Abstract
The arachidonic acid preferred long-chain acyl-CoA synthetase 4 (ACSL4) is a key enzyme for fatty acid metabolism in various metabolic tissues. In this study, we utilized hamsters fed a normal chow diet, a high-fat diet or a high cholesterol and high fat diet (HCHFD) as animal models to explore novel transcriptional regulatory mechanisms for ACSL4 expression under hyperlipidemic conditions. Through cloning hamster ACSL4 homolog and tissue profiling ACSL4 mRNA and protein expressions we observed a selective upregulation of ACSL4 in testis and liver of HCHFD fed animals. Examination of transcriptional activators of the ACSL family revealed an increased hepatic expression of PPARδ but not PPARα in HCHFD fed hamsters. To explore a role of PPARδ in dietary cholesterol-mediated upregulation of ACSL4, we administered a PPARδ specific agonist L165041 to normolipidemic and dyslipidemic hamsters. We observed significant increases of hepatic ACSL4 mRNA and protein levels in all L165041-treated hamsters as compared to control animals. The induction of ACSL4 expression by L165041 in liver tissue in vivo was recapitulated in human primary hepatocytes and hepatocytes isolated from hamster and mouse. Moreover, employing the approach of adenovirus-mediated gene knockdown, we showed that depletion of PPARδ in hamster hepatocytes specifically reduced ACSL4 expression. Finally, utilizing HepG2 as a model system, we demonstrate that PPARδ activation leads to increased ACSL4 promoter activity, mRNA and protein expression, and consequently higher arachidonoyl-CoA synthetase activity. Taken together, we have discovered a novel PPARδ-mediated regulatory mechanism for ACSL4 expression in liver tissue and cultured hepatic cells.
Keywords: ACSL4, PPARδ, Hamsters, Dietary cholesterol, Dyslipidemia, NAFLD
1. Introduction
The liver plays a central role in fatty acid (FA) homeostasis by regulating the uptake, synthesis, oxidation and export of FA based upon different nutritional conditions. Dysregulation of long chain FA (LCFA) has been associated with metabolic diseases such as non-alcoholic fatty liver disease (NAFLD) [1], type 2 diabetes mellitus [2,3] and cancer [4].
Long chain acyl-CoA synthetase (ACSL) is a family of five enzymes (ACSL1, ACSL3, ACSL4, ACSL5 and ACSL6) which esterifies a LCFA into acyl-CoA in the process of fatty acid activation [5,6]. Because LCFA must be esterified to LCFA-CoA in order for entry into various metabolic pathways including FA β-oxidation, triglyceride generation and phospholipid synthesis, ACSL is considered a rate determining enzyme in FA metabolism [4,7–10]. ACSL4, in particular, has a substrate preference for arachidonic acid (AA) and plays an important role in the cellular metabolism of this polyunsaturated FA in different tissues [5,11]. Human ACSL4 has two isoforms with the longer one of 711 amino acids expressed in the brain, while the shorter isoform of 670 amino acids is expressed in other adult tissues at different levels [12]. Clinical studies have reported that ACSL4 gene mutation causes developmental and neuronal problems such as X-linked mental retardation [12]. Based on these observations, new mechanistic studies in zebrafish demonstrated that ACSL4 regulates Bmp-Smad signaling and that it can influence the developmental process significantly [13].
Recently, multiple in vitro studies described various functions of ACSL4 in different cell lines. For example, exogenous overexpression of ACSL4 in rat smooth muscle cell inhibited IL-1β induced PGE2 secretion [14]. In breast cancer cells exogenous expression of ACSL4 promoted the cell invasiveness by regulating AA availability for enzymes such as COX, LOX and CYP450s to generate eicosanoids [15]. In pancreatic cell lines, diminished ACSL4 expression by siRNA mediated knockdown reduces glucose-stimulated insulin secretion [16]. Studies also have shown ACSL4 being capable of channeling fatty acid towards phosphatidylinositol production in COS-7 cells [17] and that it also affects AA incorporation into phospholipids in rat fibroblasts [18]. Besides being involved in fatty acid metabolism, ACSL4 is reported to regulate steroidogenic acute regulatory protein [19], which is the rate determining enzyme in steroidogenesis. Thus, ACSL4 regulates steroid hormone production in testis and adrenal gland indirectly.
Despite ACSL4's diverse functions, there are limited literature reports about its regulation. ACSL4 expression in steroidogenic cells was shown to be regulated by hormones such as EGF, ACTH, 8Br-cAMP and NGF [19]. Also, apolipoprotein O depletion in HepG2 cells causes ACSL4 mRNA to upregulate [20]. ACSL4 gene expression in mouse liver is regulated by circadian clock [21]. In addition, the transcription of ACSL4 gene in mouse steroidgenic MA-10 cells was induced by cAMP via a CRE sequence embedded in the proximal promoter region of mouse ACSL4 gene [22]. At the post-transcriptional level, ACSL4 mRNA has been identified as target for several microRNAs. It was reported that miR-347 upregulates ACSL4 in neuronal cells [23] and miR-224 downregulates ACSL4 mRNA during adipocyte differentiation [24]. ACSL4 protein is also subject to post-translational modification. Our laboratory has recently identified an AA-induced and proteasome-mediated degradation mechanism to downregulate ACSL4 protein expression in liver cells [25], and another group reported that ACSL4 forms homodimers which enables PKA and PKC to induce or reduce, respectively, its activity in Y1 cells through phosphorylation [19].
While above described studies in different cell lines have provided some understanding of the regulatory mechanisms that modulate ACSL4 expression and activity in cultured cells, to date, no precedent studies have examined the in vivo regulation of ACSL4 in different metabolic organs under normal and dyslipidemic conditions.
The golden Syrian hamster has been increasingly utilized to study lipoprotein metabolism and to evaluate the effects of hypolipidemic agents such as PPAR agonists [26,27], statins [28–30] and CETP inhibitors [31] because hamsters share more characteristic features in lipid metabolism with those found in humans than mouse or rat [32,33]. In particular, hamsters rapidly respond to dietary cholesterol and high-fat diet with elevated plasma lipid levels and hepatic accumulations of cholesterol and neutral lipids in a fashion comparable to humans. Therefore, the aims of this current study were to employ hamsters as an in vivo model to profile the mRNA and protein expressions of ACSL4 in different tissues of animals fed a normal chow diet (NCD), a high-fat diet (HFD) or a high-cholesterol and high-fat diet (HCHFD), and to further identify critical transcriptional regulatory mechanisms that modulate ACSL4 gene expression and activity under hyperlipidemic conditions in a tissue-specific fashion.
2. Materials and methods
2.1. Cells and reagents
Human hepatoma HepG2 cells and mouse hepatoma AML12 cells were obtained from ATCC. HEK293A cell line was obtained from Invitrogen. Hamster primary hepatocytes were isolated from male golden Syrian hamsters in our laboratory. Human primary hepatocytes were obtained from Invitrogen. Mouse primary hepatocytes were isolated from male C57BL/6 J mice at San Francisco General Hospital Liver Center. L165041 and GW0742 were purchased from TOCRIS Biosciences (Bristol, UK). Cholesterol, 25-hydroxylcholesterl and WY-14643 were purchased from Sigma, and 6α-ethyl-chenodeoxycholic acid (6ECDCA) and 24(S)-hydroxylcholesterol (24-OHC) were purchased from Abcam (Cambridge, MA).
2.2. Culture of primary hepatocytes
Primary hepatocytes were seeded on collagen coated plates at a density of 1 × 105 cells/well in a 24-well plate in Williams E Medium supplemented with a Cell Maintenance Cocktail (Cell Maintenance Supplement Pack, Invitrogen). After overnight seeding, cells were treated with different compounds or cytokine oncostatin M (OM) for 24 h before isolation of total RNA or cell lysates.
2.3. Cloning of hamster ACSL4 complete coding sequence
To clone the hamster ACSL4 complete coding sequence, as previously described for hamster ACSL3 cloning [34], we first compared mRNA sequence from human, mouse and rat ACSL4 gene and selected highly conserved regions across these species for primer sets design. Using primers identified by this approach, hamster ACSL4 coding region was amplified from a hamster liver cDNA library, cloned into pCR2.1-Topo vectors (Invitrogen, Carlsbad, CA, USA) and sequenced. The primers are listed in Supplemental Table 1. The hamster ACSL4 coding region was subcloned into pcDNA4.0-HisMax-TOPO vector, resulting in the plasmid pHis-HamACSL4 for hamster ACSL4 expression in mammalian cells. Recently, the genomic sequence of a female golden Syrian hamster (Mesocricetus auratus) became available on NCBI. The NCBI reference sequence (XP_005082531.1) predicted ACSL4X5 protein sequence differs from our ACSL4 protein sequence at amino acid position 311 with a proline substituted by serine.
2.4. Animals and diets
All animal experiments were performed according to procedures approved by the VA Palo Alto Health Care System Animal Care and Use Committee (IACUC). Ten-week old male golden Syrian hamsters were purchased from Harlan. Hamsters were housed (2 animals/cage) under controlled temperature (72 °F) and lighting (12 h light/dark cycle). Animals had free access to autoclaved water and food. Hamsters were fed a NCD (Teklad Global 18% Protein Rodent Diet 2018; 0% cholesterol), a HCHFD (Research Diets D12336; 35% calories from fat; 1.25% cholesterol) or a HFD (Harlan TD.88137; ~42% of total calories from fat; 0.15% cholesterol) for two weeks. At the experimental termination, hamsters were fasted overnight before euthanization for serum and tissue collections.
2.5. Drug treatments
For studying PPARδ activation in normolipidemic hamsters, eighteen hamsters fed NCD were divided into two groups. One group was given a daily dose of 10 mg/kg of L165041 intraperitoneally and the other group was given equal volume of vehicle [35]. The treatment lasted for 7 days. Hamsters were fasted overnight before euthanization for serum and liver tissue collections.
To study PPARδ activation under hyperlipidemic conditions, hamsters were fed the HFD for two months to induce hyperlipidemia and hepatosteatosis. After 8-weeks on HFD, overnight fasting blood samples were taken to randomize hamsters into homogenous treatment groups according to total cholesterol (TC) levels. Hamsters were then maintained on the HFD and were treated orally with vehicle (0.5% hydroxypropyl methylcellulose in PBS) or L165041 (10 mg/kg, once a day) for 1 week. At the experimental termination, hamsters were fasted overnight before euthanization for serum and tissue collections.
2.6. RNA isolation and quantitative real-time PCR (qPCR)
Total RNA isolation, generation of cDNA, and qPCR were conducted as previously reported [36]. Each cDNA sample was run in duplicate. The correct size of the PCR product and the specificity of each primer pair were validated by examination of PCR products on an agarose gel. Species-specific primer sequences used in qPCR are listed in Supplemental Table 1. Target mRNA expression in each sample was normalized to the housekeeping gene GAPDH. The 2−ΔΔCt method was used to calculate relative mRNA expression levels.
2.7. Western blot analysis
Approximately 50 mg of frozen tissue was homogenized in RIPA buffer containing 1 mM PMSF and protease inhibitor cocktail (Roche). After protein quantitation using BCA protein assay reagent (Pierce), 50 µg of homogenate proteins from individual tissue samples was resolved by SDS-PAGE and transferred to nitrocellulose membranes. The anti-ACSL4 antibody recognizes a 15 AA peptide of N-terminal sequences of ACSL4 of human, mouse, rat and hamster origins [25]. Rabbit anti-hamster ACSL3 antibody was previously generated in our laboratory that recognizes the C-terminal sequences of ACSL3 of human, mouse, rat and hamster origins [34]. The anti-human ACSL1 (ab76702) was obtained from Abcam, Cambridge, MA. The membranes were reprobed with anti-β-actin (Sigma), anti-GAPDH (Sigma) or anti-transferrin receptor (TFR) antibodies. Immunoreactive bands of predicted molecular mass were visualized using an ECL plus kit (GE Healthcare life Sciences, Piscataway, NJ) and quantified with the Alpha View Software with normalization by signals of β-actin or GAPDH.
2.8. Measurement of tissue acyl-CoA synthetase activity
Approximately 50 mg frozen tissue were dounce-homogenized 15 times in a buffer containing 250 mM sucrose, 10 mM Tris, pH 7.4, 1 mM EDTA, 1 mM dithiothreitol (DTT), protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Sigma). Homogenates were centrifuged at 16,000 ×g for 30 min at 4 °C, protein concentrations in supernatants were determined by BCA assay, and aliquots were stored at −80 °C. Initial rates of total ACSL activity in tissue homogenate were measured using 5 µg of homogenate at 37 °C in the presence of 175 mM Tris (pH 7.4), 8 mM MgCl2, 5 mM DTT, 10 mM ATP, 250 µM CoA, 50 µM [3H]arachidonic acid (AA) in 0.5 mM Triton X-100, and 10 µM EDTA in a total volume of 0.1 ml. The reaction was initiated by adding the homogenized sample and terminated by adding 1 ml Dole's reagent as previously described [36]. Generated [3H]AA-CoA were extracted, and the radioactivity was determined in a scintillation counter. The radioactivity in the reaction that contained all components but omitted homogenate was included as a negative control.
2.9. Construction of Ad-ham-shPPARδ adenoviral vector for PPARδ knockdown
A U6 promoter-based vector (pSH-PPARδ) that expresses a shRNA targeting hamster PPARδ coding region (5′-GCAAGCCCTTCAGTGACATCA-3′) was generated using Invitrogen's BLOCK-iT™ U6 RNAi Entry Vector Kit following the manufacturer's instruction. The sequence identity and orientation of PPARδ shRNA in pSH-PPARδ were confirmed by sequencing. The pSH-PPARδ plasmid was then recombined with pAD/BLOCK-iT DEST vector to generate Ad-shPPARδ viral vector and transduced it into HEK293A cells. The crude viral stocks were further amplified and purified.
2.10. Adenoviral transduction
Hamster primary hepatocytes were seeded at 1 × 105 cells/well in 24-well plates overnight. The next day, cells were transduced with the adenovirus at various multiplicity of infection (MOI) in 1 ml medium containing 0.5% FBS for 8 h. Then viral-containing medium was replaced by fresh complete medium and cells were cultured for 72 h before harvesting.
2.11. Constructions of human ACSL4 promoter luciferase reporters
For generation of ACSL4 promoter reporter, a DNA fragment of 3026 bp covering human ACSL4 proximal promoter region from −2997 to +29 relative to the 5′ end of exon 1 was amplified from HepG2 genomic DNA and was cloned into Topo 2.1 vector, followed by subcloning into pGL3-basic at the Sac1 and Xho1 sites to yield the promoter reporter pGL3-ACSL4-3Kb. Likewise, the plasmid pGL3-ACSL4-0.7Kb was made that covered the genomic region of −622 to +29. After transformation and propagation in E. coli, two independent clones per reporter constructs were sequenced to verify the sequence and orientation of the promoter fragment.
2.12. Luciferase reporter assay
Luciferase reporter assay was performed by using Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Promoter luciferase reporters were individually transfected into HepG2 cells along with plasmid pRL-SV40, a renilla luciferase reporter as an internal transfection efficiency control. One day after transfection, cells were switched to low serum medium for overnight before the addition of PPARδ agonist L165041 and cells were lysed 24 h later. Cells were lysed with 50 µl of lysis buffer followed by measurements of firefly luciferase and renilla luciferase activities. The firefly luciferase activity was normalized to renilla activity. Four wells were assayed for each condition.
2.13. Statistical analysis
Values are presented as the mean ± SEM. Significant differences between diet or treatment groups were assessed by two-tailed Student t-test (nonparametric Mann Whitney test) or one-way ANOVA with Dunnett's posttest. Statistical significance is displayed as p < 0.05 (one asterisk), p < 0.01 (two asterisks) or p < 0.001 (three asterisks).
3. Results
3.1. Cloning of the hamster ACSL4 coding region
To examine the dietary effects on ACSL4 expression in hamster tissues, we first cloned the ACSL4 coding region from a male golden Syrian hamster liver cDNA. The hamster ACSL4 cDNA coding sequence is 2013 base pairs in length that encodes the hamster ACSL4 protein of 670 amino acids with a calculated molecular weight of 74.4 kD. The overall homology of hamster ACSL4 amino acids to rat, mouse and human is 98, 97, and 97%, respectively (Supplemental Fig. 1). The hamster ACSL4 expressing plasmid (pHis-HamACSL4) was transfected into HepG2 and HEK293A cells. Western blot analysis using anti-His antibody detected the expression of His-tagged hamster ACSL4 as the predicted molecular mass of ~75 kD protein in HepG2 cells and anti-human ACSL4 antibody recognized the hamster ACSL4 in HEK293A cells transfected with pHis-HamACSL4 but not in mock transfected cells (Supplemental Fig. 2A,B).
3.2. Tissue-selective induction of ACSL4 expression in hamsters by hyperlipidemic diets
Next, we conducted a tissue profiling of ACSL4 mRNA and protein expressions in hamsters fed three different diets, a NCD, a HCHFD and a HFD for two weeks. Feeding hamsters HCHFD and HFD increased serum TC levels by 3.7-fold and 2.0-fold respectively as compared to that of NCD (Supplemental Fig. 3A). Serum TG levels were increased by HCHFD and HFD to comparable levels of 3.2- and 3.7-fold of the control diet (Supplemental Fig. 3B). The cholesterol-enriched HCHFD produced a more pronounced increase in hepatic TC and TG as compared to the HFD feeding (Supplemental Fig. 3C, D).
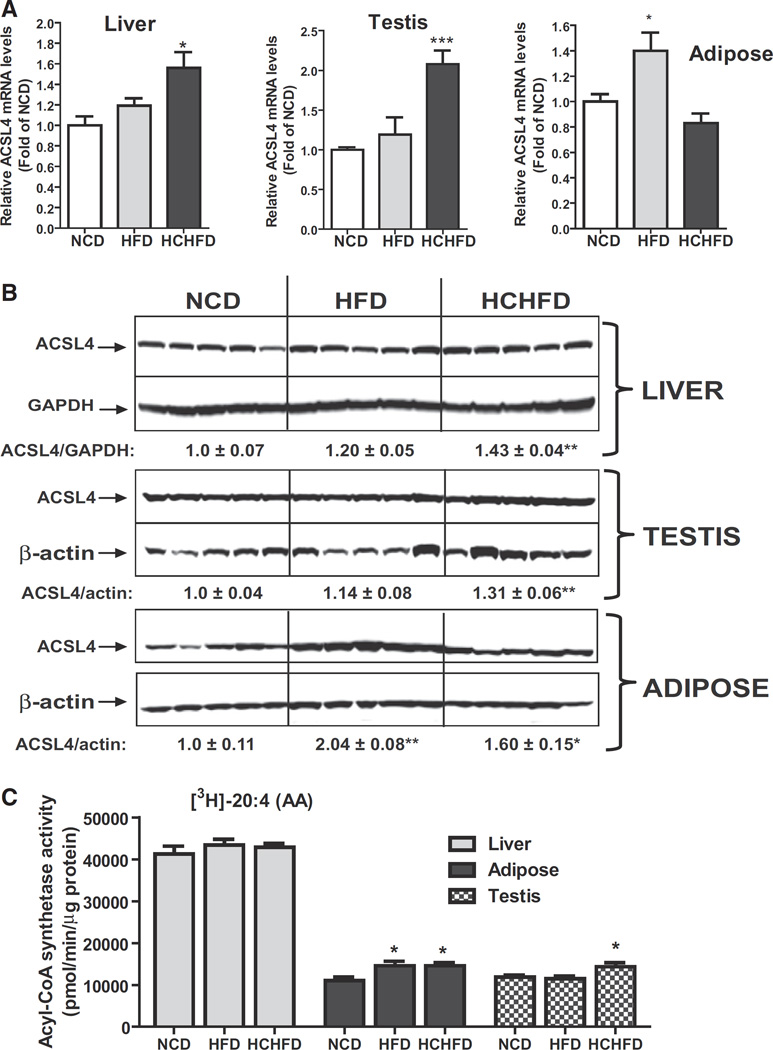
We performed qPCR to assess ACSL4 mRNA expression levels in various tissues of the three diet groups (Fig. 1A). ACSL4 mRNA levels were significantly increased by 1.6-fold (p < 0.05) in liver and 2.1-fold in testis (p < 0.001) of HCHFD fed animals and by 1.4-fold in adipose tissue (p < 0.05) of HFD fed animals as compared to that of NCD. Western blot analysis confirmed the significant increases in ACSL4 protein expressions in liver and testis of HCHFD group. In adipose, ACSL4 protein levels were elevated 2-fold (p < 0.01) by HFD and 1.6-fold (p < 0.05) by HCHFD feeding (Fig. 1B). Both hyperlipidemic diets did not alter ACSL4 mRNA or protein levels in heart, kidney, muscle and brain (Supplemental Fig. 4). In hamster brain tissues, in addition to the 75 kDa band, a brain-specific higher molecular mass species around 80 kDa was also detected by Western blotting. Again, levels of the 80 kDa isoform of ACSL4 were similar among the diet groups.
Fig. 1.
Tissue-specific inductions of ACSL4 expression by HCHFD and HFD. (A) Quantitative real-time RT-PCR analysis of ACSL4 mRNA levels in liver, adipose and testis of hamsters of three diet groups. Hamsters were sacrificed and tissues were isolated after two weeks on the indicated diets. Total RNA was isolated from each tissue sample and relative mRNA abundance of ACSL4 was determined by conducting q-PCR and normalized to GAPDH. *p < 0.05, ***p < 0.001 compared to NCD group. (B) Western blot analysis of ACSL4 protein abundance. Individual tissue protein extracts were prepared and protein concentrations were determined. 50 µg of homogenate proteins of individual samples was resolved by SDS-PAGE and ACSL4 protein was detected by immunoblotting using anti-ACSL4 antibody. The membrane was reprobed with anti-β-actin or anti-GAPDH antibody. The protein abundance of ACSL4 was quantified with Alpha View Software with normalization by signals of β-actin or GAPDH. Values are mean ± SEM of 5 samples per group. * p < 0.05 and ** p < 0.01 compared to the NCD group. (C) Measurement of tissue ACSL enzymatic activity. Initial rates of total ACSL activity of individual tissue homogenates from three diet groups were measured using 5 µg of homogenate per sample at 37 °C in the presence of [3H] labeled AA. *p < 0.05 compared to NCD group.
To compare ACSL4 protein levels among different tissues of hamsters we pooled equal amount of tissue protein homogenates from the same tissue of each diet group and 50 µg of pooled protein samples was analyzed by Western blotting (Supplemental Fig. 5). ACSL4 protein level was most abundant in hamster liver and modest levels of ACSL4 were detected in other metabolic tissues including adipose, testis, heart and muscle. The pooled samples also confirmed the elevated ACSL4 protein levels in liver and testis of HCHFD group and in adipose of HFD and HCHFD groups.
We next measured ACSL activity of individual homogenates of liver, adipose and testis from NCD, HCHFD and HFD groups in the presence of 3H-arachidonic acid (20:4), the preferred substrate of ACSL4. Fig. 1C shows that liver homogenates had approximately 4-fold higher arachidonoyl-CoA synthetase activity as compared to adipose or testis and the hepatic enzyme activities did not further increase by hyperlipidemic diets. In adipose samples, the arachidonoyl-CoA synthetase activity was increased by ~32% (p < 0.05) in HFD and HCHFD fed hamsters. Likewise, we detected a ~21% increase (p < 0.05) of arachidonoyl-CoA synthetase activity in testis of HCHFD fed animals. Taken together, our dietary studies have discovered for the first time that ACSL4 expression and the arachidonoyl-CoA synthetase activity are upregulated by hyperlipidemic diets in a tissue-specific manner.
3.3. Hepatic ACSL4 gene expression is upregulated by PPARδ agonist
The higher expression level of ACSL4 in the liver relative to other tissues suggests that ACSL4 plays an important role in hepatic FA metabolism. Therefore, it is of interest to understand how dietary cholesterol regulates its hepatic expression. Our previous studies have observed an increased hepatic expression of ACSL3 in hamsters fed a HCHFD [34]. We subsequently demonstrated that ACSL3 is a molecular target of peroxisome proliferator-activated receptor δ (PPARδ) [35]. In addition to PPARδ, PPARα has been demonstrated to transactivate ACSL1 gene expression in mouse and rat liver cells [37–39]. Thus, we examined hepatic mRNA levels of PPARδ and PPARα along with ACSL3 and observed that HCHFD did not affect PPARα gene expression, but increased PPARδ mRNA levels in liver tissue by approximately 50% (p < 0.05) as compared to the liver of hamsters fed a NCD or a HFD (Supplemental Fig. 6). In adipose and testis PPARδ mRNA levels were unchanged among diet groups.
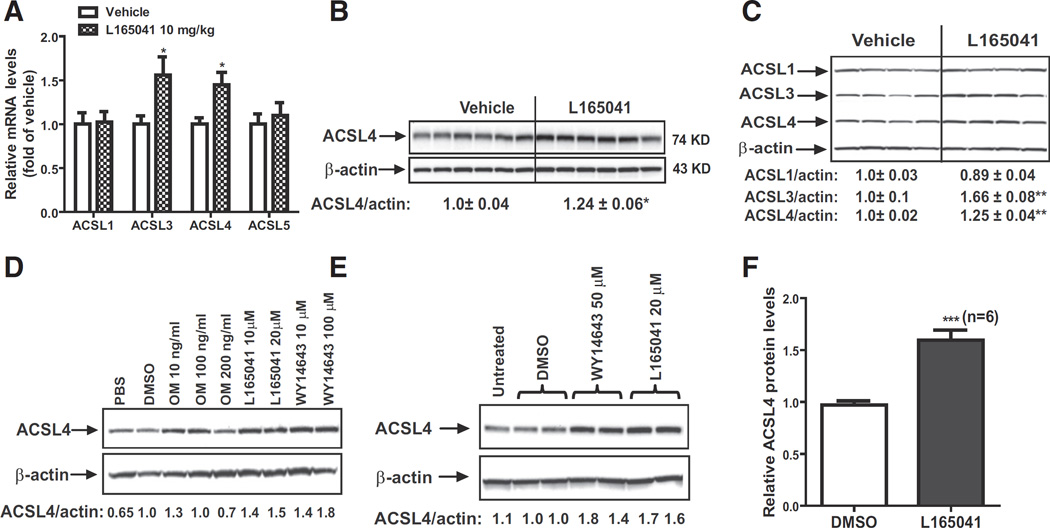
To explore a functional role of PPARδ in ACSL4 gene expression in liver tissues, we examined ACSL mRNA levels in NCD fed hamsters that were treated with vehicle or PPARδ specific ligand L165041 at a daily dose of 10 mg/kg for 7 days [35]. Real-time qPCR analysis (Fig. 2A) shows that ACSL4 and ACSL3 mRNA levels were increased to ~45% (p < 0.05) and ~56% (p < 0.05) of vehicle control, respectively by L165041 treatment. In contrast, L165041 treatment had no effect on ACSL1 or ACSL5 mRNA expression in hamster liver. Western blotting of individual liver homogenates shows that ACSL4 protein levels were 24% (p < 0.05) higher in agonist-treated livers as compared to the control (Fig. 2B). These results provide the first direct evidence to support a positive role of PPARδ in hepatic ACSL4 expression in addition to its known target ACSL3, another ACSL family member.
Fig. 2.
Elevation of ACSL4 mRNA and protein expressions in hamster liver tissue and primary hepatocytes by L165041 treatment. (A, B) Hamsters fed a NCD were treated with 10 mg/kg of L165041 (n = 9) or with vehicle (n = 9) for 7 days. At the end of treatment, animals were sacrificed for liver tissue collection. In A, total RNA was isolated from each liver sample and relative mRNA abundances of ACSL family members were determined by q-PCR and normalized to GAPDH. In B, total protein extracts were individually prepared from 6 randomly chosen liver samples of vehicle group or the treatment group. Equal amounts of homogenate proteins (50 µg) were resolved by SDS-PAGE and ACSL4 protein was detected by immunoblotting. The membrane was reprobed with anti-β-actin antibody. The protein abundance of ACSL4 was quantified and normalized by signals of β-actin. Values are mean ± SEM of 6 samples per group. (C) Effects of activation of PPARδ on hepatic expressions of ACSLs in dyslipidemic hamsters. Ten hamsters were fed HFD for eight weeks. Five hamsters were treated with L165041 at a dose of 10 mg/kg or with vehicle for 7 days. Hamsters were sacrificed and liver tissues were isolated at the end of treatment. Total protein extracts were individually prepared from 4 randomly chosen liver samples of each group. Hepatic ACSL protein levels were assessed by Western blotting with anti-ACSL1, anti-ACSL3 and anti-ACSL4 antibodies and signals were quantified. Values are mean ± SEM of 4 samples per group. (D) Hamster primary hepatocytes were cultured in HepatoZYME-SFM medium overnight. Cytokine OM, L165041 or WY14643 were added to the cells for 24 h. Total cell lysates were isolated for Western blotting of ACSL4 or β-actin. PBS was a vehicle control for OM and DMSO was vehicle control for PPAR agonists. (E) Mouse primary hepatocytes were cultured in HepatoZYME-SFM medium overnight. L165041 or WY14643 were added to the cells for 24 h. Total cell lysates were isolated for Western blotting of ACSL4 or β-actin. The relative ACSL4 protein levels were quantified. (F) The relative increase of ACSL4 protein levels over control mouse primary hepatocytes assessed by Western blot analysis is presented as the mean ± SEM of 6 separate treatments. * p < 0.05, ** p < 0.01 and *** p < 0.001 compared to the vehicle or DMSO.
Ligand-induced activation of PPARδ has been shown to improve hepatic lipid metabolism in various dyslipidemic animal models [40–43]. Thus, we fed ten hamsters a HFD for eight weeks to induce dyslipidemia which was evident by 2.7-fold increase in serum TC relative to hamsters fed a NCD (NCD, 117.8 ± 27.4 mg/dl; HFD, 326.6 ± 50.7 mg/dl). While continued on HFD, hamsters were orally dosed once a day for 7 days with L165041 (10 mg/kg) or vehicle. Analysis of circulating and hepatic lipids showed that L165051 treatment did not change serum and hepatic cholesterol levels but it reduced serum and hepatic TG levels significantly (Supplemental Fig. 7).
Western blot analysis of individual liver homogenates (Fig. 2C) shows that ACSL1 protein levels were not affected by the drug treatment. ACSL4 protein levels were elevated to 25% (p < 0.05) in the liver of L165041 treated hamsters as compared to vehicle group, which further suggests that ACSL4 gene expression in liver tissue is under the regulation of PPARδ. In line with previous studies, L165041 also increased hepatic ACSL3 protein levels [35].
To examine the direct effect of PPARδ activation on ACSL4 expression in hepatic cells, we isolated primary hepatocytes from hamster liver and treated these cells with various doses of L165041, WY14643, a PPARα agonist and oncostatin M (OM), a cytokine that induces PPARδ expression [35]. The result of Western blot analysis in Fig. 2D shows that ACSL4 protein levels were increased by all treatments with one exception of a high dose of OM that caused cytotoxicity in these primary hepatic cells. We also isolated primary hepatocytes from mouse liver and examined the effects of L165041 and WY14643. ACSL4 protein levels (Fig. 2E) were upregulated by L165043 as well as WY14643. Fig. 2F shows the summarized result of six separate experiments in which ACSL4 protein levels were increased by 64% (p < 0.001) in mouse primary hepatocytes upon L165141 treatment.
3.4. Induction of ACSL4 expression in human primary hepatocytes by PPAR ligands
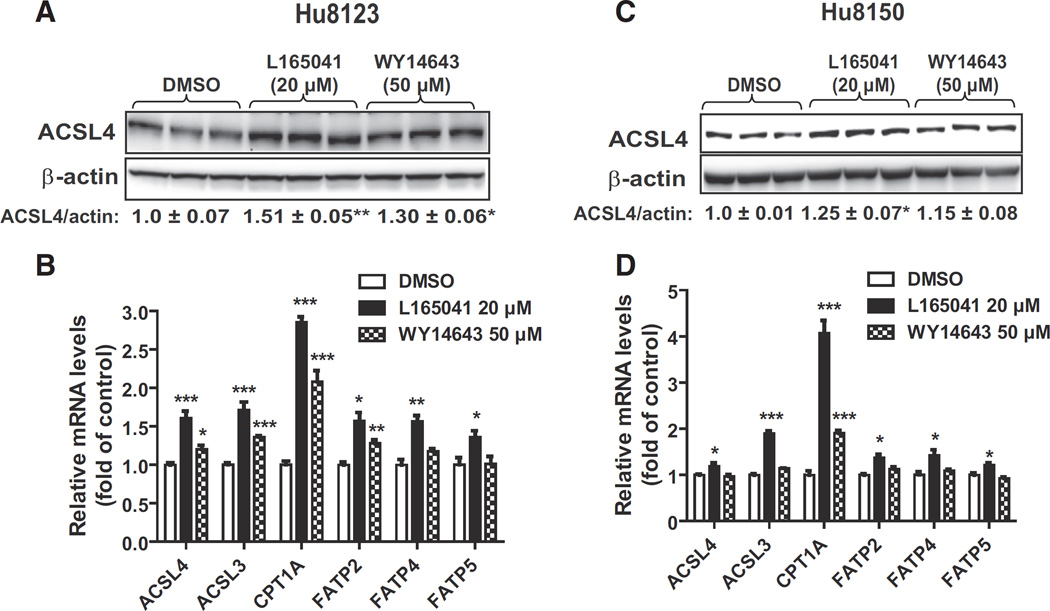
To determine whether activation of PPARs affects ACSL4 expression in normal human liver cells we treated human primary hepatocytes derived from two individual donors (Hu8123, Hu8150) with L165041 and WY14643. Western blot analysis of Hu8123 total cell lysates showed that ACSL4 protein levels were increased 51% (p < 0.01) by L165043 and 30% (p < 0.05) by WY14643 compared to control (Fig. 3A). Gene expression analysis by qRT-PCR confirmed that the mRNA levels of ACSL4 and two known PPARδ target genes (ACSL3 and CPT1A) were all significantly elevated by activators of PPARδ/α in Hu8123 cells (Fig. 3B). Additionally, we measured mRNA levels of three FA transport proteins (FATPs) in control and treated cells to learn whether ACSL4 could be co-regulated with hepatic FATPs [44,45] by PPAR activators. We observed that activation of PPARδ had stronger positive effects on FATP gene expression than activation of PPARα in Hu8123 cells.
Fig. 3.
Induction of ACSL4 protein expression in human primary hepatocytes by PPAR agonists. Human primary hepatocytes of two different donors (Hu8123, Hu8150) were treated with L165041 (20 µM) orWY14643 (50 µM) for 24 h. Triplicate wells were used in each condition. ACSL4 protein abundance was analyzed by Western blotting and q-PCR was performed to measure relative mRNA levels of indicated genes. * p < 0.05, ** p < 0.01 and *** p < 0.001 compared to the DMSO control.
In Hu8150 cells, ACSL4 protein (Fig. 3C) and mRNA levels (Fig. 3D) were slightly increased by L165043 with statistical significances. Interestingly, the inducing effects of L165041 on ACSL3 and CPT1A mRNA levels in Hu8150 cells were as strong as in Hu8123 cells. Thus, we compared mRNA levels of ACSL4, ACSL3 and CPT1A in un-induced cells of Hu8123 and Hu8150 (Supplemental Fig. 8A) and found that while ACSL3 and CPT1A mRNA levels were the same between these two donors, Hu8150 cells had ~3.5-fold more ACSL4 transcript than Hu8123 cells, which could explain its lower induction by PPAR activators. Additionally, we compared relative mRNA levels of ACSL isoforms in these human primary hepatocytes and in liver tissue of normolipidemic hamsters (NCD group) (Supplemental Fig. 8B). It is interesting to note that among ACSL family members, ACSL4 mRNA levels showed the most variations. Nevertheless, these data produced from human primary hepatocytes are consistent with the data generated in hamster and mouse hepatocytes, indicating that activation of PPARδ leads to increased ACSL4 gene expression in primary hepatocytes of human, hamster and mouse origins.
3.5. Transcriptional activation of ACSL4 gene expression by L165041 in human hepatic cell line HepG2
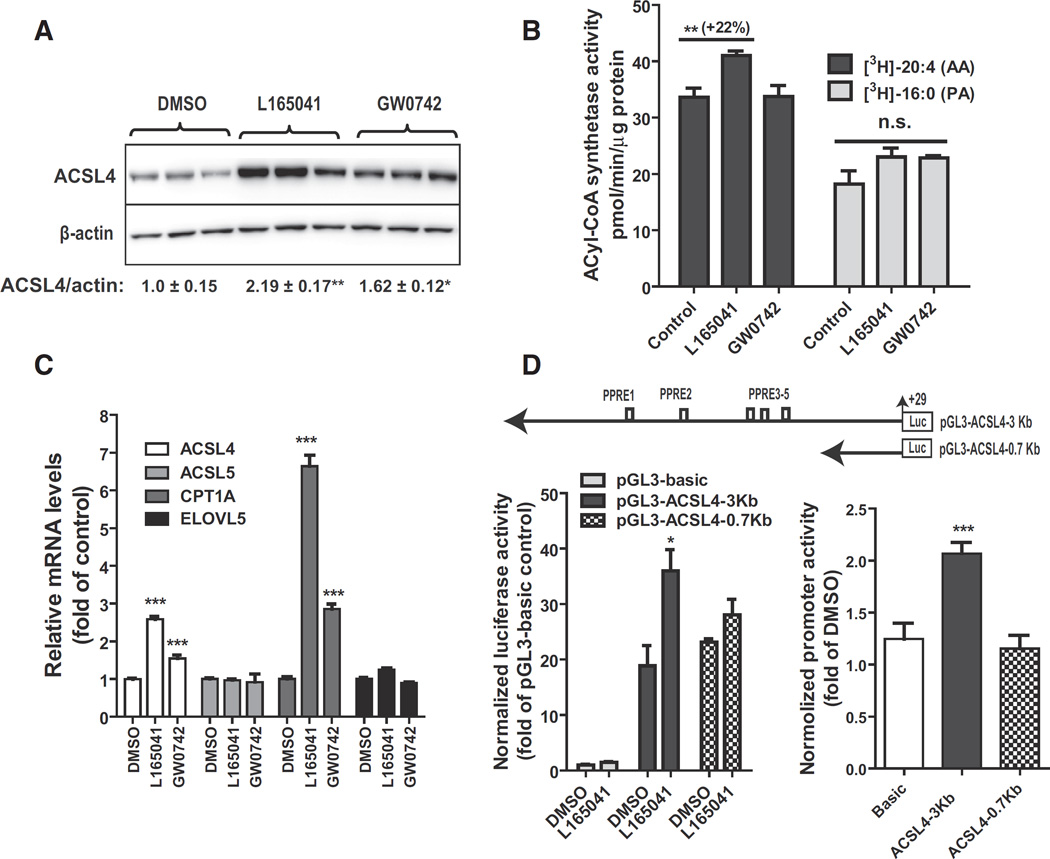
To further characterize the functional role of PPARδ in hepatic ACSL4 expression, we utilized human hepatic cell line HepG2 as a model system. HepG2 cells were treated with L165041 or another PPARδ specific ligand GW0742 for 24 h. Western blot analysis showed that ACSL4 protein levels were increased 2.2-fold (p < 0.01) by L165041 and 1.6-fold (p < 0.05) by GW0742 (Fig. 4A). The stronger induction of ACSL4 protein levels by L165041 over GW0742 was corroborated by a significant increase in arachidonoyl-CoA synthetase activity in HepG2 cells treated with this PPARδ ligand (Fig. 4B). We did not detect a statistically significant increase in palmitoyl-CoA synthetase activity upon PPARδ activation.
Fig. 4.
Activation of ACSL4 gene transcription by PPARδ agonists in HepG2 cells. HepG2 cells in triplicate wells were cultured overnight in culture medium containing 0.5% FBS, followed by treatment of L165041 (20 µM) or GW0742 (1 µM) for 24 h. In A, total protein lysates were isolated and 30 µg protein per sample was used for SDS-PAGE and Western blotting. In B, initial rates of total ACSL activity in 5 µg of cell homogenate of HepG2 were measured at 37 °C in the presence of [3H] labeled AA or [3H] labeled palmitic acid (PA). In C, total RNA was isolated and gene expression analysis was conducted by q-PCR. In D, HepG2 cells were transfected with pGL3-ACSL4-3Kb or pGL3-ACSL4-0.7Kb. The plasmid pRL-SV40 was cotransfected with ACSL4 promoter constructs. One day post-transfection, cells were treated with 25 µM L165041 for 24 h. Cell lysates were isolated to measure dual luciferase activities. In the left panel, the normalized luciferase activity of pGL3-basic in DMSO treated control cells is expressed as 1. In the right panel, the relative luciferase of each vector in ligand treated cells was compared to that in the control. Each value represents the mean ± SEM of five independent transfection experiments in which triplicate wells were assayed. * p < 0.05 and *** p < 0.001 compared to the DMSO control.
In accord with the results of Western blot and enzyme activity assays, gene expression analysis by qRT-PCR showed that ACSL4 mRNA levels were increased 2.6- and 1.5-fold by L165041 and GW0742, respectively. The stronger activity of L165041 over GW0742 was also reflected by the higher induction of PPARδ target gene CPT1A in cells treated with L165041 as compared to that of GW0742. Consistent with the hamster in vivo data, ACSL5 mRNA levels were not induced by PPARδ activation. We also did not observe changes in mRNA levels of ELOVL5, a known PPARα target gene. Altogether, these data demonstrate that ACSL4 gene expression is increased by PPARδ agonists in HepG2 cells to similar magnitudes as that in liver tissue and in various primary hepatocytes.
Analysis of the nucleotide sequence upstream of exon 1 of human ACSL4 gene by the MatInspector software revealed the presence of five putative PPAR response element (PPRE) motifs residing in the region of −2418 to −1243 upstream of the 5′ end of exon 1. Thus, we constructed two ACSL4 promoter luciferase reporter vectors (Fig. 4D upper panel). The reporter pGL3-ACSL4-3Kb includes all putative PPRE sites and the reporter pGL3-ACSL4-0.7Kb devoid these regulatory sequences. These reporters were transfected into HepG2 cells along with pRL-SV40 for the normalization. Fig. 4D left panel shows one representative transfection results and the right panel shows summarized results of 5 separate transfections. These data clearly demonstrated that while the 3 Kb and 0.7 Kb constructs have similar basal promoter activities, L165041 treatment only increases the promoter activity of 3 Kb ACSL4 construct containing the PPRE sequences. Collectively, these new results from HepG2 cells strongly suggest that ACSL4 gene transcription in liver cells is positively regulated by PPARδ.
3.6. Hepatic depletion of PPARδ leads to the suppression of ACSL4 gene expression
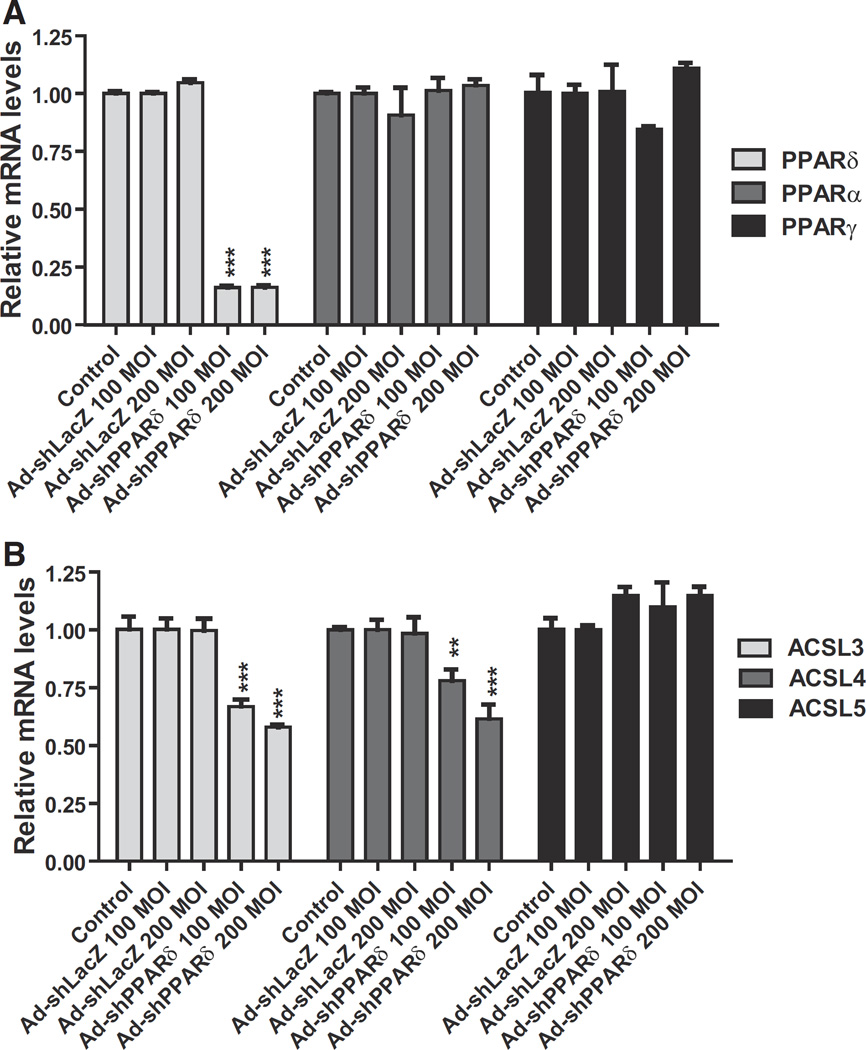
As aforementioned, we observed the correlated upregulation of ACSL4 and PPARδ in liver tissue in HCHFD fed hamsters. Here we performed ex vivo experiments to examine the impact of depleting PPARδ on hepatic ACSL4 mRNA expression. We made an Ad-shPPARδ adenoviral construct that expresses a shRNA targeting hamster PPARδ coding sequence. Hamster primary hepatocytes were transduced with different MOIs of Ad-shPPARδ or a shRNA control virus Ad-shLacZ for 3 days, and subsequently gene expression was analyzed by q-PCR. Fig. 5A shows that Ad-shPPARδ transduction specifically reduced PPARδ mRNA levels without affecting PPARα or PPARγ mRNA expression, thus confirming the specific targeting effect of the shRNA to hamster PPARδ. Importantly, both hamster ACSL3 and ACSL4 mRNA levels were significantly attenuated by the depletion of PPARδ whereas mRNA levels of ACSL5 in hamster primary hepatocytes were not affected by Ad-shPPARδ infection (Fig. 5B). These results further demonstrate that hepatic ACSL4 gene transcription is under the control of PPARδ.
Fig. 5.
Knockdown efficiency and suppression of ACSL4 expression by Ad-shPPARδ transduction. Hamster primary hepatocytes were transduced with indicated MOIs of Ad-shPPARδ or Ad-shLacZ for 3 days. The mRNA levels of PPARs (A) and ACSL isoforms (B) in transduced cells were measured by q-PCR.
3.7. Hepatic PPARδ gene expression is not regulated by the nuclear sterol-activated receptors LXR and FXR
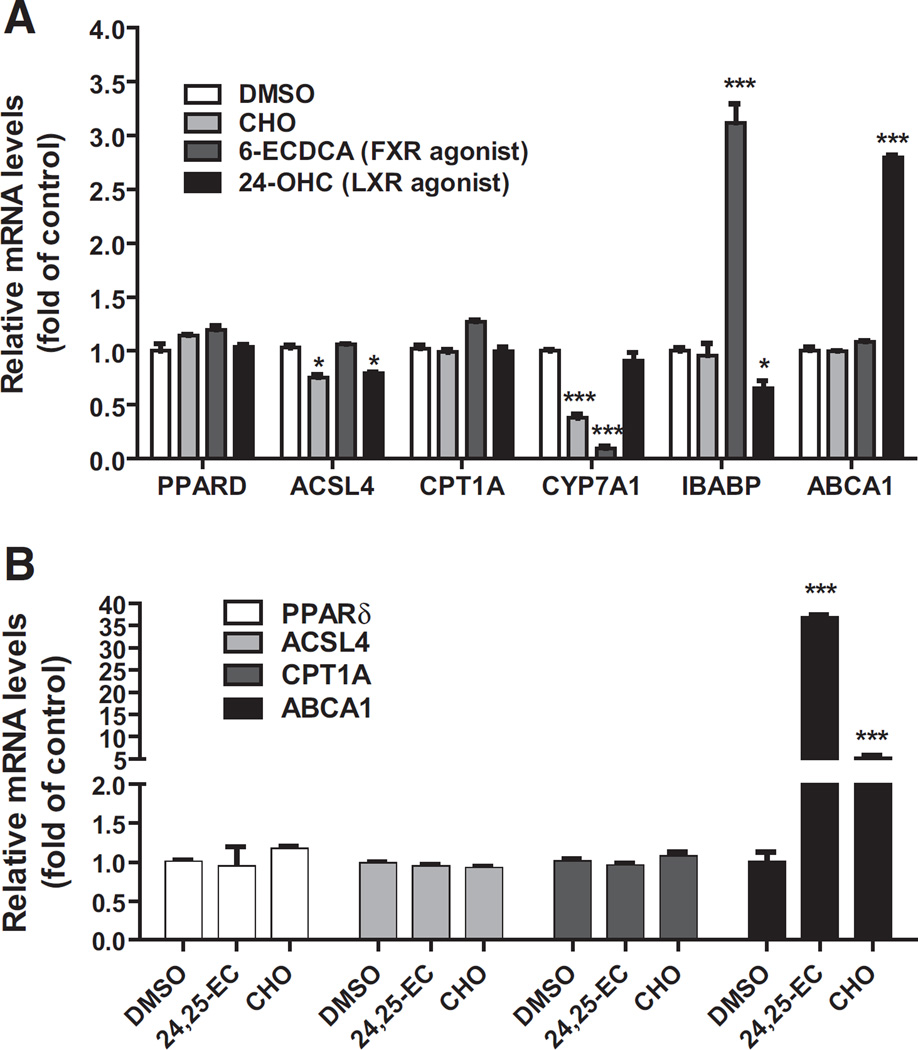
The LXRs and farnesoid X receptors (FXRs) are nuclear receptors that are activated by oxysterols and bile acids in liver tissue. Our observation of elevated hepatic PPARδ mRNA levels in hamsters fed the cholesterol enriched diet (HCHFD) raised an interesting question of whether PPARδ gene transcription is upregulated by LXR or FXR. To address this question, we treated HepG2 cells with cholesterol (CHO), a strong LXR agonist 24-OHC or a FXR agonist 6-ECDCA for 24 h. Gene expression analysis (Fig. 6A) showed that mRNA level of LXR target gene ABCA1 (ATP-binding cassette transporter A1) was greatly elevated in cells treated with 24-OHC; FXR suppressive gene CYP7A1 (cytochrome P450 7A1) mRNA level was inhibited by cholesterol and 6-ECDCA whereas IBABP (ileal bile acid binding protein) mRNA expression was increased 3.2-fold of control by the FXR agonist. However, in contrast to these known LXR and FXR target genes, activation of these nuclear receptors did not affect the mRNA levels of PPARδ and its target gene CPT1A. We observed a small decrease in ACSL4 mRNA levels in HepG2 cells treated with cholesterol and 24-OHC. To further validate these findings, we performed similar experiments in mouse hepatic AML12 cells (Fig. 6B) and hamster primary hepatocytes (Supplemental Fig. 9). Again, we did not detect any increases in PPARδ or ACSL4 mRNA levels after LXR activation. Altogether, these data suggest that hepatic PPARδ transcription is not directly activated by LXR or FXR. The precise molecular mechanism of dietary cholesterol-induced elevation of PPARδ mRNA levels in liver tissue remained elusive.
Fig. 6.
Examination of the effects of LXR and FXR ligands on PPARδ mRNA expression in cultured hepatic cell lines. HepG2 cells (A) or AML12 cells (B) were cultured in medium supplemented with 10% LPDS overnight. The next day, DMSO (as vehicle), CHO (10 µg/ml cholesterol + 1 µg/ml 25-hydroxycholesterol), 24-OHC (20 µM) or 6-ECDCA (20 µM) was added to HepG2 cells for 24 h prior to RNA isolation; cholesterol or 24,25-EC (20 µM) was added to AML12 cells. Hepatic mRNA levels were assessed by q-PCR with triplicate measurement of each cDNA sample. After normalization with GAPDH mRNA levels, the relative levels are presented. The data shown are representative results of two separate experiments in which duplicate wells were used in each condition. * p < 0.05 and *** p < 0.001 compared to the DMSO control.
4. Discussion
ACSL4 is one of the five members of the ACSL family that are essential for LCFA metabolism and utilization in mammalian cells. It has been well recognized that whereas ACSL enzymes catalyze similar enzymatic reactions to esterify free FA to acyl-CoA, they exhibit variable cellular functions and generate distinct metabolic outcomes in an isoform specific and tissue/cell type specific manner [10,46]. It is generally believed that the substrate specificity, subcellular localization, tissue specific expression and upstream signaling pathways all contribute to the unique functions of the individual ACSLs. In particular, how an ACSL isozyme is transcriptionally activated might affect its cellular functions under that stimulated condition.
When ACSL4 was first cloned and characterized, its mRNA expression in adult human tissues was analyzed by Northern blot [12,47]. The ACSL4 expression was detected at high levels in brain, placenta, testis, ovary, spleen and adrenal cortex; low levels were detected in liver samples. A subsequent study of Western blot using anti-ACSL4 antibody also confirmed its low expression in adult liver sample as compared to the intestine and brain [48]. However, recent studies have revealed elevated ACSL4 mRNA expression in human liver samples of NAFLD [49–51]. The interesting and unanswered questions are 1) how ACSL4 expression is elevated in steatotic human liver? And 2) is heightened ACSL4 a causal factor for the development of NAFLD or it is needed to compensate the demand of liver to metabolize excessive FAs due to oversupply of nutrients?
In this current study, we tissue profiled ACSL4 mRNA and protein expressions in hamsters fed three different diets. Through this process, we have uncovered for the first time that ACSL4 expression and arachidonoyl-CoA synthetase activity are upregulated by hyperlipidemic diets in a tissue-specific manner: HCHFD feeding upregulated ACSL4 mRNA and protein expressions in liver and testis, and ACSL4 protein abundance in adipose tissue was increased by both hyperlipidemic diets.
PPARs are transcriptional activators of genes encoding enzymes to metabolize FA in the liver as well as other metabolic tissues [52]. Both PPARα and PPARγ have been reported to activate ACSL1 transcription in different cell lines of mouse or rat origins and to increase ACSL1 expression in animal models [37–39]. PPARδ was demonstrated to bind to ACSL3 gene promoter and increase its transcription in HepG2 cells and in the liver of hamsters treated with PPARδ ligand L165041 [35]. In this current study, PPARα mRNA expressions in liver, testis and adipose of hamsters of the three diet groups did not change, we observed an increased PPARδ mRNA levels in liver tissues but not in adipose or testis.
The co-induction of PPARδ with ACSL4 as well as with ACSL3, a known PPARδ target gene by HCHFD, suggested a positive role of PPARδ in ACSL4 gene expression in liver tissue. Indeed, treating hamsters with L165041 for 7 days increased ACSL4 mRNA and protein levels in the liver. This induction occurred under normolipidemic and dyslipidemic conditions. It is worthy to note that ACSL1 and ACSL5 are abundant isoforms of ACSL family in hamster liver tissue; their expressions were not responsive to PPARδ activation under in vivo conditions.
It has been demonstrated that the increased PPARδ expression level alone in liver by adenovirus-mediated over expression and by direct ligand activation without PPARδ overexpression both led to its target gene expression [40–43]. It is also known that some fatty acids and fatty acid derivatives are endogenous ligands for PPARs [52]. Thus, it is conceivable that HCHFD feeding or HFD feeding increased endogenous PPARδ ligands in liver, testis or adipose tissues of hamsters that led to an increased PPARδ-mediated upregulation of ACSL4 expression with or without changes in PPARδ expression levels.
In addition to the in vivo study, we examined the effects of specific PPARδ and PPARα ligands on ACSL4 expression in human primary hepatocytes as well as hepatocytes isolated from hamster and mouse. Our results clearly indicate that activations of PPARs, particularly PPARδ, elevate ACSL4 protein and mRNA expressions regardless of the hepatocyte origin. Based upon these consistent results from hamster liver tissue and various hepatocytes, we opted to use HepG2 cells as a model system to further elucidate the underlying mechanism of PPARδ-dependent upregulation of ACSL4 expression. Unlike liver tissue and primary hepatocytes in which ACSL1 and ACSL5 are dominant isoforms, HepG2 cells express abundant ACSL4 in a level higher than ACSL1 according to mRNA analysis [53]. Thus, changes in its expression level could affect ACSL enzyme activity, in particular, when labeled AA is used as the substrate. Indeed, we showed that treating HepG2 cells with L165041 produced approximately a 2-fold increase in ACSL4 protein abundance that was accompanied by a 22% increase in arachidonoyl-CoA synthetase activity.
Our sequence analysis suggested five putative PPRE motifs residing in the 5′ flanking region of the human ACSL4 gene from −2418 to −1243 upstream of the 5′ end of exon 1. To firmly demonstrate the transcriptional induction of ACSL4 mRNA expression by PPARδ agonist, initially we made two luciferase constructs with one that contains the PPRE clusters (pGL3-ACSL4-3Kb) and one without (pGL3-ACSL4-0.7Kb). Multiple transfection experiments in HepG2 cells clearly demonstrated that L165041 treatment induced the promoter activity of the former without an effect on the latter. These results establish that activation of PPARδ increases ACSL4 gene expression by inducing its promoter transcriptional activity.
To substantiate the critical role of PPARδ in ACSL4 transcription, we utilized the approach of adenovirus-mediated depletion of hamster PPARδ in ex vivo experiments. Transduction of Ad-shPPARδ did not affect PPARα or PPARγ mRNA levels but markedly lowered PPARδ mRNA abundance, which was accompanied with significant reductions of ACSL4 and ACSL3 mRNA levels without lowering ACSL1 (data not shown) or ACSL5 mRNA abundance, thereby further support the selective regulation of PPARδ to ACSL4 and ACSL3 isoforms.
We were puzzled by the observation of increased PPARδ mRNA levels in the liver of HCHFD fed animals. It is well known that over accumulation of hepatic sterols activates the LXR and FXR pathways to accelerate the cholesterol execration from the body by inducing target gene expression [54]. We used HepG2 and AML12 cells as well as hamster primary hepatocytes to examine the possibility of FXR or LXR-mediated upregulation of PPARδ in liver cells. We demonstrate that treating these cells with a LXR agonist or a FXR agonist strongly impacted the expression of their canonical target genes without any inducing effects on PPARδ and PPARδ regulated gene CPT1A or ACSL4, which largely rule out the possibility of a direct effect of LXR or FXR on PPARδ gene expression. These data are also consistent with our in vivo finding that treating hamsters with LXR agonist did not affect liver PPARδ mRNA expression [36].
Currently, little is known about the transcriptional networks that regulate PPARδ gene expression. A recent study by Chistyakov et al. [55] reported that in astrocytes PPARδ expression and activity were induced by LPS. The LPS-induced kinetics of PPARδ expression was similar to that of the proinflammatory gene cyclooxynase 2. Other studies of PPARδ agonists in diabetic rats [56] and in LDLR−/− mice [41] have suggested that PPARδ activation attenuates hepatic steatosis in part by anti-inflammatory mechanism. Thus, it is conceivable to speculate that the elevated levels of PPARδ in HCHFD-fed liver could reflect the liver response to lessen the low-grade chronic inflammatory status caused by excessive cholesterol accumulation. Further studies to examine the signaling pathways linked to inflammatory response might aid to elucidate the mechanism of dietary cholesterol induced PPARδ gene expression in liver tissue.
Our current understandings about the ACSL family with regard to their tissue expression patterns and responses to nutritional status are largely gained from animal studies, particularly from rats and mice [57–59]. In this study, by utilizing human primary hepatocytes we showed that activation of PPARδ increased ACSL4 gene expression to different degrees in hepatocytes derived from two individual donors and the magnitude of induction was reversely correlated to the basal ACSL4 expression levels. It is tempting to postulate that hepatic ACSL4 expression in humans is highly regulated and this ACSL isoform has important roles in hepatic lipid metabolism in humans.
In conclusion, we have demonstrated that hyperlipidemic diets upregulate ACSL4 expression in a tissue-specific manner. Importantly, combined the in vivo upregulation of ACSL4 in liver tissue by PPARδ specific agonist with the direct inducing effects of PPARδ activation on ACSL4 promoter activity, protein expression and arachidonoyl-CoA synthetase activity in HepG2 cells, our comprehensive studies have identified PPARδ as a novel transcriptional activator for ACSL4 expression in liver cells. This work further supports our hypothesis that the transcription of each ACSL isoformis controlled by specific members of the PPAR family, and this intrinsic relationship may endow individual ACSLs with unique cellular functions under an induced condition.
Supplementary Material
Acknowledgments
This study was supported by the Department of Veterans Affairs (Office of Research and Development, Medical Research Service) and by grants (1R01 AT002543-01A1 and 1R01AT006336-01A1) from the National Center of Complementary and Alternative Medicine.
Abbreviations
- AA
arachidonic acid
- ACSL
long-chain acyl-CoA synthetase
- 24,25-EC
24,25-epoxycholesterol
- 6-ECDCA
6α-ethyl-chenodeoxycholic acid
- FA
fatty acid
- FATP
fatty acid transport protein
- FXR
farnesoid X receptor
- HCHFD
high-cholesterol and high-fat diet
- HFD
high-fat diet
- LXR
liver X receptor
- MOI
multiplicity of infection
- NAFLD
non-alcoholic fatty liver disease
- NCD
normal chow diet
- 24-OHC
24S-hydroxycholesterol
- PPAR
peroxisome proliferator-activated receptor
- PPRE
PPAR response element
Footnotes
Conflict of interest
All authors have no conflict of interest for this study.
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbalip.2015.01.008.
References
- 1.Scorletti E, Byrne CD. Omega-3 Fatty acids, hepatic lipid metabolism, and nonalcoholic fatty liver disease. Annu. Rev. Nutr. 2013;33:231–248. doi: 10.1146/annurev-nutr-071812-161230. [DOI] [PubMed] [Google Scholar]
- 2.Ruderman NB, Saha AK, Vavvas D, Witters LA. Malonyl-CoA, fuel sensing, and insulin resistance. Am. J. Physiol. 1999;276:E1–E18. doi: 10.1152/ajpendo.1999.276.1.E1. [DOI] [PubMed] [Google Scholar]
- 3.Laybutt DR, Schmitz-Peiffer C, Saha AK, Ruderman NB, Biden TJ, Kraegen EW. Muscle lipid accumulation and protein kinase C activation in the insulin-resistant chronically glucose-infused rat. Am. J. Physiol. 1999;277:E1070–E1076. doi: 10.1152/ajpendo.1999.277.6.E1070. [DOI] [PubMed] [Google Scholar]
- 4.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soupene E, Kuypers FA. Mammalian long-chain acyl-CoA synthetases. Exp. Biol. Med. (Maywood) 2008;233:507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopes-Marques M, Cunha I, Reis-Henriques MA, Santos MM, Castro LF. Diversity and history of the long-chain acyl-CoA synthetase (Acsl) gene family in vertebrates. BMC Evol. Biol. 2013;13:271. doi: 10.1186/1471-2148-13-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis JM, Frahm JL, Li LO, Coleman RA. Acyl-coenzyme A synthetases in metabolic control. Curr. Opin. Lipidol. 2010;21:212–217. doi: 10.1097/mol.0b013e32833884bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman RA, Van Horn CG, GonzalezBaro MR. Do acyl-coA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J. Nutr. 2002;132:2123–2126. doi: 10.1093/jn/132.8.2123. [DOI] [PubMed] [Google Scholar]
- 9.Watkins PA, Ellis JM. Peroxisomal acyl-CoA synthetases. Biochim. Biophys. Acta. 2012;1822:1411–1420. doi: 10.1016/j.bbadis.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grevengoed TJ, Klett EL, Coleman RA. Acyl-CoA metabolism and partitioning. Annu. Rev. Nutr. 2014;34:1–30. doi: 10.1146/annurev-nutr-071813-105541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang MJ, Fujino T, Sasano H, Minekura H, Yabuki N, Nagura H, Iijima H, Yamamoto TT. A novel arachidonate-preferring acyl-CoA synthetase is present in steroidogenic cells of the rat adrenal, ovary, and testis. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piccini M, Vitelli F, Bruttini M, Pober BR, Jonsson JJ, Villanova M, Zollo M, Borsani G, Ballabio A, Renieri A. FACL4, a new gene encoding long-chain acyl-CoA synthetase 4, is deleted in a family with Alport syndrome, elliptocytosis, and mental retardation. Genomics. 1998;47:350–358. doi: 10.1006/geno.1997.5104. [DOI] [PubMed] [Google Scholar]
- 13.Miyares RL, Stein C, Renisch B, Anderson JL, Hammerschmidt M, Farber SA. Long-chain Acyl-CoA synthetase 4A regulates Smad activity and dorsoventral patterning in the zebrafish embryo. Dev. Cell. 2013;27:635–647. doi: 10.1016/j.devcel.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golej DL, Askari B, Kramer F, Barnhart S, Vivekanandan-Giri A, Pennathur S, Bornfeldt KE. Long-chain acyl-CoA synthetase 4 modulates prostaglandin E(2) release from human arterial smooth muscle cells. J. Lipid Res. 2011;52:782–793. doi: 10.1194/jlr.M013292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orlando UD, Garona J, Ripoll GV, Maloberti PM, Solano AR, Avagnina A, Gomez DE, Alonso DF, Podesta EJ. The functional interaction between Acyl-CoA synthetase 4, 5-lipooxygenase and cyclooxygenase-2 controls tumor growth: a novel therapeutic target. PLoS One. 2012;7:e40794. doi: 10.1371/journal.pone.0040794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klett EL, Chen S, Edin ML, Li LO, Ilkayeva O, Zeldin DC, Newgard CB, Coleman RA. Diminished acyl-CoA synthetase isoform 4 activity in INS 832/13 cells reduces cellular epoxyeicosatrienoic acid levels and results in impaired glucose-stimulated insulin secretion. J. Biol. Chem. 2013;288:21618–21629. doi: 10.1074/jbc.M113.481077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuch EM, Vellaramkalayil R, Zhang I, Lehnen D, Brugger B, Stremmel W, Ehehalt R, Poppelreuther M, Fullekrug J. Differentially localized acyl-CoA synthetase 4 isoenzymes mediate the metabolic channeling of fatty acids towards phosphatidylinositol. Biochim. Biophys. Acta. 2013;1841:227–239. doi: 10.1016/j.bbalip.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Kuwata H, Yoshimura M, Sasaki Y, Yoda E, Nakatani Y, Kudo I, Hara S. Role of long-chain acyl-coenzyme A synthetases in the regulation of arachidonic acid metabolism in interleukin 1beta-stimulated rat fibroblasts. Biochim. Biophys. Acta. 2013;1841:44–53. doi: 10.1016/j.bbalip.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Smith ME, Saraceno GE, Capani F, Castilla R. Long-chain acyl-CoA synthetase 4 is regulated by phosphorylation. Biochem. Biophys. Res. Commun. 2013;430:272–277. doi: 10.1016/j.bbrc.2012.10.138. [DOI] [PubMed] [Google Scholar]
- 20.Wu CL, Zhao SP, Yu BL. Microarray analysis provides new insights into the function of apolipoprotein O in HepG2 cell line. Lipids Health Dis. 2013;12:186. doi: 10.1186/1476-511X-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudo T, Tamagawa T, Kawashima M, Mito N, Shibata S. Attenuating effect of clock mutation on triglyceride contents in the ICR mouse liver under a high-fat diet. J. Biol. Rhythm. 2007;22:312–323. doi: 10.1177/0748730407302625. [DOI] [PubMed] [Google Scholar]
- 22.Orlando U, Cooke M, Cornejo MF, Papadopoulos V, Podesta EJ, Maloberti P. Characterization of the mouse promoter region of the acyl-CoA synthetase 4 gene: role of Sp1 and CREB. Mol. Cell. Endocrinol. 2013;369:15–26. doi: 10.1016/j.mce.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Gubern C, Camos S, Ballesteros I, Rodriguez R, Romera VG, Canadas R, Lizasoain I, Moro MA, Serena J, Mallolas J, Castellanos M. miRNA expression is modulated over time after focal ischaemia: up-regulation of miR-347 promotes neuronal apoptosis. FEBS J. 2013;280:6233–6246. doi: 10.1111/febs.12546. [DOI] [PubMed] [Google Scholar]
- 24.Peng Y, Xiang H, Chen C, Zheng R, Chai J, Peng J, Jiang S. MiR-224 impairs adipocyte early differentiation and regulates fatty acid metabolism. Int. J. Biochem. Cell Biol. 2013;45:1585–1593. doi: 10.1016/j.biocel.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 25.Kan CF, Singh AB, Stafforini DM, Azhar S, Liu J. Arachidonic acid downregulates acyl-CoA synthetase 4 expression by promoting its ubiquitination and proteasomal degradation. J. Lipid Res. 2014;55:1657–1667. doi: 10.1194/jlr.M045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava RA. Evaluation of anti-atherosclerotic activities of PPAR-alpha, PPAR-gamma, and LXR agonists in hyperlipidemic atherosclerosis-susceptible F(1)B hamsters. Atherosclerosis. 2011;214:86–93. doi: 10.1016/j.atherosclerosis.2010.10.033. [DOI] [PubMed] [Google Scholar]
- 27.Srivastava RA, He S. Anti-hyperlipidemic and insulin sensitizing activities of fenofibrate reduces aortic lipid deposition in hyperlipidemic Golden Syrian hamster. Mol. Cell. Biochem. 2010;345:197–206. doi: 10.1007/s11010-010-0573-8. [DOI] [PubMed] [Google Scholar]
- 28.Mangaloglu L, Cheung RC, Van Iderstine SC, Taghibiglou C, Pontrelli L, Adeli K. Treatment with atorvastatin ameliorates hepatic very-low-density lipoprotein over production in an animal model of insulin resistance, the fructose-fed Syrian golden hamster: evidence that reduced hypertriglyceridemia is accompanied by improved hepatic insulin sensitivity. Metabolism. 2002;51:409–418. doi: 10.1053/meta.2002.30954. [DOI] [PubMed] [Google Scholar]
- 29.Dong B, Wu M, Li H, Kraemer FB, Adeli K, Seidah NG, Park SW, Liu J. Strong induction of PCSK9 gene expression through HNF1alpha and SREBP2: mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J. Lipid Res. 2010;51:1486–1495. doi: 10.1194/jlr.M003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naples M, Federico LM, Xu E, Nelken J, Adeli K. Effect of rosuvastatin on insulin sensitivity in an animal model of insulin resistance: evidence for statin-induced hepatic insulin sensitization. Atherosclerosis. 2008;198:94–103. doi: 10.1016/j.atherosclerosis.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 31.Castro-Perez J, Briand F, Gagen K, Wang SP, Chen Y, McLaren DG, Shah V, Vreeken RJ, Hankemeier T, Sulpice T, Roddy TP, Hubbard BK, Johns DG. Anacetrapib promotes reverse cholesterol transport and bulk cholesterol excretion in Syrian golden hamsters. J. Lipid Res. 2011;52:1965–1973. doi: 10.1194/jlr.M016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohtani H, Hayashi K, Hirata Y, Dojo S, Nakashima K, Nishio E, Kurushima H, Saeki M, Kajiyama G. Effects of dietary cholesterol and fatty acids on plasma cholesterol level and hepatic lipoprotein metabolism. J. Lipid Res. 1990;31:1413–1422. [PubMed] [Google Scholar]
- 33.Sullivan MP, Cerda JJ, Robbins FL, Burgin CW, Beatty RJ. The gerbil, hamster, and guinea pig as rodent models for hyperlipidemia. Lab. Anim. Sci. 1993;43:575–578. [PubMed] [Google Scholar]
- 34.Wu M, Liu H, Chen W, Fujimoto Y, Liu J. Hepatic expression of long-chain acyl-CoA synthetase 3 is upregulated in hyperlipidemic hamsters. Lipids. 2009;44:989–998. doi: 10.1007/s11745-009-3341-3. [DOI] [PubMed] [Google Scholar]
- 35.Cao A, Li H, Zhou Y, Wu M, Liu J. Long chain acyl-CoA synthetase-3 is a molecular target for peroxisome proliferator-activated receptor delta in HepG2 hepatoma cells. J. Biol. Chem. 2010;285:16664–16674. doi: 10.1074/jbc.M110.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong B, Kan CF, Singh AB, Liu J. High-fructose diet downregulates long-chain acyl-CoA synthetase 3 expression in liver of hamsters via impairing LXR/RXR signaling pathway. J. Lipid Res. 2013;54:1241–1254. doi: 10.1194/jlr.M032599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoonjans K, Watanabe M, Suzuki H, Mahfoudi A, Krey G, Wahli W, Grimaldi P, Staels B, Yamamoto T, Auwerx J. Induction of the acyl-coenzyme A synthetase gene by fibrates and fatty acids is mediated by a peroxisome proliferator response element in the C promoter. J. Biol. Chem. 1995;270:19269–19276. doi: 10.1074/jbc.270.33.19269. [DOI] [PubMed] [Google Scholar]
- 38.Martin GK, Schoonjans A-M, Lefebvre B, Auwerx J. Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARα and PPARγ activators. J. Biol. Chem. 1997;272:28210–28217. doi: 10.1074/jbc.272.45.28210. [DOI] [PubMed] [Google Scholar]
- 39.Rakhshandehroo M, Hooiveld G, Muller M, Kersten S. Comparative analysis of gene regulation by the transcription factor PPARalpha between mouse and human. PLoS One. 2009;4:e6796. doi: 10.1371/journal.pone.0006796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Hatano B, Zhao M, Yen CC, Kang K, Reilly SM, Gangl MR, Gorgun C, Balschi JA, Ntambi JM, Lee CH. Role of peroxisome proliferator-activated receptor δ/β in hepatic metabolic regulation. J. Biol. Chem. 2011;286:1237–1247. doi: 10.1074/jbc.M110.138115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bojic LA, Telford DE, Fullerton MD, Ford RJ, Sutherland BG, Edwards JY, Sawyez CG, Gros R, Kemp BE, Steinberg GR, Huff MW. PPARdelta activation attenuates hepatic steatosis in Ldlr−/− mice by enhanced fat oxidation, reduced lipogenesis, and improved insulin sensitivity. J. Lipid Res. 2014;55:1254–1266. doi: 10.1194/jlr.M046037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham TL, Mookherjee C, Suckling KE, Palmer CN, Patel L. The PPARdelta agonist GW0742 reduces atherosclerosis in LDLR(−/−) mice. Atherosclerosis. 2005;181:29–37. doi: 10.1016/j.atherosclerosis.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 43.Salvado L, Serrano-Marco L, Barroso E, Palomer X, Vazquez-Carrera M. Targeting PPARbeta/delta for the treatment of type 2 diabetes mellitus. Expert Opin. Ther. Targets. 2012;16:209–223. doi: 10.1517/14728222.2012.658370. [DOI] [PubMed] [Google Scholar]
- 44.Krammer J, Digel M, Ehehalt F, Stremmel W, Fullekrug J, Ehehalt R. Overexpression of CD36 and acyl-CoA synthetases FATP2, FATP4 and ACSL1 increases fatty acid uptake in human hepatoma cells. Int. J. Med. Sci. 2011;8:599–614. doi: 10.7150/ijms.8.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kazantzis M, Stahl A. Fatty acid transport proteins, implications in physiology and disease. Biochim. Biophys. Acta. 2012;1821:852–857. doi: 10.1016/j.bbalip.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mashek DG, Li LO, Coleman RA. Long-chain acyl-CoA synthetases and fatty acid channeling. Futur. Lipidol. 2007;2:465–476. doi: 10.2217/17460875.2.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y, Traer E, Zimmerman GA, McIntyre TM, Prescott SM. Cloning, expression, and chromosomal localization of human long-chain fatty acid-CoA ligase 4 (FACL4) Genomics. 1998;49:327–330. doi: 10.1006/geno.1998.5268. [DOI] [PubMed] [Google Scholar]
- 48.Cao Y, Murphy KJ, McIntyre TM, Zimmerman GA, Prescott SM. Expression of fatty acid-CoA ligase 4 during development and in brain. FEBS Lett. 2000;467:263–267. doi: 10.1016/s0014-5793(00)01159-5. [DOI] [PubMed] [Google Scholar]
- 49.Westerbacka J, Kolak M, Kiviluoto T, Arkkila P, Siren J, Hamsten A, Fisher RM, Yki-Jarvinen H. Genes involved in fatty acid partitioning and binding, lipolysis, monocyte/macrophage recruitment, and inflammation are overexpressed in the human fatty liver of insulin-resistant subjects. Diabetes. 2007;56:2759–2765. doi: 10.2337/db07-0156. [DOI] [PubMed] [Google Scholar]
- 50.Kotronen A, Yki-Jarvinen H, Aminoff A, Bergholm R, Pietilainen KH, Westerbacka J, Talmud PJ, Humphries SE, Hamsten A, Isomaa B, Groop L, Orho-Melander M, Ehrenborg E, Fisher RM. Genetic variation in the ADIPOR2 gene is associated with liver fat content and its surrogate markers in three independent cohorts. Eur. J. Endocrinol. 2009;160:593–602. doi: 10.1530/EJE-08-0900. [DOI] [PubMed] [Google Scholar]
- 51.Stepanova M, Hossain N, Afendy A, Perry K, Goodman ZD, Baranova A, Younossi Z. Hepatic gene expression of Caucasian and African–American patients with obesity-related non-alcoholic fatty liver disease. Obes. Surg. 2010;20:640–650. doi: 10.1007/s11695-010-0078-2. [DOI] [PubMed] [Google Scholar]
- 52.Michalik L, Auwerx J, Berger JP, Chatterjee VK, Glass CK, Gonzalez FJ, Grimaldi PA, Kadowaki T, Lazar MA, O'Rahilly S, Palmer CNA, Plutzky J, Reddy JK, Spiegelman BM, Staels B, Wahli W. International Union of Pharmacology. LXI. Peroxisome Proliferator-Activated Receptors. Pharmacol. Rev. 2006;58:726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- 53.Sandoval A, Fraisl P, Rias-Barrau E, DiRusso CC, Singer D, Sealls W, Black PN. Fatty acid transport and activation and the expression patterns of genes involved in fatty acid trafficking. Arch. Biochem. Biophys. 2008;477:363–371. doi: 10.1016/j.abb.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 54.Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chistyakov DV, Aleshin S, Sergeeva MG, Reiser G. Regulation of peroxisome proliferator-activated receptor beta/delta expression and activity levels by toll-like receptor agonists and MAP kinase inhibitors in rat astrocytes. J. Neurochem. 2014;130:563–574. doi: 10.1111/jnc.12757. [DOI] [PubMed] [Google Scholar]
- 56.Lee MY, Choi R, Kim HM, Cho EJ, Kim BH, Choi YS, Naowaboot J, Lee EY, Yang YC, Shin JY, Shin YG, Chung CH. Peroxisome proliferator-activated receptor delta agonist attenuates hepatic steatosis by anti-inflammatory mechanism. Exp. Mol. Med. 2012;44:578–585. doi: 10.3858/emm.2012.44.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mashek DG, Li LO, Coleman RA. Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J. Lipid Res. 2006;47:2004–2010. doi: 10.1194/jlr.M600150-JLR200. [DOI] [PubMed] [Google Scholar]
- 58.Lewin TM, de Jong H, Schwerbrock NJ, Hammond LE, Watkins SM, Combs TP, Coleman RA. Mice deficient in mitochondrial glycerol-3-phosphate acyltransferase-1 have diminished myocardial triacylglycerol accumulation during lipogenic diet and altered phospholipid fatty acid composition. Biochim. Biophys. Acta. 2008;1781:352–358. doi: 10.1016/j.bbalip.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li LO, Ellis JM, Paich HA, Wang S, Gong N, Altshuller G, Thresher RJ, Koves TR, Watkins SM, Muoio DM, Cline GW, Shulman GI, Coleman RA. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. J. Biol. Chem. 2009;284:27816–27826. doi: 10.1074/jbc.M109.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.