Abstract
Large-scale quantitative proteomics can provide a near-global view of cellular protein abundance. Yet, the time, effort, and expertise required to achieve reasonable protein coverage and reliable quantification have limited the broad application of this technology. To fully leverage mass spectrometry for the elucidation of biological systems requires sufficient throughput to monitor dynamic changes across conditions and to enable replicate analysis to provide statistical power. We report a straightforward approach to increase the multiplexing capacity of quantitative mass spectrometry, which provides a platform for the analysis of cellular signaling pathways. Using triplex metabolic labeling and six-plex isobaric tags, we monitored changes in protein abundance from 18 samples simultaneously, performing biological triplicates of a six-point time course of rapamycin-stimulated yeast. The data set provides temporal abundance profiles for thousands of yeast proteins, highlighting the complex cellular roles of the TOR (target of rapamycin) pathway.
INTRODUCTION
By providing a means to identify proteins from complex mixtures without prior knowledge of the components, mass spectrometry (MS) has transformed modern molecular biology; it is routinely used to identify cellular proteins, identify members of protein complexes, and map sites of post-translational modifications. Although the enormous dynamic range of protein abundance still limits detection of the least abundant proteins, the depth of quantitative proteomic analysis continues to increase. In the budding yeast Saccharomyces cerevisiae, a few reports have demonstrated relative quantification of the bulk of predicted proteins (1–3), bringing one of the coveted goals of proteomics, comprehensive protein quantification, tantalizingly close. Despite these advances, such analysis is not yet routine and access to these methods is limited.
Large-scale proteomics experiments typically use enzymatic digestion of cellular proteins to peptides followed by a fractionation step to simplify the peptide mixture. Each fraction is then analyzed by liquid chromatography–tandem MS (LC-MS/MS). As peptides elute from a reversed-phase high-performance liquid chromatography (RP-HPLC) column, they are ionized and introduced into the mass spectrometer for analysis. The combination of intact peptide masses, measured in full scans (MS or MS1), and fragment ion masses for each peptide measured in subsequent tandem mass spectra (MS/MS or MS2) after collisional dissociation, enables efficient algorithmic matching to theoretical spectra of peptide sequences predicted from the genome sequence. Quantification is commonly achieved through the use of stable isotope labels, such as 13C or 15N, incorporated either metabolically or by chemical addition. These labels alter the mass, but not the chemical characteristics, of peptides. Differentially labeled but same-sequence peptides behave identically during all separation steps and enter the mass spectrometer together; however, their mass differences make them distinguishable by the instrument. The relative height of the corresponding peaks in a mass spectrum provides quantitative information.
Stable isotope labeling of amino acids in cell culture (SILAC) (4) is a common technique for quantitative proteomics entailing the addition of stable isotope–enriched essential amino acids to the culture medium. Labeled amino acids are metabolically incorporated into all proteins. Samples from labeled and unlabeled cells are combined and processed together. Peptide quantification proceeds through the comparison of peaks representing intact peptide ions in MS1 spectra (Fig. 1A). Although a powerful and effective technique, SILAC is not easily multiplexed and is most often used only for binary (4) and, occasionally, ternary (5) comparisons. Quantitative comparisons of multiple conditions or time points by SILAC require repeated analyses for each additional sample, creating a bottleneck that limits its application in such experiments. In contrast to SILAC, chemically incorporated isobaric mass-tagging reagents, such as “isobaric tags for relative and absolute quantification” (iTRAQ) and “tandem mass tags” (TMT), provide label-based quantification that is measurable only after peptides are fragmented in the mass spectrometer and the fragments are detected in MS/MS. Unlike metabolic labeling, the different isobaric labels bear identical intact masses. Same-sequence peptides labeled with any of them co-elute from RP-HPLC, enter the mass spectrometer together, and appear as a single composite peak in a mass spectrum of the intact peptides (Fig. 1B). However, upon isolation and fragmentation, peptides labeled with each reagent generate unique reporter ions that can be observed in a subsequent MS/MS scan. The intensities of the reporter ions provide a relative quantitative measure of the peptides derived from the different samples. The great advantage of this method is that it can be readily multiplexed. Commercial reagents are available for four-, six-, and eight-plex analysis (6).
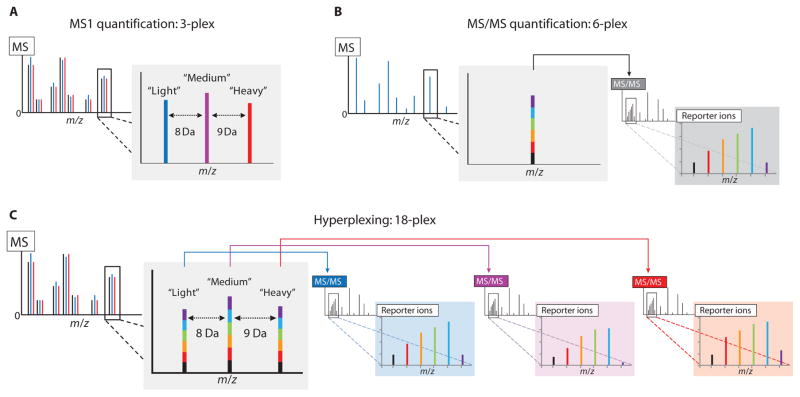
Fig. 1.
Combining labels for MS1 and MS/MS analysis for higher-order proteomic multiplexing. (A) MS1-based quantification, such as SILAC, relies on the incorporation of isotopic labels to separate co-eluting unfragmented peptides with the same sequence in an MS1 scan, enabling their simultaneous detection and quantification. (B) MS/MS-based methods use isobaric tags to label multiple samples with tags of identical intact mass. Unfragmented, same-sequence peptides from the different samples appear as a single composite peak in an MS1 scan. Upon isolation and fragmentation, the different tags produce unique reporter ions that can be detected and quantified in an MS/MS scan. (C) Hyperplexing uses a combination of metabolically and chemically incorporated stable isotope labels to achieve increases in multiplexing capacity. Metabolic labels provide intact mass differences distinguishable in an MS1 scan of intact peptide ions. Upon isolation and fragmentation of the light, medium, and heavy versions of a peptide, the isobaric labels provide separate multiplexed quantitative measurements for each in the MS/MS spectra. A schematic representation of a 3 × 6 hyperplexing experiment using triplex SILAC and six-plex TMT to simultaneously quantify 18 samples is shown.
Seeking to further expand the multiplexing capacity of isobaric tagging, we posited that a mass separation of intact peptides in an MS1 scan could be exploited to allow the simultaneous quantification of multiple sets of isobaric labels in a single run, in a technique that we call “hyperplexing.” For example, a “3 × 6” experiment using three MS1-separable SILAC metabolic labels and six MS/MS-distinguishable TMT isobaric labels enables 18 samples to be monitored in a single run (Fig. 1C). We rigorously tested this hyperplexing method in a biologically meaningful system, using the yeast response to the immunosuppressant drug rapamycin, which inhibits the kinase target of rapamycin (TOR).
The highly conserved TOR pathway acts as a nutrient sensor in all eukaryotes, regulating complex and diverse cellular functions including cell growth and proliferation, ribosomal biogenesis, protein synthesis, autophagy, and aging. By inhibiting the kinase activity of TOR and downstream signaling through this pathway, rapamycin has widespread effects on cellular metabolism (7). Rapamycin has been used for its immunosuppressant activity in organ transplant patients, and its antiproliferative properties have been explored for the treatment of human cancers (8). Whereas several groups have examined the transcriptional response of yeast cells to rapamycin by means of DNA microarrays (9–12), a thorough proteomic analysis has not been reported. Here, we present the details of the hyperplexing MS method that increases the multiplexing capacity of quantitative MS with a combination of metabolic and chemical labels. We demonstrate its efficacy and utility by profiling the yeast response to rapamycin, performing three biological replicate time-course analyses (18 samples) simultaneously.
RESULTS
To test the efficacy of our strategy, we first performed a “2 × 6” experiment, combining pairwise SILAC labels with six-plex TMT labels to compare six biological replicates of rapamycin- or dimethyl sulfoxide (DMSO)–treated yeast samples (Fig. 2A and fig. S1A). Twelve separate cultures of wild-type yeast (NY107) were grown to mid-log phase in synthetic medium containing either naturally occurring lysine (“light”) or [13C615N2]lysine (“heavy”). Three each of the light and heavy cultures were treated with 200 nM rapamycin and the remaining cultures with DMSO for 60 min. The cells were lysed, and proteins were digested with lysyl endopeptidase (lysC). The resulting peptides were labeled with six-plex TMT reagents and mixed in equal amounts before separation by strong cation exchange (SCX) chromatography. We used a reported method using MS2 scans for peptide identification and a subsequent round of isolation and fragmentation with quantification of the reporter ions in the resultant MS3 scans to overcome interference from co-isolated peptides that compromises the accuracy of quantification (13) (fig. S1B).
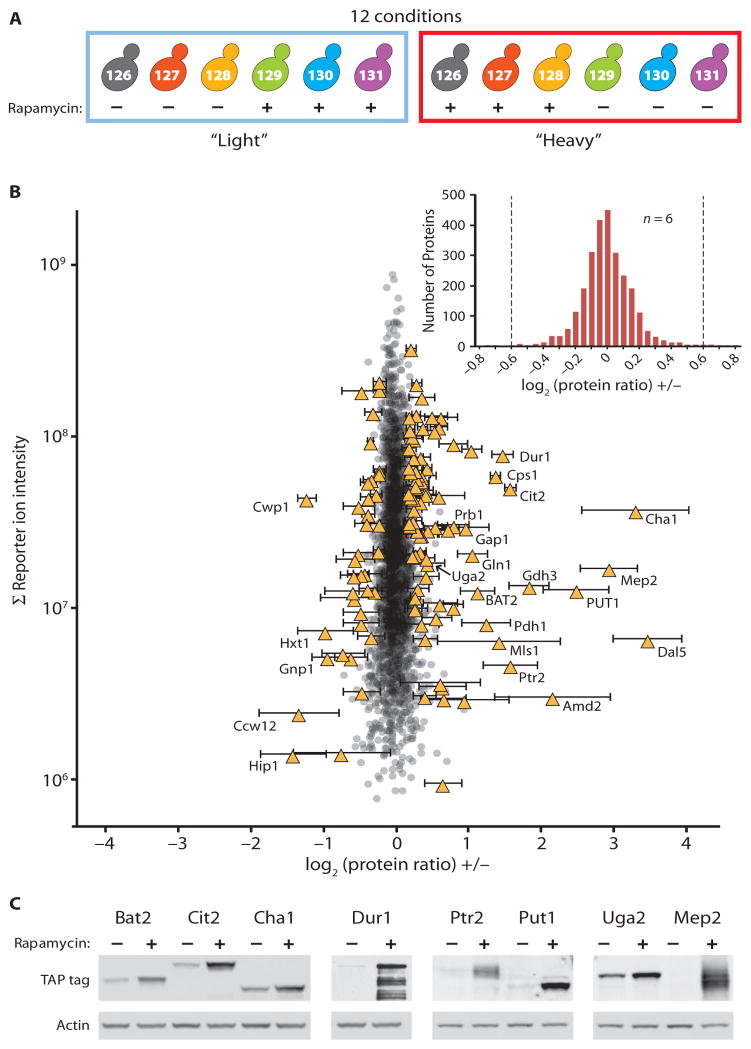
Fig. 2.
Simultaneous quantitative analysis of 12 conditions by 2 × 6 hyperplexing. (A) Six biological replicates of rapamycin- or DMSO-treated cells were analyzed using duplex metabolic labeling (heavy and light) and six-plex isobaric tags (represented by the different colors). The numbers indicate the m/z ratios of the reporter ions generated from each of the six different labeling reagents. (B) A total of 2666 proteins were quantified in all six samples. Each point represents a single protein plotted showing the average log2 (protein ratios) (rapamycin treated: DMSO treated) on the x axis and the summed reporter ion intensities (abundant proteins generate greater ion signals) on the y axis. Significantly changing proteins (121) are shown as yellow triangles with error bars (SD). Select regulated proteins are labeled. The inset shows a histogram of the same data. Dashed lines indicate the position of 1.5-fold changes between rapamycin-treated and control samples. (C) Rapamycin-regulated changes were tested by immunoblotting extracts from epitope-tagged strains. Blots were reprobed for actin as a loading control. Shown are representative results from biological duplicate (Ptr2, Put1, Uga2, Mep2) or triplicate (Bat2, Cit2, Cha1, Dur1) analyses.
We detected and quantified 2981 proteins at a 1% false discovery rate (FDR), 2666 of which were quantified in all six replicates (table S1). Most of the proteins exhibited no change upon rapamycin treatment (Fig. 2B). The log2 SD of the complete set of 2981 proteins was 0.26, less than a 1.2-fold change compared to the control. For the 2666 proteins detected in all six replicates, the log2 SD was 0.23. The width of this distribution was narrower still for proteins that generated higher total reporter ion counts (Fig. 2B). Because we analyzed multiple replicates (n = 6), we could apply stringent statistical cutoffs and confidently assign the significance of small but consistent protein changes. We used Welch’s t test for unequal variances with Benjamini-Hochberg multiple hypothesis testing (14) to assign a significance score (p1) to all protein abundance changes. For each protein, we also calculated a second significance score (p2) from the fold change over control, using a nonparametric outlier significance score (see Materials and Methods). Proteins with both p1 and p2 ≤ 0.05 were considered significantly changed or regulated. We identified 121 regulated proteins, 84 of which increased in abundance and 37 that decreased (table S1 and Fig. 2B, yellow triangles). To test the response of individual proteins to rapamycin treatment, we repeated the experiment in epitope-tagged yeast strains from the TAP tag collection (15) and immunoblotted for the tag. We confirmed the rapamycin-induced increase in the abundance of Cha1, Cit2, Dur1, Bat2, Put1, Ptr2, Mep2, and Uga2 (Fig. 2C). Apparent quantitative disparities between the immunoblot and the MS data may reflect the difference in strain background between the 2 × 6 experiment, K699, and the TAP tag library strain, S288c. Although several papers have reported hundreds of genes with changes at the transcriptional level in response to rapamycin (9–12), our observation of a much smaller number of changes at the protein level (121) after a 60-min treatment is consistent with an earlier proteomic analysis of rapamycin-treated yeast (16).
Most of the identified proteins that were increased in abundance were encoded by known rapamycin-responsive genes; 77 of 84 exhibited equal or greater increases at the mRNA level than those observed at the protein level in either of two independent experiments (10, 11), and 62 of those were equal or greater in both experiments. Many regulated proteins have functions related to the cellular response to nitrogen starvation. Microarray analysis has defined a core group of 91 nitrogen catabolite repression (NCR)–sensitive genes induced by growth on poor nitrogen sources or rapamycin treatment (12). The corresponding gene products were heavily overrepresented in the set of changing proteins (13.8-fold enrichment, P = 2.6 × 10−19); 20 of the 46 NCR gene products measured in our data set were among the 84 proteins increased in abundance, including many of those undergoing the largest changes (table S1). The cellular response to low amounts of nitrogen was also revealed as enriched by Gene Ontology (GO) analysis of the regulated proteins. “Carbon-nitrogen lyase activity” (21.5-fold statistical overenrichment, P = 10−4.9), “transaminase activity” (9.5-fold enrichment, P = 10−3.9), and “amino acid transmembrane transporter activity” (13.8-fold enrichment, P = 10−3.0) were among the most enriched terms (table S2).
To evaluate the reproducibility available through each metabolic labeling channel, we compared the measured fold change over control for the 121 regulated proteins, considering each set of three biological replicates separately. We observed a strong correlation (R2 = 0.87), demonstrating that similar results were obtained from samples grown in both light and heavy SILAC medium (fig. S2A). Finally, as further validation of the accuracy of the hyperplexing method, we repeated the experiment, using SILAC to compare rapamycin- and DMSO-treated yeast (fig. S2B). From a single, binary comparison (n = 1), we detected and quantified 3073 proteins (1% FDR) with SD = 0.28 (table S1 and fig. S3). A comparison of SILAC and hyperplexing ratios for 23 regulated proteins showed excellent agreement in direction and magnitude of change (fig. S2B). Although SILAC analysis provided similar values, a single replicate provided no measure of reproducibility and no opportunity for the kind of statistical analysis that allowed us to identify small but significant changes. For example, Idp1p, a mitochondrial isocitrate dehydrogenase, increased by <25%, but had p1 = 7.9 × 10−4 and p2 = 7.1 × 10−3 (table S1). Acquiring comparable data by duplex SILAC would require six separate analyses, each requiring nearly comparable time and effort to a single hyperplexed analysis.
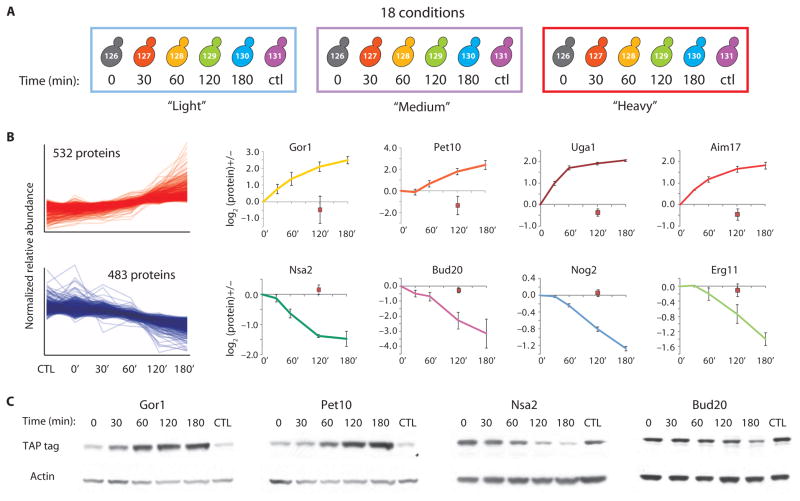
To further demonstrate the utility of the hyperplexing method, we examined the time-resolved response of yeast cells (BY4742) to rapamycin by sampling stimulated cells at 0, 30, 60, 120, and 180 min along with a DMSO-treated control sample at 120 min (Fig. 3A and fig. S4). Using a third metabolic label, [13C615N2D9]lysine, we performed this experiment in triplicate, in a 3 × 6 experiment, measuring all 18 conditions simultaneously. Peptides from all samples were labeled and mixed in equal amounts before separation by SCX and LC-MS/MS analysis. An example of this analysis for a single regulated peptide sequence from Uga1p found in light, medium, and heavy samples appears in fig. S5. We detected and quantified 2745 proteins (1% FDR), 2217 of which were quantified in all three replicates (table S3). In contrast to the fixed-point assay at 60 min, where only a handful of proteins changed, analysis of variance (ANOVA) revealed that more than half of the measured proteins changed (P ≤ 0.05). Log2 protein ratio distributions at each time point revealed that most of these changes were not observed until the 120 min time point (fig. S6). We used the “cluster affinity search technique” (17) to identify patterns within the set of regulated proteins. More than 80% (1015) of the regulated proteins fell into two clusters, one group with proteins that increased in abundance (532 “up-regulated” proteins) and the other with proteins that decreased in abundance (483 “down-regulated” proteins) (Fig. 3B). The data describe the dynamic changes in the abundance of thousands of proteins in response to rapamycin. Representative plots for four proteins from the up-regulated cluster, Gor1 (a glyoxylate reductase), Pet10 (an uncharacterized lipid particle protein), Uga1 (a γ-aminobutyrate trans-aminase), and Aim17 (a protein involved in mitochondrial inheritance), are shown in Fig. 3B along with those for four proteins from the down-regulated cluster, Nsa2 (a component of 66S pre-ribosomal particles), Bud20 (a protein involved in bud-site selection), Nog2 [a putative nucleolar guanosine triphosphatase (GTPase)], and Erg11 (an ergosterol biosynthesis protein). Immunoblotting extracts from epitope-tagged strains subjected to identical rapamycin treatment revealed similar patterns of regulation to those observed by hyperplexing for Gor1, Pet10, Nsa2, and Bud20 (Fig. 3C).
Fig. 3.
Simultaneous quantitative analysis of 18 conditions by 3 × 6 hyperplexing. (A) Three separate cultures grown in light, medium, or heavy medium were treated with 200 nM rapamycin, and samples were removed at 0, 30, 60, 120, and 180 min. A single 120-min sample was taken from parallel cultures treated with DMSO. Equal amounts of peptides from each sample were labeled with six-plex TMT reagents, mixed, and separated by SCX before LC-MS/MS analysis. Significantly changing proteins were determined by ANOVA and clustered using the clustering algorithm search technique (17). (B) More than 80% of changing proteins fell into two dominant clusters, one up-regulated and the other down-regulated upon rapamycin stimulation. Individual protein plots are shown for select proteins. The DMSO control sample appears as a red square. Error bars represent SD. (C) Epitope-tagged strains from the TAP tag collection were treated with rapamycin and analyzed by immunoblotting. Blots were reprobed for actin as a loading control.
GO analysis of the proteins that increased in abundance was dominated by terms related to mitochondrial functions and components (table S2), including “cellular respiration” (2.7-fold enrichment, P = 10−8.1), “proton-transporting adenosine triphosphate (ATP)–synthase complex” (4.2-fold enrichment, P = 10−8.1), “tricarboxylic acid cycle” (3.4-fold enrichment, P = 10−6.2), and “mitochondrial inner membrane” (2.5-fold enrichment, P = 10−11.2). In addition, a number of uncharacterized or minimally characterized mitochondrial proteins were also in the up-regulated cluster. These included seven FMP (“found in mitochondrial proteome”) gene products and six AIM (“altered inheritance rate of mitochondria”) genes. Analysis of the proteins in the down-regulated cluster reflected the well-characterized rapamycin-induced attenuation of protein translation and ribosome biogenesis. Enriched terms included “cytosolic ribosome” (3.4-fold enrichment, P = 10−33.6), “ribosome biogenesis” (2.9-fold enrichment, P = 10−56.2), and “translational initiation” (2.8-fold enrichment, P = 10−7.1). Additional GO analysis results appear in table S2.
DISCUSSION
The most comprehensive proteomic analysis to date of the yeast response to rapamycin quantified fewer than 800 proteins at any of seven time points and only slightly more than half of these at all time points (16). One of the advantages of using isobaric tags is that peptides from all time points are isolated and quantified together; thus, measured peptides are almost always quantified from all samples. We quantified >2500 proteins from four or more time points in each biological replicate and >2200 of these in all three biological replicates. Whereas the previous analysis required weeks of MS, performing this analysis with the hyperplexing method, at about threefold greater protein coverage depth, was completed in about 2 days. The biological triplicate analysis of the time-dependent response to rapamycin provided data for thousands of proteins at a quality similar to that expected of directed studies of individual proteins.
Here, we have described and validated the hyperplexing technique, which combines metabolic labeling and isobaric tags to increase the multiplexing capacity of quantitative proteomics. The increased multiplexing capacity enabled us to perform complex experimentation and do so with biological replicates, providing the statistical power required to identify significant trends. Monitoring 18 separate samples simultaneously, we generated time-resolved protein abundance profiles in response to drug treatment for thousands of proteins. Similar analysis of only a few proteins by immunoblotting could take nearly as much time as an entire hyperplexing experiment and in most systems would require the generation of antibodies or the introduction of an epitope tag for each new protein of interest. When other label combinations are used, even greater channel capacity is possible. With the use of available eight-plex iTRAQ reagents, 24 channels could be monitored, and with additional MS1 labels or new isobaric labeling reagents, even higher capacities could be achieved. Although we have demonstrated hyperplexing in the context of large-scale shotgun proteomics, it is equally applicable to targeted quantitative assays where a single or small number of targets isolated from many different conditions could be quantified without SCX separation in a single short LC-MS/MS analysis. With improvements to instrumentation and the generation of new labeling reagents, hyperplexing can generate global protein abundance data for potentially dozens of samples in parallel. We expect that the increase in proteomic throughput enabled by our method will enhance both the access to and the quality of quantitative proteomic analysis.
MATERIALS AND METHODS
Yeast strains
Strain NY107 (MAT a, ade2-1, trp1-1, can1-100, leu2-3,112, his3, lys1::URA3) was created from a wild-type K699 strain by single-step deletion of Lys1. Strain BY4742 (MAT α, his3Δ1, leu2Δ0, lys2Δ0, ura3Δ0) is the MAT α, wild-type S288c strain. All TAP tag strains are derived from the TAP tag collection (15) made in a BY4741 background (MAT a, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0), the MAT a, wild-type S288c strain through single-step insertion of the TAP tag cassette at the C terminus of each target gene.
Cell growth and lysis
For the fixed-point assay, 12 separate cultures of wild-type yeast (NY107) were grown to mid-log phase at 30°C in synthetic complete medium containing either naturally occurring lysine (light) or [13C615N2]lysine (heavy) (Cambridge Isotopes). Three each of light and heavy cultures were treated with 200 nM rapamycin (LC Laboratories), and the remaining six cultures with an equal volume of DMSO for 60 min at 30°C. Samples were normalized to cell number on the basis of optical density and mixed pairwise such that each pair consisted of one rapamycin-treated and one untreated sample, as well as one light and one heavy sample. The combined cell pellets were washed with ice-cold water, aspirated, frozen in liquid nitrogen, and stored at −80°C. Cells were thawed on ice and then resuspended in 700 μl of urea lysis buffer containing protease inhibitors [8 M urea, 75 mM NaCl, 50 mM tris-HCl (pH 8.0), 1 mM AEBSF [4-(2-aminoethyl) benzene-sulfonyl fluoride hydrochloride], 2.5 mM benzamidine, leupeptin (1 μg/ml), and pepstatin (1 μg/ml)]. Acid-washed glass beads (1.5 ml; BioSpec) were added, and the cells were ruptured by bead-beating on a BioSpec Mini-Beadbeater using 3 × 60-s cycles with 3-min rests on ice between cycles. Lysates were transferred to fresh tubes and cleared by centrifugation at 21,000g for 10 min at 4°C. The supernatants were removed to fresh tubes, and the protein concentration was measured by dye-binding assay (Bio-Rad). Disulfide bonds were reduced by addition of dithiothreitol to a final concentration of 2.5 mM and incubating at 56°C for 40 min. The extract was allowed to cool to room temperature, and the reduced cysteines were alkylated by addition of iodoacetamide to 7.5 mM and incubation for 40 min in the dark at room temperature. Alkylation was quenched with an additional 5 mM dithiothreitol. For time-course experiments, three cultures of wild-type yeast (BY4742) were grown in medium containing natural lysine (light), [13C615N2]lysine (medium), or [13C615N2D9]lysine (heavy) (Cambridge Isotopes). Cells were treated with 200 nM rapamycin or DMSO. Equal cell numbers were harvested at 0, 30, 60, 120, and 180 min from the rapamycin-treated sample and at 120 min from the DMSO-treated sample. Cells were lysed and prepared for digestion as described above.
Peptide digestion and labeling
Proteins were diluted 2.5-fold into 25 mM (final concentration) tris-HCl (pH 8.8) and digested by the addition of lysC (Wako) to a final concentration of 10 ng/μl with gentle agitation overnight at room temperature. Digested peptides were acidified by the addition of neat formic acid to 1%, and the resultant precipitate was pelleted by centrifuging for 2 min at 21,000g. The supernatants were desalted on 200-mg tC18, reversed-phase solid-phase extraction cartridges (Waters) with 1% formic acid as aqueous loading and wash buffer, eluted with 750 μl of 70% acetonitrile (CH3CN)/1% formic acid, and dried in a SpeedVac (Thermo Fisher). Dried peptides were resuspended in 100 μl of 1% formic acid. Peptides were normalized on the basis of the protein assay, taking off 100 μg total from each sample into a fresh tube, and were dried once again in a SpeedVac. Dried peptides were resuspended in 100 μl of 0.2 M Hepes (pH 8.5). TMT six-plex reagents (0.8 mg per vial) (Thermo Fisher) were resuspended in 41 μl of anhydrous CH3CN and 10 μl of each reagent was added to each sample. Reactions were allowed to proceed at room temperature for 1 hour, after which they were quenched by the addition of 8 μl of 5% hydroxylamine for 15 min and then acidified by the addition of 16 μl of neat formic acid. Reaction products were combined and 1 ml of 1% formic acid was added before desalting on a 100-mg tC18 Sep-Pak. Eluted peptides were dried in a SpeedVac and stored at −20°C.
SCX chromatography
SCX was performed as described (18) with some changes. Briefly, 600 μg of labeled peptide mixture from all samples was resuspended in 300 μl of SCX buffer A [7 mM KH2PO4 (pH 2.65) and 30% CH3CN] and separated on a 4.6 mm × 20 cm PolySULFOETHYL A column (5-μm particle size, 200 Å pore size) (PolyLC) with a two-stage gradient of 8 to 20% SCX buffer A to SCX buffer B [7 mM KH2PO4 (pH 2.65), 30% CH3CN, and 350 mM KCl] at 0.5 ml/min for 16 min followed by a 20 to 40% step at 1.0 ml/min over 20 min. Fractions were collected at 1.5-min intervals in a 96-well plate and desalted on an Empore C-18, 96-well plate (3M).
MS analysis
SCX fractions were resuspended in 50 μl of 5% formic acid, and 1 μl was analyzed on an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with an Agilent 1100 binary pump (Agilent Technologies) or an Accela 600 quaternary pump (Thermo Fisher Scientific) and a Famos Microautosampler (LC Packings). Nanospray tips were hand-pulled with 100-μm inside diameter fused-silica tubing and packed with 0.5 cm of Magic C4 resin (5 μm, 100 Å; Michrom Bioresources) followed by 20 cm of Maccel C18AQ resin (3 μm, 200 Å; Nest Group). Peptides were separated with a gradient of 6 to 24% CH3CN in 0.125% formic acid over 150 min (2 × 6 experiment) or 120 min (3 × 6 experiment) at a flow rate of ~500 nl/min. Peptides were detected in a hybrid dual-cell quadrupole linear ion trap–orbitrap mass spectrometer (LTQ Orbitrap Velos, Thermo Fisher) by means of a data-dependent Top10-MS2/MS3 method (13). For each cycle, one full MS scan of mass/charge ratio (m/z) = 400 to 1800 was acquired in the Orbitrap at a resolution of 30,000 or 60,000 at m/z = 400 with automatic gain control (AGC) target of 2 × 106. Each full scan was followed by the selection of the most intense ions, up to 10, for collision-induced dissociation (CID) using the wide-band activation feature and MS2 analysis in the linear ion trap for peptide identification and subsequent higher-energy collisional dissociation (HCD) and MS3 analysis in the Orbi-trap for quantification of the TMT reporter ions. AGC targets of 4 × 103 and 2 × 104 were used for MS2 and MS3 scans, respectively. Ions selected for MS2 analysis were excluded from reanalysis for 120 s (2 × 6) or 90 s (3 × 6). Ions with a charge of 1 or unassigned were also excluded from further analysis. A single MS3 scan was performed for each MS2 scan selecting the most intense ion from the MS2 falling in the range of 110 to 160% of the mass of the MS1-detected precursor ion for fragmentation in the HCD cell. The resultant fragment ions were detected in the orbitrap at a resolution of 7500. Maximum ion accumulation times were 1000 ms for each full MS scan, 150 ms for MS2 scans, and 250 ms for MS3 scans. Lockmass, with atmospheric polydimethylsiloxane (m/z = 371.1012) as an internal standard, was used in all runs to calibrate orbitrap MS precursor masses. SILAC-only samples were analyzed as described above for the hyperplexing experiments with the following changes. Samples were separated with a 90-min gradient. SCX fractions were analyzed on an LTQ Orbitrap Velos with a data-dependent Top20-MS2 method. For each cycle, one full MS scan of m/z = 300 to 1500 was acquired in the orbitrap. Each full scan was followed by the selection of the most intense ions, up to 20, for CID and MS2 analysis in the linear ion trap. Ions selected for MS2 analysis were excluded from reanalysis for 90 s.
Peptide identification and filtering
MS2 spectra were searched using SEQUEST v.28 (rev. 13) against a composite database containing the translated sequences of all predicted open reading frames of S. cerevisiae (http://downloads.yeastgenome.org, downloaded 30 October 2009) and its reversed complement with the following parameters: a precursor mass tolerance of ±20 parts per million (ppm); 1.0-dalton product ion mass tolerance; lysC digestion; up to two missed cleavages; static modifications of carbamidomethylation on cysteine (+57.0214), TMT reagent adducts (+229.1629) on lysine and peptide amino termini; and dynamic modifications of methionine oxidation (+15.9949), 13C615N2-lysine (+8.0142), and for the 3 × 6 experiment, 13C615N2D9-lysine (+17.0706). SILAC-only searches were performed in the same manner, omitting the TMT (+229.1639) static modification. Peptide spectral matches were filtered to 1% FDR using the target-decoy strategy (19) combined with linear discriminant analysis (LDA) (20) using several different parameters including the SEQUEST Xcorr and ΔCn′ scores, precursor mass error, observed ion charge state, and predicted solution charge state. Linear discriminant models were calculated for each LC-MS/MS run with peptide matches to forward and reversed protein sequences as positive and negative training data. Peptide spectral matches within each run were sorted in descending order by discriminant score and filtered to a 1% FDR as revealed by the number of decoy sequences remaining in the data set. The data were further filtered to control protein-level FDRs. Peptides from all fractions in each experiment were combined and assembled into proteins. Protein scores were derived from the product of all LDA peptide probabilities, sorted by rank, and filtered to 1% FDR as described for peptides. The FDR of the remaining peptides fell markedly after protein filtering. Remaining peptide matches to the decoy database were removed from the final data set.
Peptide quantification
Raw reporter ion intensities were denormalized by multiplying with the ion accumulation times for each MS3 scan and corrected for isotopic overlap between reporter ions by using empirically derived values. We required each peptide to have denormalized reporter ion intensities ≥20 for the zero time point and at least four of six TMT channels. In cases where a protein was completely absent from either the stimulated or the unstimulated sample, this filter would exclude highly regulated proteins from consideration. Manual inspection of the disqualified peptides identified only one regulated protein, Asp3-4, the product of an NCR-sensitive gene (12), that was omitted from three of six replicates in the 2 × 6 experiment. SILAC ratios were calculated automatically with the VISTA program (21). We required a signal-to-noise ratio of ≥2. For the 2 × 6 experiment, 14 SCX fractions were analyzed on an LTQ Orbitrap Velos, leading to the identification and quantification of 54,749 peptides (0.2% FDR) and 2981 proteins (1.0% FDR). Of the identified peptides, 68% were found in both light and heavy SILAC channels. For the 3 × 6 time-course experiment, 20 SCX fractions were analyzed as described, leading to the identification and quantification of 76,341 peptides (0.2% FDR) and 2745 proteins (1.0% FDR). For all identified peptides in this data set, 53% were found in all three SILAC channels, 77% in at least two channels, and 23% in only a single channel.
Protein quantification
Peptides assigned to light and heavy, or light, medium, and heavy labeled samples were analyzed separately. C-terminal peptides lacking lysine cannot be assigned to either light, medium, or heavy channels and were removed from the data set. Reporter ion intensities were normalized by scalar transformation to equalize the sum intensity of all observed signals for each channel across all peptides in each metabolic labeling channel, effectively recentering the distribution of all peptide ratios at 1:1. Relative protein abundances were calculated as a weighted average from the ratio of the summed reporter ion intensities of all peptides for each protein in the rapamycin- and DMSO-treated samples and log2-transformed for all subsequent analysis. Protein ratios from the SILAC experiment were calculated as described (2) by means of the median log2 ratio of all peptides for each protein. For proteins represented by a single quantified peptide, a minimum signal-to-noise ratio of ≥5 was required for both light and heavy signals. The ratios were normalized to recenter the distribution at 1:1 (log2 = 0).
Statistical analysis
For the 2 × 6 experiment, two significance values were calculated for proteins quantified in all six biological replicates. p1 was calculated from the reporter ion intensities with a two-tailed Welch’s t test with unequal variance and was adjusted for multiple hypothesis testing with the method of Benjamini-Hochberg (14). We generated a second significance score on the basis of the magnitude of the average log2 protein ratio relative to its position in the total population as described (22). Proteins were grouped into bins of about 300 by their summed reporter ion intensities, and a robust z score was calculated for each protein using only those proteins within each bin. Significance score p2 was calculated as , where erfc(z) is the complementary error function. Proteins with p1 and p2 ≤0.05 were classified as significantly changed (regulated) upon rapamycin treatment. The significance of overrepresentation of NCR genes in the set of up-regulated proteins was calculated from the hypergeometric distribution with WolframAlpha (http://www.wolframalpha.com/). ANOVA and clustering were performed with the multiexperiment viewer (23) using the default parameters for each. Raw P values for ANOVA were corrected with Benjamini-Hochberg (14) multiple hypothesis testing. Proteins with adjusted P ≤ 0.05 were clustered with multiexperiment viewer and the clustering affinity search technique (17).
Immunoblotting
Immunoblotting was performed by standard methods. Each strain from the TAP tag collection (15) was grown to mid-log phase, split into two cultures of equal volumes, and treated with 200 nM rapamycin or DMSO for the indicated times. Cells were lysed by boiling in 0.1 M NaOH and 2% SDS (24). Extracts were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Transfer efficiency was confirmed by Ponceau S staining. The membranes were immunoblotted with an antibody recognizing the calmodulin-binding peptide (GenScript) followed by horseradish peroxidase–coupled anti-rabbit immunoglobulin G (Jackson Immunoresearch Laboratories) and developed using chemiluminescent substrate (Thermo Scientific). Membranes were stripped with Restore Western Blot Stripping Buffer (Thermo Scientific) and re-blotted for actin (Abcam).
GO analysis
GO analysis was performed with High-Throughput GoMiner (25). For the 2 × 6 experiment, the set of 2666 proteins quantified in all six replicates was used as the background, and the sets of 84 and 37 up- and down-regulated proteins as the query sets. For the 3 × 6 experiment, the set of 2217 proteins observed in all replicates served as the background, and the 532 up-regulated and 483 down-regulated proteins were the query sets. Categories listed in table S2 were selected to remove redundancy and highlight the most significant findings.
Supplementary Material
Fig. S1. Proteomic analysis of rapamycin-stimulated yeast by hyperplexing.
Fig. S2. Quantitative reproducibility and accuracy of the hyperplexing method.
Fig. S3. Protein quantification by SILAC.
Fig. S4. “3 × 6” hyperplexed analysis of rapamycin-stimulated yeast.
Fig. S5. Workflow for identification and quantification of a representative protein from the “3 × 6” experiment.
Fig. S6. Delayed response of the yeast proteome to 200 nM rapamycin.
Table S1. Log2 ratios of the proteins from the “2 × 6” experiment. (Excel file)
Table S2. Gene ontology analysis of the regulated proteins from the “2 × 6” and “3 × 6” experiments. (Excel file)
Table S3. Log2 ratios of the proteins from the “3 × 6” experiment of rapamycin-treated yeast. (Excel file)
Acknowledgments
We thank J. Rogers at Thermo Fisher for helpful suggestions and E. Huttlin for helpful comments and discussion.
Funding: Supported by grants from the U.S. NIH to S.P.G. (HG003456 and GM067945).
Footnotes
Author contributions: N.D. and S.P.G. designed the research; N.D. performed all experiments and data analysis; and N.D. and S.P.G. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All proteomic raw data have been deposited in the Proteome Commons Tranche Repository in three sets with the following hashes: lL+VbZyU5EbEir6/HbrNnxg/zD0vzH3XoycRfDHQ3tqlGOvCY4Shl2C7Z8L7Knu7rR85-RZp4ZH1JxXsmOzo9hOsqKy8AAAAAAAAIdA==, Viph3LfvM44fYdJQI9ak9wSFBJ4xtyeicCfqkfg+ ZyqvRPxVuYPk/+PJOW393oE0oNuNWQ+a6bGTjhdbQ4eviaoVvuQAAAAAAAAG6Q==, and uoaJP4NSizi8fXTWhXMh/fN/bDJPkrztOlbxoxFk4H1TbLQU2qcDK0NPqiH9wlX3oeM1-FNGrXr2Q8FqyfVD0kXUeusMAAAAAAAALDA==.
REFERENCES AND NOTES
- 1.de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 2.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu R, Dephoure N, Haas W, Huttlin EL, Zhai B, Sowa ME, Gygi SP. Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol Cell Proteomics. 2011;10:M111.009654. doi: 10.1074/mcp.M111.009654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 6.Walsh GM, Rogalski JC, Klockenbusch C, Kast J. Mass spectrometry-based proteomics in biomedical research: Emerging technologies and future strategies. Expert Rev Mol Med. 2010;12:e30. doi: 10.1017/S1462399410001614. [DOI] [PubMed] [Google Scholar]
- 7.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabatini DM. mTOR and cancer: Insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 9.Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Zhu H, Haggarty SJ, Spring DR, Hwang H, Jin F, Snyder M, Schreiber SL. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc Natl Acad Sci USA. 2004;101:16594–16599. doi: 10.1073/pnas.0407117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, Edenberg HJ, Wek RC. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J Biol Chem. 2010;285:16893–16911. doi: 10.1074/jbc.M110.121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scherens B, Feller A, Vierendeels F, Messenguy F, Dubois E. Identification of direct and indirect targets of the Gln3 and Gat1 activators by transcriptional profiling in response to nitrogen availability in the short and long term. FEMS Yeast Res. 2006;6:777–791. doi: 10.1111/j.1567-1364.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 13.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 15.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 16.Fournier ML, Paulson A, Pavelka N, Mosley AL, Gaudenz K, Bradford WD, Glynn E, Li H, Sardiu ME, Fleharty B, Seidel C, Florens L, Washburn MP. Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol Cell Proteomics. 2010;9:271–284. doi: 10.1074/mcp.M900415-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Dor A, Shamir R, Yakhini Z. Clustering gene expression patterns. J Comput Biol. 1999;6:281–297. doi: 10.1089/106652799318274. [DOI] [PubMed] [Google Scholar]
- 18.Villén J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 20.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakalarski CE, Elias JE, Villén J, Haas W, Gerber SA, Everley PA, Gygi SP. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J Proteome Res. 2008;7:4756–4765. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 23.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 24.von der Haar T. Optimized protein extraction for quantitative proteomics of yeasts. PLoS One. 2007;2:e1078. doi: 10.1371/journal.pone.0001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, Burt SK, Elnekave E, Hari DM, Wynn TA, Cunningham-Rundles C, Stewart DM, Nelson D, Weinstein JN. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Proteomic analysis of rapamycin-stimulated yeast by hyperplexing.
Fig. S2. Quantitative reproducibility and accuracy of the hyperplexing method.
Fig. S3. Protein quantification by SILAC.
Fig. S4. “3 × 6” hyperplexed analysis of rapamycin-stimulated yeast.
Fig. S5. Workflow for identification and quantification of a representative protein from the “3 × 6” experiment.
Fig. S6. Delayed response of the yeast proteome to 200 nM rapamycin.
Table S1. Log2 ratios of the proteins from the “2 × 6” experiment. (Excel file)
Table S2. Gene ontology analysis of the regulated proteins from the “2 × 6” and “3 × 6” experiments. (Excel file)
Table S3. Log2 ratios of the proteins from the “3 × 6” experiment of rapamycin-treated yeast. (Excel file)