Abstract
BACKGROUND
For more than a decade, risk stratification for hypertrophic cardiomyopathy has been enhanced by targeted genetic testing. Using sequencing results, clinicians routinely assess the risk of hypertrophic cardiomyopathy in a patient’s relatives and diagnose the condition in patients who have ambiguous clinical presentations. However, the benefits of genetic testing come with the risk that variants may be misclassified.
METHODS
Using publicly accessible exome data, we identified variants that have previously been considered causal in hypertrophic cardiomyopathy and that are overrepresented in the general population. We studied these variants in diverse populations and reevaluated their initial ascertainments in the medical literature. We reviewed patient records at a leading genetic-testing laboratory for occurrences of these variants during the near-decade-long history of the laboratory.
RESULTS
Multiple patients, all of whom were of African or unspecified ancestry, received positive reports, with variants misclassified as pathogenic on the basis of the understanding at the time of testing. Subsequently, all reported variants were recategorized as benign. The mutations that were most common in the general population were significantly more common among black Americans than among white Americans (P<0.001). Simulations showed that the inclusion of even small numbers of black Americans in control cohorts probably would have prevented these misclassifications. We identified methodologic shortcomings that contributed to these errors in the medical literature.
CONCLUSIONS
The misclassification of benign variants as pathogenic that we found in our study shows the need for sequencing the genomes of diverse populations, both in asymptomatic controls and the tested patient population. These results expand on current guidelines, which recommend the use of ancestry-matched controls to interpret variants. As additional populations of different ancestry backgrounds are sequenced, we expect variant reclassifications to increase, particularly for ancestry groups that have historically been less well studied. (Funded by the National Institutes of Health.)
Although hypertrophic cardiomyopathy is best known as a fatal disease of young athletes, it causes considerable morbidity and mortality among patients of all ages and lifestyles.1, 2 The defining feature of hypertrophic cardiomyopathy is unexplained left ventricular hypertrophy, but its clinical presentation is variable; it can manifest as severe heart failure in some patients yet be asymptomatic in others.3 In more than one third of patients, causal genetic lesions are identified, which enables clinicians to assess risk among the patient’s relatives4 and, in rare circumstances, to tailor therapy for a patient who is found to have a tractable disorder, such as Fabry’s disease.5 In addition, in patients who have clinical features but who do not have a definitive diagnosis of hypertrophic cardiomyopathy, the identification of a pathogenic variant may be used to help establish a diagnosis.
The provision of false genetic information to a patient, such as when a patient is incorrectly informed that one of his or her variants is causal when in fact it is benign, can have far-reaching adverse consequences within the family. First, relatives with the noncausal variant receive prolonged at-risk screening, are advised about lifestyle modifications (e.g., cessation of certain sports and activities), and may have stress and economic burden consequent to an incorrect diagnosis. Second, relatives who do not have the noncausal variant are given false reassurance that further surveillance is unnecessary. Third, for patients with clinical features but without a definitive diagnosis of hypertrophic cardiomyopathy, such as young athletes with modest hypertrophy and a family history of sudden cardiac death, misclassification of a benign variant as pathogenic may lead to overestimation of the benefits of implantation of a cardioverter–defibrillator to prevent sudden cardiac death. In addition, when the status of a variant is changed from pathogenic to benign, the sequencing laboratory often (but not obligatorily) recontacts the referring physician, who in turn recontacts the patient and their tested family members, engendering confusion and distrust.
Much effort has gone into developing standards for the correct interpretation of genetic variants.6–10 The principal challenge is to separate truly pathogenic variants from the historically underappreciated amount of background variant noise in the genome.8, 11 Expert guidelines generally recommend classifying variants with the use of sequence data from ancestry-matched controls.4, 6
When large-scale control sequence data from the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project12 were systematically reviewed for hypertrophic cardiomyopathy– associated variants labeled “disease-causing” or “pathogenic” in an expert-curated database,13, 14 many more variants were found than were expected in the general population (given the prevalence of hypertrophic cardiomyopathy), which implied that penetrance might have been lower than previously thought or that misclassification errors might have been present in previous studies of the association between the variants and the condition. We found that five high-frequency variants (i.e., variants with a minor-allele frequency >1% in either black Americans or white Americans in the Exome Sequencing Project) accounted for the majority of this overabundance of misclassified variation and that these variants occurred disproportionately among black Americans.
We hypothesized that the identification of high-frequency variants associated with hypertrophic cardiomyopathy in the general population would indicate the presence of historical errors in patient reports and that most or all persons affected would be of African ancestry. We further posited that these variant associations stemmed from ascertainment bias and other methodologic shortcomings in the original studies. To test these hypotheses, we searched patient records for occurrences of these variants at an established genetic testing laboratory and reviewed the literature implicating the variants. We describe here a cautionary tale of broad relevance to genetic diagnosis. Although this tale is well known to geneticists, clinicians may be less familiar with it.
Methods
Study Populations
We used publicly accessible sequence data from the NHLBI Exome Sequencing Project,12 1000 Genomes Project,15 and Human Genome Diversity Project.16 For the estimation of minor-allele frequency, the NHLBI Exome Sequencing Project had exome data for 4300 white Americans and 2203 black Americans, the 1000 Genomes Project phase 1 had whole-genome data for 1092 persons from 14 populations worldwide, and the Human Genome Diversity Project had whole-genome single-nucleotide polymorphism (SNP) data for 938 persons from 51 populations worldwide. Clinical records for patients with hypertrophic cardiomyopathy were reviewed at the Laboratory for Molecular Medicine, Partners HealthCare, Boston. This population has been described in detail previously.17 All patient reports that originally included a variant status of “pathogenic,” “presumed pathogenic,” “unknown significance,” or “pathogenicity debated” were included in the analysis (Table 1). Demographic data were acquired from the genetic-testing requisition form. The Laboratory for Molecular Medicine patient population, which included patients who were referred to the laboratory for testing, was a mixed population of 64% white American patients and 8% black American patients, with the remaining patients of other or unspecified ancestry. 17 This study was conducted in accordance with a protocol that was approved by the institutional review board at Partners HealthCare, which waived the requirement for informed consent.
Table 1.
Clinical Findings for High-Frequency Variants Associated with Hypertrophic Cardiomyopathy.
| Originally Reported Status of Variant* |
Patient’s Age |
Patient’s Ethnic Background |
Report Year |
Report Result |
Variant | Most Significant Pathogenic Variant† |
Indication for Test |
|---|---|---|---|---|---|---|---|
| Pathogenic | 46 yr | Unavailable | 2005 | Positive | TNNI3 (P82S) | Yes | Clinical diagnosis of hypertrophic cardiomyopathy |
| Pathogenic | 75 yr | Unavailable | 2005 | Positive | TNNI3 (P82S) | Yes | Family history and clinical symptoms of hypertrophic cardiomyopathy |
| Presumed pathogenic | 32 yr | African ancestry | 2005 | Positive | TNNI3 (P82S) | No | Clinical diagnosis of hypertrophic cardiomyopathy |
| Pathogenicity debated | 34 yr | African ancestry | 2005 | Positive | TNNI3 (P82S) | No | Clinical diagnosis and family history of hypertrophic cardiomyopathy |
| Unknown significance | 12 yr | African ancestry | 2006 | Inconclusive | TNNI3 (P82S) | Yes | Family history of hypertrophic cardiomyopathy |
| Unknown significance | 40 yr | African ancestry | 2007 | Inconclusive | TNNI3 (P82S) | Yes | Clinical diagnosis of hypertrophic cardiomyopathy |
| Unknown significance | 45 yr | African ancestry | 2007 | Inconclusive | TNNI3 (P82S) | Yes | Clinical features of hypertrophic cardiomyopathy |
| Unknown significance | 16 yr | Asian ancestry | 2008 | Positive | TNNI3 (P82S) | No | Clinical diagnosis and family history of hypertrophic cardiomyopathy |
| Presumed pathogenic | 59 yr | African ancestry | 2006 | Positive | MYBPC3 (G278E) | Yes | Clinical features of hypertrophic cardiomyopathy |
| Presumed pathogenic | 15 yr | African ancestry | 2007 | Positive | MYBPC3 (G278E) | Yes | Clinical diagnosis of hypertrophic cardiomyopathy |
| Presumed pathogenic | 16 yr | African ancestry | 2007 | Positive | MYBPC3 (G278E) | Yes | Clinical diagnosis of hypertrophic cardiomyopathy |
| Presumed pathogenic | 22 yr | African ancestry | 2007 | Positive | MYBPC3 (G278E) | No | Clinical diagnosis and family history of hypertrophic cardiomyopathy |
| Unknown significance | 48 yr | African ancestry | 2008 | Positive | MYBPC3 (G278E) | No | Clinical diagnosis of hypertrophic cardiomyopathy |
All variants subsequently have been reclassified as benign.
Information in this column indicates whether the variant was unequivocally the most pathogenic variant in the original report that was provided to the patient.
Ascertainment of Variants
Using PubMed, we performed a targeted search for initial disease–variant associations for all high-frequency variants associated with hypertrophic cardiomyopathy in the medical literature. All Human Genome Variation Society names for the variants (e.g., K247R and Lys247Arg) were used, as well as all possible transcript variants obtained from the National Center for Biotechnology Information SNP database (dbSNP), build 140.18 All original reports of disease–variant associations were in agreement with those listed in the Human Gene Mutation Database Professional Version 2016.1.14 High-frequency variants associated with hypertrophic cardiomyopathy were defined as variants with a minor-allele frequency greater than 1% in either NHLBI subpopulation. The “penetrance” of a genetic variant was defined as the proportion of persons with the variant who had hypertrophic cardiomyopathy.
Statistical Analysis
P values were computed with the chi-square test and Mann–Whitney U-test. The Human Genome Diversity Project Selection Browser19 was used to display allele frequencies in worldwide populations. Unless otherwise specified, all analyses were performed with the R statistical package, version 3.2.1.20
Results
Hypertrophic Cardiomyopathy Gene Variation in the General Population
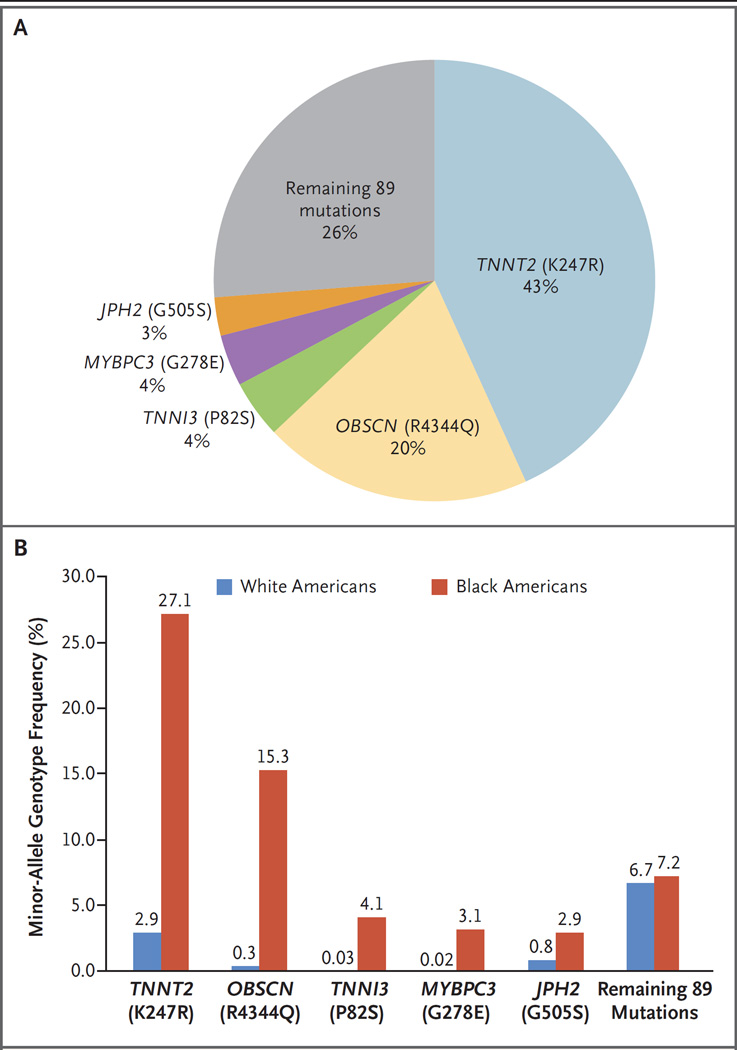
The NHLBI Exome Sequencing Project has previously been searched for any variant labeled a “Disease causing mutation” for hypertrophic cardiomyopathy in the Human Gene Mutation Database, version 2012.2.13, 14 Although 94 distinct variants that had previously been characterized as associated with hypertrophic cardiomyopathy were found in the Exome Sequencing Project data, we found that relatively few variants accounted for the bulk of the genotype prevalence signal (Fig. 1A). Of the 94 Human Gene Mutation Database hypertrophic cardiomyopathy– associated variants identified in the Exome Sequencing Project data, 5 met our threshold for being classified as high-frequency variants (minor-allele frequency of >1% in either NHLBI subpopulation), and these variants accounted for nearly 75% of the overall genotype prevalence signal.
Figure 1. Genetic Variants Associated with Hypertrophic Cardiomyopathy.
Panel A shows variants associated with hypertrophic cardiomyopathy that are overrepresented in the general population. The five highest-frequency variants account for 74% of the misclassified variation in the general population. Panel B shows the minor-allele genotype frequencies for high-frequency variants associated with hypertrophic cardiomyopathy; all variants are significantly more common among black Americans than among white Americans (P<0.001 by the chi-square test for all five variants).
High-Frequency Variants Associated with Hypertrophic Cardiomyopathy in Black Americans
All five high-frequency variants associated with hypertrophic cardiomyopathy had significantly greater frequencies among black Americans than among white Americans in the NHLBI Exome Sequencing Project data set (P<0.001 for each comparison by the chi-square test) (Fig. 1B). The minor-allele frequencies for these five variants ranged from 1.5 to 14.9% among black Americans, from 0.01 to 1.5% among white Americans, and from 0.5 to 6.0% in the combined population. The genotype frequency, defined as the number of persons with at least one copy of the minor allele divided by the total number of persons, ranged from 2.9 to 27.1% among black Americans, from 0.02 to 2.9% among white Americans, and from 1.0 to 11.1% in the combined population. The summed genotype frequency of the remaining 89 variants did not differ significantly between black Americans and white Americans.
Penetrance of High-Frequency Variants Associated with Hypertrophic Cardiomyopathy in Black Americans
We computed the penetrance for each variant across several clinical contexts (Fig. S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Because hypertrophic cardiomyopathy is rare in the general population, with a prevalence of 1 case in 500 persons,2 even variants with a minor-allele frequency as low as 1% have a theoretical maximal penetrance of 0.2; however, the penetrance is probably much lower, given the high allelic heterogeneity of hypertrophic cardiomyopathy4 and the fact that most alleles are probably benign. Even if the TNNT2 K247R variant were present in all black Americans with hypertrophic cardiomyopathy (which is certain not to be the case), K247R would have a penetrance of less than 1%. Penetrance may take on rather different values in other clinical contexts. It is notable that for first-degree relatives, benign high-frequency variants may have high apparent penetrance owing to the high prior probability of disease in this population.
Classification of High-Frequency Variants Associated with Hypertrophic Cardiomyopathy
Applying the clinical classification algorithm in use at the Laboratory for Molecular Medicine, Partners HealthCare Personalized Medicine,9 we classified all high-frequency variants unambiguously as “benign,” given their high frequency in control populations and given the mix of patient and functional data available for these variants. In contrast, in the Human Gene Mutation Database database, version 2016.1, four of the five variants remain classified in the most pathogenic category, “disease-causing mutation.” Only one variant (OBSCN R4344Q) was downgraded from “disease-causing” to “disease-causing?” (in September 2012).
Benign Variants Misclassified as Pathogenic in Genetic Reports
Seven patients, all of African or unspecified ancestry, received reports between 2005 and 2007 that they had one of two variants, TNNI3 P82S or MYBPC3 G278E, that were classified as “pathogenic” or “presumed pathogenic” (Table 1); these variants were later reclassified as benign. In five of the seven reports, P82S or G278E was the most pathogenic variant reported to the patient. Six additional patients with inconclusive or positive genetic-testing results that were reported later were listed as having one of these two variants, which were characterized as being of “unknown significance” or “pathogenicity debated.” Among the 13 patients, 9 had a clinical diagnosis of hypertrophic cardiomyopathy, 2 had clinical features of hypertrophic cardiomyopathy, and 1 had clinical symptoms of hypertrophic cardiomyopathy. Five of the 13 patients had a documented family history of hypertrophic cardiomyopathy. From the records available, it was not possible to determine whether the families affected by the reclassification of these variants were recontacted.
Sample Sizes and Representativeness of Original Studies
All high-frequency variants were examined for their initial association in the medical literature (Table 2).21–25 In the initial studies of TNNI3 P82S and MYBPC3 G278E, the control sample sizes were 85 and 100, respectively, which are below and equal to, respectively, the minimum currently accepted (but probably still inadequate) standard needed to corroborate pathogenicity.4 Furthermore, none of the studies implicating these variants involved persons of African ancestry explicitly; however, several studies might have involved sequencing or genotyping of persons of African ancestry during the discovery stage (Table 2). In general, the original study that established the association between a variant and hypertrophic cardiomyopathy consisted of three steps. First, a handful of genes previously connected with hypertrophic cardiomyopathy were sequenced in DNA samples obtained from patients with hypertrophic cardiomyopathy. Second, the discovered variants were examined in ostensibly ancestry-matched unrelated controls and, when available, in family members of the patients. Third, functional analyses were conducted in a subset of studies to assess the mechanism by which the variant may cause disease.
Table 2.
Studies That Initially Implicated High-Frequency Variants Associated with Hypertrophic Cardiomyopathy.
| Gene (Variant) | Reference | Discovery Phase | No. of Cases |
No. of Controls |
Variant Assessment |
Country | Included in LMM Clinical Panel |
|
|---|---|---|---|---|---|---|---|---|
| In Vitro |
In Vivo |
|||||||
| TNNT2 (K247R) | García-Castro et al.21 | Targeted gene sequencing of unrelated cases and controls from Asturias |
30 | 200 | No | No | Spain | Yes |
| OBSCN* (R4344Q) | Arimura et al.22 | Targeted gene sequencing of unrelated Japanese cases and controls |
144 | 288 | Yes | No | Japan | No |
| TNNI3 (P82S) | Niimura et al.23 | Targeted gene sequencing of unrelated cases and controls† |
31 | 85 | No | No | United States |
Yes |
| MYBPC3 (G278E) | Richard et al.24 | Targeted gene sequencing of unrelated cases and controls‡ |
197 | 100 | No | No | France | Yes |
| JPH2* (G505S) | Matsushita et al.25 | Targeted gene sequencing of Japanese cases and controls |
195 | 236 | Yes | No | Japan | No |
OBSCN and JPH2 have never been included in cardiomyopathy testing at the Laboratory for Molecular Medicine (LMM).
No specific ethnic background was provided, but “informed consent was obtained in accordance with human subject committee guidelines at Brigham and Women’s Hospital, St. George’s Hospital Medical School [U.K.], and Minneapolis Heart Institute Foundation.”23
“Patients were recruited in France, and most of them were of European origin.”24 The sample of patients included persons of African ancestry (Richard P: personal communication).
Variation in MYBPC3 and TNNI3 among Black Americans
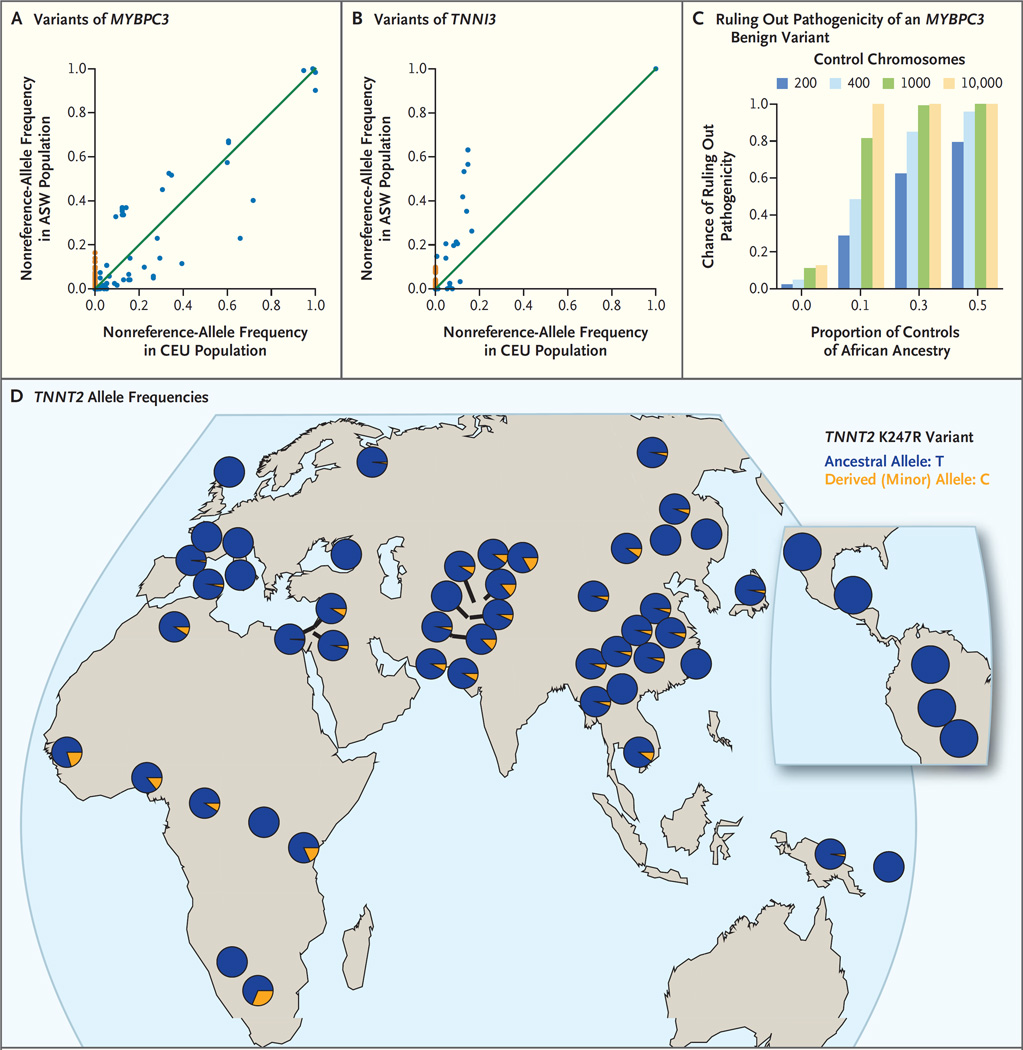
We used the data from the 1000 Genomes Project to compare sequence variation between black Americans and white Americans, using as proxies the ASW (African Ancestry in Southwest USA) and CEU (Utah Residents [CEPH] with Northern and Western European ancestry) populations, respectively. As shown in Figure 2A and 2B, black Americans harbored significantly more segregating loci (i.e., variable genomic sites within a population) in both genes than did white Americans. These “private sites,” at which the frequency of the nonreference allele (i.e., the allele that is not present in the reference sequence of the human genome) is nonzero in one population but zero in the other population, are shown in Figure 2A and 2B. For MYBPC3, there were 66 private sites in the ASW population and 15 private sites in the CEU population; for TNNI3, there were 45 private sites in the ASW population and 6 private sites in the CEU population.
Figure 2. Prevention of Misclassification of Variants with the Use of Data from Diverse Populations.
Panels A and B show frequencies of the nonreference allele (i.e., the allele that is not present in the reference sequence of the human genome) for the 1000 Genome Project populations ASW (African Ancestry in Southwest USA) (61 persons) and CEU (Utah Residents [CEPH] with Northern and Western European ancestry) (85 persons) for the genes MYBPC3 (Panel A) and TNNI3 (Panel B). Each point represents a distinct variant. There are significantly more private sites (sites for which the nonreference-allele frequency is nonzero in one population but zero in the other population) among black Americans (nonreference-allele frequency, 0% in CEU and >0% in ASW, with ASW private sites shown in orange) than among white Americans (nonreference-allele frequency, 0% in ASW and >0% in CEU). Panel C shows the chance of correctly ruling out pathogenicity for a truly benign variant that is found predominantly in one ancestry group; the probability generally increases with the fraction of the control cohort that is made up of that ancestry group and with the number of controls (numbers of control chromosomes are shown in the key). These simulations use the allele frequencies of the MYBPC3 G278E variant, which has a minor-allele frequency of 0.0157 among black Americans and 0.000122 among white Americans. Panel D shows a map of TNNT2 allele frequencies; the K247R variant of TNNT2 (rs3730238 in dbSNP) was genotyped in the Human Genome Diversity Project. Most populations around the world have a nonzero minor-allele frequency.
Reducing the Risk of False Positives with Genetically Diverse Populations
As shown in Figure 2C, even small studies of diverse populations are comparatively well-powered to avoid misclassification of the five high-frequency variants associated with hypertrophic cardiomyopathy. Conservatively, we used the lower-frequency variant of the two that were misclassified in patients (MYBPC3 G278E; minor-allele frequency, 0.0157 in black Americans and 0.000122 in white Americans). At these frequencies, even if black Americans constituted just 10% of the control cohort, we would have a 50% chance of correctly ruling out pathogenicity with a control cohort of only 200 persons.
We documented the way in which the hypertrophic cardiomyopathy–associated high-frequency variants could be studied in worldwide populations (Fig. 2D, and Table S1 in the Supplementary Appendix). For example, the highest-frequency hypertrophic cardiomyopathy variant (TNNT2 K247R) was a locus in the Human Genome Diversity Project16 (Fig. 2D) and has a nonzero minor-allele frequency in many worldwide populations.
Errors from a Paucity of Diverse Control Data
Table 3 shows the probability of ruling out pathogenicity for truly benign variants with the use of several sequencing resources that have been in use for the past several years. For example, using the 1000 Genomes Project population Mexican Ancestry from Los Angeles (MXL), which consists of 66 persons, we have only a 48% chance of ruling out pathogenicity when the minor-allele frequency is 0.5%. If the minor-allele frequency is 0.1%, such as for a rare variant discovered by high-coverage exome sequencing, the probability of ruling out pathogenicity is only 12% when the MXL population is used. In contrast, if new variant association studies use a resource like the recently created Exome Aggregation Consortium database,26 they are well-powered for ruling out pathogenicity even for rarer variation.
Table 3.
Control Sequencing Resources for Ruling Out Pathogenicity in Several Worldwide Populations.
| Database and Cohort | No. of Persons |
Probability of Ruling out Pathogenicity of a Benign Variant | ||
|---|---|---|---|---|
| Minor-Allele Frequency of 1% |
Minor-Allele Frequency of 0.5% |
Minor-Allele Frequency of 0.1% |
||
| percent | ||||
| National Heart, Lung, and Blood Institute Exome Sequencing Project |
||||
| White Americans | 4,300 | 100 | 100 | 100 |
| Black Americans | 2,203 | 100 | 100 | 99 |
| 1000 Genomes Project | ||||
| Mexican Ancestry from Los Angeles | 66 | 73 | 48 | 12 |
| Han Chinese in Beijing | 97 | 86 | 62 | 18 |
| Southern Han Chinese | 100 | 87 | 63 | 18 |
| Exome Aggregation Consortium | ||||
| African or black American | 5,203 | 100 | 100 | 100 |
| Non-Finnish European | 33,370 | 100 | 100 | 100 |
| East Asian | 4,327 | 100 | 100 | 100 |
Discussion
We hypothesized that genetic variants that are common among black Americans had previously been misclassified in patients undergoing genetic testing for hypertrophic cardiomyopathy. We identified multiple persons, all of African or unspecified ancestry, who had received positive reports that were based on misclassified benign variants. Such misclassifications invalidate risk assessments undertaken in relatives, requiring a chain of amended reports and management plans. Our findings suggest that false positive reports are an important and perhaps underappreciated component of the “genotype-positive– phenotype-negative” subgroup of tested persons.27 These findings show how health disparities may arise from genomic misdiagnosis. Disparities may result from errors that are related neither to access to care nor to posited “physiological differences” but, rather, to the historical dearth of control populations that include persons of diverse racial and ethnic backgrounds. Future work is needed to assess the extent to which this pattern holds across other variants, types of misclassifications, and diseases.
Minimizing misclassifications by sifting through genomic noise for causal variants is closely related to assessing penetrance, the proportion of persons with the variant who have the disease. However, estimating penetrance is often difficult because it is sensitive to clinical context (Fig. S1 in the Supplementary Appendix) and because many studies are designed for the discovery of variants rather than for an unbiased estimation of true effect sizes, a pattern that is not limited to hypertrophic cardiomyopathy.28 This approach is due, in part, to historically limited sequencing data. Fortunately, several large-scale sequencing efforts are mitigating this aspect of the variant annotation challenge,12, 15 although they are also introducing an unprecedented scale of novel variants and genes to consider. 8, 29 Even when penetrance cannot be computed precisely, we may still be able to determine the penetrance to within a certain range in order to infer that a variant is benign, given a sufficiently high minor-allele frequency in the general population.9 Although the NHLBI Exome Sequencing Project is a powerful resource in its supply of exome sequence data for black Americans and white Americans, analyses using similar resources for genomic data from Native Americans and Asian Americans are urgently needed (Table 3). Large-scale sequencing resources such as the NHLBI Exome Sequencing Project and the Exome Aggregation Consortium not only are well-powered to “rule out” benign variants and reduce false positives (Fig. 2C), but they also allow pathogenicity to be corroborated for truly pathogenic variants (i.e., they help “rule in” variants in addition to reducing false negatives). Indeed, sequencing data from diverse populations are necessary to find ancestry-specific pathogenic variants.30 In addition, sequencing data from diverse populations have been used successfully in “admixture mapping” to find risk loci by detecting deviations in local ancestry.31
Large-scale sequencing data from the general population also enable systematic reassessments of previous disease–variant associations.13, 32, 33 For the interpretation of variants in hypertrophic cardiomyopathy, expert guidelines generally recommend the use of ancestry-matched controls.4 However, an insistence on using only ancestry-matched controls may delay proper annotation if the matching is imperfect. As an example, consider MYBPC3 G278E, a high-frequency hypertrophic cardiomyopathy–associated variant that was first discovered in a Parisian cohort,24 was also identified in subsequent studies,34 and was misclassified in several persons of African ancestry (Table 1). We were able to verify that although most of the patients in the original study were of European origin,24 the sample also included patients of African ancestry (Richard P: personal communication). If only or primarily persons of European ancestry were included in the control sample in the original study or in follow-up studies, analyses would be underpowered to classify the variant as nonpathogenic (Fig. 2C). These findings suggest how current guidelines might be extended: sequencing data from diverse ancestry groups may be used to refute the possibility that novel variants are pathogenic, as well as to reevaluate the status (with respect to pathogenicity) of known variants.9, 35
We believe that with several steps, care could be improved going forward. First, sequencing data from diverse populations could be used to evaluate novel variants and reevaluate known variants. Second, if researchers and genetic testing laboratories adopted a probabilistic framework by computing relative risks explicitly, the confusion in pathogenicity assessments would probably be reduced. Third, reevaluation of the fragmented literature regarding disease variants depends on continued data-sharing and standardization of reporting, which are the aims of centralized databases like ClinVar.36 In line with President Barack Obama’s Precision Medicine Initiative, we support the development of an information commons37 that stores deidentified genetic-testing results from patients and their family members.38 This effort would show the value of responsible data sharing, as detailed in a recent Institute of Medicine report.39, 40 Finally, as variant annotations are updated, agile clinical systems would ideally make this information available in near-real time to physicians and genetic-testing laboratories.41 Many physicians express a desire for additional guidance on genetic testing.42 Point-of-care support for physicians and genetic-testing professionals, such as risk calculators for each hypertrophic cardiomyopathy variant, incorporating family history and ethnic background, would assist in decision making and help minimize misclassification errors. We expect that many “variants of uncertain significance”8 will be recategorized in the near future with ongoing efforts to sequence the DNA of persons from diverse populations. Far from being a clear binary decision, variant classification is an evolving process. A synergy of clinical, genetic, statistical, and political perspectives is needed to ensure that genomic medicine benefits all populations equally.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health (NIH) National Human Genome Research Institute (5T32HG002295-9), the NIH National Institute of Mental Health (P50MH094267), and the NIH National Center for Biomedical Computing (5U54-LM-008748).
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults: echocardiographic analysis of 4111 subjects in the CARDIA Study. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 3.Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet. 2013;381:242–255. doi: 10.1016/S0140-6736(12)60397-3. [DOI] [PubMed] [Google Scholar]
- 4.Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60:705–715. doi: 10.1016/j.jacc.2012.02.068. [DOI] [PubMed] [Google Scholar]
- 5.Weidemann F, Niemann M, Breunig F, et al. Long-term effects of enzyme replacement therapy on Fabry cardiomyopathy: evidence for a better outcome with early treatment. Circulation. 2009;119:524–529. doi: 10.1161/CIRCULATIONAHA.108.794529. [DOI] [PubMed] [Google Scholar]
- 6.Richards CS, Bale S, Bellissimo DB, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 7.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013;14:295–300. doi: 10.1038/nrg3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duzkale H, Shen J, McLaughlin H, et al. A systematic approach to assessing the clinical significance of genetic variants. Clin Genet. 2013;84:453–463. doi: 10.1111/cge.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norton N, Robertson PD, Rieder MJ, et al. Evaluating pathogenicity of rare variants from dilated cardiomyopathy in the exome era. Circ Cardiovasc Genet. 2012;5:167–174. doi: 10.1161/CIRCGENETICS.111.961805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacArthur DG, Balasubramanian S, Frankish A, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NHLBI GO Exome Sequencing Project (ESP) Exome variant server. ( http://evs.gs.washington.edu/EVS/)
- 13.Andreasen C, Nielsen JB, Refsgaard L, et al. New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet. 2013;21:918–928. doi: 10.1038/ejhg.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stenson PD, Mort M, Ball EV, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 17.Alfares AA, Kelly MA, McDermott G, et al. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet Med. 2015;17:880–888. doi: 10.1038/gim.2014.205. [DOI] [PubMed] [Google Scholar]
- 18.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickrell JK, Coop G, Novembre J, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 21.García-Castro M, Reguero JR, Batalla A, et al. Hypertrophic cardiomyopathy: low frequency of mutations in the beta-myosin heavy chain (MYH7) and cardiac troponin T (TNNT2) genes among Spanish patients. Clin Chem. 2003;49:1279–1285. doi: 10.1373/49.8.1279. [DOI] [PubMed] [Google Scholar]
- 22.Arimura T, Matsumoto Y, Okazaki O, et al. Structural analysis of obscurin gene in hypertrophic cardiomyopathy. Biochem Biophys Res Commun. 2007;362:281–287. doi: 10.1016/j.bbrc.2007.07.183. [DOI] [PubMed] [Google Scholar]
- 23.Niimura H, Patton KK, McKenna WJ, et al. Sarcomere protein gene mutations in hypertrophic cardiomyopathy of the elderly. Circulation. 2002;105:446–451. doi: 10.1161/hc0402.102990. [DOI] [PubMed] [Google Scholar]
- 24.Richard P, Charron P, Carrier L, et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 25.Matsushita Y, Furukawa T, Kasanuki H, et al. Mutation of junctophilin type 2 associated with hypertrophic cardiomyopathy. J Hum Genet. 2007;52:543–548. doi: 10.1007/s10038-007-0149-y. [DOI] [PubMed] [Google Scholar]
- 26.Exome Aggregation Consortium (ExAC) ExAC browser (beta) 2016 ( http://exac.broadinstitute.org)
- 27.Maron BJ, Yeates L, Semsarian C. Clinical challenges of genotype positive (+)-phenotype negative (−) family members in hypertrophic cardiomyopathy. Am J Cardiol. 2011;107:604–608. doi: 10.1016/j.amjcard.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 28.Beutler E, Felitti VJ, Koziol JA, Ho NJ, Gelbart T. Penetrance of 845G→A (C282Y) HFE hereditary haemochromatosis mutation in the USA. Lancet. 2002;359:211–218. doi: 10.1016/S0140-6736(02)07447-0. [DOI] [PubMed] [Google Scholar]
- 29.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Struewing JP, Hartge P, Wacholder S, et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 31.Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat Rev Genet. 2011;12:523–528. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 33.Bick AG, Flannick J, Ito K, et al. Burden of rare sarcomere gene variants in the Framingham and Jackson Heart Study cohorts. Am J Hum Genet. 2012;91:513–519. doi: 10.1016/j.ajhg.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morita H, Rehm HL, Menesses A, et al. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899–1908. doi: 10.1056/NEJMoa075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ioannidis JPA, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 36.Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. Washington, DC: National Academies Press; 2011. [PubMed] [Google Scholar]
- 38.Manrai AK, Ioannidis JPA, Kohane IS. Clinical genomics: from pathogenicity claims to quantitative risk estimates. JAMA. 2016;315:1233–1234. doi: 10.1001/jama.2016.1519. [DOI] [PubMed] [Google Scholar]
- 39.Lo B. Sharing clinical trial data: maximizing benefits, minimizing risk. JAMA. 2015;313:793–734. doi: 10.1001/jama.2015.292. [DOI] [PubMed] [Google Scholar]
- 40.Drazen JM. Sharing individual patient data from clinical trials. N Engl J Med. 2015;372:201–202. doi: 10.1056/NEJMp1415160. [DOI] [PubMed] [Google Scholar]
- 41.Wilcox AR, Neri PM, Volk LA, et al. A novel clinician interface to improve clinician access to up-to-date genetic results. J Am Med Inform Assoc. 2014;21(e1):e117–e1 21. doi: 10.1136/amiajnl-2013-001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klitzman R, Chung W, Marder K, et al. Attitudes and practices among internists concerning genetic testing. J Genet Couns. 2013;22:90–100. doi: 10.1007/s10897-012-9504-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.