Abstract
Background
Rapid dose assessment using biological dosimetry methods is essential to increase the chance of survival of exposed individuals in radiation accidents.
Objectives
We compared the expression levels of the FDXR and RAD51 genes at 6 and 18 MV beam energies in human peripheral blood lymphocytes. The results of our study can be used to analyze radiation energy in biological dosimetry.
Methods
For this in vitro experimental study, from 36 students in the medical physics and virology departments, seven voluntary, healthy, non-smoking male blood donors of Khuzestan ethnicity with no history of exposure to ionization radiation were selected using simple randomized sampling. Sixty-three peripheral blood samples were collected from the seven healthy donors. Human peripheral blood was then exposed to doses of 0, 0.2, 0.5, 2, and 4 Gy with 6 and 18 MV beam energies in a Linac Varian 2100C/D (Varian, USA) at Golestan hospital in Ahvaz, Iran. After RNA extraction and cDNA synthesis, the expression levels of FDXR and RAD51 were determined 24 hours post-irradiation using the gel-purified reverse transcription polymerase chain reaction (RT-PCR) technique and TaqMan strategy (by real-time PCR).
Results
The expression level of FDXR gene was significantly increased at doses of 2 Gy and 4 Gy in the 6 - 18 MV energy range (P < 0.001 and P < 0.008, respectively). The medians with interquartile ranges (IQRs) of the copy numbers of the FDXR gene at 2 Gy and 4 Gy doses under 6 and 18 MV beam energies were 2393.59 (1798.21, 2575.37) and 2983.00 (2199.48, 3643.82) and 3779.12 (3051.40, 5120.74) and 5051.26 (4704.83, 5859.17), respectively. However, RAD51 gene expression levels only showed a significant difference between samples at a dose of 2 Gy with 6 and 18 MV beam energies, respectively (P < 0.040). The medians with IQRs of the copy numbers of the RAD51 gene were 2092.77 (1535.78, 2705.61) and 3412.57 (2979.72, 4530.61) at beam energies of 6 and 18 MV, respectively.
Conclusions
The data suggest that the expression analysis of the FDXR gene, contrary to that of the RAD51 gene, may be suitable for assessment of high-energy X-ray. In addition, RAD51 is not a suitable gene for dose assessment in biological dosimetry.
Keywords: Lymphocytes, Dose-Response Relationship, X-Rays
1. Background
Biological dosimetry involves biological and biophysical methods to estimate the level of radiation exposure of an accident victim in the absence of physical dosimetry or after large-scale radiological events like the radiation accident in Chernobyl (1, 2). Progress in biological dosimetry via rapid dose assessment is essential to increase survival through medical intervention (3-5). The effect of partial body exposure, dose rate, and radiation energy on radiation biological dosimetry has not been studied in the context of radiological or nuclear accidents. Thus, it is of great importance to design studies for the prediction of the absorbed dose in exposed people (6, 7).
Cytogenetic techniques, such as dicentric assay, are time-consuming and cannot be used on the scale that would be necessary in a mass casualty situation (7); however, gene expression biodosimetry is not a laborious approach. The change in the quantity of RNA transcripts in cells exposed to ionizing radiation suggests that gene expression analysis may enable quantitative dose assessment (8, 9). Up to now, many studies have been conducted to develop a gene expression profiling strategy, but there have been few studies on different energies of ionization radiation (10, 11).
Ionization radiations, such as megavoltage X-ray and neutron radiation, interact with DNA and generate several double-strand breaks (DSBs), cause mitotic death, and change the copy number of mRNA in some genes (12). DSBs are considered to represent important radiation-induced DNA damage that can result in cell death. DNA damage is critically affected by radiation type and energy (13).
It is clear that accurate dose estimation can be achieved via the analysis of the genes involved in different pathways, such as cell cycle regulation, DNA repair, and apoptosis (14, 15). Suitable genes show a linear dose-response relationship between the RNA transcription level and the absorbed dose. Microarray is used to find these genes, and the results are verified by real-time polymerase chain reaction (PCR) (4, 8, 16, 17). The FDXR gene is a suitable gene in biological dosimetry with a linear dose response, while RAD51 is the most important gene for DNA repair in the homologous recombination (HR) mechanism.
2. Objectives
In this study, the response of the FDXR and RAD51 genes to different doses (0, 0.2, 0.5, 2, and 4 Gy) with beam energies of 6 and 18 MV was recorded to draw the dose-response curve in human peripheral blood.
3. Methods
3.1. Blood Collection and Ethics
In this in vitro experimental study, from 36 students in the medical physics and virology departments, 7 healthy non-smoking male voluntary blood donors of Khuzestan ethnicity who had no history of exposure to ionization radiation were selected using simple randomized sampling. Sixty-three peripheral blood samples were collected from the healthy donors (aged 30 - 35 years). An informed consent form was signed by all participants from Khuzestan Province of Iran. The samples were transferred into K3 EDTA-coated tubes (FL, Italy). This work was approved by the medical ethics committee of Ahvaz Jundishapur University of Medical Sciences (ethical code no. U-92133), Iran. The study was conducted in the virology department at this university from 2013 to 2014.
3.2. Irradiation Conditions and Cell Culture
All samples were irradiated in a Linac Varian 2100C/D (Varian, USA) in Golestan hospital in Ahvaz (Ahvaz Jundishapur University of Medical Sciences) with a dose rate of 0.3 Gy/min. Sixty-three samples (7 samples per group) were treated and conditioned for the in vitro calibration curve of total body exposure with 0, 0.2, 0.5, 2, and 4 Gy doses at 6 and 18 MV of energy. Dosimetry conditions of all samples included a source-to-surface distance of 100 cm and a field size of 20 × 20 cm.
3.3. RNA Extraction
RNA was prepared from 500 µL of peripheral blood using a High Pure RNA Isolation Kit (Roche, Korea) following the manufacturer’s recommendations (for 500 µL of peripheral blood). The quantification of RNA was performed using a NanoDrop-2000 spectrophotometer (Thermo, USA) by analyzing A260/A280 and A260/A230.
3.4. cDNA Synthesis
Total RNA (0.7 µg) was reverse transcribed to cDNA using a High-Capacity cDNA Archive Kit (Thermo, US) according to the manufacturer’s instructions. The quality of cDNA synthesis was evaluated by reverse transcription (RT)-PCR and agarose gel electrophoresis. Samples with a sharp band of expected size in the gel were used for amplification by real-time PCR.
3.5. Homemade Standard Construction and Gel Purification for a Standard Curve
Standard samples were prepared using PCR amplification of total cDNA from lymphocytes using FDXR and RAD51 primers. The PCR products were loaded onto 2% agarose gels with 50 and 100 bp markers for RAD51 and FDXR, respectively. Then, size bands for FDXR (85 bp) and RAD51 (150 bp) were excised from the agarose gels and purified (Bioneer, Daejoon, South Korea). The final concentration of standards was determined using Nanodrop. Ten serial dilutions were prepared (ranging from 102 to 106 FDXR and RAD51 mRNA copies per µL). The PCR reaction volume was 20 µL, and the range of copy numbers/reactions was 5 to 2 × 105.
The concentrations of gel purification products for FDXR and RAD51 genes were 13 ng/µL and 17 ng/µL, respectively. The molecular weights of each copy number from the FDXR gene were 52567.77 (8.729035912 × 10-11 ng) and 68003.02 daltons (1.13 × 10-10 ng), respectively. The schematic representation of the experiment is shown in Appendix 1 in Supplementary File Figure 1.
Figure 1. Typical Gel Electrophoresis Data and an Amplification curve for the RAD51 Gene.
The specific PCR products were confirmed using a 50 bp molecular weight marker for RAD51 (lane 1). RAD51-specific target products were used as the first to fifth concentrations from serial dilutions of standards (lanes 2 - 7).
3.6. Quantitative Real-Time PCR
PCR amplification was scored as successful using a step one calibrated real-time PCR machine (Applied Bio Systems). The cycling conditions for the FDXR gene were as follows: 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 20 seconds and 60°C for 1 minute. Primers and probes were purchased from Bioneer Inc. (Daejeon, South Korea). The cycling conditions for RAD51 gene were as follows: 48°C for 10 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 20 seconds and 60°C for 1 minute. The probes were synthesized using 6-carboxyfluorescein (FAM) at the 5ˊ end and a TAMRA quencher at the 3ˊ end.
All reactions were carried out in duplicate using Takara master mix (Japan) with the primer and probe set for FDXR and RAD51 genes at a concentration of 200 nM and 1 µL of cDNA in a 20 µL reaction volume. The designed primers and probes are shown in Table 1. Cycle threshold (Ct) values were converted to transcript quantity using standard curves obtained by serial dilution of PCR-amplified cDNA fragments of the FDXR and RAD51 genes. The internal control gene was 18S. 18S and GAPDH have been identified as the most suitable endogenous control genes for gene expression in biological dosimetry, and the latter was used in this study (18). Briefly, the changes in quantity of the RNA transcripts were measured in the cells exposed to ionizing radiation by real-time PCR.
Table 1. Oligonucleotide Primers and Probes.
| Gene | Accession. No | PCR Primers | Probe | Amplicon Length, bp |
|---|---|---|---|---|
| FDXR | NM-024417 | GTCCGTCTGAGTGGGACTTT (Fwd) | CACCTCTGCTGATCCCG | 85 |
| GAGGAGAGACGCTGGAAGAG (Rev) | GCC | |||
| RAD51 | NM-024153 | GAGGTGAAGGAAAGGCCATGT (Fwd) | TGGCTGAGAGGTATGG | 150 |
| GGGTCTGGTGGTCTGTGTTGA (Rev) | TCTCTCTGGCA | |||
| 18S | NR-046237 | CCTGCCAAGTATGATGACATCAAGA (Fwd) | TGGTGAAGCAGGCGGC | 187 |
| GTAGCCCAGGATGCCCTTTAGT (Rev) | CGAG |
3.7. Statistical Analysis
Curve fitting was carried out to fit the mean experimental values using standard regression analysis programs in Microsoft Excel 2007 (Microsoft Corporation, Redmond, WA, USA), and all the statistical tests were done using the Statistical Package for Social Sciences, version 16.0 (SPSS, Inc., Chicago, IL, USA). The Mann-Whitney non-parametric U test was then applied to present the gene expression levels of the FDXR and RAD51 genes under 6 and 18 MV beam energies.
4. Results
4.1. Quantitative Real-Time PCR Data
Demographic data on healthy donors are listed in Table 2. Quantitative real-time PCR was conducted for all samples and plotted as an average response ratio. The results are indicated as copy number/reaction. The PCR product was confirmed by size using electrophoretic separation on 2% agarose gel (Figure 1). The linear dynamic range of standard curves included five orders of magnitude (from × 102 to × 106 standard cDNA copy number). For all runs, the PCR efficiency was between 89% and 106% for FDXR and RAD51 genes, R2 > 0.98. The Ct for FDXR and RAD51 genes was started at 18 and 21 cycles for the first concentration of standard samples and was finished at 32 and 34 cycles for the last concentration. Typical data from a real-time PCR run are shown in Appendix 2 in Supplementary File, Figure 2.
Table 2. Demographic Characteristics of the Participants (n = 7)a,b.
| Variables | Value (n = 7) |
|---|---|
| Weight, kg | 81.58 ± 3.83 |
| Age, y | 33.02 ± 2.32 |
| Ethnicity | Khuzestan province of Iran |
| Body Mass Index (BMI) | 23.5 ± 1.5 |
| Gender | Male |
aValues are expressed as mean ± standard deviation (SD) or No. (%).
bThe inclusion criteria in this study included healthy donors with no history of exposure to ionization radiation and who were non-smokers; The gene expression level in human peripheral blood was compared before and after irradiation; therefore, there were no confounding factors in this experimental study; The blood samples were collected from students in the Medical Physics and Virology departments at the Ahvaz Jundishapour University of Medical Sciences, Ahvaz, Iran.
Figure 2. Real-Time PCR Data.
A, The specific PCR products were confirmed using a 100 bp molecular weight marker (lane 6); five samples with dose of 0.5 Gy at both energies are shown (lanes 1 - 5); B, PCR amplification curve for the RAD51 gene with five concentrations from tenfold serial dilutions.
4.2. In Vitro Dose-Response Curve of Blood Irradiation Under 6 and 18 MV Energy Beams
The gene expression levels of FDXR and RAD51 were determined for samples 24 hours after irradiation with 0, 0.2, 0.5, 2, and 4 Gy doses with 6 and 18 MV energy beams. The median and IQR of the sample responses for FDXR and RAD51 at all doses are shown in Table 3. The best fits for the dose-response curves of both genes were polynomial equations (Figures 3 and 4). The results of regression analysis as polynomial equation were fitted to the dose-response curves of the FDXR and RAD51 genes, and goodness of fit (R2) is presented in Table 4. Moreover, the p-values for significant differences are shown for the same doses for 6 and 18 MV beam energies.
Table 3. Real-Time PCR Data From the Irradiated Samples with a 6 MV Energy Beam: Cycle Thresholds (Cts) for FDXR and 18Sa.
| Gene/Energy | Median (IQR) | ||||
|---|---|---|---|---|---|
| 0 Gy | 0.2 Gy | 0.5 Gy | 2 Gy | 4 Gy | |
| FDXR/6 MV | 581.11 (369.39, 1018.30) | 830.65 (610.13, 1190.82) | 1203.05 (738.38, 1785.42) | 2393.59 (1798.21, 2575.37) | 2983.00 (2199.48, 3643.82) |
| FDXR/18 MV | 581.11 (369.39, 1018.30) | 1301.09 (924.40, 1490.32) | 1332.34 (1091.79, 2131.46) | 3779.12 (3051.40, 5120.74) | 5051.26 (4704.83, 5859.17) |
| RAD51/6 MV | 788.30 (573.98, 929.09) | 805.38 (507.65, 1280.51) | 1195.34 (778.96, 1300.37) | 2092.77 (1535.78, 2705.61) | 2006.59 (1325.34, 2634.10) |
| RAD51/18 MV | 788.30 (573.98, 929.09) | 803.95 (641.30, 1221.25) | 1761.08 (1311.64, 2465.03) | 3412.57 (2979.72, 4530.61) | 3035.60 (2409.63, 4177.00) |
aThe median with the interquartile range (IQR) of the copy number of RNA was calculated from the molecular weight of each RNA (Nano grams).
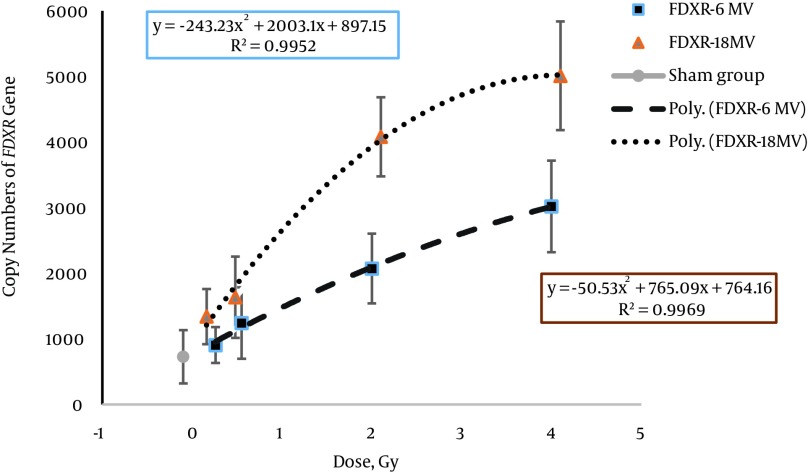
Figure 3. Comparison of the Expression Levels of the FDXR Gene in Human Peripheral Blood Lymphocytes at Doses of 0, 0.2, 0.5, 2, and 4 Gy with 6 and 18 MV Photon Beams by Absolute Quantification.
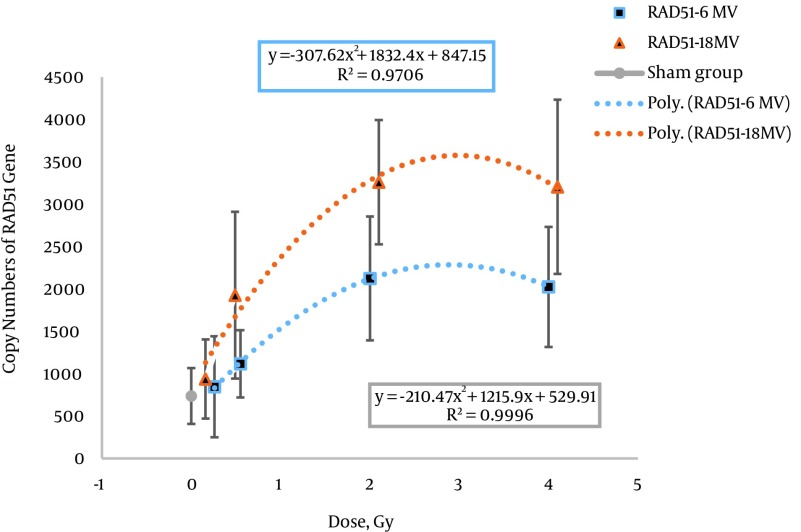
Figure 4. Comparison of the Expression levels of the RAD51 Gene in Human Peripheral Blood Lymphocytes at Doses of 0, 0.2, 0.5, 2, and 4 Gy With 6 and 18 MV Photon Beams by Absolute Quantification.
A non-significant increase in gene expression level was observed at different doses of X-ray (except 2 Gy) with 6 and 18 MV beam energies.
Table 4. Results of the Regression Analysis for the Dose-Response Curve of the FDXR and RAD51 Genes and P Values for Significant Differences Between the 6 and 18 MV Beam Energies (1, 2).
| Gene/Energy | Equation | R2 | P Value | |||
|---|---|---|---|---|---|---|
| 0.2 Gy | 0.5 Gy | 2 Gy | 4 Gy | |||
| FDXR/6 MV | y = -51.198 x2 + 773.2 x + 743.75 | 0.9987 | 0.2 70a | 0.51 3a | 0.001b | 0.008b |
| FDXR/18 MV | y = -243.23 x2 + 2003.1 x + 897.15 | 0.9952 | ||||
| RAD51/6 MV | y = -210.47 x2 + 1215.9 x + 529.91 | 0.9996 | 827a | 0.2 75a | 0.04 0b | 0.12 7b |
| RAD51/18 MV | y = -307.62 x2 + 1832.4 x + 847.15 | 0.9706 | ||||
aNon-significant differences between the copy numbers of genes at the same doses at 6 and 18 MV photon beam energies.
bSignificant differences between the copy number of genes at the same doses at 6 and 18 MV photon beam energies (P < 0.05).
In the dose-response curve for 6 MV, significant increases were shown in the copy number of the FDXR gene at doses of 2 Gy and 4 Gy doses (P < 0.001 and P < 0.008, respectively). The dose estimation curve for 18 MV showed the plateau phase for doses higher than 2 Gy.
For the dose response of the RAD51 gene, an increasing level of gene expression was shown up to 2 Gy at 6 and 18 MV energies of photon beams. After this dose, the descending part of the dose-response curve was well characterized for both energies. A significant increase in the copy number of RAD51 was revealed at a dose of 2 Gy (P < 0.040).
5. Discussion
In this study, peripheral blood samples were collected from seven healthy donors. Human peripheral blood was then exposed to different doses in 6 and 18 MV beam energies in Linac. After RNA extraction and cDNA synthesis, the expression levels of FDXR and RAD51 were determined at 24 hours post-irradiation. We used the PCR amplification product of total cDNA from lymphocytes using RAD51 and FDXR primers as a standard for absolute quantification in gene expression biological dosimetry. This has the advantages of being a comparatively simple, rapid method. In this study, for the first time, absolute quantification was performed in gene expression biological dosimetry using a gel-purified RT-PCR product as the standard sample. The PCR product was confirmed by size using the gel electrophoresis method. In previous studies, the expression level of FDXR was reported in human lymphocytes in radiation response using relative quantification or absolute quantification real-time PCR (9, 12, 19-21).
These genes may be considered as biomarkers of ionization radiation because of increasing expression levels with an increase in dose (22). FDXR is a proper gene for biological dosimetry, so we selected it as a radiation biomarker to compare the results of our quantitative study with previous studies (19). RAD51 was also selected because of its important role in the DNA repair mechanism. DNA repair is a biological pathway affected by ionization radiation. In previous studies, the expression level of RAD51 was reported in human lymphocytes in response to radiation (15, 23, 24); however, there has been no in vivo biological dosimetry study for the RAD51 gene at different radiation doses using real-time PCR.
The best fits for the dose-response curves of the FDXR gene included linear and polynomial equations for 6 and 18 MV beam energies. The polynomial equation was the best fit for the RAD51 dose-response curve. The best fit equation and goodness of fit (R2) for both beam energies are given in Table 4. In conventional radiotherapy, patients are routinely treated with 6 and 18 MV beam energies, as used in this study. However, there has been no in vivo biological dosimetry study on gene expression with different radiation energies using real-time PCR. The difference in copy number of the FDXR and RAD51 genes under 6 and 18 MV beam energies is related to two components, namely the effect of higher energy (18 MV versus 6 MV) and neutron contamination (photo neutron) at 18 MV. Rapid neutron contamination is caused by X-ray beams over 10 MV (25, 26). Figures 3 and 4 show that the DNA damage in cells was increased with increasing dose in low and intermediate doses (0.2, 0.5, and 2 Gy). Consequently, the copy number of both genes increased with increasing DNA damage.
The saturation part of the dose-response curve (Figures 3 and 4) was predictable in the presence of sub-lethal damage repair and cellular repopulation. The complicated issue was the plateau of the curve, which was not shown in the dose-response curve of the FDXR gene at 6 MV of photon energy. However, the plateau was observed in the dose–response curve at 18 MV of energy after a 2 Gy dose. Manning et al. investigated the FDXR dose-response curve under different irradiation conditions. They reported a plateau in the dose-response curve of the FDXR gene. This was related to the in vitro conditions of cell culture or cell death in high-dose irradiation in Manning et al.’s study, while the plateau was observed after a 2 Gy dose at 18 MV of energy in our study. We suggest that cell death is the most important reason for the plateau in the dose-response curve.
In the dose-response curve of the high-dose group at 6 MV of photon energy, the expression level of the FDXR gene was increased with an increasing photon dose. A possible explanation for this is that the megavoltage photons create noticeable sub-lethal DNA damage, and the expression level of the FDXR gene is increased in response to this damage. Sub-lethal DNA damage is then repaired by the DNA repair mechanisms, and cell survival increases.
Photo-neutrons created lethal DNA damage at high doses of the dose-response curve and 18 MV of energy. After radiation, lethal damage was induced and cell death mechanisms (apoptosis and mitotic death) led to a decrease in the measured level of mRNA transcripts. This plateau may be related to a high expression level of the FDXR gene at high doses. In contrast, there was significant cell death within 24 hours after irradiation. The photo-neutron dose equivalent was reported to be a 1 - 3 mSv/Gy dose on the skin with an 18 MV photon beam (mSv per unit photon Gy delivered to isocenter) (27). Thus, it is understandable why the Mann-Whitney test showed significant differences (P < 0.001 and P > 0.008, respectively) between 6 and 18 MV for 2 Gy and 4 Gy doses in the FDXR gene (Table 4). However, we think that analysis of FDXR expression may be a good biomarker for discrimination of radiation at different energy levels with intermediate and high doses.
The dose-response curve of the RAD51 gene only showed a significant difference (P < 0.040) at 2 Gy with 6 and 18 MV beams. The dose-response curve of the RAD51 gene at 6 and 18 MV of energy showed increasing expression levels up to 2 Gy and then a constant expression level that was similar to the dose-response curve of the FDXR gene (as shown in Figure 3); however, differences and more complications were observed. The great variation in the number of RAD51 copies compared to FDXR and the lower effectiveness of radiation energy on the expression of the RAD51 gene compared to the FDXR gene may be related to the DNA repair mechanisms.
Peripheral blood lymphocytes are usually in the resting G0 phase. Homologous recombination (HR) is frequently initiated when a DSB occurs soon after DNA has been replicated, namely in the S or G2 phases, and non-homologous end-joining (NHEJ) is the dominant mechanism of DNA repair after DSBs in G1 and G0. The HR and NHEJ mechanisms are differentially regulated depending on two factors, namely the nature of the DSB and the cell cycle phase in mammalian cells (28, 29). The variation in the copy number of RAD51 and the less prominent effect on radiation quality can be related to this mechanism. It is possible to simultaneously measure the expression level of principal genes in the NHEJ mechanism in association with the RAD51 gene for use in biological dosimetry.
Briefly, the increasing expression level of FDXR gene in the dose response compared to RAD51 was consistent with the hypothesis that the FDXR gene is superior to RAD51 for quantitative dose and energy assessment in gene expression biological dosimetry. In future, several genes should be checked in relation to different pathways, including the cell cycle and apoptosis. The low sample size was another weak point of our study. A larger sample size could improve the accuracy of the study. The study of well-known genes, such as CDKN1, GADD45, and so on, in response to different heterogeneous high-energy X-rays is suggested.
5.1. Conclusion
In contrast to the RAD51 gene, FDXR is a suitable gene for quantitative dose and energy assessment in biological dosimetry, even in high energy X-rays.
Supplements
Supplementary material(s) is available |http://ircmj.com/?page=download&file_id=64909|.
Acknowledgments
This paper was based on the thesis of Ehsan Khodamoradi, and financial support was provided by the Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. We would like to thank Jafar Fattahi Asl and Reza Maskani for assistance in the irradiation of samples. We acknowledge the Deputy of Research Affairs at Ahvaz Jundishapur University of Medical Sciences for financial support for this research.
Footnotes
Authors’ Contribution:All of the authors approved the content of the manuscript and contributed significantly to the research involved in the writing of the manuscript.
Funding/Support:This study was supported by Ahvaz Jundishapur University of Medical Sciences.
References
- 1.Dorr H, Goulko G, Meineke V. Clinical Aspects of Radiation Accident Medical Management. Radiation Emergency Med. 2014;3(2):13–8. [Google Scholar]
- 2.Voisin P. Standards in biological dosimetry: A requirement to perform an appropriate dose assessment. Mutation Research. Genetic Toxicol and Environment Mutagenesis. 2015;739:115–22. doi: 10.1016/j.mrgentox.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Ghandhi SA, Smilenov LB, Elliston CD, Chowdhury M, Amundson SA. Radiation dose-rate effects on gene expression for human biodosimetry. BMC Med Genomics. 2015;8:22. doi: 10.1186/s12920-015-0097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brzoska K, Kruszewski M. Toward the development of transcriptional biodosimetry for the identification of irradiated individuals and assessment of absorbed radiation dose. Radiat Environ Biophys. 2015;54(3):353–63. doi: 10.1007/s00411-015-0603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsuyama N, Mizuno H, Katafuchi A, Abe Y, Kurosu Y, Yoshida M, et al. Identification of low-dose responsive metabolites in X-irradiated human B lymphoblastoid cells and fibroblasts. J Radiat Res. 2015;56(1):46–58. doi: 10.1093/jrr/rru078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Redon CE, Nakamura AJ, Gouliaeva K, Rahman A, Blakely WF, Bonner WM. Q(gamma-H2AX), an analysis method for partial-body radiation exposure using gamma-H2AX in nonhuman primate lymphocytes. Radiat Meas. 2011;46(9):877–81. doi: 10.1016/j.radmeas.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meineke V, Fliedner TM. Radiation-induced multi-organ involvement and failure: challenges for radiation accident medical management and future research. British J Radiol. 2005;27(1):196–200. doi: 10.1259/bjr/25654769. [DOI] [Google Scholar]
- 8.Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, et al. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007;4(4):106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 2008;71(4):1236–44. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee KF, Weng JTY, Hsu PWC, Chi YH, Chen CK, Liu IY, et al. Gene expression profiling of biological pathway alterations by radiation exposure. BioMed Res Inter. 2014;2014 doi: 10.1155/2014/834087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavakoli H, Manoochehri M, Modarres Mosalla SM, Ghafori M, Karimi AA. Dose-dependent and gender-related radiation-induced transcription alterations of Gadd45a and Ier5 inhuman lymphocytes exposed to gamma ray emitted by (60)Co. Radiat Prot Dosimetry. 2013;154(1):37–44. doi: 10.1093/rpd/ncs164. [DOI] [PubMed] [Google Scholar]
- 12.Omaruddin RA, Roland TA, Wallace H3, Chaudhry MA. Gene expression as a biomarker for human radiation exposure. Hum Cell. 2013;26(1):2–7. doi: 10.1007/s13577-013-0059-6. [DOI] [PubMed] [Google Scholar]
- 13.Cannan WJ, Pederson DS. Mechanisms and Consequences of Double‐Strand DNA Break Formation in Chromatin. J Cellular Physiol. 2016;231(1):3–14. doi: 10.1002/jcp.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boldt S, Knops K, Kriehuber R, Wolkenhauer O. A frequency-based gene selection method to identify robust biomarkers for radiation dose prediction. Int J Radiat Biol. 2012;88(3):267–76. doi: 10.3109/09553002.2012.638358. [DOI] [PubMed] [Google Scholar]
- 15.Knops K, Boldt S, Wolkenhauer O, Kriehuber R. Gene expression in low- and high-dose-irradiated human peripheral blood lymphocytes: possible applications for biodosimetry. Radiat Res. 2012;178(4):304–12. doi: 10.1667/rr2913.1. [DOI] [PubMed] [Google Scholar]
- 16.Tucker JD, Joiner MC, Thomas RA, Grever WE, Bakhmutsky MV, Chinkhota CN, et al. Accurate gene expression-based biodosimetry using a minimal set of human gene transcripts. Int J Radiat Oncol Biol Phys. 2014;88(4):933–9. doi: 10.1016/j.ijrobp.2013.11.248. [DOI] [PubMed] [Google Scholar]
- 17.Nosel I, Vaurijoux A, Barquinero JF, Gruel G. Characterization of gene expression profiles at low and very low doses of ionizing radiation. DNA Repair (Amst). 2013;12(7):508–17. doi: 10.1016/j.dnarep.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 18.Vaiphei ST, Keppen J, Nongrum S, Chaubey RC, Kma L, Sharan RN. Evaluation of endogenous control gene(s) for gene expression studies in human blood exposed to 60Co gamma-rays ex vivo. J Radiat Res. 2015;56(1):177–85. doi: 10.1093/jrr/rru074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manning G, Kabacik S, Finnon P, Bouffler S, Badie C. High and low dose responses of transcriptional biomarkers in ex vivo X-irradiated human blood. Int J Radiat Biol. 2013;89(7):512–22. doi: 10.3109/09553002.2013.769694. [DOI] [PubMed] [Google Scholar]
- 20.Kabacik S, Manning G, Raffy C, Bouffler S, Badie C. Time, dose and ataxia telangiectasia mutated (ATM) status dependency of coding and noncoding RNA expression after ionizing radiation exposure. Radiation Res. 2015;183(3):325–37. doi: 10.1667/RR13876.1. [DOI] [PubMed] [Google Scholar]
- 21.Chauhan V, Howland M, Wilkins R. Identification of gene-based responses in human blood cells exposed to alpha particle radiation. BMC Med Genomics. 2014;7:43. doi: 10.1186/1755-8794-7-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Badie C, Kabacik S, Balagurunathan Y, Bernard N, Brengues M, Faggioni G, et al. Laboratory intercomparison of gene expression assays. Radiat Res. 2013;180(2):138–48. doi: 10.1667/RR3236.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budworth H, Snijders AM, Marchetti F, Mannion B, Bhatnagar S, Kwoh E, et al. DNA repair and cell cycle biomarkers of radiation exposure and inflammation stress in human blood. PLoS One. 2012;7(11):48619. doi: 10.1371/journal.pone.0048619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li MJ, Wang WW, Chen SW, Shen Q, Min R. Radiation dose effect of DNA repair-related gene expression in mouse white blood cells. Med Sci Monitor. 2011;17(10):290–7. doi: 10.12659/msm.881976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brockstedt S, Holstein H, Jakobsson L, Tomaszewicz A, Knoos T. Be aware of neutrons outside short mazes from 10-MV linear accelerators X-rays in radiotherapy facilities. Radiat Prot Dosimetry. 2015;165(1-4):464–7. doi: 10.1093/rpd/ncv046. [DOI] [PubMed] [Google Scholar]
- 26.Alem Bezoubiri A, Bezoubiri F, Badreddine A, Mazrou H, Lounis Mokrani Z. Monte Carlo estimation of photoneutrons spectra and dose equivalent around an 18MV medical linear accelerator. Radiation Physics Chem. 2014;97:381–92. doi: 10.1016/j.radphyschem.2013.07.013. [DOI] [Google Scholar]
- 27.Howell RM, Hertel NE, Wang Z, Hutchinson J, Fullerton GD. Calculation of effective dose from measurements of secondary neutron spectra and scattered photon dose from dynamic MLC IMRT for 6 MV, 15 MV, and 18 MV beam energies. Med Phys. 2006;33(2):360–8. doi: 10.1118/1.2140119. [DOI] [PubMed] [Google Scholar]
- 28.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst). 2006;5(9-10):1021–9. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011;30(6):1079–92. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]