Abstract
Autophagy is a dynamic and catabolic process that results in the breakdown and recycling of cellular components through the autophagosomal-lysosomal pathway. Many autophagy genes identified in yeast and mammals have orthologs in C. elegans. In recent years, gene inactivation, by RNAi and/or chromosomal mutations, has been useful to probe the functions of autophagy in C. elegans, and a role for autophagy has been shown in multiple processes such as, the adaptation to stress, longevity, cell death, cell growth control, clearance of aggregate prone proteins, degradation of P granules during embryogenesis, and apoptotic cell clearance. Here we discuss some of these roles and describe methods that can be used to study autophagy in C. elegans. Specifically, we summarize how to visualize autophagy in embryos, larva, or adults, how to detect the lipidation of LGG-1 by western blot, and how to inactivate autophagy genes by RNAi.
INTRODUCTION
Autophagy in C. elegans
Autophagy is a lysosomal-mediated pathway resulting in the degradation and recycling of long-lived proteins, protein aggregates, as well as damaged and old organelles (Klionsky, 2004). It is highly conserved and has been shown to be a fundamental catabolic process in eukaryotes that is required for key developmental and pathological events. Autophagy was first described in mammals, through morphological studies of rat liver cells (Deter et al., 1967). However, it was in yeast where many autophagy genes (atg) were discovered, by screening for mutations that decreased the survival of yeast cells under starvation, as well as mutations that disrupted the cytoplasm-to-vacuole targeting (cvt) process (Harding, 1996; Harding et al., 1995; Hutchins and Klionsky, 2001; Klionsky et al., 2003; Thumm et al., 1994; Tsukada and Ohsumi, 1993).
The process of autophagy is composed of several distinct steps: formation of a phagophore (also referred to as an isolation membrane or preautophagosomal structure); elongation and closure of the phagophore to form the double membrane autophagosome; transport and fusion of the autophagosome with a lysosome; and finally, degradation of the autophagosomal contents, and recycling of degraded material (FIG. 1) (Mizushima, 2007; Nakatogawa et al., 2009; Xie and Klionsky, 2007). In addition to fusing with a lysosome, an autophagosome may also fuse with an endosome to form a hybrid organelle called the amphisome (Jing and Tang, 1999; Liou et al., 1997). When an amphisome or autophagosome fuses with a lysosome, it is referred to as an autophagolysosome (or an autolysosome).
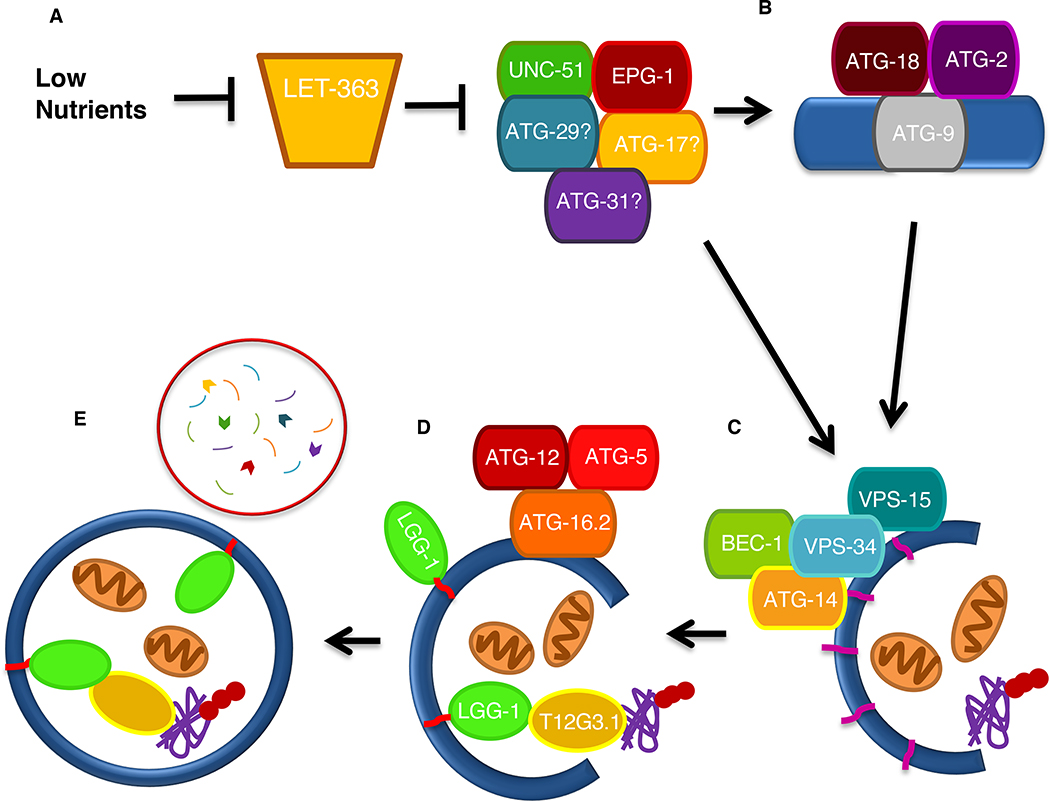
Figure 1.
Autophagy in C. elegans A. The process of autophagy has been delineated by studies in yeast and mammalian cells. We presume that induction of autophagy begins with the activation of UNC-51. B. Autophagosome formation requires the integral protein ATG-9, thought to contribute membrane to the developing autophagosome. C. Nucleation requires the Class III PI3K complex, which recruits downstream autophagy proteins to the isolation membranes (IM) in mammals or pre-autophagosomal structure (PAS) in yeast, through the production of PI3P (light purple). D. Two conjugation complexes (LGG-1 and ATG-12) are required for elongation of the isolation membranes and completion of the developing autophagosome. LGG-1 conjugated to phospatidylethanolamine (PE, red) binds to both the inner and outer membranes of the autophagosome. LGG-1 also has the ability to bind to the autophagic adaptor proteins, such as SQST-1 which bind poly-ubiquitinated aggregates. E. The complete autophagosome eventually fuses with the lysosome leading to the degradation of cargo within the autophagosome.
The evolutionary conservation of autophagy genes between yeast and C. elegans allowed for the identification of genes that encode core components of the autophagic machinery in C. elegans, on the basis of genomic sequence homology (Table 1) (Meléndez and Levine, 2009; Meléndez et al., 2003). Genetic screens for mutations that disrupt the degradation of P granules, has recently discovered autophagy genes not previously identified in C. elegans on the basis of sequence homology, including: epg-1, the ortholog of yeast ATG13, and epg-8, the ortholog of yeast ATG14 (Table 1) (Tian et al., 2009; Yang and Zhang, 2011). Although, the similarities between S. cerevisiae, mammals, and C. elegans autophagy proteins suggest that the molecular mechanisms of autophagosome formation may be conserved (FIG. 1 and Table 1) (Meléndez and Levine, 2009), genes recently identified in C. elegans that do not have a yeast ortholog may indicate that autophagy involves more complex membrane dynamics in higher eukaryotes. It is important to uncover further details about the roles of autophagy genes in autophagosome formation and maturation in C. elegans, and the role of these genes in different settings where autophagy is required.
Table 1:
C. elegans autophagy genes
| C. elegans Atg gene | Allele | Yeast/Mammalian ortholog | Phenotype in C. elegans | Reference |
|---|---|---|---|---|
| let-363R | h98 | TOR1,2/mTOR | Let, LL | (Brown et al. 1994; Noda and Ohsumi 1998; Vellai et al. 2003; Jia et al. 2004; Hansen et al. 2007; Hansen et al. 2008) |
| unc-51R | e369 | ATG1/Ulk1/2 | Unc, AbD, Pg, Egl | (Hedgecock et al. 1985; Ogura et al. 1994; Matsuura et al. 1997; Kuroyanagi et al. 1998; Meléndez et al. 2003; Zhang et al. 2009) |
| epg-1R | bp414 | ATG13/Atg13 | Dv, Pg | (Funakoshi et al. 1997; Chan et al. 2009; Tian et al. 2009) |
| bec-1R | ok691 | ATG6,VPS30/beclin 1 | Let, AbD, St, SL, Pg pQ | (Seaman et al. 1997; Kametaka et al. 1998; Kihara et al. 2001; Meléndez et al. 2003; Takacs-Vellai et al. 2005; Jia et al. 2007; Hansen et al. 2008; Zhao et al. 2009; Ruck et al. 2011) |
| let-512/vps-34R | h797 | VPS34/Vps34 | Let, SL, Pg | (Seglen and Gordon 1982; Volinia et al. 1995; Roggo et al. 2002; Zhao et al. 2009; Ruck et al. 2011) |
| ZK930.1R | ok3132 | VPS15/p150 | ND | (Panaretou et al. 1997; Kihara et al. 2001; Kovács et al. 2003) |
| epg-8R | bp251 | ATG14/Atg14L, Barkor | Dv, Pg | (Kihara et al. 2001; Obara et al. 2006; Sun et al. 2008); (Fan et al. 2011; Yang and Zhang 2011) |
| epg-6 | bp242 | -/WIPI4 | Dv, Pg | (Lu et al. 2011) |
| epg-3R | bp405 | -/VMP1 | Dv, Pg | (Tian et al. 2010) |
| epg-4R | bp425 | -/EI24,PIG8 | Dv, Pg | (Tian et al. 2010) |
| atg-3 | bp412 | ATG3/Atg3 | Pg | (Tanida et al. 2002; Zhang et al. 2009) |
| atg-4.1*R | tm3949 | ATG4/Atg4 | Pg | (Kirisako et al. 2000; Tanida et al. 2004; Zhang et al. 2009) |
| atg-4.2*R | tm3948 | ATG4/Atg4 | ND | (Kirisako et al. 2000; Tanida et al. 2004; Zhang et al. 2009) |
| atg-5 | bp545 | ATG5/Atg5 | ND | (Mizushima et al. 1998; Mizushima et al. 2001; Tian et al. 2010) |
| atg7/M7.5R | tm831 | ATG7/Atg7 | AbD, SL, Pg, pQ | (Kim et al. 1999; Tanida et al. 2001; Meléndez et al. 2003) |
| Igg-1R | bp500, tm3489 | ATG8/LC3 | Let, Dv, AbD, SL, Pg | (Kirisako et al. 2000; He et al. 2003; Meléndez et al. 2003); (Zhang et al. 2009; Alberti et al. 2010) |
| Igg-2R | - | ATG8/LC3 | Let, Dv, AbD, SL, Pg | (Kirisako et al. 2000; He et al. 2003; Meléndez et al. 2003; Zhang et al. 2009; Alberti et al. 2010) |
| atg-10R | bp588 | ATG10/Atg10 | ND | (Takahiro Shintani 1999; Mizushima et al. 2002; Meléndez et al. 2003; Tian et al. 2010) |
| Igg-3R | gk1857 | ATG12/Atg12 | SL, Pg | (Mizushima et al. 1998; Meléndez et al. 2003; Hars et al. 2007) |
| atg-16.1*R | - | ATG16/Atg16L1 | ND | (Kuma et al. 2002; Mizushima et al. 2003; Tian et al. 2010) |
| atg-16.2*R | ok3224 | ATG16/Atg16L1 | ND | (Kuma et al. 2002; Mizushima et al. 2003; Tian et al. 2010) |
| atg-2 | bp576 | ATG2/Atg2 | (Shintani 2001; Wang et al. 2001; Lu et al. 2011) | |
| atg-9R | bp564 | ATG9/Atg9 | (Noda et al. 2000; Yamada et al. 2005; Reggiori 2006) | |
| atg-18R | gk378 | ATG18/WIPI1/2 | Let, AbD, Pg, pQ | (Barth 2001; Meléndez et al. 2003; Jia et al. 2007); (Polson et al. 2010; Tian et al. 2010) |
| epg-2 | bp287 | -/- | Pg | (Tian et al. 2010) |
| epg-5R | bp450 | -/ KIAA1632 | Dv, Pg | (Tian et al. 2010) |
| sepa-1 | bp402 | -/- | Pg | (Zhang et al. 2009) |
| T12G3.1 | ok2892 | -/p62(SQSTM1) | (Tian et al. 2010; Lu et al. 2011) |
Let= Lethal; Unc= uncoordinated; Dv = Decreased viability of L1s during starvation; AbD= Abnormal Dauer; St= Sterile; LL= Long lifespan; SL= Short Lifespan; Pg= P granule accumulation; Egl= Egg laying defective; pQ= polyQ expansion susceptibility; ND= Not determined
Paralogs in C. elegans
RNAi clone available
Alberti A, Michelet X, Djeddi A, Legouis R. 2010. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 6: 622–633.
Barth H, Meiling-Wesse, K., Epple, U. D., and Thumm, M. 2001. Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS letters 508: 23–28.
Brown EJ, Albers MW, Bum Shin T, Ichikawa K, Keith CT, Lane WS, Schreiber SL. 1994. A mammalian protein targeted by Gl-arresting rapamycin–receptor complex. Nature 369: 756–758.
Chan EY, Longatti A, McKnight NC, Tooze SA. 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Molecular and cellular biology 29: 157–171.
Fan W, Nassiri A, Zhong Q. 2011. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). PNAS 108.
Funakoshi T, Matsuura A, Noda T, Ohsumi Y. 1997. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192: 207–213.
Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. 2008. A Role for Autophagy in the Extension of Lifespan by Dietary Restriction in C. elegans. PLoS genetics 4: e24.
Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. 2007. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging cell 6: 95–110.
Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF. 2007. Autophagy regulates ageing in C. elegans. Autophagy 3: 93–95.
He H, Dang Y, Dai F, Guo Z, Wu J, She X, Pei Y, Chen Y, Ling W, Wu C et al. 2003. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. The Journal of biological chemistry 278: 29278–29287.
Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. 1985. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Developmental biology 111: 158–170.
Jia K, Chen D, Riddle DL. 2004. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131: 3897–3906.
Jia K, Hart AC, Levine B. 2007. Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy 3: 21–25.
Kametaka S, Okano T, Ohsumi M, Ohsumi Y. 1998. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. The Journal of biological chemistry 273: 22284–22291.
Kihara A, Noda T, Ishihara N, Ohsumi Y. 2001. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. The Journal of cell biology 152: 519–530.
Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. 1999. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Molecular biology of the cell 10: 1337–1351.
Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. The Journal of cell biology 151: 263–276.
Kovács AL, Vellai T, Müller F. 2003. Autophagy in Caenorhabditis elegans. In Autophagy, D. Klionsky. 219–225.
Kuma A, Mizushima N, Ishihara N, Ohsumi Y. 2002. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. The Journal of biological chemistry 277: 18619–18625.
Kuroyanagi H, Yan J, Seki N, Yamanouchi Y, Suzuki Y, Takano T, Muramatsu M, Shirasawa T. 1998. Human ULK1, a Novel Serine/Threonine Kinase Related to UNC-51 Kinase of Caenorhabditis elegans: cDNA Cloning, Expression, and Chromosomal Assignment. Genomics 51: 76–85.
Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovacs AL, Yu L, Zhang H. 2011. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell 21: 343–357.
Matsuura A, Miki T, Wada Y, Ohsumi Y. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192: 245–250.
Meléndez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301: 1387–1391.
Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. 2003. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. Journal of Cell Science 116: 1679–1688.
Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. 1998. A New Protein Conjugation System in Human THE COUNTERPART OF THE YEAST Apg12p CONJUGATION SYSTEM ESSENTIAL FOR AUTOPHAGY*. The Journal of biological chemistry 273: 33889–33892.
Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. 2001. Dissection of Autophagosome Formation using Apg5-deficient Mouse Embryonic Stem Cells. Journal of Cell Biology 152: 657–667.
Mizushima N, Yoshimori T, Ohsumi Y. 2002. Mouse Apg10 as an Apg12-conjugating enzyme: analysis by the conjugation-mediated yeast two-hybrid Method. FEBS letters 532: 450–454.
Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. 2000. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. The Journal of cell biology 148: 465–480.
Noda T, Ohsumi Y. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. The Journal of biological chemistry 273: 3963–3966.
Obara K, Sekito T, Ohsumi Y. 2006. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Molecular biology of the cell 17: 1527–1539.
Ogura K, Wicky C, Magnenat L, Tobler H, Mori I, Muller F, Ohshima Y. 1994. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes & development 8: 2389–2400.
Panaretou C, Domin J, Cockcroft S, Waterfield MD. 1997. Characterization of p150, an Adaptor Protein for the Human Phosphatidylinositol (PtdIns) 3-Kinase. Journal of Biological Chemistry 272: 2477–2485.
Polson HE, de Lartigue J, Rigden D, Reedijk M, Urbé S, Clague M, Tooze S. 2010. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 6: 506–522.
Reggiori F, and Klionsky, D. J. 2006. Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae. Journal of Cell Science 119: 2903–2911.
Roggo L, Bernard V, Kovacs AL, Rose AM, Savoy F, Zetka M, Wymann MP, Muller F. 2002. Membrane transport in Caenorhabditis elegans: an essential role for VPS34 at the nuclear membrane. The EMBO journal 21: 1673–1683.
Ruck A, Attonito J, Garces KT, Nunez L, Palmisano NJ, Rubel Z, Bai Z, Nguyen KC, Sun L, Grant BD et al. 2011. The Atg6/Vps30/Beclin1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7.
Seaman MNJ, Marcusson EG, Cereghino JL, Emr SD. 1997. Endosome to Golgi Retrieval of the Vacuolar Protein Sorting Receptor, Vps10p, Requires the Function of the VPS29, VPS30, and VPS35Gene Products. Journal of Cell Biology 137: 79–92.
Seglen PO, Gordon PB. 1982. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proceedings of the National Academy of Sciences of the United States of America 79: 1889–1892.
Shintani T, Suzuki, K., Kamada, Y., Noda, T., and Ohsumi, Y. 2001. Apg2p functions in autophagosome formation on the perivacuolar structure. Journal of Biological Chemistry 276: 30452–30460.
Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. 2008. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proceedings of the National Academy of Sciences of the United States of America 105: 19211–19216.
Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, Kovacs AL, Muller F. 2005. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Current biology : CB 15: 1513–1517.
Takahiro Shintani NM, Yoko Ogawa, Akira Matsuura, Takeshi Noda and Yoshinori Ohsumi. 1999. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. The EMBO journal 18: 5234–5241.
Tanida I, Sou YS, Ezaki J, Minematsu-Ikeguchi N, Ueno T, Kominami E. 2004. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. The Journal of biological chemistry 279: 36268–36276.
Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. 2002. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. The Journal of biological chemistry 277: 13739–13744.
Tanida I, Tanida-Miyake E, Ueno T, Kominami E. 2001. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. The Journal of biological chemistry 276: 1701–1706.
Tian E, Wang F, Han Ja, Zhang H. 2009. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy 5: 608–615.
Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X et al. 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141: 1042–1055.
Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. 2003. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426: 620.
Volinia S, Dhand R, Vanhaesebroeck B, MacDougall LK, Stein R, Zvelebil MJ, Domin J, Panaretou C, Waterfield MD. 1995. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. The EMBO journal 14: 3339–3348.
Wang CW, Kim J, Huang WP, Abeliovich H, Stromhaug PE, Dunn WA, Jr., Klionsky DJ. 2001. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. The Journal of biological chemistry 276: 30442–30451.
Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. 2005. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. The Journal of biological chemistry 280: 18283–18290.
Yang P, Zhang H. 2011. The coiled-coil domain protein EPG-8 plays an essential role in the autophagy pathway in C. elegans. Autophagy 7: 159–165.
Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 136: 308–321.
Zhao Y, Tian E, Zhang H. 2009. Selective autophagic degradation of maternally-loaded germline P granule components in somatic cells during C. elegans embryogenesis. Autophagy 5: 717–719.
The role of autophagy genes in C. elegans development has emerged from studies using chromosomal mutations or RNA interference against autophagy genes. Chromosomal mutations exist for many of the autophagy genes found in C. elegans and many RNAi clones are available (Table 1).
Autophagy in C. elegans Development and Aging
L1 arrest after starvation:
Autophagy plays a role mediating the developmental changes associated with survival during extracellular and/or intracellular stress, such as starvation (Levine and Klionsky, 2004). In the absence of food, L1 larvae undergo a reversible developmental arrest and can survive for 1–2 weeks (Johnson et al., 1984). The insulin/IGF-1 signaling pathway, composed of the insulin-like/IGF-1 receptor daf-2 and the FOXO transcription factor daf-16, is involved in regulating L1 arrest triggered by starvation (Baugh and Sternberg, 2006; Fukuyama et al., 2006; Gems, 1998). Interestingly, reduced levels of autophagy have been shown to greatly decrease the survival of starved L1 larvae, emphasizing the importance of autophagy during early stages of development (Alberti et al., 2010; Kang et al., 2007; Lu et al., 2011; Tian et al., 2009; Tian et al., 2010; Yang and Zhang, 2011).
Dauer development:
During the first larval molt, animals that are exposed to a limited food supply develop into an alternate L3 larval stage termed dauer (Albert et al., 1981). Dauer development is associated with morphological and behavioral changes that allow for survival under harsh conditions and stress (Cassada and Russell, 1975; Golden and Riddle, 1984). The regulation of dauer development has been well characterized and requires the IGF-1/insulin-like, guanylyl cyclase, and TGF-β signaling pathways, as mutations in any of these pathways can result in a dauer constitutive phenotype (Daf-c) or a dauer defective phenotype (Daf-d) (Birnby et al., 2000; da Graca et al., 2004; Estevez et al., 1993; Gottlieb and Ruvkun, 1994; Inoue and Thomas, 2000; Patterson et al., 1997; Ren, 1996; Schackwitz, 1996; Thomas et al., 1993). Dauer development is associated with an increase in autophagy, which appears to be required for the cell remodeling associated with proper dauer formation (Meléndez et al., 2003).
Longevity pathways:
In C. elegans, aging is controlled by multiple longevity pathways, such as insulin-like growth factor signaling, TOR signaling, dietary restriction, mitochondrial activity, and germline signaling (Antebi, 2007). Recent genetic studies suggest that autophagy interacts with many of these longevity signals to regulate C. elegans aging (Hansen et al., 2008; Lapierre et al., 2011; Meléndez et al., 2003; Toth et al., 2008). Insulin/IGF-1R/daf-2 mutants display an increase in autophagy, as detected by an increase in the number of punctate structures labeled by the autophagy marker, GFP::LGG-1, in hypodermal seam cells, a cell type commonly used to visualize autophagy in C. elegans (FIG. 2) (Hansen et al., 2008; Meléndez et al., 2003). A reduction in autophagy during development, or only during adulthood, shortens the long lifespan of daf-2 mutants (Hansen et al., 2008; Hars et al., 2007; Meléndez et al., 2003). Reduced food intake without malnutrition, otherwise referred to as dietary restriction, occurs in eat-2 mutants (Avery, 1993). These animals lack a nicotinic acetylcholine receptor specific to the pharynx, thereby exhibiting reduced pharyngeal pumping, and have an extended lifespan phenotype (Lakowski and Hekimi, 1998; Raizen et al., 1995). Consistent with a role for TOR in dietary restriction, eat-2 mutants have reduced TOR signaling, display an increase in autophagy, and require autophagy for their long-lived phenotype (Hansen et al., 2008; Jia and Levine, 2007; Toth et al., 2008). The reduction in mitochondrial respiration in isp-1 mutants extends lifespan (Dillin et al., 2002; Lee et al., 2003), and this phenotype is also dependent on autophagy (Toth et al., 2008). Finally, glp-1/Notch germline-less mutants induce autophagy, and require autophagy for lifespan extension (Lapierre et al., 2011). Interestingly, HLH-30, the ortholog of the mammalian TFEB transcription factor, is required for the lifespan extension associated with the longevity pathways described above, and also regulates autophagy (Lapierre et al., 2013). In conclusion, autophagy is required as part of most longevity pathways in C. elegans, the only exception thus far being the longevity associated with a reduction in protein translation (Hansen et al., 2008; Pan et al., 2007).
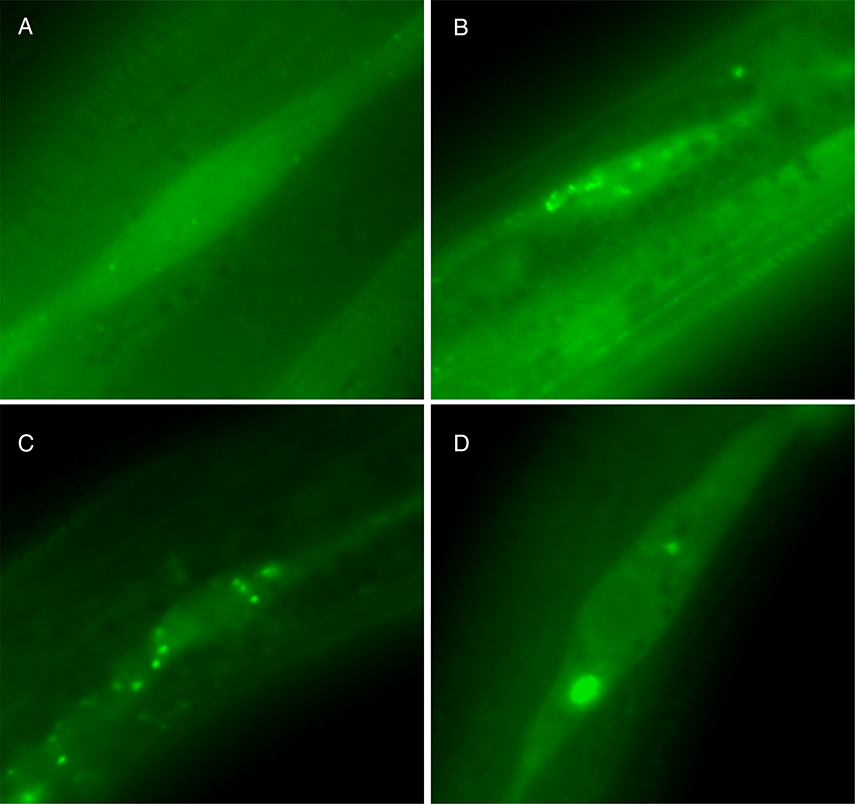
Figure 2.
GFP::LGG-1 expression in hypodermal seam cells of daf-2(e1370) mutants A. daf-2(e1370) mutants grown on OP50 E. coli, at 15°C, display a diffuse localization of GFP::LGG-1. B. daf-2(e1370) mutants grown on OP50 E. coli, at 25°C, display an increase in GFP::LGG-1 positive puncta (up to 12 GFP::LGG-1 positive puncta/seam cell) that represent early autophagic structures or autophagosomes. C. daf-2(e1370) mutants grown on control RNAi E. coli (transformed with empty vector, L4440), at 25°C, display the characteristic GFP::LGG-1 positive punctate structures. D. daf-2(e1370) mutants fed bec-1 RNAi, and raised at 25°C, display an increase in GFP::LGG-1 expression and large GFP::LGG-1 positive aggregates
Degradation of paternal mitochondria:
Directly after fertilization, autophagy is induced resulting in the elimination of spermatozoon specific organelles, including paternal mitochondria (Al Rawi et al., 2011; Sato and Sato, 2011). Whether autophagy also acts in higher eukaryotes to degrade paternal mitochondria is not known, however, an increase in ubiquitination and the localization of LC3 near the sperm mid-piece at the point of entry, may suggest that this is the case in fertilized mouse zygotes (Al Rawi et al., 2011).
Autophagy in apoptosis, necrosis and cell clearance:
Although autophagy has a role in homeostasis as an important pro-survival mechanism in response to stress, an excess in autophagy may result in cell death (Kang et al., 2007). Autophagy is also required for necrotic cell death, a type of cell death characterized by the loss of plasma membrane integrity (Samara et al., 2008; Toth et al., 2007). Additionally, similar to mammals, BEC-1, a component of the class III Phosphatidylinositol 3-Kinase (PI3K) complex (FIG. 1), interacts with the anti-apoptotic ortholog of Bcl-2, CED-9, suggesting cross-talk between autophagy and apoptosis (Erdelyi et al., 2011; Takacs-Vellai et al., 2005). Autophagy proteins have been shown to play a role in the proper degradation of apoptotic cell corpses in C. elegans, since in autophagy deficient animals, apoptotic cells are internalized, but not properly degraded (Li et al., 2012; Ruck et al., 2011). Interestingly, rescue experiments indicate that autophagy genes are required within the engulfing cell to promote apoptotic cell degradation (Li et al., 2012).
Detecting autophagy in C. elegans:
Autophagy can be monitored by transmission electron microscopy (TEM), fluorescent image analysis of the GFP::LGG-1 reporter or other autophagy reporters (Table 2), and by western blot, evaluating LGG-1 lipidation. It should be noted that an increase in the number of autophagosomes does not necessarily reflect an induction of autophagy (Klionsky, 2012), and is therefore important to distinguish between induction of autophagy, an increase in autophagic flux, and the accumulation of autophagosomes due to inefficient or blocked autophagy (Klionsky, 2012). Usually, it is useful to infer the turnover of autophagosomes in the presence and absence of lysosomal degradation. In C. elegans, this may be achieved by RNAi knockdown of genes with lysosomal function, such as cup-5 (Fares and Greenwald, 2001; Kostich et al., 2000; Sun et al., 2011), or the addition of inhibitors such as bafilomycin A1, or chloroquine, routinely used in mammalian cells, which have also been successful in C. elegans (Ji et al., 2006; Oka and Futai, 2000; Pivtoraiko et al., 2010). Clearly, the use of multiple assays to verify an increase in functional autophagy is recommended. A comprehensive list of guidelines was recently reported (Klionsky et al., 2012). Here we describe four protocols for the basic study of autophagy in C. elegans: detection of autophagy using GFP::LGG-1, autophagy in embryos, western blotting to evaluate lipidation of LGG-1, and RNAi as a method to target the knockdown of autophagy genes.
Table 2.
Fluorescent reporters for monitoring Autophagy in C. elegans
| Protein | Function | Tissue Expression¥ | TransgenesΦ | References |
|---|---|---|---|---|
| LGG-1 | Microtubule – associated protein-1/Ubiquitin-like protein | Intestine, Hypodermis, Muscle, Pharynx, Neurons, Vulva, Somatic Gonad, Germline | adIs2122[Plgg-1::GFP::LGG-1; rol-6(su1006)] izEx1[Plgg-1::GFP::LGG-1; rol-6(su1006)] izEx5[Plgg-1::GFP::LGG-1; Podr-1::RFP] vkEx1093[Pnhx-2::mCherry::LGG-1] dkIs399[Ppie-1::GFP::lgg-1, unc-119 (+)] Is(Ppie-1::GFP::mCherry::LGG-1; unc-119(+)) Ex[Plgg-1::DsRED::LGG-1; Pmyo-2::GFP] | (Melendez et al. 2003; Kang et al. 2007; Samara et al. 2008; Gosai et al. 2010; Manil-Segalen et al. 2014) |
| LGG-2 | Ubiquitin-like protein | Hypodermis, Intestine, Vulva, Pharynx, Neurons, Muscle | RD108 Ex[Plgg-2::GFP::LGG-2; rol-6(su1006)] RD217 unc119(ed3)III; Ex[unc-119(+); Ppie-1::gfp::mcherry::lgg-1] VIG9 unc119(ed3)III; Is[unc-119(+); Plgg-2::gfp::lgg-2] | (Alberti et al. 2010; Manil-Segalen et al. 2014) |
| DFCP1 | Double FYVE-Containing Protein | Head, Tail, Vulva, Neurons | bpIs168[Pnfya-1::DFCP1::GFP; unc-76(+)] | (Derubeis et al. 2000; Cheung et al. 2001; Axe et al. 2008; Tian et al. 2010) |
| ATG-16.1 | WD repeat-containing protein | Intestine, Head, Pharynx, Muscle, Neurons | [Patg-16.1::ATG-16.1::GFP; rol-6(su10006)] | (Zhang et al. 2013) |
| ATG-16.2 | WD repeat-containing protein | Intestine, Head, Pharynx, Muscle, Neurons | [Patg-16.2::ATG-16.2::GFP; rol-6(su10006)] | (Zhang et al. 2013) |
| ATG-9 | Integral Membrane Protein | Head, Tail, Vulva, Neurons | bpIs211[Pnfya-1::ATG-9::GFP; unc-76(+)] | (Noda et al. 2000; Lu et al. 2011; Liang et al. 2012; Lin et al. 2013) |
| EPG-1 | Atg13 homolog | Neurons, Pharynx, Muscle | bpIs175[Pepg-1::EPG-1::GFP; rol-6(su1006)] | (Tian et al. 2009) |
| EPG-9 | Atg101 homolog | Intestine, Pharynx, Neurons | bpIs214[Pepg-9::EPG-9::GFP; unc-76(+)] | (Liang et al. 2012) |
| BEC-1 | Coiled-Coil protein | Intestine, Hypodermis, Vulva, Neurons, Somatic Gonad | swEx520 [Pbec-1::BEC-1::GFP; rol-6(su1006)] grEx129[Pbec-1::BEC-1::mRFP; lin-15(+)] Ex[Pced-1::mCherry::BEC-1; rol-6(su1006)] Ex[Pegl1-::mCherry::BEC-1; rol-6(su1006)] | (Takacs-Vellai et al. 2005; Rowland et al. 2006; Ruck et al. 2011; Huang et al. 2012) |
| SQST-1 | p62/Autophagy adaptor protein | Hypodermis, Neurons, Intestine, Vulva, Muscle | bpIs151[Psqst-1::SQST-1::GFP; unc-76(+)] | (Hunt-Newbury et al. 2007; Pankiv et al. 2007; Tian et al. 2010) |
| SEPA-1 | Autophagy adaptor protein | Intestine, Head, Tail | bpIs131[Psepa-1::SEPA-1::GFP; unc-76(+)] | (Zhang et al. 2009; Tian et al. 2010) |
| PGL-1 | RNA-binding protein/P-granule component | P-granules, Intestine | bnIs1[Ppie-1::GFP::PGL-1; unc-119(+)] bnIs26[Pelt-2::PGL-1::GFP; Pmyo-2::mCherry] | (Cheeks et al. 2004; Zhang et al. 2009; Updike et al. 2011) |
Tissue expression may vary depending on the specific promoter used
Transgenes shown are those found in autophagy studies or those which may be beneficial in autophagy studies; Additional transgenes may be available for each gene
Alberti A, Michelet X, Djeddi A, Legouis R. 2010. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 6: 622–633.
Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of cell biology 182: 685–701.
Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. 2004. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Current biology : CB 14: 851–862.
Cheung PC, Trinkle-Mulcahy L, Cohen P, Lucocq JM. 2001. Characterization of a novel phosphatidylinositol 3-phosphate-binding protein containing two FYVE fingers in tandem that is targeted to the Golgi. The Biochemical journal 355: 113–121.
Derubeis AR, Young MF, Jia L, Robey PG, Fisher LW. 2000. Double FYVE-containing protein 1 (DFCP1): isolation, cloning and characterization of a novel FYVE finger protein from a human bone marrow cDNA library. Gene 255: 195–203.
Gosai SJ, Kwak JH, Luke CJ, Long OS, King DE, Kovatch KJ, Johnston PA, Shun TY, Lazo JS, Perlmutter DH et al. 2010. Automated high-content live animal drug screening using C. elegans expressing the aggregation prone serpin alpha1-antitrypsin Z. PloS one 5: e15460.
Huang S, Jia K, Wang Y, Zhou Z, Levine B. 2012. Autophagy genes function in apoptotic cell corpse clearance during C. elegans embryonic development. Autophagy 9.
Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A et al. 2007. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS biology 5: e237.
Kang C, You YJ, Avery L. 2007. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes & development 21: 2161–2171.
Liang Q, Yang P, Tian E, Han J, Zhang H. 2012. The C. elegans ATG101 homolog EPG-9 directly interacts with EPG-1/Atg13 and is essential for autophagy. Autophagy 8: 1426–1433.
Lin L, Yang P, Huang X, Zhang H, Lu Q, Zhang H. 2013. The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. The Journal of cell biology 201: 113–129.
Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovacs AL, Yu L, Zhang H. 2011. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Developmental cell 21: 343–357.
Manil-Segalen M, Lefebvre C, Jenzer C, Trichet M, Boulogne C, Satiat-Jeunemaitre B, Legouis R. 2014. The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Developmental cell 28: 43–55.
Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301: 1387–1391.
Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. 2000. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. Journal of Cell Biology 148: 465–480.
Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. The Journal of biological chemistry 282: 24131–24145.
Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA. 2006. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. The Journal of neuroscience : the official journal of the Society for Neuroscience 26: 1711–1720.
Ruck A, Attonito J, Garces KT, Nunez L, Palmisano NJ, Rubel Z, Bai Z, Nguyen KC, Sun L, Grant BD et al. 2011. The Atg6/Vps30/Beclin1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7.
Samara C, Syntichaki P, Tavernarakis N. 2008. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell death and differentiation 15: 105–112.
Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, Kovacs AL, Muller F. 2005. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Current biology : CB 15: 1513–1517.
Tian E, Wang F, Han Ja, Zhang H. 2009. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy 5: 608–615.
Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X et al. 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141: 1042–1055.
Updike DL, Hachey SJ, Kreher J, Strome S. 2011. P granules extend the nuclear pore complex environment in the C. elegans germ line. The Journal of cell biology 192: 939–948.
Zhang H, Wu F, Wang X, Du H, Wang X, Zhang H. 2013. The two C. elegans ATG-16 homologs have partially redundant functions in the basal autophagy pathway. Autophagy 9: 1965–1974.
Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 136: 308–321.
References
- Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V, 2011. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 334, 1144–1147. [DOI] [PubMed] [Google Scholar]
- Albert PS, Brown SJ, Riddle DL, 1981. Sensory control of dauer larva formation in Caenorhabditis elegans. J Comp Neurol 198, 435–451. [DOI] [PubMed] [Google Scholar]
- Alberti A, Michelet X, Djeddi A, Legouis R, 2010. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 6, 622–633. [DOI] [PubMed] [Google Scholar]
- Antebi A, 2007. Genetics of aging in Caenorhabditis elegans. PLoS genetics 3, 1565–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, 1993. The genetics of feeding in Caenorhabditis elegans. Genetics 133, 897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Sternberg PW, 2006. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Current Biology 16, 780–785. [DOI] [PubMed] [Google Scholar]
- Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, Thomas JH, 2000. A Transmembrane Guanylyl Cyclase (DAF-11) and Hsp90 (DAF-21) Regulate a Common Set of Chemosensory Behaviors in Caenorhabditis elegans. Genetics 155, 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada RC, Russell RL, 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol 46, 326–342. [DOI] [PubMed] [Google Scholar]
- da Graca LS, Zimmerman KK, Mitchell MC, Kozhan-Gorodetska M, Sekiewicz K, Morales Y, Patterson GI, 2004. DAF-5 is a Ski oncoprotein homolog that functions in a neuronal TGF beta pathway to regulate C. elegans dauer development. Development 131, 435–446. [DOI] [PubMed] [Google Scholar]
- Deter RL, Baudhuin P, De Duve C, 1967. Participation of Lysosomes in Cellular Autophagy Induced in Rat Liver by Glucagon. Journal of Cell Biology 35, C11–C16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C, 2002. Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401. [DOI] [PubMed] [Google Scholar]
- Erdelyi P, Borsos E, Takacs-Vellai K, Kovacs T, Kovacs AL, Sigmond T, Hargitai B, Pasztor L, Sengupta T, Dengg M, Pecsi I, Toth J, Nilsen H, Vertessy BG, Vellai T, 2011. Shared developmental roles and transcriptional control of autophagy and apoptosis in Caenorhabditis elegans. J Cell Sci 124, 1510–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez M, Attisano L, Wrana JL, Albert PS, Massague J, Riddle DL, 1993. The daf-4 gene encodes a bone morphogenetic protein receptor controlling C. elegans dauer larva development. Nature 365, 644–649. [DOI] [PubMed] [Google Scholar]
- Fares H, Greenwald I, 2001. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nature genetics 28, 64–68. [DOI] [PubMed] [Google Scholar]
- Fukuyama M, Rougvie A, E., Rothman JH, 2006. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Current Biology 16, 773–779. [DOI] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, and Riddle DL, 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150, 129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden JW, Riddle DL, 1984. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol 102, 368–378. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G, 1994. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics 137, 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C, 2008. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4, e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding TM, Hefner-Gravink A, Thumm M, and Klionsky DJ, 1996. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. Journal of Biological Chemistry 271, 17621–17624. [DOI] [PubMed] [Google Scholar]
- Harding TM, Morano KA, Scott SV, Klionsky DJ, 1995. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol 131, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hars ES, Qi H, Ryazanov AG, Jin S, Cai L, Hu C, Liu LF, 2007. Autophagy Regulates Ageing in C. elegans. Autophagy 3, 93–95. [DOI] [PubMed] [Google Scholar]
- Hutchins MU, Klionsky DJ, 2001. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem 276, 20491–20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Thomas JH, 2000. Suppressors of transforming growth factor-beta pathway mutants in the Caenorhabditis elegans dauer formation pathway. Genetics 156, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji YJ, Choi KY, Song HO, Park BJ, Yu JR, Kagawa H, Song WK, Ahnn J, 2006. VHA-8, the E subunit of V-ATPase, is essential for pH homeostasis and larval development in C. elegans. FEBS Lett 580, 3161–3166. [DOI] [PubMed] [Google Scholar]
- Jia K, Levine B, 2007. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy 3, 597–599. [DOI] [PubMed] [Google Scholar]
- Jing Y, Tang XM, 1999. The convergent point of the endocytic and autophagic pathways in leydig cells. Cell research 9, 243–253. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Mitchell DH, Kline S, Kemal R, Foy J, 1984. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mechanisms of ageing and development 28, 23–40. [DOI] [PubMed] [Google Scholar]
- Kang C, You YJ, Avery L, 2007. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev 21, 2161–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky B.L.a.D.J., 2004. Development by Self-Digestion: Molecular Mechanisms and Biological Functions of Autophagy. Developmental cell 6, 463–477. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, 2012. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 1–100.22082964 [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA Jr., Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y, 2003. A unified nomenclature for yeast autophagy-related genes. Dev Cell 5, 539–545. [DOI] [PubMed] [Google Scholar]
- Kostich M, Fire A, Fambrough DM, 2000. Identification and molecular-genetic characterization of a LAMP/CD68-like protein from Caenorhabditis elegans. J Cell Sci 113 ( Pt 14), 2595–2606. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S, 1998. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A 95, 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, Gelino S, Ong B, Davis AE, Irazoqui JE, Dillin A, Hansen M, 2013. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun 4, 2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre LR, Gelino S, Melendez A, Hansen M, 2011. Autophagy and Lipid Metabolism Coordinately Modulate Life Span in Germline-less C. elegans. Current biology : CB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Clough EA, Yellon P, Teslovich TM, Stephan DA, Baehrecke EH, 2003. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Current biology : CB 13, 350–357. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ, 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6, 463–477. [DOI] [PubMed] [Google Scholar]
- Li W, Zou W, Yang Y, Chai Y, Chen B, Cheng S, Tian D, Wang X, Vale RD, Ou G, 2012. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J Cell Biol 197, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou W, Geuze HJ, Geelen MJ, Slot JW, 1997. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. The Journal of cell biology 136, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Yang P, Huang X, Hu W, Guo B, Wu F, Lin L, Kovacs AL, Yu L, Zhang H, 2011. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell 21, 343–357. [DOI] [PubMed] [Google Scholar]
- Meléndez A, Levine B, 2009. Autophagy in C. elegans. WormBook, 1–26. [DOI] [PubMed] [Google Scholar]
- Meléndez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B, 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387–1391. [DOI] [PubMed] [Google Scholar]
- Mizushima N, 2007. Autophagy: process and function. Genes Dev 21, 2861–2873. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y, 2009. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nature Reviews Molecular Cell Biology 10, 458–467. [DOI] [PubMed] [Google Scholar]
- Oka T, Futai M, 2000. Requirement of V-ATPase for ovulation and embryogenesis in Caenorhabditis elegans. The Journal of biological chemistry 275, 29556–29561. [DOI] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P, 2007. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GI, Koweek A, Wong A, Liu Y, Ruvkun G, 1997. The DAF-3 Smad protein antagonizes TGF-beta-related receptor signaling in the Caenorhabditis elegans dauer pathway. Genes & development 11, 2679–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pivtoraiko VN, Harrington AJ, Mader BJ, Luker AM, Caldwell GA, Caldwell KA, Roth KA, Shacka JJ, 2010. Low-dose bafilomycin attenuates neuronal cell death associated with autophagy-lysosome pathway dysfunction. Journal of neurochemistry 114, 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen DM, Lee RY, Avery L, 1995. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics 141, 1365–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, and Riddle DL, 1996. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274, 1389–1391. [DOI] [PubMed] [Google Scholar]
- Ruck A, Attonito J, Garces KT, Nunez L, Palmisano NJ, Rubel Z, Bai Z, Nguyen KC, Sun L, Grant BD, Hall DH, Melendez A, 2011. The Atg6/Vps30/Beclin1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara C, Syntichaki P, Tavernarakis N, 2008. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ 15, 105–112. [DOI] [PubMed] [Google Scholar]
- Sato M, Sato K, 2011. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334, 1141–1144. [DOI] [PubMed] [Google Scholar]
- Schackwitz WS, Inoue T, and Thomas JH, 1996. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17, 719–728. [DOI] [PubMed] [Google Scholar]
- Sun T, Wang X, Lu Q, Ren H, Zhang H, 2011. CUP-5, the C. elegans ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, is required for proteolytic degradation in autolysosomes. Autophagy 7, 1308–1315. [DOI] [PubMed] [Google Scholar]
- Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, Kovacs AL, Muller F, 2005. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol 15, 1513–1517. [DOI] [PubMed] [Google Scholar]
- Thomas JH, Birnby DA, Vowels JJ, 1993. Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134, 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH, 1994. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett 349, 275–280. [DOI] [PubMed] [Google Scholar]
- Tian E, Wang F, Han J.a., Zhang H, 2009. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy 5, 608–615. [DOI] [PubMed] [Google Scholar]
- Tian Y, Li Z, Hu W, Ren H, Tian E, Zhao Y, Lu Q, Huang X, Yang P, Li X, Wang X, Kovacs AL, Yu L, Zhang H, 2010. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 141, 1042–1055. [DOI] [PubMed] [Google Scholar]
- Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, Vellai T, 2008. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 4, 330–338. [DOI] [PubMed] [Google Scholar]
- Toth ML, Simon P, Kovacs AL, Vellai T, 2007. Influence of autophagy genes on ion-channel-dependent neuronal degeneration in Caenorhabditis elegans. J Cell Sci 120, 1134–1141. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y, 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333, 169–174. [DOI] [PubMed] [Google Scholar]
- Xie Z, Klionsky DJ, 2007. Autophagosome formation: core machinery and adaptations. Nature cell biology 9, 1102–1109. [DOI] [PubMed] [Google Scholar]
- Yang P, Zhang H, 2011. The coiled-coil domain protein EPG-8 plays an essential role in the autophagy pathway in C. elegans. Autophagy 7, 159–165. [DOI] [PubMed] [Google Scholar]