Abstract
Heat shock transcription factors (Hsfs) are important transcription factors (TFs) in protecting plants from damages caused by various stresses. The released whole genome sequences of wild peanuts make it possible for genome-wide analysis of Hsfs in peanut. In this study, a total of 16 and 17 Hsf genes were identified from Arachis duranensis and A. ipaensis, respectively. We identified 16 orthologous Hsf gene pairs in both peanut species; however HsfXs was only identified from A. ipaensis. Orthologous pairs between two wild peanut species were highly syntenic. Based on phylogenetic relationship, peanut Hsfs were divided into groups A, B, and C. Selection pressure analysis showed that group B Hsf genes mainly underwent positive selection and group A Hsfs were affected by purifying selection. Small scale segmental and tandem duplication may play important roles in the evolution of these genes. Cis-elements, such as ABRE, DRE, and HSE, were found in the promoters of most Arachis Hsf genes. Five AdHsfs and two AiHsfs contained fungal elicitor responsive elements suggesting their involvement in response to fungi infection. These genes were differentially expressed in cultivated peanut under abiotic stress and Aspergillus flavus infection. AhHsf2 and AhHsf14 were significantly up-regulated after inoculation with A. flavus suggesting their possible role in fungal resistance.
Keywords: heat shock transcription factor, peanut, abiotic stress, selective pressure, purifying selection
Introduction
Abiotic stresses, including heat, cold, drought, and salinity, affect plant growth and development and cause serious loss of crop production (Wang et al., 2004; Al-Whaibi, 2011; Qiao et al., 2015). As sessile organisms, plants could not change their locations when facing such stress conditions (Guo et al., 2016). However, plants have evolved adaptation strategies to these stresses (Scharf et al., 2012; Guo et al., 2016). Transcription factors play a crucial role in stress tolerance by regulating the expression of thousands of genes under unfavorable conditions (Schwechheimer and Bevan, 1998; Kreps et al., 2002; Shinozaki and Yamaguchi-Shinozaki, 2007; Wang et al., 2014). Plant heat shock transcription factors (Hsfs) are important transcription factors (TFs) in protecting plants from heat stress and other stresses, including cold, salinity, and drought (Kotak et al., 2007a; Swindell et al., 2007; Hu et al., 2015). Hsfs were found in eukaryotes from yeast to humans (Ritossa, 1962; Tanabe et al., 1997; Akerfelt et al., 2010). Hsfs could protect cells from extreme proteotoxic damage via the activation of related genes (Dalton et al., 2000; Akerfelt et al., 2010; Yang et al., 2014; Jaeger et al., 2016). Studies showed that Hsfs are also involved in plant growth and development (Almoguera et al., 2002; Díaz-Martín et al., 2005; Kotak et al., 2007b).
Hsfs regulate heat shock response via activating the expression of heat shock protein (HSP) genes by binding to the heat shock elements (HSEs) (Pelham, 1982; Akerfelt et al., 2010). The sequences and geometrys of HSEs (5′-AGAAnnTTCT-3′) are variable (Guertin et al., 2012; Mendillo et al., 2012; Vihervaara et al., 2013). Hsfs could also bind to SatIII repeat element, 5′-cgGAAtgGAAtg-3′ (Grady et al., 1992). Like many other transcription factors, Hsfs have an N-terminal DNA binding domain (DBD) and followed by an oligomerization domain (OD). OD is composed of two hydrophobic heptad repeats (HR-A/B) which allows homo- and hetero-multimerization (Peteranderl et al., 1999; Nover et al., 2001; Baniwal et al., 2004). Certain Hsfs contained nuclear location signal (NLS) domain, nuclear export signal (NES), and C-terminal activation (AHA) domain (Döring et al., 2000; Hsu et al., 2003; Maere et al., 2005). Based on the structural characteristics of HR-A/B domain and the phylogenetic relationship, plant Hsfs are divided into A, B, and C groups (Von Koskull-Döring et al., 2007; Wang et al., 2014; Yang et al., 2014). Additional sequences were found in HR-A/B domain of group A and C, but not in group B Hsfs (Nover et al., 2001; Schmidt et al., 2012; Wang et al., 2014; Yang et al., 2014).
Only the active Hsfs are capable of recognizing and binding to the promoters of target genes. The inactive monomer could be converted into active oligomer under variety of stress conditions (Hartl and Hayer-Hartl, 2002; Wang et al., 2012; Li et al., 2014). There are only a few Hsf genes in yeast and animals, while 20–50 Hsf genes were found in plants (Scharf et al., 2012; Lin et al., 2014; Qiao et al., 2015). Hsf genes were identified in many plants and expressed in various tissues at different developmental stages during different stress conditions (Giorno et al., 2012; Chung et al., 2013; Xue et al., 2014).
Peanut (Arachis hypogaea L.) is an important oil crop in the world. In developing countries, peanuts were rain-fed, so it is important to study the drought stress tolerance of peanut (Ramu et al., 2015). Aspergillus flavus produces potent mycotoxins known as aflatoxins that could cause serious health concerns (Zhang et al., 2015). It is unknown on the role of Hsf genes in peanut response to abiotic stresses and A. flavus infection.
Cultivated peanut is an allotetraploid (AABB, 4n = 4x = 40) originated from a single hybridization and genome duplication event between two wild type diploid peanuts (AA and BB genomes) (Kochert et al., 1996; Freitas et al., 2007; Moretzsohn et al., 2013; Wang et al., 2016). Recently, the whole genome sequencing of the two ancestral species (A. duranensis and A. ipaensis) have been completed (Bertioli et al., 2016; http://peanutbase.org/). Here, we genome-widely identified and analyzed the Hsf genes from two wild peanuts species: A. duranensis (AA genome) and A. ipaensis (BB genome), respectively. We analyzed the gene duplication events in the wild peanut species, the difference of selection pressure in A, B, and C group of Arachis Hsfs, and the structures of these proteins. Our results provide basic information for further understanding the functional divergence and evolution of Arachis Hsfs. We also applied the knowledge gained from wild species to cultivated one to understand their possible functions on peanut response to abiotic and biotic stress.
Materials and methods
Data collection and identification of Hsf genes
The genome sequence data of two wild peanut species (AA and BB genomes) were obtained from the peanut genome database (http://peanutbase.org/). The conserved domains of Hsfs are Hsf-type DBD domain. The HMM ID of this domain is PF00447 in the pfam database (http://pfam.xfam.org/). The amino acid sequences of HMMs were used as queries to identify all possible Hsf protein sequences in AA and BB genome database using BLASTP (E < 0.001). SMART software (http://smart.embl-heidelberg.de/) was used to identify integrated DBD domain and (HR-A/B) domain in the putative peanut Hsfs. Candidate proteins without integrated DBD domain and HR-A/B domain were removed. NLS domains in peanut Hsfs were predicted using cNLS Mapper software (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). NES domains were predicted using NetNES 1.1 server software (http://www.cbs.dtu.dk/services/NetNES/). AHA domains were predicted based on the conserved-type AHA motif sequence FWxxF/L, F/I/L (Kotak et al., 2004). Protein isoelectric point (pI) and molecular weight (Mw) were analyzed using Expasy software (http://web.expasy.org/compute_pi/).
The genome, protein, and cDNA sequences were collected from the related genome databases for the following additional plant species: Arabidopsis thaliana (http://www.plantgdb.org/AtGDB/), Glycine max (http://www.plantgdb.org/GmGDB/), Lotus japonicus (http://www.plantgdb.org/LjGDB/), Medicago truncatula (http://www.plantgdb.org/MtGDB/), Cajanus cajan (http://gigadb.org/dataset/100028) and Cicer arietinum (http://nipgr.res.in/CGAP/home.php).
Orthologous gene identification and structure analysis
Orthologous gene pairs were identified according to (1) the best-hit between A.duranensis and A. ipaensis, (2) the position in the phylogenetic tree (bootstrap value >50), and (3) identity between ortholougs gene pairs (>90%). Circos software was used to plot the chromosomal location (Krzywinski et al., 2009). Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/) was used to plot the gene structure.
Analysis of synteny
Intraspecies synteny analysis of AA or BB genome and interspecies synteny analysis between AA and BB genomes were based on comparison of 100 kb blocks of chromosome containing Hsf genes according to previous reports (Sato et al., 2008; Zhang et al., 2011; Lin et al., 2014). Hsf genes were set as anchor points according to their chromosome locations. Blocks were identified by local all-vs-all BLASTN (E < 10−20). In intraspecies analysis, when four or more homology genes were detected, these two blocks were considered to be originated from a large-scale duplication event (Zhang et al., 2011; Lin et al., 2014). In interspecies analysis, when three or more conserved homology genes were detected, these two blocks were considered syntenic blocks (Sato et al., 2008; Lin et al., 2014; Wang et al., 2016).
Multiple sequence alignment and phylogenetic analysis
Protein multiple sequence alignment was performed using online software Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo). Neighbor-Joining (NJ) trees were constructed using MEGA 6.0 with protein sequences. To support the calculated relationship, 1000 bootstrap samples were generated. A total of 21 A. thaliana Hsfs (Scharf et al., 2012), 11 M. truncatula Hsfs, 10 L. japonicus Hsfs, 16 C. cajan Hsfs (Lin et al., 2014), 11 C. arietinum Hsfs, and 40 G. max Hsfs were included in the phylogenetic analysis (Poisson correction, pairwise deletion, and bootstrap = 1000 replicates; Xue et al., 2014). All Hsfs used in this study was listed in the Table S1.
Gene duplication analysis
Two standards for duplication gene identification were used. High-stringency standard: coding protein pair with ≥50% identity and covering ≥90% protein length. Low-stringency standard: protein pair with ≥30% identity and covering ≥70% protein length (Rizzon et al., 2006). Tandem duplication of genes was marked according to the previously described method (Yuan et al., 2015). Chromosome segmental or large scale duplication of genes was identified based on the intraspecies synteny (Zhang et al., 2011; Lin et al., 2014; Qiao et al., 2015).
Protein structure analysis and homology modeling
SWISS-MODLE (http://www.swissmodel.expasy.org/interactive) was used to calculate secondary structure and build three-dimensional structure of proteins. The templates for building protein 3D model were selected in PDB database based on the best identity. Protein 3D models were selected based on the best global model quality estimation (GMQE). Homology modeling templates included 5d5v.1 (monomer of DBD domain), 5d5v.1 (homo-dimer of DBD domain interacted with SalIII), 5d5u.1 (homo-dimer DBD domain interacted with HSE), 4r0r.1.A (monomer of HR-A/B domain) and 4r0r.1 (homo-trimer of HR-A/B).
Analysis of selective pressure
Codeml program under PAML (phylogenetic analysis maximum likelihood) version 4.7 software (Yang, 2007) was used to detect whether the Hsf genes underwent positive selection. In PAML, six site models, M0 (one ratio), M1a (neutral), M2a (positive selection), M3 (discrete), M7 (beta) and M8 (beta and ω) could be applied to selection pressure analysis. Positive selection sites could be identified by the comparison of M0-M3, M1a-M2a, and M7-M8 (Yang et al., 2000).
Analysis of cis-acting regulatory elements in promoter
Plantcare software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict cis-acting regulatory elements.
Plant materials, stress treatments, and RNA isolation
Cultivated peanut cv. Luhua-14 was used in this study. Eleven-day-old peanut seedlings were subjected to drought (removed from wet medium and kept in air on filter paper), cold (4°C) and high temperature (42°C) treatment. Leaf samples were collected at 0, 1, and 6 h after treatment and immediately frozen in liquid nitrogen. Leaf samples without treatment were used as control. Peanut seeds inoculated with A. flavus for 3 days were collected and seeds without A. flavus inoculation were used as control according to a previous report (Zhang et al., 2015). RNAs were isolated by CTAB method according to a previous method (Wang et al., 2016). For reverse transcription, the first-strand cDNA was synthesized with an oligo (dT) primer using a PrimeScript™ first-strand cDNA synthesis kit (TaKaRa). Three technical replicates were carried out in this study.
Gene expression analysis
Quantitative real time PCR (qRT-PCR) was performed using the FastStart Universal SYBR Green Master (ROX) with ABI™ 7500. The qRT-PCR program was set as the following: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s. Relative gene expression levels were calculated using the ΔΔCT method. The primers for qRT-PCR were provided in the Table S2. T-test was used to analyze the significance.
Results
Identification of Hsf genes in wild peanut species
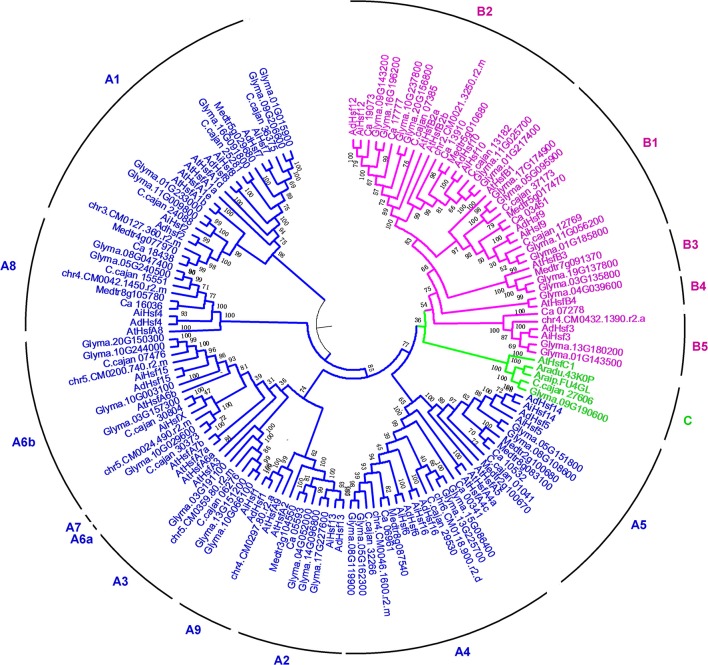
The amino acid sequences of Hsfs were extracted from AA and BB wild peanut genome database using the BLASTP program. The amino acid sequences of Hsf DBD domains (Pfam: PF00447) were used as queries. From AA and BB genomes, we identified 16 and 17 Hsf genes, respectively. The polypeptide lengths of Hsfs varied from 209 to 656 aa in A. duranensis and from 282 to 514 aa in A. ipaensis. A. thaliana Hsf family were often employed as reference to classify Hsf family in other plant species (Scharf et al., 2012; Li et al., 2014; Wang et al., 2014; Qiao et al., 2015). We employed Hsfs from A. thaliana and other species to construct phylogenetic tree together with Hsfs in two wild peanut species. In this study, 21 A. thaliana Hsfs, 11 M. truncatula Hsfs, 10 L. japonicus Hsfs, 16 C. cajan Hsfs, 11 C. arietinum Hsfs, and 40 G. max Hsfs were used for phylogenetic tree construction (Figure 1). These Hsfs were divided into A, B, and C groups that was consistent with previous studies (Scharf et al., 2012; Li et al., 2014; Lin et al., 2014; Wang et al., 2014; Qiao et al., 2015). Group A was divided into 10 clusters, group B was divided into five clusters, and group C contained only one cluster. Clusters in the group A were named as A1–A5, A6a, A6b, A7–A9. Clusters in the group B were named as B1–B5. B5 cluster was not presented in Arabidopsis; however, B5 cluster was identified in many leguminous species including wild peanut species. In wild peanut species, A3, A6a, A7, B3, and B4 clusters were absent (Figure 1). Orthologous of all 16 AA genome Hsfs were found in the BB genome with >90% identity (Table S3).
Figure 1.
Neighbor-joining phylogenic analysis of Hsfs.
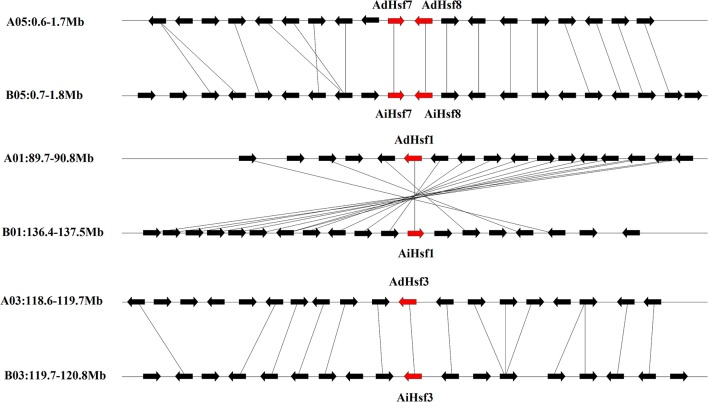
Interspecies synteny analysis showed that high level synteny was maintained between AA and BB genomes (Figure 2). This synteny analysis supported the identification of orthologous-pairs of Hsfs between AA and BB genomes. The nomenclature of AA genome Hsfs was based on their chromosome location order, AdHsf1-16. BB genome Hsfs were named based on their orthologous genes in AA genome AiHsf1-16 and AiHsfX. The orthologous gene of AiHsfX (Araip. A5C77) was not found in AA genome. The gene IDs and physical locations information of wild peanut Hsf genes were showed in Table 1, Figure 3.
Figure 2.
Syntenic chromosomal segments between genes flanking AA genome Hsfs and their orthologous genes in BB genome. Red arrows represent Hsf, black arrows represent flanking genes.
Table 1.
Hsfs identified in wild type peanuts.
| Gene name | Chromosome location | Gene ID |
|---|---|---|
| AdHsf12 | Aradu.A05:106,432,571.106,434,237 | Aradu.H5REB |
| AdHsf9 | Aradu.A05:7,998,864.8,000,946 | Aradu.S4DGV |
| AdHsf10 | Aradu.A05:14,375,242.14,377,500 | Aradu.NTQ7W |
| AdHsf1 | Aradu.A01:90,085,291.90,088,131 | Aradu.5S8J3 |
| AdHsf2 | Aradu.A03:8,559,905.8,562,946 | Aradu.N3N49 |
| AdHsf16 | Aradu.A10:1,011,979.1,014,302 | Aradu.H8ZFN |
| AdHsf15 | Aradu.A09:110,970,626.110,972,271 | Aradu.MA5WH |
| AdHsf6 | Aradu.A03:134,564,420.134,565,759 | Aradu.RVF1V |
| AdHsf7 | Aradu.A05:1,337,984.1,342,330 | Aradu.G1NHG |
| AdHsf4 | Aradu.A03:127,573,853.127,576,100 | Aradu.N1DAK |
| AdHsf5 | Aradu.A03:133,622,480.133,625,670 | Aradu.R0U9B |
| AdHsf11 | Aradu.A05:91,450,473.91,451,748 | Aradu.43K0P |
| AdHsf14 | Aradu.A09:108,105,828.108,110,202 | Aradu.RFN6Q |
| AdHsf8 | Aradu.A05:1,343,331.1,346,309 | Aradu.95F2Q |
| AdHsf3 | Aradu.A03:119,139,289.119,141,972 | Aradu.X3DNX |
| AdHsf13 | Aradu.A06:108,820,015.108,822,104 | Aradu.RY508 |
| AiHsf7 | Araip.B05:1,259,505.1,263,848 | Araip.T98BR |
| AiHsf12 | Araip.B05:146,768,456.146,770,030 | Araip.8V58J |
| AiHsf9 | Araip.B05:8,412,092.8,414,222 | Araip.90YM8 |
| AiHsf10 | Araip.B05:15,070,998.15,073,228 | Araip.K80UC |
| AiHsf1 | Araip.B01:137,050,298.137,053,902 | Araip.DCD5Q |
| AiHsf2 | Araip.B03:11,777,014.11,780,056 | Araip.U7I8R |
| AiHsf16 | Araip.B10:2,967,059.2,969,380 | Araip.4A18K |
| AiHsf15 | Araip.B09:145,996,176.145,998,569 | Araip.24AK5 |
| AiHsfX | Araip.B06:1,771,852.1,780,911 | Araip.A5C77 |
| AiHsf6 | Araip.B03:135,626,726.135,628,064 | Araip.2D8LN |
| AiHsf4 | Araip.B03:128,239,020.128,240,481 | Araip.3P0PJ |
| AiHsf5 | Araip.B03:889,869.893,296 | Araip.Z1XB6 |
| AiHsf11 | Araip.B05:137,352,977.137,354,281 | Araip.FU4GL |
| AiHsf3 | Araip.B03:120,142,696.120,145,308 | Araip.UGV6F |
| AiHsf8 | Araip.B05:1,265,312.1,268,368 | Araip.SMI4I |
| AiHsf14 | Araip.B09:130,155,536.130,159,602 | Araip.G2FC5 |
| AiHsf13 | Araip.B06:133,072,149.133,074,221 | Araip.B0RQS |
These gene ID could be searched on web (http://peanutbase.org/keyword_search).
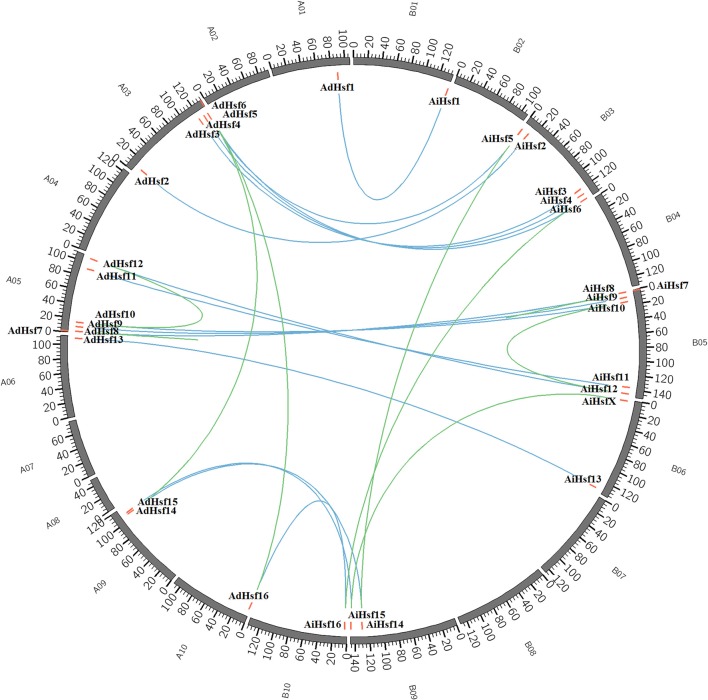
Figure 3.
Genome location of Hsfs in AA and BB genome peanuts. AA and BB wild peanut Hsfs were marked with red short lines on their genome location. Duplicated gene-pairs linked with green long curves and orthologous gene-pairs linked with blue curves.
Duplication of Hsf genes in peanut
Duplicated gene-pairs were found in both AA and BB genomes, including high-stringency standard duplicated gene-pairs AdHsf5-AdHsf14, AdHsf6-AdHsf16 in AA genome and AiHsf5-AiHsf14, AiHsf6-AiHsf16, AiHsf7-AiHsf8 in BB genome, low-stringency standard duplicated gene-pairs AdHsf7-AdHsf8 in AA genome and AiHsf15-AiHsfX in BB genome. Intraspecies synteny analysis showed that the duplicated gene-pair blocks were not collinear. No chromosome segmental or large scale duplication gene pairs were identified. AiHsf7-AiHsf8 and AdHsf7-AdHsf8 were identified as tandem duplicated gene-pairs.
Features of Hsfs in wild peanut species
Most members of Hsf gene families in both AA and BB genomes contained one intron and two exons. However, AdHsf7 contained three exons and AdHsf14 contained four exons in the AA genome, AiHsf15 contained three exons, AiHsf14, and AiHsfX contained four exons in the BB genome. AdHsf14 contained four exons, while its duplicated gene AdHsf5 contained only two exons. Intronless Hsfs were also found in both AA and BB genomes (Figure S1).
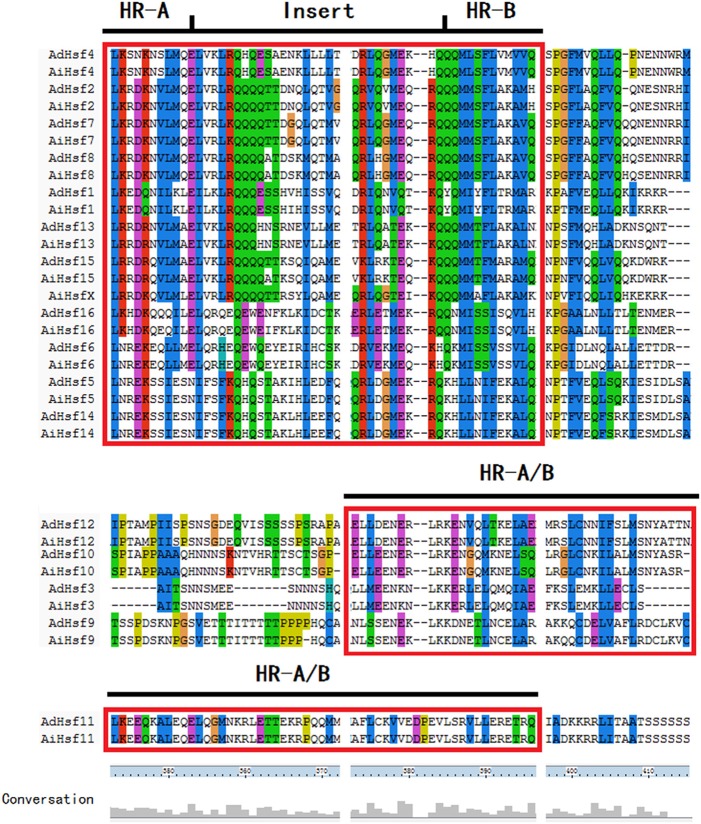
HR-A/B domain is critical for one Hsf interacting with other Hsfs to form trimer through a helical coiled-coil structure (Scharf et al., 2012; Jaeger et al., 2016; Neudegger et al., 2016). Similar to other plant Hsfs, group A Hsfs have an insertion between HR-A and HR-B regions in peanut. However, this insertion was not found in the group B Hsfs. In Arachis, the sequence of group B Hsf HR-A/B was not conserved compare with that in group A (Figure 4). The DBD domains were conserved in two wild peanut species. The most conserved motif of DBD domains were “FSSFI/VRQLNT/I” in peanut (Figure S2).
Figure 4.
HR-A/B domain in peanut Hsfs.
The 3D structure of Hsfs in wild peanut species
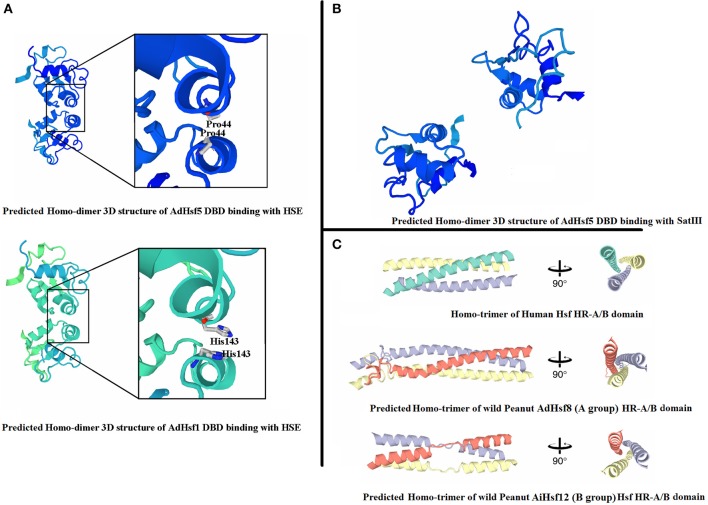
The predicted 3D structures of BDB domain of all AA and BB Hsfs were similar to that of human Hsf BDB (Figure 5A). The predicted 3D structures of HR-A/B domain of AA and BB Hsfs were also similar to the human Hsfs (Figure 5C). The 3D structures of BDB domain of peanut orthologous were highly conserved.
Figure 5.
3D structure of peanut Hsfs. (A) Represents 3D structures of peanut Hsf DBD domains binding with HSE. (B) Represents 3D structures of peanut Hsf DBD domains binding with SatIII. (C) Represents homo-trimer of Hsf HR-A/B domain.
When adjacent DBD molecules bound to HSE element, two DBD molecules formed symmetrical protein-protein interaction involving the helix α2. The closest intermolecular contact occurred between the Gly50 residues located at the N-terminal end of the α2 helices in chordate Hsfs. Gly50 is conserved and is surrounded by Gln49 and Gln51 in chordate Hsfs (Neudegger et al., 2016). In peanut, we predicted that the closest intermolecular contact residues by homologous comparison and 3D model comparison. The results showed that the closest contact residues were not conserved between chordate Hsfs and peanut Hsfs. For example, in AdHsf1 and AdHsf5, the predicted closest intermolecular contact occurred between the residues His143 (Figure 5A). We also built models that DBD domain of AA and BB wild peanut Hsfs bound to SatIII element. The result showed that the predicted dimer structures of DBD-DBD interaction for binding to SatIII element and HSE element were distinct (Figure 5B).
Selective pressure analysis of Hsfs in wild peanut species
Site models were used to detect whether different groups of Hsfs were under different selective pressure in peanuts. Group C Hsfs contained only one gene, it could not be analyzed. M0 showed that both AdHsfs and AiHsfs in group A underwent strong purifying selection (ω = 0.31723 in AA genome and ω = 0.40488 in BB genome; Table S4). Interestingly, in group B, both AdHsfs and AiHsfs were underwent positive selection (ω = 1.69713 in AA genome and ω = 1.95226 in BB genome). M0 vs. M3, M1a vs. M2a and M7 vs. M8 comparisons detected 399 positive selection sites in group B AdHsfs (P < 0.05) and 382 positive selection sites in group B AiHsfs (P < 0.001; Table S4). The identification of these positive selection sites in group B Hsfs indicated extensive functional diversity and structural variation (Wang et al., 2016).
Cis-acting regulatory element analysis of peanut Hsf promoter
In silico survey of the putative cis-acting regulatory elements in the 1500 kb promoter region of Hsfs was performed. The majority of Hsf promoters contained HSE elements. HSE was not found in AdHsf3, AdHsf12, AdHsf14, AiHsf1, or AdHsf11 promoters. Many Hsf promoters except AiHsf2 contained abiotic stress responsive element such as MBS (drought inducible), LTR (low temperature responsive), and ARE elements (anaerobic induction). RNA-seq data showed that two A. duranensis Hsf genes (Aradu.X3DNX, AdHsf3, and Aradu.5S8J3, AdHsf1) were up-regulated significantly under drought stress (log2 FC > 2, FDR < 0.05) (Guimarães et al., 2012; Brasileiro et al., 2015). Phytohormone-induced elements, such as ERE element (ethylene-responsive element), AuxRR-core or TGA-element (auxin responsive), GARE-motif, or P-box element (gibberellin-responsive), ABRE element (ABA responsive), TCA-element (salicylic acid responsive), and TGACG-motif or CGTCA-motif element (MeJA-responsive) were found in some Hsf promoters. Five AdHsfs (AdHsf2, AdHsf4, AdHsf16, AdHsf8, and AdHsf6) and two AiHsfs (AdHsf11 and AdHsf10) contained fungal elicitor responsive elements. Promoters of orthologous genes between AA and BB genomes were similar (Table S5).
Expression of Hsfs in various tissues in cultivated peanut
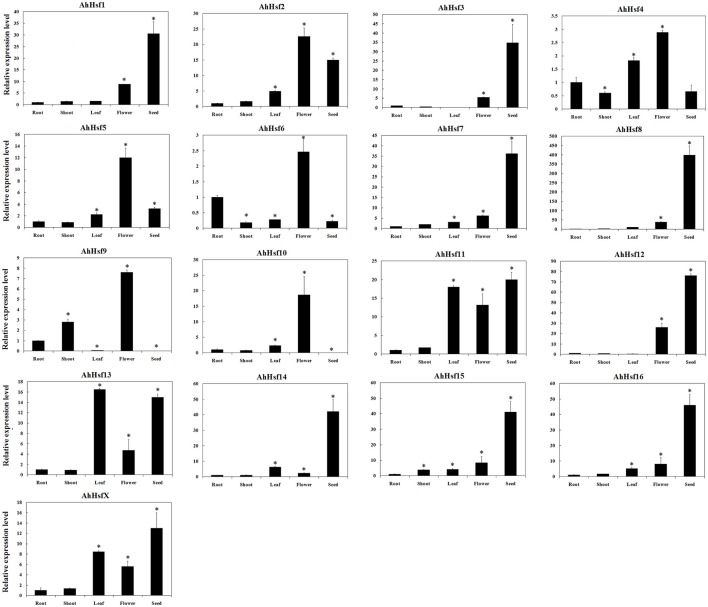
We used Hsfs of wild peanut species as queries to identify Hsfs in cultivated peanut species from transcriptome and genomic sequences (unpublished data). Totally, 17 Hsfs were identified in cultivated peanut species and named as AhHsf1- AhHsf16 and AhHsfX. The sequences of these genes were similar to their orthologous genes in wild peanut species (Table S6). To predict the possible function of these genes in cultivated peanut, the expression of these genes was investigated by qRT-PCR. Results showed that AhHsf1, 3, 7, 8, 11, 12, 14, 15, 16, and X were expressed predominantly in seeds, while the expression of AhHsf9 and 10 was not detected in seeds. AhHsf2, 4, 5, 6, 9, and 10 were highly expressed in flower. The expression of AhHsf1, 7, 12, 15, and 16 was higher in flower than that in root, shoot or leaf. The expression of AhHsf11, 13, and X was higher in leaf than that in root, shoot or flower. The expression of AhHsf4 and AhHsf6 was higher in root than that in shoot or seed. The expression level of AhHsf9 was higher in shoot than that in leaf or seed (Figure 6).
Figure 6.
Relative expression levels of Hsfs in different tissues in cultivated peanut. T-test was used to perform analysis of significance. * represents significantly difference (P < 0.05) compared with control (0 h).
Hsf expression in response to various stresses in cultivated peanut
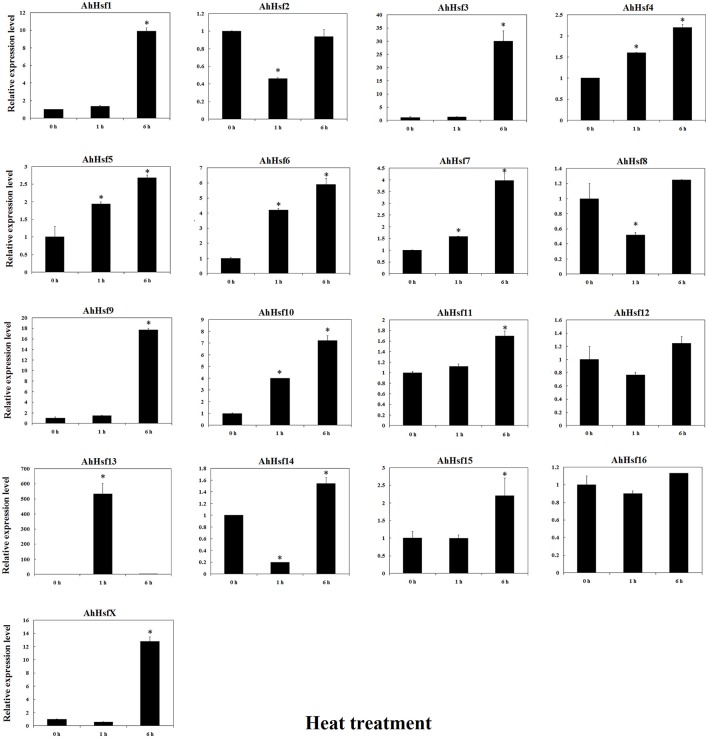
The expression of AhHsf was analyzed under high temperature, drought and low temperature by qRT-PCR. The expression levels of most Hsfs (AhHsf1, 3, 4, 5, 6, 7, 9, 10, 11, 13, 14, 15, and X) were up-regulated under high temperature. The expression of AhHsf1, 3, 9, 15, and X was up-regulated up to ~9-folds after 6 h treatment with 42°C. AhHsf4, 5, 6, 10, 11, and 13 could response rapidly to high temperature, and up-regulated after 1 h treatment. The expression of AhHsf4, 5, 6, 10, and 11 was continuously increased during 1–6 h of 42°C treatment. The expression of AhHsf13 was decreased at 6 h after 42°C treatment (Figure 7).
Figure 7.
Relative expression levels of Hsfs under heat stress in cultivated peanut. T-test was used to perform analysis of significance. * represents significantly difference (P < 0.05) compared with control (0 h).
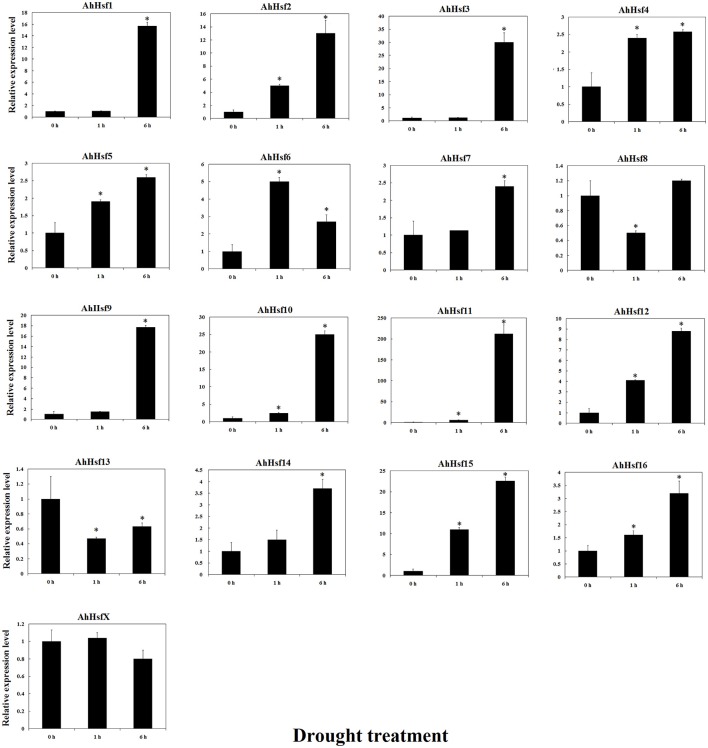
The expression of most AhHsfs was up-regulated under drought stress. The expression levels of AhHsf2, 4, 5, 7, 12, 14, 15, and 16 were increased after 1 h of drought treatment. The expression of AhHsf2, 5, 12, 14, 15, and 16 was continuously increased during the first 6 h of drought treatment. The expression of AhHsf1, 3, 9, 10, and 11 was up-regulated after 6 h of drought stress (~15-folds). AhHsfX didn't respond much to drought stress (Figure 8).
Figure 8.
Relative expression levels of Hsfs under drought stress in cultivated peanut. T-test was used to perform analysis of significance. * represents significantly difference (P < 0.05) compared with control (0 h).
The expression of most AhHsfs was up-regulated after 1 h of 4°C treatment, and then down-regulated at 6 h after treatment. The expression of AhHsf 12 was continuously up-regulated during 6 h of cold treatment. The expression of AhHsf14 was decreased at 1 h and then increased at 6 h after 4°C treatment (Figure S3).
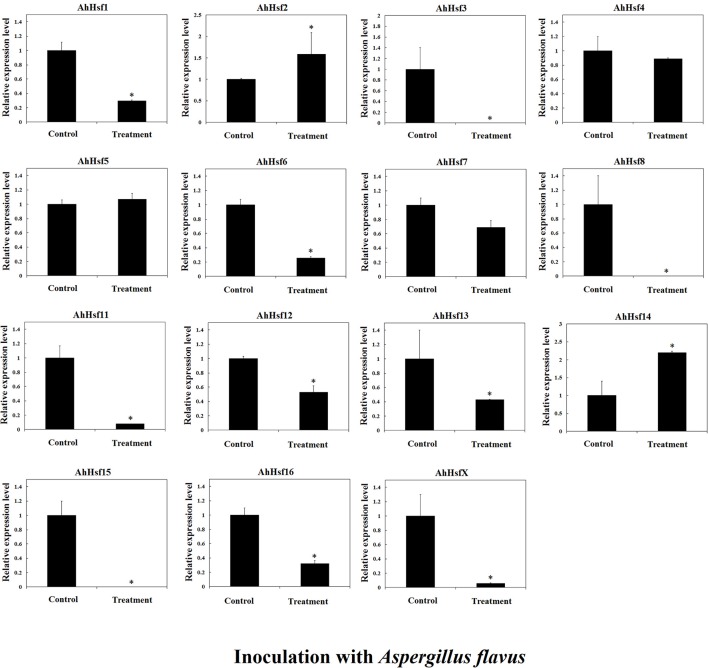
Previous study showed that Hsfs may be involved in disease resistance (Pick et al., 2012). In this study, we analyzed the expression of AhHsfs in peanut seeds after A. flavus infection. The expression of most AhHsfs was down-regulated in seed after A. flavus inoculation, while the expression of AhHsf2 and 14 was up-regulated (~1.5-fold; Figure 9).
Figure 9.
Relative expression variation of Hsfs in seeds inoculated with Aspergillus flavus in cultivated peanut. T-test was used to perform analysis of significance. * represents significantly difference (P < 0.05) compared with control (0 h).
Discussions
Leguminous contained different Hsf clusters
B5 cluster was not presented in Arabidopsis Hsfs, while B5 cluster was identified in most leguminous species, such as C. cajan, L. japonicus wild peanuts, and G. max. B5 Hsf cluster were not detected in Medicago truncatula. Phylogenetic tree showed that the leguminous plants contained different Hsf group members. Both in AA and BB wild peanut species, A3, A6a, A7, B3, and B4 cluster members were not found. Only soybean and M. truncatula contained the B3 members. A6a and A7 Hsf cluster was not found in leguminous. A3 cluster was not found in wild peanuts and M. truncatula. Group C Hsfs were not found in L. japonicus and M. truncatula. Soybean contained most clusters but not A6a and A7. The number of Hsfs from wild peanut species was relative small to compare with cotton, soybean and rosaceae (Li et al., 2014; Wang et al., 2014; Qiao et al., 2015). Phylogenetic tree showed that A. duranensis is the closer relative of A. ipaensis compared with other Leguminous.
WGD may not the major driving force of Hsfs large scale expansion in Arachis
Our results showed that Hsf gene duplication occurred in both AA and BB peanut genomes. The majority pf Hsf duplication events were similar between AA and BB genomes. For example, the duplicated gene pair AdHsf7-AdHsf8 was located on chromosome 5. The distance between AdHsf7 and AdHsf8 was about 1 kb. The duplicated gene pair AiHsf7-AiHsf8 was located on chromosome 5, and the distance between these two genes was about 2 kb. However, AiHsfX was located on chromosome 6 of BB peanut and its duplicated gene AiHsf15 was on chromosome 9 of BB peanut. It is possible that AdHsf15 didn't undergo duplication or the orthologous of AdHsfX was lost during the evolution (Figure 3). We only found one tandem duplication gene pairs in A. duranensis and A. ipaensis, respectively. Both AA and BB genomes or their common ancestor were underwent the early papilionoid whole-genome duplication (WGD) about 58 million year ago (Ks = 0.65) (Bertioli et al., 2016). Intraspecies synteny analysis showed that Hsf duplication in wild peanut species was not originated from a large scale duplication event, because no intraspecies synteny blocks containing Hsfs was found. However, the recent WGD could be a driving force for the expansion of Hsf gene family in Chinese white pear and apple (Qiao et al., 2015). That may be the reason why peanut has less Hsfs than that in cotton, soybean and rosaceae (Li et al., 2014; Wang et al., 2014; Qiao et al., 2015).
Hsfs is different in Group B from that in Group A
Group B Hsfs underwent positive selection (Table S4). Positive selection could contribute to adaptive evolution, functional diversity, and neofunctionalization (Beisswanger and Stephan, 2008). Study on barley showed that many gene families involved in adaptation to environment were under positive selection. Positive selection may lead to the expansion of these gene families (Zeng et al., 2015). However, group A Hsfs underwent purifying selection. Purifying selection may generate genes with conserved functions or pseudogenization (Zhang, 2003). These results indicated that the function of Arachis group A Hsfs may be more conserved and the function of group B Hsfs may be more diverged. The sequences of Hsf group B HR-A/B were not conserved compare with group A HR-A/B which was in agreement with the differential selection they experienced (Figure 4). The 3D structure of peanut group B Hsfs was different from group A and C Hsfs. The 3D structure of group A and C HR-A/B was a continuous helix, while group B HR-A/B 3D structure contained helixes which were linked by a linear part (Figure 5).
The possible roles of Arachis Hsfs in abiotic and biotic stresses
Hsfs play a central role in protecting plants from high temperature or other stresses (Nishizawa-Yokoi et al., 2009; Scharf et al., 2012). Many Hsfs could regulate a set of heat-shock protein genes to enhance the thermo-tolerance in plants. Some Hsfs could be regulated by DREB genes as part of drought stress signaling pathway, and enhance drought tolerance (Scharf et al., 2012; Ma et al., 2015; Guo et al., 2016). Some Hsfs could regulate WRKY transcription factors which are involved in response to abiotic stresses, such as drought and cold (Ren et al., 2010; Zou et al., 2010; Jiang et al., 2012; Shen et al., 2015). Arabidopsis HsfA9 could be activated by ABI3 to enhance seed desiccation tolerance and longevity (Verdier et al., 2013).
In our study, the majority of Hsf promoters contained HSE elements (Table S5) suggesting that peanut Hsfs could be regulated by other Hsfs. Many peanut Hsf promoters contained MYB binding sites which are involved in drought response (Table S5). It indicated that peanut Hsfs could be regulated by MYB transcription factors under drought stress. Many Arachis Hsf promoters contained ABRE and DRE elements which are involved in ABA-dependent or independent stress tolerance (Chen et al., 2012). Therefore, Hsfs could play important roles for gene regulation in response to different stresses in peanuts. Some Arachis Hsf promoters contained salicylic acid responsive, MeJA-responsive or fungal elicitor responsive elements, suggesting their roles in response to pathogen infection.
In cultivated peanut cultivars, the expression level of AhHsf13 was approximately 500-folds as high as the control after 1 h of heat treatment, and then the expression was decreased after 6 h of treatment. Expression levels of AhHsf1, 3, 9, and AhHsfX were up-regulated by about 10-folds after 6 h of heat treatment to compare with the control. The expression of these Hsfs kept at a high level under continuous heat stress (Figure 7). Group A1a Hsfs were master regulators for acquired thermo tolerance in tomato and Arabidopsis (Scharf et al., 2012). However, we found that the expression of AhHsf2 (group A1) did not respond to heat and cold, but to drought stress. In cultivated peanut, expression levels of AhHsf1, 2, 3, 9, 10, 11, 15 were about 10-folds as high to compare with the control after 6 h of drought stress (Figure 8). The expression of some Hsfs was altered after Podosphaera aphanis inoculation in woodland strawberry (Hu et al., 2015). Aspergillus flavus produces potent mycotoxin known as aflatoxin which is a key issue of food safety in peanut (Zhang et al., 2015). We detected whether peanut Hsf genes were involved in the response to A. flavus infection. The results showed that the expression of AhHsf2 and AhHsf14 were significantly up-regulated after A. flavus inoculation. The expression of some AhHsfs was down-regulated by A. flavus infection.
Hsf gene family were highly expressed in peanut seed
Some Hsfs play key roles in plant seed development (Wang et al., 2014). In sunflower and Arabidopsis, HsfA9 was expressed specifically in seeds and the expression of Hsps was changed during seed development (Almoguera et al., 2002; Kotak et al., 2007b). In rice, HsfA7 was expressed specifically in seed under normal condition (Chauhan et al., 2011). In peanut, expression levels of more than half of the AhHsfs were higher in seeds than that in other tissues. These expression patterns may suggest their roles in peanut seed development.
Conclusions
Genome-wide identification and comparison of peanut Hsfs with other plant species revealed that peanut contained a small number of Hsfs. Phylogenetic tree showed that B5 cluster Hsfs might present only in leguminous. Small scale segmental and tandem duplication but not WGD played important roles in Hsfs expansion in Arachis. The sequences of group B Hsf HR-A/B were not conserved compare with group A HR-A/B which was in agreement with the different selection pressure they experienced. We built the 3D structures of peanut Hsfs with the newly submitted templates and found the difference between group A and B members. Peanut Hsfs may play important roles in abiotic and biotic stress tolerance based on their expression responses to these stresses.
Author contributions
XW designed the study, wrote the manuscript and finalized the figures and tables. PW and LH carried out the majority of experiments, data analysis, and wrote the method section of the manuscript. HS, CL, PL, AL, and HG performed experiments.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by the National grants (31500217; 31471526; 2013AA102602; 31500257), Shandong provincial grants (BS2013SW006; BS2014SW017; ZR2015CQ012; ZR2015YL061), Agricultural scientific, and technological innovation project (CXGC2016C08) and Young Talents Training Program of Shandong Academy of Agricultural Sciences.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00106/full#supplementary-material
Structure of peanut Hsfs.
DBD domain in peanut Hsfs.
Relative expression levels of Hsfs under cold stress in cultivated peanut. T-test was used to perform analysis of significance. “*” represents significantly difference (P < 0.05) compared with control (0 h).
Hsfs identified in other species.
Primers of cultivated peanut Hsfs for qRT-PCR.
Identity of wild peanut Hsf orthologous.
Likelihood values and parameter estimate for wild peanut Hsfs.
Cis-acting regulatory elements of wild peanut Hsf promoters.
Identity of wild and cultivated peanut Hsfs.
References
- Akerfelt M., Morimoto R. I., Sistonen L. (2010). Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 11, 545–555. 10.1038/nrm2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almoguera C., Rojas A., Diaz-Martin J., Prieto-Dapena P., Carranco R., Jordano J. (2002). A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. J. Biol. Chem. 277, 43866–43872. 10.1074/jbc.M207330200 [DOI] [PubMed] [Google Scholar]
- Al-Whaibi M. H. (2011). Plant heat-shock proteins: a mini review. J. King Saud Univ. Sci. 23, 139–150. 10.1016/j.jksus.2010.06.022 [DOI] [Google Scholar]
- Baniwal S. K., Bharti K., Chan K. Y., Fauth M., Ganguli A., Kotak S., et al. (2004). Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29, 471–487. 10.1007/BF02712120 [DOI] [PubMed] [Google Scholar]
- Beisswanger S., Stephan W. (2008). Evidence that strong positive selection drives neofunctionalization in the tandemly duplicated polyhomeotic genes in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 5447–5452. 10.1073/pnas.0710892105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertioli D. J., Cannon S. B., Froenicke L., Huang G., Farmer A. D., Cannon E. K., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. 10.1038/ng.3517 [DOI] [PubMed] [Google Scholar]
- Brasileiro A. C., Morgante C. V., Araujo A. C., Leal-Bertioli S. C., Silva A. K., Martins A. C., et al. (2015). Transcriptome profiling of wild arachis from water-limited environments uncovers drought tolerance candidate genes. Plant Mol. Biol. Rep. 33, 1876–1892. 10.1007/s11105-015-0882-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan H., Khurana N., Agarwal P., Khurana P. (2011). Heat shock factors in rice (Oryza sativa L.): genome-wide expression analysis during reproductive development and abiotic stress. Mol. Genet. Genomics 286, 171–187. 10.1007/s00438-011-0638-8 [DOI] [PubMed] [Google Scholar]
- Chen L., Chen Y., Jiang J., Chen S., Chen F., Guan Z., et al. (2012). The constitutive expression of Chrysanthemum dichrum ICE1 in Chrysanthemum grandiflorum improves the level of low temperature, salinity and drought tolerance. Plant Cell Rep. 31, 1747–1758. 10.1007/s00299-012-1288-y [DOI] [PubMed] [Google Scholar]
- Chung E., Kim K. M., Lee J. H. (2013). Genome-wide analysis and molecular characterization of heat shock transcription factor family in Glycine max. J. Genet. Genomics 40, 127–135. 10.1016/j.jgg.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Dalton T. P., Solis W. A., Nebert D. W., Carvan M. J. (2000). Characterization of the MTF-1 transcription factor from zebrafish and trout cells. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 126, 325–335. 10.1016/S0305-0491(00)00180-2 [DOI] [PubMed] [Google Scholar]
- Díaz-Martín J., Almoguera C., Prieto-Dapena P., Espinosa J. M., Jordano J. (2005). Functional interaction between two transcription factors involved in the developmental regulation of a small heat stress protein gene promoter. Plant Physiol. 139, 1483–1494. 10.1104/pp.105.069963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring P., Treuter E., Kistner C., Lyck R., Chen A., Nover L. (2000). The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12, 265–278. 10.2307/3870927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas F. O., Moretzsohn M. C., Valls J. F. (2007). Genetic variability of Brazilian Indian landraces of Arachis hypogaea L. Genet. Mol. Res. 6, 675–684. [PubMed] [Google Scholar]
- Giorno F., Guerriero G., Baric S., Mariani C. (2012). Heat shock transcriptional factors in Malus domestica: identification, classification and expression analysis. BMC Genomics 13:639. 10.1186/1471-2164-13-639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady D. L., Ratliff R. L., Robinson D. L., McCanlies E. C., Meyne J., Moyzis R. K. (1992). Highly conserved repetitive DNA sequences are present at human centromeres. Proc. Natl. Acad. Sci. U.S.A. 89, 1695–1699. 10.1073/pnas.89.5.1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin M. J., Martins A. L., Siepel A., Lis J. T. (2012). Accurate prediction of inducible transcription factor binding intensities in vivo. PLoS Genet. 8:e1002610. 10.1371/journal.pgen.1002610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães P. M., Brasileiro A. C., Morgante C. V., Martins A. C., Pappas G., Silva O. B., et al. (2012). Global transcriptome analysis of two wild relatives of peanut under drought and fungi infection. BMC Genomics 13:387. 10.1186/1471-2164-13-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Liu J., Ma X., Luo D., Gong Z., Lu M. (2016). The plant heat stress transcription factors (HSFs): structure, regulation and function in response to abiotic stresses. Front. Plant Sci. 7:114. 10.3389/fpls.2016.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U., Hayer-Hartl M. (2002). Molecular chaperones in the cytosol: from nascent chain to foldedprotein. Science 295, 1852–1858. 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- Hsu A. L., Murphy C. T., Kenyon C. (2003). Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142–1145. 10.1126/science.1083701 [DOI] [PubMed] [Google Scholar]
- Hu Y., Han Y.-T., Wei W., Li Y.-J., Zhang K., Gao Y.-R., et al. (2015). Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 6:736. 10.3389/fpls.2015.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger A. M., Pemble C. W., Sistonen L., Thiele D. J. (2016). Structures of HSF2 reveal mechanisms for differential regulation of human heat-shock factors. Nat. Struct. Mol. Biol. 23, 147–154. 10.1038/nsmb.3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. J., Liang G., Yu D. Q. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5, 1375–1388. 10.1093/mp/sss080 [DOI] [PubMed] [Google Scholar]
- Kochert G., Stalker H. T., Gimenes M., Galgaro L., Romero-Lopes C., Moore K. (1996). RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut, Arachis hypogaea (Leguminosae). Am. J. Bot. 83, 1282–1291. [Google Scholar]
- Kotak S., Larkindale J., Lee U., von Koskull-Döring P., Vierling E., Scharf K.-D. (2007a). Complexity of the heat stress response in plants. Curr. Opin. Plant Biol. 10, 310–316. 10.1016/j.pbi.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Kotak S., Port M., Ganguli A., Bicker F., von Koskull-Döring P. (2004). Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 39, 98–112. 10.1111/j.1365-313X.2004.02111.x [DOI] [PubMed] [Google Scholar]
- Kotak S., Vierling E., Bäumlein H., von Koskull-Döring P. (2007b). A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19, 182–195. 10.1105/tpc.106.048165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps J. A., Wu Y., Chang H. S., Zhu T., Wang X., Harper J. F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130, 2129–2141. 10.1104/pp.008532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. S., Yu T. F., He G. H., Chen M., Zhou Y. B., Chai S. C., et al. (2014). Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genomics 15:1009. 10.1186/1471-2164-15-1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Cheng Y., Jin J., Jin X., Jiang H., Yan H., et al. (2014). Genome duplication and gene loss affect the evolution of heat shock transcription factor genes in legumes. PLoS ONE 9:e102825. 10.1371/journal.pone.0102825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Wang C., Yang B., Cheng H., Wang Z., Mijiti A., et al. (2015). CarHSFB2, a Class B heat shock transcription factor, is involved in different developmental processes and various stress responses in chickpea (Cicer Arietinum L.). Plant Mol. Biol. Rep. 34, 1–14. 10.1007/s11105-015-0892-8 [DOI] [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459. 10.1073/pnas.0501102102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo M. L., Santagata S., Koeva M., Bell G., Rong H., Tamimi R. M., et al. (2012). HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562. 10.1016/j.cell.2012.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretzsohn M. C., Gouvea E. G., Inglis P. W., Leal-Bertioli S. C. M., Valls J. F. M., Bertioli D. J. (2013). A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers. Ann. Bot. 111, 113–126. 10.1093/aob/mcs237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudegger T., Verghese J., Hayer-Hartl M., Hartl F. U., Bracher (2016). A. Structure of human heat-shock transcription factor 1 in complex with DNA. Nat. Struct. Mol. Biol. 23, 140–146. 10.1038/nsmb.3149 [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A., Yoshida E., Yabuta Y., Shigeoka S. (2009). Analysis of the regulation of target genes by an Arabidopsis heat shock transcription factor, HsfA2. Biosci. Biotechnol. Biochem. 73, 890–895. 10.1271/bbb.80809 [DOI] [PubMed] [Google Scholar]
- Nover L., Bharti K., Döring P., Mishra S. K., Ganguli A., Scharf K.-D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R. B. (1982). A regulatory upstream promoter element in the Drosophila hsp70 heat-shock gene. Cell 30, 517–528. 10.1016/0092-8674(82)90249-5 [DOI] [PubMed] [Google Scholar]
- Peteranderl R., Rabenstein M., Shin Y. K., Liu C. W., Wemmer D. E., King D. S., et al. (1999). Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry 38, 3559–3569. 10.1021/bi981774j [DOI] [PubMed] [Google Scholar]
- Pick T., Jaskiewicz M., Peterhansel C., Conrath U. (2012). Heat shock factor HsfB1 primes gene transcription and systemic acquired resistance in Arabidopsis. Plant Physiol. 159, 52–55. 10.1104/pp.111.191841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X., Li M., Li L., Yin H., Wu J., Zhang S. (2015). Genome-wide identification and comparative analysis of the heat shock transcription factor family in Chinese white pear (Pyrus bretschneideri) and five other Rosaceae species. BMC Plant Biol. 15:12. 10.1186/s12870-014-0401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu V. S., Swetha T. N., Sheela S. H., Babitha C. K., Rohini S., Reddy M. K., et al. (2015). Simultaneous expression of regulatory genes associated with specific drought-adaptive traits improves drought adaptation in peanut. Plant Biotechnol. J. 14, 1008–1020. 10.1111/pbi.12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. Z., Chen Z. Z., Liu Y., Zhang H. R., Zhang M., Liu Q. A., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. 10.1111/j.1365-313X.2010.04248.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa F. (1962). A new puffing pattern induced by temperature shock and DNP in Drosophila. Experimentia 18, 571–573. 10.1007/BF02172188 [DOI] [Google Scholar]
- Rizzon C., Ponger L., Gaut B. S. (2006). Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput. Biol. 2:e115. 10.1371/journal.pcbi.0020115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Nakamura Y., Kaneko T., Asamizu E., Kato T., Nakao M., et al. (2008). Genome structure of the legume, Lotus japonicus. DNA Res. 15, 227–239. 10.1093/dnares/dsn008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K. D., Berberich T., Ebersberger I., Nover L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 1819, 104–119. 10.1016/j.bbagrm.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Schmidt R., Schippers J. H., Welker A., Mieulet D., Guiderdoni E., Mueller-Roeber B. (2012). Transcription factor OsHsfC1b regulates salt tolerance and development in Oryza sativa ssp. japonica. AoB Plants 2012:pls011. 10.1093/aobpla/pls011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C., Bevan M. (1998). The regulation of transcription factor activity in plants. Trends Plant Sci. 3, 378–383. 10.1016/S1360-1385(98)01302-8 [DOI] [Google Scholar]
- Shen Z., Yao J., Sun J., Chang L., Wang S., Ding M., et al. (2015). Populus euphratica HSF binds the promoter of WRKY1 to enhance salt tolerance. Plant Sci. 235, 89–100. 10.1016/j.plantsci.2015.03.006 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. 10.1093/jxb/erl164 [DOI] [PubMed] [Google Scholar]
- Swindell W. R., Huebner M., Weber A. P. (2007). Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8:125. 10.1186/1471-2164-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M., Nakai A., Kawaoe Y., Nagata K. (1997). Different thresholds in the responses of two heat shock transcription factors, HSF1 and HSF3. J. Biol. Chem. 272, 15389–15395. 10.1074/jbc.272.24.15389 [DOI] [PubMed] [Google Scholar]
- Verdier J., Lalanne D., Pelletier S., Torres-Jerez I., Righetti K., Bandyopadhyay K., et al. (2013). A regulatory networkbased approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 163, 757–774. 10.1104/pp.113.222380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A., Sergelius C., Vasara J., Blom M. A. H., Elsing A. N., Roos-Mattjus P., et al. (2013). Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc. Natl. Acad. Sci. U.S.A. 110, E3388–E3397. 10.1073/pnas.1305275110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Koskull-Döring P., Scharf K.-D., Nover L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. 10.1016/j.tplants.2007.08.014 [DOI] [PubMed] [Google Scholar]
- Wang F., Dong Q., Jiang H., Zhu S., Chen B., Xiang Y. (2012). Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol. Biol. Rep. 39, 1877–1886. 10.1007/s11033-011-0933-9 [DOI] [PubMed] [Google Scholar]
- Wang J., Sun N., Deng T., Zhang L., Zuo K. (2014). Genome-wide cloning, identification, classification and functional analysis of cotton heat shock transcription factors in cotton (Gossypium hirsutum). BMC Genomics 15:961. 10.1186/1471-2164-15-961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Gao C., Bian X., Zhao S., Zhao C., Xia H., et al. (2016). Genome-wide identification and comparative analysis of cytosine-5 DNA methyltransferase and demethylase families in wild and cultivated peanut. Front. Plant Sci. 7:7. 10.3389/fpls.2016.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Vinocur B., Shoseyov O., Altman A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. 10.1016/j.tplants.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Xue G. P., Sadat S., Drenth J., McIntyre C. L. (2014). The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 65, 539–557. 10.1093/jxb/ert399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yang Z., Nielsen R., Goldman N., Pedersen A. M. (2000). Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wang Y., Gao Y., Zhou Y., Zhang E., Hu Y., et al. (2014). Adaptive evolution and divergent expression of heat stress transcription factors in grasses. BMC Evol. Biol. 14:147. 10.1186/1471-2148-14-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S. X., Xu B., Zhang J., Xie Z. N., Cheng Q., Yang Z., et al. (2015). Comprehensive analysis of CCCH-type zinc finger family genes facilitates functional gene discovery and reflects recent allopolyploidization event in tetraploid switchgrass. BMC Genomics 16:129. 10.1186/s12864-015-1328-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Long H., Wang Z., Zhao S., Tang Y., Huang Z., et al. (2015). The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau. Proc. Natl. Acad. Sci. U.S.A. 112, 1095–1100. 10.1073/pnas.1423628112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Scharfenstein L., Zhang D., Chang P., Montalbano B. G., Guo B., et al. (2015). Peanut resistance gene expression in response to Aspergillus flavus infection during seed germination. J Phytopathol. 163, 212–221. 10.1111/jph.12311 [DOI] [Google Scholar]
- Zhang J. Z. (2003). Evolution by gene duplication: an update. Trends Ecol. 18, 292–298. 10.1016/s0169-5347(03)00033-8 [DOI] [Google Scholar]
- Zhang X., Feng Y., Cheng H., Tian D., Yang S. (2011). Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol. Genet. Genomics 285, 79–90. 10.1007/s00438-010-0587-7 [DOI] [PubMed] [Google Scholar]
- Zou C. S., Jiang W. B., Yu D. Q. (2010). Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 61, 3901–3914. 10.1093/jxb/erq204 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure of peanut Hsfs.
DBD domain in peanut Hsfs.
Relative expression levels of Hsfs under cold stress in cultivated peanut. T-test was used to perform analysis of significance. “*” represents significantly difference (P < 0.05) compared with control (0 h).
Hsfs identified in other species.
Primers of cultivated peanut Hsfs for qRT-PCR.
Identity of wild peanut Hsf orthologous.
Likelihood values and parameter estimate for wild peanut Hsfs.
Cis-acting regulatory elements of wild peanut Hsf promoters.
Identity of wild and cultivated peanut Hsfs.