | 1 | 2 | 2 | ND | ND |
| Levofloxacin | 0.5 | 1 | ND | <1 | 0.5 |
| Rifampin | 0.03 | 0.06 | 0.5 | <0.125 | 0.03 |
| Doxycycline | 0.03 | 0.06 | 0.25 | <0.125 | 0.03 |
| Cotrimoxazole | >32 | >32 | >16 | ND | 50 |
| Thiamphenicol | 2 | 4 | ND | ND | ND |
| Chloramphenicol | ND | ND | >32 | >16 | 2-8e |
| Telithromycin | >1 | >1 | ND | ND | ND |
| Erythromycin | >8 | >8 | >8 | >8 | >16 |
DQ, Diff-Quick assay; PCR, real-time PCR assay.
One strain each from Minnesota, New York, and Wisconsin.
Isolates 6008 and 6003, cultured in 1996; isolates 7013, 7019, and NY13, cultured in 1997; and isolate NY18, cultured in 1998.
ND, not done.
The MIC ranged from 2 to 8 μg/ml according to the strain tested.
Sequence alignment of the 23S rRNA genes.
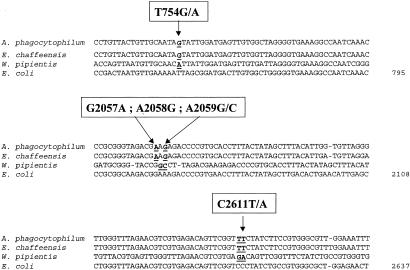
In Fig. 2, we present a sequence alignment of E. chaffeensis, A. phagocytophilum, W. pipientis, and E. coli 23S rDNAs. We found specific bases at positions that have been implicated in macrolide resistance in other bacteria: transitions A2058G, G2057A, A2059G, C2611T, and T754G (E. coli numbering) (44, 48) (Fig. 2).
FIG. 2.
DNA sequence alignment of the 23S rRNA genes of A. phagocytophilum, E. chaffeensis, W. pipientis, and E. coli.
DISCUSSION
The experimental evaluation of antibiotic therapy of Ehrlichia and Anaplasma infections is difficult because these bacteria are fastidious intracellular bacteria, requiring cell culture systems for their propagation. Moreover, conventional phenotypic assays using staining methods cannot allow a precise quantification of bacteria because evaluation of positive cells may vary according to the operators. Brouqui and Raoult performed a blind evaluation of uninfected cells stained by Diff-Quick and found a false-positive rate (cells interpreted as infected) of 3.6% (7). The quantitative PCR we describe in this report enabled us to precisely and objectively determine the number of DNA copies of the bacteria. The specificity of the primers used in this study was previously assessed by using 19 different genera belonging to the different subgroups of Proteobacteria (35).
The MICs we found by the two methods (Tables 1 and 2) were similar to those reported previously (7, 8, 23, 28, 33, 40). Doxycycline and rifampin were the most active drugs in vitro against E. chaffeensis, E. canis, and A. phagocytophilum, and similar findings have been reported previously (Tables 1 and 2). Tetracyclines are considered to be the first-line antibiotics for treatment of ehrlichial diseases (14). They have the advantage of being active against Borrelia burgdorferi, the agent of Lyme disease, which is transmitted by the same tick vector as A. phagocytophilum (12). Alternatives to tetracyclines are needed for children under the age of 8 years because of the potential for tooth discoloration; for pregnant women because of possible bone toxicity in the fetus; for allergic patients; and for patients with gastric intolerance of the antibiotic. Also, although tetracyclines are effective during the acute phase of canine ehrlichiosis, chronically infected dogs do not respond to treatment with tetracyclines (22, 26). This is probably due to a lack of bactericidal activity of these compounds.
Rifampin can be used for pregnant women or children, and there are anecdotal reports of successful treatment of infections using this antibiotic (9, 14, 24, 29).
Ampicillin, ceftriaxone, and cotrimoxazole were not active in vitro, as was previously reported (7, 8, 28). Ehrlichiae are highly pleomorphic and are enveloped in a rippled thin outer membrane which lacks thickening of the inner or outer leaflet and shows no sign of a peptidoglycan layer or lipopolysaccharide (38), which is the β-lactam target site. Recently, analysis of E. chaffeensis and A. phagocytophilum genome sequences revealed that these bacteria lack the genes for biosynthesis of lipid A and most genes for biosynthesis of peptidoglycan, both of which confer structural strength on gram-negative bacteria (30). This situation is very similar to that of mycoplasmas, which are also naturally resistant to all β-lactam compounds (34).
Our study confirms that macrolides and telithromycin (a ketolide derivative) lack antimicrobial activity against these bacteria (6, 7, 28, 33, 40). Many cases of macrolide resistance in clinical isolates can be linked to alteration of specific nucleotides in the 23S rRNA gene within the large ribosomal subunit (44). We report, for the first time, numerous specific nucleotides known to confer resistance to macrolides in the E. chaffeensis and A. phagocytophilum 23S rRNA genes (Fig. 2): T754(G/A), G2057A, A2058G, A2059(G/C), and C2611(T/A). In addition, we found as many as 4 of these specific nucleotides occurring together, which has never been reported before. W. pipientis, a closely related organism, also possesses mutations A2058G, A2059C, C2611G, and T754A, which may explain the intrinsic resistance of this bacterium to macrolides (18). The role of each of these mutations is not clear, and correlation between resistance to macrolides and specific nucleotide sequences in the 23S rDNA should be investigated in the future in order to better understand the molecular mechanism of resistance in this group of bacteria. Mutations at position 2058 (using the E. coli numbering system [GenBank accession number AF278710]) are the only substitutions to confer high resistance to all the macrolides (44). Mutations at position 2059 have been found in vivo in mycobacteria, propionibacteria, Helicobacter pylori, Streptococcus pneumoniae, and Mycoplasma spp. (36, 44). Low-level drug resistance is provided by mutations at positions 2057, 2452, and 2611 and has been described for an E. coli laboratory strain with a mutation at position 754 in hairpin 35 (48). Mutations at position 2057 in clinical isolates have been found in erythromycin-resistant Propionibacterium spp. (42), chloramphenicol- and erythromycin-resistant E. coli (44), and a clarithromycin-resistant double-mutant strain of H. pylori (44).
We found that thiamphenicol appeared to be more active than chloramphenicol against E. canis (8) and E. chaffeensis (7). However, Klein et al. reported 75% growth inhibition of A. phagocytophilum with chloramphenicol at 32 μg/ml after 3 days (28), and Maurin et al. reported variable activity among strains (33). In practice, chloramphenicol is not widely used because of hematological side effects and inconsistent activity (1, 11, 16, 17, 19, 21). Both chloramphenicol and thiamphenicol inhibit protein synthesis by decreasing peptidyl transferase activity of the 70S ribosome subunit, but chloramphenicol binds to the target site with higher affinity and dramatically decreases polynuclear phagocytic function (10). Thiamphenicol would have the advantage of being less toxic (43), but further clinical data are needed to define the usefulness of this drug.
Ehrlichia and Anaplasma are not usually susceptible to aminoglycosides. However, amikacin is the only drug that is tested often. Brouqui and Raoult reported a decrease in the number of infected cells after 9 days of incubation with gentamicin at 32 μg/ml (7). Klein et al. reported inhibitory activity of gentamicin against A. phagocytophilum exposed for 3 days to 50 μg/ml, a level not achievable in human blood without severe toxicity (28). Gentamicin MICs found in this study were 2.5 to 10 μg/ml for E. chaffeensis and E. canis and 20 μg/ml for A. phagocytophilum.
Fluoroquinolones are more active in vitro against A. phagocytophilum than against E. chaffeensis and E. canis (Tables 1 and 2). gyrA-mediated resistance has recently been found in E. canis and E. chaffeensis, and this might explain the differences in sensitivity that we found (32).
Conclusion.
Ehrlichioses and anaplasmoses are emerging infectious diseases that can cause severe disease and are of major medical concern. Clinical data on the treatment of these diseases are limited, and Ehrlichia and Anaplasma are regarded as highly resistant organisms, because only two drugs are always effective in treatment: doxycycline and rifampin. The new quantitative PCR assay that we describe proved very useful for determination of the antibiotic susceptibilities of the organisms. Because this method is very sensitive and reproducible, it could easily be standardized for quantification of the bacteria with a low possibility of contamination. Antibiotic selective forces in ticks and animals are probably minimal, but more data with regard to new isolates may be important in tracking these emerging infections and their resistance patterns. Since these bacteria are extremely fastidious, routine evaluation of antibiotic susceptibility is not possible and should not dictate therapy.
Acknowledgments
We thank Pat Kelly and Esther Platt for careful review of the manuscript.
REFERENCES
- 1.Barton, L. L., and T. M. Foy. 1989. Ehrlichia canis infection in a child. Pediatrics 84:580-582. [PubMed] [Google Scholar]
- 2.Blanco, J. R., and J. A. Oteo. 2002. Human granulocytic ehrlichiosis in Europe. Clin. Microbiol. Infect. 8:763-772. [DOI] [PubMed] [Google Scholar]
- 3.Boulos, A., J. M. Rolain, M. Maurin, and D. Raoult. 2004. Evaluation of antibiotic susceptibilities against Coxiella burnetii by real time PCR. Int. J. Antimicrob. Agents 23:169-174. [DOI] [PubMed] [Google Scholar]
- 4.Boulos, A., J. M. Rolain, and D. Raoult. 2004. Antibiotic susceptibility of Tropheryma whipplei in MRC5 cells. Antimicrob. Agents Chemother. 48:747-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan, R. E., and J. E. Samuel. 2003. Evaluation of Coxiella burnetii antibiotic susceptibilities by real-time PCR assay. J. Clin. Microbiol. 41:1869-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brouqui, P., and D. Raoult. 1990. In vitro susceptibility of Ehrlichia sennetsu to antibiotics. Antimicrob. Agents Chemother. 34:1593-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouqui, P., and D. Raoult. 1992. In vitro antibiotic susceptibility of the newly recognized agent of ehrlichiosis in humans, Ehrlichia chaffeensis. Antimicrob. Agents Chemother. 36:2799-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouqui, P., and D. Raoult. 1993. Susceptibilities of Ehrlichiae to antibiotics, p. 179-199. In D. Raoult (ed.), Antimicrobial agents and intracellular pathogens. CRC Press, Boca Raton, Fla.
- 9.Buitrago, M. I., J. W. Ijdo, P. Rinaudo, H. Simon, J. Copel, J. Gadbaw, R. Heimer, E. Fikrig, and F. J. Bia. 1998. Human granulocytic ehrlichiosis during pregnancy treated successfully with rifampin. Clin. Infect. Dis. 27:213-215. [DOI] [PubMed] [Google Scholar]
- 10.Cannon, M., S. Harford, and J. Davies. 1990. A comparative study on the inhibitory actions of chloramphenicol, thiamphenicol and some fluorinated derivatives. J. Antimicrob. Chemother. 26:307-317. [DOI] [PubMed] [Google Scholar]
- 11.Doran, T. I., R. T. Parmley, P. C. Logas, and S. Chamblin. 1989. Infection with Ehrlichia canis in a child. J. Pediatr. 114:809-812. [DOI] [PubMed] [Google Scholar]
- 12.Dumler, J. S. 1997. Is human granulocytic ehrlichiosis a new Lyme disease? Review and comparison of clinical, laboratory, epidemiological, and some biological features. Clin. Infect. Dis. 25:S43-S47. [DOI] [PubMed] [Google Scholar]
- 13.Dumler, J. S., and J. S. Bakken. 1995. Ehrlichial diseases of humans: emerging tick-borne infections. Clin. Infect. Dis. 20:1102-1110. [DOI] [PubMed] [Google Scholar]
- 14.Dumler, J. S., and J. S. Bakken. 1998. Human ehrlichioses: newly recognized infections transmitted by ticks. Annu. Rev. Med. 49:201-213. [DOI] [PubMed] [Google Scholar]
- 15.Dumler, J. S., A. F. Barbet, C. P. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 16.Dumler, J. S., P. Brouqui, J. Aronson, J. P. Taylor, and D. H. Walker. 1991. Identification of Ehrlichia in human tissue. N. Engl. J. Med. 325:1109-1110. [DOI] [PubMed] [Google Scholar]
- 17.Eng, T. R., D. B. Fishbein, J. E. Dawson, C. R. Greene, and M. Redus. 1990. Surveillance of human ehrlichiosis in the United States: 1988. Ann. N. Y. Acad. Sci. 590:306-307. [DOI] [PubMed] [Google Scholar]
- 18.Fenollar, F., M. Maurin, and D. Raoult. 2003. Wolbachia pipientis growth kinetics and susceptibilities to 13 antibiotics determined by immunofluorescence staining and real-time PCR. Antimicrob. Agents Chemother. 47:1665-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fishbein, D. B., J. E. Dawson, and L. E. Robinson. 1994. Human ehrlichiosis in the United States, 1985 to 1990. Ann. Intern. Med. 120:736-743. [DOI] [PubMed] [Google Scholar]
- 20.Goodman, J. L., C. Nelson, B. Vitale, J. E. Madigan, J. S. Dumler, T. J. Kurtti, and U. G. Munderloh. 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 334:209-215. [DOI] [PubMed] [Google Scholar]
- 21.Harkess, J. R., S. A. Ewing, T. Brumit, and C. R. Mettry. 1991. Ehrlichiosis in children. Pediatrics 87:199-203. [PubMed] [Google Scholar]
- 22.Harrus, S., T. Waner, I. Aizenberg, and H. Bark. 1998. Therapeutic effect of doxycycline in experimental subclinical canine monocytic ehrlichiosis: evaluation of a 6-week course. J. Clin. Microbiol. 36:2140-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horowitz, H. W., T.-C. Hsieh, M. E. Aguero-Rosenfeld, F. Kalantarpour, I. Chowdhury, G. P. Wormser, and J. M. Wu. 2001. Antimicrobial susceptibility of Ehrlichia phagocytophila. Antimicrob. Agents Chemother. 45:786-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horowitz, H. W., E. Kilchevsky, S. Haber, M. Aguero-Rosenfeld, R. Kranwinkel, E. K. James, S. J. Wong, F. Chu, D. Liveris, and I. Schwartz. 1998. Perinatal transmission of the agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 339:375-378. [DOI] [PubMed] [Google Scholar]
- 25.Hulinska, D., J. Votypka, J. Plch, E. Vlcek, M. Valesova, M. Bojar, V. Hulinsky, and K. Smetana. 2002. Molecular and microscopical evidence of Ehrlichia spp. and Borrelia burgdorferi sensu lato in patients, animals and ticks in the Czech Republic. New Microbiol. 25:437-448. [PubMed] [Google Scholar]
- 26.Iqbal, Z., and Y. Rikihisa. 1994. Reisolation of Ehrlichia canis from blood and tissues of dogs after doxycycline treatment. J. Clin. Microbiol. 32:1644-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlsson, U., A. Bjoersdorff, R. F. Massung, and B. Christensson. 2001. Human granulocytic ehrlichiosis—a clinical case in Scandinavia. Scand. J. Infect. Dis. 33:73-74. [DOI] [PubMed] [Google Scholar]
- 28.Klein, M. B., C. M. Nelson, and J. L. Goodman. 1997. Antibiotic susceptibility of the newly cultivated agent of human granulocytic ehrlichiosis: promising activity of quinolones and rifamycins. Antimicrob. Agents Chemother. 41:76-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause, P. J., C. L. Corrow, and J. S. Bakken. 2003. Successful treatment of human granulocytic ehrlichiosis in children using rifampin. Pediatrics 112:e252-e253. [DOI] [PubMed] [Google Scholar]
- 30.Lin, M., and Y. Rikihisa. 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun. 71:5324-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masselot, F., A. Boulos, M. Maurin, J. M. Rolain, and D. Raoult. 2003. Molecular evaluation of antibiotic susceptibility: the Tropheryma whipplei paradigm. Antimicrob. Agents Chemother. 47:1658-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maurin, M., C. Abergel, and D. Raoult. 2001. DNA gyrase-mediated natural resistance to fluoroquinolones in Ehrlichia spp. Antimicrob. Agents Chemother. 45:2098-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maurin, M., J. S. Bakken, and J. S. Dumler. 2003. Antibiotic susceptibilities of Anaplasma (Ehrlichia) phagocytophilum strains from various geographic areas in the United States. Antimicrob. Agents Chemother. 47:413-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormack, W. M. 1993. Susceptibility of mycoplasmas to antimicrobial agents: clinical implications. Clin. Infect. Dis. 17(Suppl. 1):S200-S201. [DOI] [PubMed] [Google Scholar]
- 35.Parola, P., V. Roux, J. L. Camicas, I. Baradji, P. Brouqui, and D. Raoult. 2000. Detection of ehrlichiae in African ticks by polymerase chain reaction. Trans. R. Soc. Trop. Med. Hyg. 94:707-708. [DOI] [PubMed] [Google Scholar]
- 36.Pereyre, S., P. Gonzalez, B. de Barbeyrac, A. Darnige, H. Renaudin, A. Charron, S. Raherison, C. Bebear, and C. M. Bebear. 2002. Mutations in 23S rRNA account for intrinsic resistance to macrolides in Mycoplasma hominis and Mycoplasma fermentans and for acquired resistance to macrolides in M. hominis. Antimicrob. Agents Chemother. 46:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remy, V., Y. Hansmann, S. De Martino, D. Christmann, and P. Brouqui. 2003. Human anaplasmosis presenting as atypical pneumonitis in France. Clin. Infect. Dis. 37:846-848. [DOI] [PubMed] [Google Scholar]
- 38.Rikihisa, Y., N. Zhi, G. P. Wormser, B. H. Wen, H. W. Horowitz, and K. E. Hechemy. 1997. Ultrastructural and antigenic characterization of a granulocytic ehrlichiosis agent directly isolated and stably cultivated from a patient in New York State. J. Infect. Dis. 175:210-213. [DOI] [PubMed] [Google Scholar]
- 39.Rolain, J. M., M. N. Mallet, P. E. Fournier, and D. Raoult. 2004. Real-time PCR for universal antibiotic susceptibility testing. J. Antimicrob. Chemother. 54:538-541. [DOI] [PubMed] [Google Scholar]
- 40.Rolain, J. M., M. Maurin, A. Bryskier, and D. Raoult. 2000. In vitro activities of telithromycin (HMR 3647) against Rickettsia rickettsii, Rickettsia conorii, Rickettsia africae, Rickettsia typhi, Rickettsia prowasekii, Coxiella burnetii, Bartonella henselae, Bartonella quintana, Bartonella bacilliformis, and Ehrlichia chaffeensis. Antimicrob. Agents Chemother. 44:1391-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rolain, J. M., L. Stuhl, M. Maurin, and D. Raoult. 2002. Evaluation of antibiotic susceptibilities of three rickettsial species including Rickettsia felis by a quantitative PCR DNA assay. Antimicrob. Agents Chemother. 46:2747-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross, J. I., E. A. Eady, J. H. Cove, C. E. Jones, A. H. Ratyal, Y. W. Miller, S. Vyakrnam, and W. J. Cunliffe. 1997. Clinical resistance to erythromycin and clindamycin in cutaneous propionibacteria isolated from acne patients is associated with mutations in 23S rRNA. Antimicrob. Agents Chemother. 41:1162-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skolimowski, I. M., R. C. Knight, and D. I. Edwards. 1983. Molecular basis of chloramphenicol and thiamphenicol toxicity to DNA in vitro. J. Antimicrob. Chemother. 12:535-542. [DOI] [PubMed] [Google Scholar]
- 44.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker, D. H., and J. S. Dumler. 1996. Emergence of the ehrlichioses as human health problems. Emerg. Infect. Dis. 2:1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Webster, P., J. W. Ijdo, L. M. Chicoine, and E. Fikrig. 1998. The agent of human granulocytic ehrlichiosis resides in an endosomal compartment. J. Clin. Investig. 101:1932-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wellman, M. L., S. Krakowka, R. M. Jacobs, and G. J. Kociba. 1988. A macrophage-monocyte cell line from a dog with malignant histiocytosis. In Vitro Cell. Dev. Biol. 24:223-229. [DOI] [PubMed] [Google Scholar]
- 48.Xiong, L., S. Shah, P. Mauvais, and A. S. Mankin. 1999. A ketolide resistance mutation in domain II of 23S rRNA reveals the proximity of hairpin 35 to the peptidyl transferase centre. Mol. Microbiol. 31:633-639. [DOI] [PubMed] [Google Scholar]