Abstract
Due to the highly restricted species-tropism of Hepatitis C virus (HCV) a limited number of animal models exist for pre-clinical evaluation of vaccines and antiviral compounds. The human-liver chimeric mouse model allows heterologous challenge with clinically relevant strains derived from patients. However, to date, the transmission and longitudinal evolution of founder viral populations in this model have not been characterized in-depth using state-of-the-art sequencing technologies. Focusing on NS3 protease encoding region of the viral genome, mutant spectra in a donor inoculum and individual recipient mice were determined via Illumina sequencing and compared, to determine the effects of transmission on founder viral population complexity. In all transmissions, a genetic bottleneck was observed, although diverse viral populations were transmitted in each case. A low frequency cloud of mutations (<1%) was detectable in the donor inoculum and recipient mice, with single nucleotide variants (SNVs) > 1% restricted to a subset of nucleotides. The population of SNVs >1% was reduced upon transmission while the low frequency SNV cloud remained stable. Fixation of multiple identical synonymous substitutions was apparent in independent transmissions, and no evidence for reversion of T-cell epitopes was observed. In addition, susceptibility of founder populations to antiviral therapy was assessed. Animals were treated with protease inhibitor (PI) monotherapy to track resistance associated substitution (RAS) emergence. Longitudinal analyses revealed a decline in population diversity under therapy, with no detectable RAS >1% prior to therapy commencement. Despite inoculation from a common source and identical therapeutic regimens, unique RAS emergence profiles were identified in different hosts prior to and during therapeutic failure, with complex mutational signatures at protease residues 155, 156 and 168 detected. Together these analyses track viral population complexity at high-resolution in the human-liver chimeric mouse model post-transmission and under therapeutic intervention, revealing novel insights into the evolutionary processes which shape viral protease population composition at various critical stages of the viral life-cycle.
Keywords: HCV protease, Transmission, Founder populations, Longitudinal evolution, RAS emergence
1. Introduction
Direct Acting antivirals (DAAs) have revolutionized treatment of chronic HCV infection (Lange et al., 2014; Polenakovik, 2015; Zhang et al., 2016). In 2003 the first anti-HCV DAA was described (BILN 2061, also known as Ciluprevir), providing proof of concept that targeting the HCV protease represents a potent therapeutic option (Lamarre et al., 2003). The non-structural protein 3 (NS3) is a multifunctional protein that contains a serine protease domain at its N-terminus. Inhibition of the NS3 protease supports viral clearance via both inhibition of viral polyprotein cleavage, reducing viral replication, in addition to restoration of MAVS and TRIF mediated innate immune signaling (Meylan et al., 2005). IFN-free DAA combination therapies including second generation NS3 PIs can achieve high clearance rates in hard-to-treat patient groups (Buti et al., 2016; Zeuzem et al., 2015). However, despite the NS3 protease domain’s importance as a therapeutic target, genetic variability in viral NS3 protease populations remains poorly characterized in HCV infection at different stages of the viral life-cycle.
Studying HCV transmission in a natural setting is challenging due to the often asymptomatic nature of infection (Irving and Brown, 2010), with limited numbers of studies characterizing samples from both donor and recipient(s). To circumvent the low availability of samples from donor/recipient pairs, permissive animal models can be used to model such scenarios (Vercauteren et al., 2015b), yielding novel insights into the transmission process (Brown et al., 2012). HCV replication in infected hepatocytes is characterized by a high rate of virus production and a correspondingly high level of genetic diversity in circulating viruses (Simmonds, 2004; Tarr et al., 2015). This is due to the lack of efficient proofreading by the HCV RNA-dependent RNA polymerase. As a result, HCV exists as a dynamic swarm of genetically related genomes within infected patients rather than a clonal population (Martell et al., 1992; Pawlotsky, 2006). While chimpanzees are permissive to infection with non-laboratory strains of HCV (Bukh, 2012) ethical concerns have resulted in research animals being retired in both the US and the EU. Consequently, the current state-of-the-art animal model for heterologous challenge with clinically relevant strains is the human-liver chimeric mouse model: the liver of Alb-uPA transgenic SCID mice can be engrafted with human adult hepatocytes rendering them permissive to HCV infection (Vercauteren et al., 2015b, 2014).
In this study, we focused on the genomic region encoding the NS3 protease domain, tracking sequence evolution upon transmission to a new host and the subsequent susceptibility of newly transmitted viral populations to antiviral therapy. We modeled a single-source outbreak to multiple recipients, infecting multiple human-liver chimeric mice with a viral inoculum derived from a single chronically infected patient. We utilized next generation sequencing (NGS) to track the evolution of viral protease populations upon viral transmission, comparing the donor viral population with founder populations in individual recipient mice. In addition, after transmission, we sought to track independent longitudinal evolution of NS3 protease populations under PI monotherapy in individual animals and the emergence of RAS at high-resolution.
2. Material and methods
2.1. Experimental design
Human liver-chimeric mice used in this study were produced as previously reported (Meuleman et al., 2005) and represent some of the monotherapy arm animals from Vercauteren et al. (2015a). Mice were infected with a serum-derived virus (titer 2 × 105 IU/ml) from a chronically HCV infected patient (genotype 1b, P05) (Fafi-Kremer et al., 2010). Patient serum was obtained with informed consent following a protocol approved by the local Ethics Committee of the University of Strasbourg Hospitals (CPP 10–17). A high-titre viremia was established in the donor mouse (DM), which was subsequently utilized to infect eight recipient animals (r1-8: 1 × 104 IU/mouse). Virus was inoculated at least four weeks prior to start of Ciluprevir monotherapy, which was administered by oral gavage twice daily at 10 mg/kg. Ciluprevir is a peptidomimetic macrocyclic compound that binds non-covalently to the active site of the HCV NS3 serine protease. Viremia was tracked as previously described (Vercauteren et al., 2015a).
2.2. Viral population sampling
RNA extraction, cDNA synthesis and amplicon generation were performed according to Vercauteren et al. (2015a). Briefly, viral RNA was extracted from EDTA-plasma using a Viral RNA MiniElute kit (Qiagen). Synthesis of cDNA was achieved using 8 μl viral RNA, a genotype 1b specific primer and Superscript III reverse transcriptase (Invitrogen). Quantification of viral cDNA input into sequencing amplicons was determined via qPCR as previously described (Irving et al., 2014). Amplicon generation was performed via nested PCR using Platinum Taq High-fidelity polymerase (Invitrogen) and genotype 1b-specific primers.
2.3. Illumina MiSeq sequencing
NGS library preparation was performed in accordance with the manufacturer’s instructions using a TruSeq DNA PCR-Free Sample Preparation Kit (Illumina Inc., San Diego, CA, USA). Quality control of NGS libraries was performed using an Agilent Bioanalyzer HS Chip (Agilent Technologies). NGS library quantification was performed via qPCR (KAPA Library Quantification Kit) and 300 cycles of sequencing performed using a MiSeq Reagent Kit v3 (Illumina Inc.).
2.4. Data analysis: read mapping and SNV calling
NGS data were analyzed as previously described (Irving et al., 2014; Vercauteren et al., 2015a) with minor modifications. The nucleotide consensus of the P05 NS3 protease population (540bp) was determined and used as a reference sequence in all subsequent assemblies. Raw Fastq files were mapped against the P05 consensus using CLC Genomics Workbench v8b (Qiagen Arhaus). BAM contigs were used to generate coverage plots and for SNV calling to track high-frequency SNVs (>1%): only SNVs exhibiting average PHRED scores >30 were included. To track ultra-low frequency SNVs (<1%) and estimate Shannon entropy in viral populations the reference-based short read aligner TANOTI was used for mapping followed by a subsequent SNV calling employing DiversiTools.
3. Results
3.1. Characterization of viral population diversity pre and post transmission
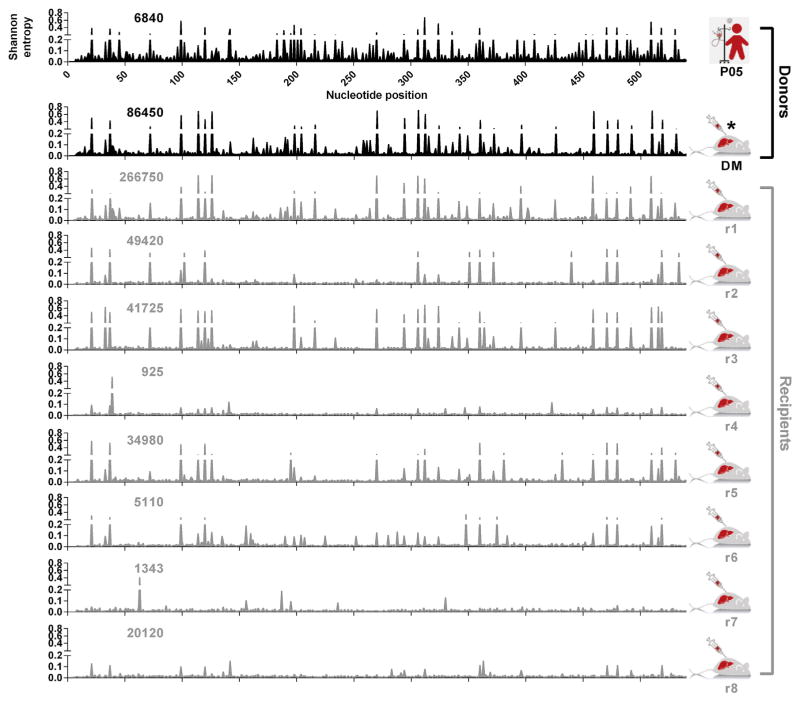
Comparative analyses of the viral population in the patient serum and donor mouse were performed to quantify any loss of population diversity, which may have occurred in the passaging process, and ensure a diverse inoculum was transmitted to recipient mice (Fig. 1). Comparative analyses of protease populations in the DM and recipient animals were then performed by calculating Shannon Entropy. Shannon Entropy is a quantitative measure of uncertainty which can be used to determine the variability in a population of sequences. Sliding window analyses enabled visualization of cross-sectional diversity in viral populations and revealed diverse and distinct viral populations in the patient sera, DM and recipients (Fig. 1). The majority of this population variability is low frequency although mutational hotspots indicating high levels of population heterogeneity are evident in most recipient animals. To ensure robust comparison of viral populations at different samplings, quantification of viral genomic input into sequencing amplicons was determined by qPCR (Irving et al., 2014). These comparisons revealed that viral population diversity was independent of genomic input. For example, population heterogeneity in patient P05 (6840 genomes sampled) was greater than the DM (86,450 genomes sampled) despite shallower depth of genome sampling. These data also revealed viral populations take different evolutionary trajectories post-transmission: despite high-titer inoculation from a common source, some founder populations exhibit greater diversity (r1) while others appear genetically more homogenous (r8).
Fig. 1. The effects of transmission on HCV population diversity.
Cross-sectional viral diversity in the genomic region encoding the NS3 protease domain in patient serum (top panel, black), donor inoculum (second panel, black) and eight recipient human-liver chimeric mice (bottom panels, grey). The x-axis represents the 540 nucleotides encoding the N-terminus of NS3, while the y-axis represents diversity in the viral population measured in Shannon entropy. Sample identifiers are presented to the right of individual plots. Numbers within individual plots represent the genomic input into respective sequencing amplicons and were determined via 5′UTR RT-qPCR (Irving et al., 2014).
3.2. Low frequency clouds of SNVs detected pre- and post-transmission
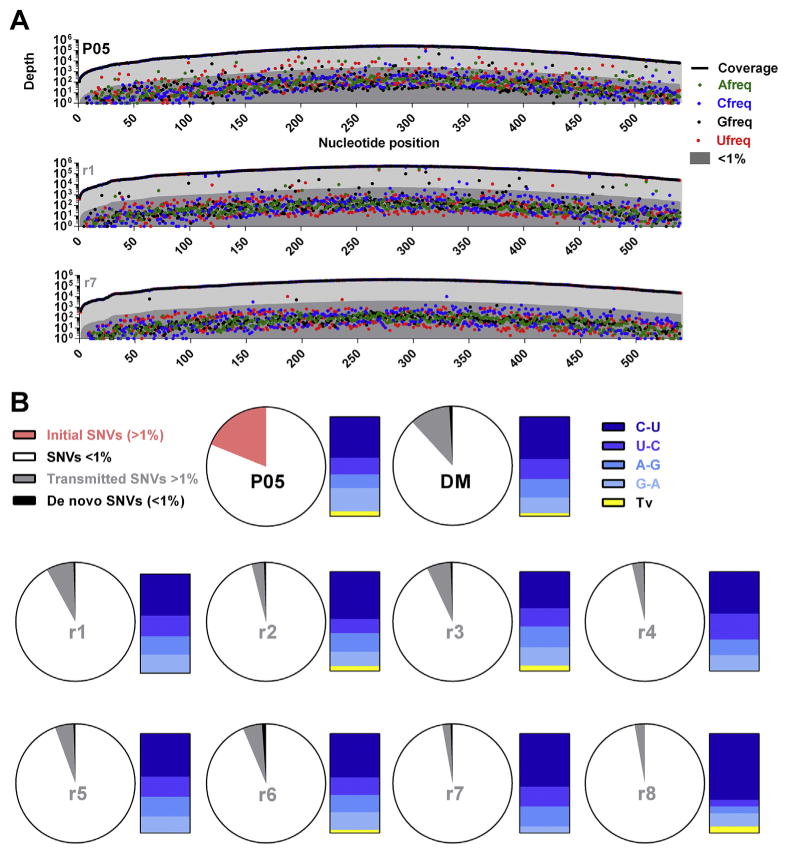
The majority of detectable variation in NS3 viral populations was low frequency (Fig. 2a). Cross-sectional analyses of the patient serum (top panel) and recipient mice with either diverse (middle panel) or more homogeneous (bottom panel) viral populations revealed a low frequency (<1%) cloud of SNVs detectable across the entire sequence length. This cloud is maintained post-transmission in all recipient animals. However, numbers of high-frequency SNVs (>1%) are reduced upon transmission and differ between individual recipient animals.
Fig. 2. SNV frequencies pre- and post-transmission.
(A) Cross-sectional detection of a low-frequency mutational cloud in the patient serum (upper panel) and two example recipient mice (lower panels). The x-axis represents the 540 nucleotides encoding the N-terminus of NS3, while the y-axis represents the depth of nucleotide coverage across this region. Sample identifiers are presented in the top right of individual plots. The thick black line represents total nucleotide coverage at a specific position whilst the frequencies of individual nucleotides are color-coded. Population frequencies above 1% are highlighted in light-grey while frequencies below 1% are highlighted in dark-grey. (B) Proportions of low- and -high frequency transmitted SNVs. Pie charts represent total nucleotides analyzed per sample (540bp). For all samples, SNVs exhibiting variation <1% (low-frequency) are highlighted in white. For the patient sera, the total SNVs exhibiting variation >1% (high-frequency) are highlighted in pink. To visualize whether there was a preference for transmission of high-frequency SNVs, for the donor inoculum and individual recipient mice, SNV >1% which were also detected in the patient inoculum at >1% are highlighted in grey. SNVs present at >1% in recipient mice that remained either undetected or had frequencies <1% in the patient serum are highlighted in black. To the left of each pie chart, high-frequency SNVs are categorized in bar charts for each sample, with different transitional substitutions highlighted in shades of blue and transversions highlighted in yellow.
3.3. High-frequency SNVs are differentially transmitted to recipient animals and reduced in number
In the initial virus population of the patient, ~19% of sites (n = 102) exhibited SNVs with >1% population frequencies. This was reduced ~12% of sites (n = 64) in the DM and ranges between ~8% and ~3% in recipient animals (Fig. 2b). The vast majority of high-frequency SNVs detected in all donor animals were transmitted (i.e. also detectable in the patient or donor inoculum at >1%). Very few high-frequency SNVs post-transmission were derived from the low-frequency SNV pool in the patient or donor inoculum, supporting the notion that restrictions exist on the mutational space that can be effectively explored by RNA virus populations (Brown et al., 2014). In all viral populations analyzed, an extreme transition/transversion bias existed, with the vast majority of high-frequency SNVs representing transitional mutations (C〈〉U or A〈〉G): this pattern was not altered post-transmission (Fig. 2b).
3.4. Synonymous mutations predominate post transmission and a subset becomes fixed
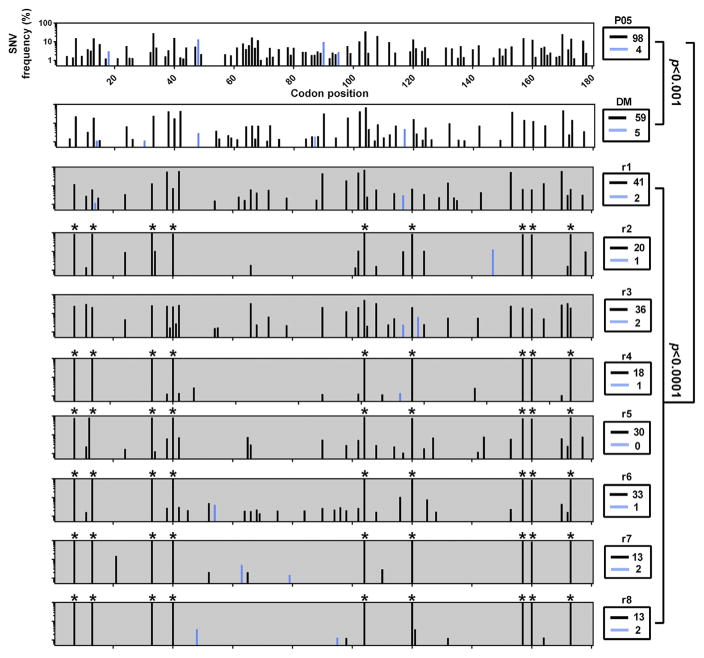
Levels of synonymous (dS) and non-synonymous (dN) substitutions were determined pre- and post-transmission (Fig. 3). Significant reductions in viral diversity were observed upon passage of the patient sera into the DM (p < 0.001) and in each separate transmission of the DM to recipients (p < 0.0001). In the patient serum, DM and recipient mice the consensus amino acid sequence remained identical. Although dN substitutions were detectable in 7/8 animals post-transmission, their population frequencies were low (<10%) and were restricted to 1–2 sites. No dN substitutions were detected at sites associated with DAA resistance or residues comprising the catalytic triad of the serine protease. Mapping against the P05 consensus enabled visualization of SNVs in the patient sera which were swept towards population fixation post-transmission. In 6/8 founder populations, nine dS SNVs exhibited evidence for an increase population frequency to become dominant (70–99% frequency: sites 21, 37, 99, 120, 312, 360, 471, 480 and 519) (Fig. 3).
Fig. 3. Synonymous and non-synonymous SNV frequencies pre- and post-transmission.
Cross-sectional quantification of silent and amino acid changing SNVs in the NS3 protease domain in patient serum (top panel, white background), donor inoculum (second panel, white background) and eight recipient human-liver mice (bottom panels, grey background). For each plot, all mutations present at greater than 1% are plotted relative to the P05 consensus master sequence. The x-axis represents the 180 codons of the NS3 protease domain. The y-axis represents the relative percentage frequencies of SNVs across the entire NS3 protease domain, with synonymous (dS, black bars) and non-synonymous (dN, blue bars) frequencies in the viral population. Sample identifiers are presented to the right of individual plots with the absolute number of sites exhibiting variation boxed below. Statistical significance was calculated via Yates corrected Chi2 tests, with adjusted p-values for multiple comparisons. SNVs which become dominant post transmission are marked with *.
3.5. Susceptibility of founder populations to antiviral therapy
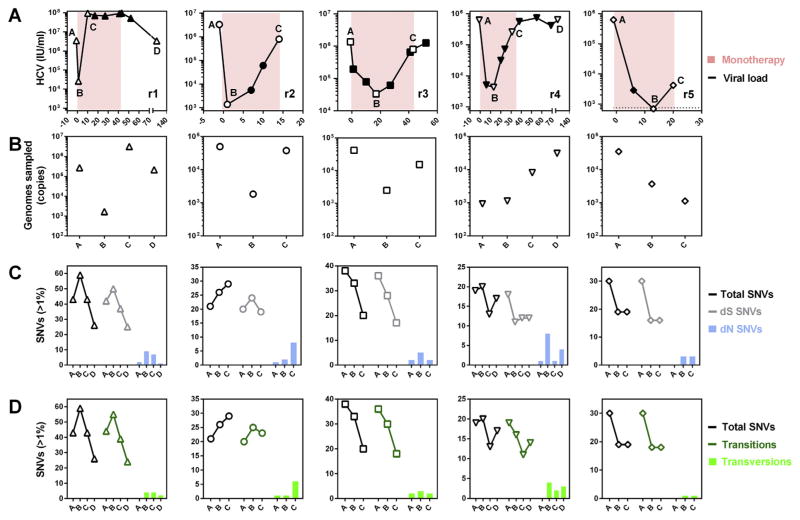
Evolution of founder populations in response to antiviral therapy was tracked in five individual mice post-transmission (Fig. 4). In all animals, viremia rapidly declined upon therapy initiation but quickly rebounded (Fig. 4a). Absolute quantification of viral genomic input into sequencing amplicons revealed good sampling depth for all analyzed populations (Fig. 4b). High frequency SNVs (>1%) were generally synonymous (Fig. 4c) and transitional (Fig. 4d) in all animals with SNV >1% declining in 4/5 animals over the course of therapy (Fig. 4C and D). Despite this decline, an increase in both dN (Fig. 4c) and tv SNV frequencies (Fig. 4d) was observed after therapy initiation.
Fig. 4. Longitudinal evolution of the protease domain during PI monotherapy.
(A) Each panel represents monotherapy duration in animals r1-r5, with days presented on the x-axis and viremia on the y-axis. Labeled open symbols represent samples which were available for NGS while the pink background indicates the duration of monotherapy. For all animals, A represents the pre-therapy sample, B represents the pre-breakthrough sample and C represents the post-breakthrough sample. For r1 and r4, an additional sample D was also available after cessation of monotherapy. (B) Viral genomic input into sequencing amplicons. (C) Quantification of total SNVs >1% (black line) and the proportions of synonymous (dS) (grey line) and non-synonymous (dN) (blue bars) SNVs in individual mice over the course of PI monotherapy. (D) Quantification of total SNVs >1% (black line) and the proportions of transitions (green line) and transversions (green bars).
3.6. Population mutational signatures underlying RAS emergence
To visualize population diversity and RAS emergence at high-resolution, mouse r1 is shown as an example (Fig. 5). Comparative analyses of viral populations over the course of therapy revealed a reduction in population diversity. After an initial increase, dS SNVs >1% declined after breakthrough. Prior to break-through, seven discrete RAS were detected at positions R155, A156 and D168 at different frequencies (Fig. 5a). To illustrate population complexity observed at susceptible sites, mutational frequencies in the viral population underlying residue 168 are visualized in Fig. 5b. These analyses reveal a homogenous population prior to therapy initiation followed by extreme population complexity prior to breakthrough. Underlying mutational complexity decreases post-breakthrough and again post therapy cessation, indicating complex population dynamics contribute to therapeutic failure which differs between hosts.
Fig. 5. Progressive evolution of the entire protease domain and complex patterns of RAS emergence under PI monotherapy.
(A) Substitutional evolution across the entire protease domain in mouse r1 pre, during and post-monotherapy. For each plot, all mutations present at greater than 1% are plotted relative to the P05 consensus master sequence. Pre and post-therapy plots have grey backgrounds while on-therapy plots have pink backgrounds. The x-axis represents the 180 codons of the NS3 protease domain. The y-axis represents the relative percentage of SNVs across the entire NS3 protease domain, with dS (black bars), dN (blue bars) and RAS (red bars) in the viral population presented. (B) Example of complex mutational signatures underlying RAS emergence. Web-logos (Crooks et al., 2004) depicting the population frequencies of nucleotides at individual codon positions underlying amino acid residue 168 in mouse r1 are presented in chronological order, from left to right.
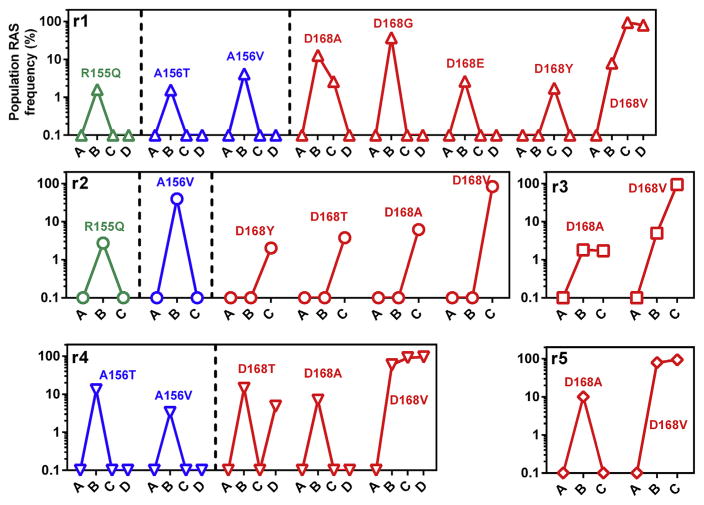
3.7. Different profiles and trajectories of RAS emergence
Patterns of RAS emergence differed between individual animals during monotherapy (Fig. 6). In all animals, no RAS were detectable prior to therapy initiation. At viral nadir, complex RAS profiles were detectable in all animals, with transient RAS at residues R155, A156 (3/5 animals) and multiple simultaneous RAS at residue D168 (5/5 animals). RAS complexity decreased in all animals after virological breakthrough with RAS at R155 and A156 becoming undetectable. D168V RAS eventually became the dominant circulating variant in all animals. Together these data reveal that despite inoculation from a common source and identical therapeutic regimens, RAS emergence takes unique trajectories in different hosts prior to and during therapeutic failure.
Fig. 6. Profiles of RAS emergence during PI monotherapy.
Each panel represents RAS emergence in animals r1 to r5 over the course of monotherapy. Within each panel, individual RAS are plotted separately to enable visualization of distinct emergence trajectories. Green lines represent RAS at R155, blue lines represent RAS at A156 and red lines representing RAS at D168, with individual RAS at these positions labeled.
4. Discussion
Previous studies which have tracked HCV transmission events from donor to recipient often focus on the envelope glycoproteins E1 and E2, which are located on the virion surface and represent the primary determinants of HCV entry (Tarr et al., 2015). These data indicate that, in specific instances, a genetic bottleneck is observed upon transmission due to initial infection with a limited number of founder viruses (D’Arienzo et al., 2013; Li et al., 2016, 2012; Wang et al., 2010). However, current antiviral therapies target the non-structural proteins and the effect of transmission on these important coding-regions was previously poorly defined. To address this shortfall, we utilized NGS to track the effect of HCV transmission on NS3 protease populations in vivo. In some scenarios of HCV transmission, such as via needle stick injury or intravenous drug use, a limited number of founder strains initiate productive infection (D’Arienzo et al., 2013; Li et al., 2016, 2012; Wang et al., 2010). These scenarios are not accurately modeled in our study as a relatively high multiplicity of infection was used to inoculate recipient animals. However, in instances of vertical transmission from mother-to-child (Honegger et al., 2013) or graft re-infection post orthotopic liver transplantation (Fafi-Kremer et al., 2010), a high multiplicity of strains can potentially initiate infection. Our study is likely to recapitulate the viral population complexity transmitted in these scenarios. Furthermore, our analyses confirm that diverse viral populations, similar to those observed in natural infection, can be transmitted in animal challenge studies for studying preclinical effectiveness of vaccine candidates and novel therapeutic regimens. However, in addition to viral load, the diversity of transmitted populations differs between individual recipients and this should be monitored and taken into consideration when interpreting results.
E1E2 evolution at transmission has been explored in multiple previous studies. Vertical transmission selects for E1E2 variants with increased replication fitness due of CTL escape mutations (Honegger et al., 2013). Experimental infection of chimeric liver mice selects for E1E2 variants with increased capacity for cellular entry (Brown et al., 2012) while in orthotopic liver transplantation, preferential transmission of neutralization resistant E1E2s with enhanced entry capacity occurs (Fafi-Kremer et al., 2010; Felmlee et al., 2016; Fofana et al., 2012). In our study focusing on the HCV protease, no evidence for T-cell reversion was observed upon transmission. However, while no selection of advantageous amino acids was observed, multiple silent transitional substitutions, which were present at >1% in the initial patient serum or donor inoculum, became dominant in the population post-transmission. Whether these fixations confer a replication or translation advantage into viral populations in a new host liver remains untested and requires further investigation. However as an identical pattern was detected in independent transmissions, this observation is unlikely due to random genetic drift. Despite high-titer inoculations, sites exhibiting variation >1% in the population were reduced in all recipients, with differential levels of viral diversity transmitted: most detectable variation was due to synonymous transitions. NS3 was surprisingly stable upon transmission, with an identical amino acid consensus in the donor and all recipients. These data indicate a combination of strong purifying selection and a genetic bottleneck shape viral protease evolution at transmission. The majority of nucleotide positions exhibit only low-frequency SNVs in the patient sera, DM and recipient animal, indicating that selective constraints limit the frequencies of SNVs that can reach high frequencies in the viral population.
While single genome amplification (SGA) has advantages over clonal sequencing to assess viral population diversity (Salazar-Gonzalez et al., 2008), sampling depth remains highly restricted and represents only a fraction of the total population. NGS allows viral populations to be scanned at unprecedented depth, orders of magnitude greater than previous approaches. Our analyses reveal the presence of a low frequency mutational cloud in all populations analyzed. The extent and diversity of this cloud, or the extreme RAS heterogeneity observed prior to virological breakthrough would not be accurately represented using clonal/SGA approaches. The virally encoded RdRp is responsible for genome replication via a negative stranded intermediate and has low fidelity. The resultant mutational spectrum generated facilitates the rapid emergence of escape mutants in response to selective pressures. Due to the limitations of current technologies, a proportion of the low frequency SNVs detected are likely to represent assay noise (Whitfield and Andino, 2016). However, the low frequency SNV cloud observed also contains a combination of defective or fitness-impaired viral genomes and represents the raw variation on which natural selection can act in viral populations, evidenced by the rapid emergence of RAS upon therapy initiation. In conclusion, these analyses provide novel insights into the dynamics of HCV protease populations at transmission and under therapeutic intervention, quantifying the evolutionary processes shaping NS3 sequence divergence and governing RAS emergence in vivo.
Acknowledgments
TFB acknowledges grant support from the European Union (ERC-AdG-2014-671231-HEPCIR, FP7 HepaMab, H2020-2015-667273-HEP-CAR), the NIH (NIAID U19AI123862) and LabEx HEP-SYS (ANR-10-LABX-0028_ HEPSYS). PM was supported by Ghent University (Concerted Action Grant 01G01712), the Agency for Innovation by Science and Technology (IWT SBO project HLIM-3D), the Belgian Science Policy Office (BELSPO; IUAP P7/47-HEPRO-2) and the European Union (FP7, HepaMab). TP was supported the German Centre for Infection Research (DZIF). Ciluprevir BILN 2061 (purity 99%) was kindly provided by Gilead (Foster City, CA).
References
- Brown RJP, Hudson N, Wilson G, Rehman SU, Jabbari S, Hu K, Tarr AW, Borrow P, Joyce M, Lewis J, Zhu LF, Law M, Kneteman N, Tyrrell DL, McKeating JA, Ball JK. Hepatitis C virus envelope glycoprotein fitness defines virus population composition following transmission to a new host. J Virol. 2012;86:11956–11966. doi: 10.1128/JVI.01079-12. http://dx.doi.org/10.1128/JVI.01079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RJP, Koutsoudakis G, Urbanowicz RA, Mirza D, Ginkel C, Riebesehl N, Calland N, Albecka A, Price L, Hudson N, Descamps V, Backx M, McClure CP, Duverlie G, Pecheur EI, Dubuisson J, Perez-del-Pulgar S, Forns X, Steinmann E, Tarr AW, Pietschmann T, Ball JK. Analysis of serine codon conservation reveals diverse phenotypic constraints on hepatitis C virus glycoprotein evolution. J Virol. 2014;88:667–678. doi: 10.1128/JVI.01745-13. http://dx.doi.org/10.1128/JVI.01745-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142:1279–1287. doi: 10.1053/j.gastro.2012.02.016. http://dx.doi.org/10.1053/j.gastro.2012.02.016e3. [DOI] [PubMed] [Google Scholar]
- Buti M, Gordon SC, Zuckerman E, Lawitz E, Calleja JL, Hofer H, Gilbert C, Palcza J, Howe AYM, DiNubile MJ, Robertson MN, Wahl J, Barr E, Forns X. Grazoprevir, Elbasvir, and Ribavirin for chronic hepatitis C virus genotype 1 infection after failure of pegylated interferon and Ribavirin with an Earlier-generation protease inhibitor: final 24-week results from C-SALVAGE. Clin Infect Dis. 2016;62:32–36. doi: 10.1093/cid/civ722. http://dx.doi.org/10.1093/cid/civ722. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. http://dx.doi.org/10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiversiTools: Beyond the consensus HTS (High-throughput sequencing) tools. http://josephhughes.github.io/btctools/
- D’Arienzo V, Moreau A, D’Alteroche L, Gissot V, Blanchard E, Gaudy-Graffin C, Roch E, Dubois F, Giraudeau B, Plantier JC, Goudeau A, Roingeard P, Brand D. Sequence and functional analysis of the envelope glycoproteins of hepatitis C virus variants selectively transmitted to a new host. J Virol. 2013;87:13609–13618. doi: 10.1128/JVI.02119-13. http://dx.doi.org/10.1128/JVI.02119-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fafi-Kremer S, Fofana I, Soulier E, Carolla P, Meuleman P, Leroux-Roels G, Patel AH, Cosset FL, Pessaux P, Doffoel M, Wolf P, Stoll-Keller F, Baumert TF. Viral entry and escape from antibody-mediated neutralization influence hepatitis C virus reinfection in liver transplantation. J Exp Med. 2010;207:2019–2031. doi: 10.1084/jem.20090766. http://dx.doi.org/10.1084/jem.20090766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felmlee DJ, Coilly A, Chung RT, Samuel D, Baumert TF. New perspectives for preventing hepatitis C virus liver graft infection. Lancet Infect Dis. 2016;16:735–745. doi: 10.1016/S1473-3099(16)00120-1. http://dx.doi.org/10.1016/S1473-3099(16)00120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fofana I, Fafi-Kremer S, Carolla P, Fauvelle C, Zahid MN, Turek M, Heydmann L, Cury K, Hayer J, Combet C, Cosset FL, Pietschmann T, Hiet MS, Bartenschlager R, Habersetzer F, Doffoel M, Keck ZY, Foung SKH, Zeisel MB, Stoll-Keller F, Baumert TF. Mutations that alter use of hepatitis C virus cell entry factors mediate escape from neutralizing antibodies. Gastroenterology. 2012;143:223–233. doi: 10.1053/j.gastro.2012.04.006. http://dx.doi.org/10.1053/j.gastro.2012.04.006e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger JR, Kim S, Price AA, Kohout JA, McKnight KL, Prasad MR, Lemon SM, Grakoui A, Walker CM. Loss of immune escape mutations during persistent HCV infection in pregnancy enhances replication of vertically transmitted viruses. Nat Med. 2013;19:1529–1533. doi: 10.1038/nm.3351. http://dx.doi.org/10.1038/nm.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving WL, Brown RJP. Acute hepatitis C virus infection: a dynamic-and challenging-concept. J Infect Dis. 2010;202:1765–1767. doi: 10.1086/657318. http://dx.doi.org/10.1086/657318. [DOI] [PubMed] [Google Scholar]
- Irving WL, Rupp D, McClure CP, Than LM, Titman A, Ball JK, Steinmann E, Bartenschlager R, Pietschmann T, Brown RJP. Development of a high-throughput pyrosequencing assay for monitoring temporal evolution and resistance associated variant emergence in the Hepatitis C virus protease coding-region. Antivir Res. 2014;110:52–59. doi: 10.1016/j.antiviral.2014.07.009. http://dx.doi.org/10.1016/j.antiviral.2014.07.009. [DOI] [PubMed] [Google Scholar]
- Lamarre D, Anderson PC, Bailey M, Beaulieu P, Bolger G, Bonneau P, Bos M, Cameron DR, Cartier M, Cordingley MG, Faucher AM, Goudreau N, Kawai SH, Kukolj G, Lagacé L, LaPlante SR, Narjes H, Poupart MA, Rancourt J, Sentjens RE, St George R, Simoneau B, Steinmann G, Thibeault D, Tsantrizos YS, Weldon SM, Yong CL, Llinás-Brunet M. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature. 2003;426:186–189. doi: 10.1038/nature02099. http://dx.doi.org/10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- Lange CM, Jacobson IM, Rice CM, Zeuzem S. Emerging therapies for the treatment of hepatitis C. EMBO Mol Med. 2014;6:4–15. doi: 10.1002/emmm.201303131. http://dx.doi.org/10.1002/emmm.201303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stoddard MB, Wang S, Blair LM, Giorgi EE, Parrish EH, Learn GH, Hraber P, Goepfert PA, Saag MS, Denny TN, Haynes BF, Hahn BH, Ribeiro RM, Perelson AS, Korber BT, Bhattacharya T, Shaw GM. Elucidation of hepatitis C virus transmission and early diversification by single genome sequencing. PLoS Pathog. 2012;8:31–33. doi: 10.1371/journal.ppat.1002880. http://dx.doi.org/10.1371/journal.ppat.1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Stoddard MB, Wang S, Giorgi EE, Blair LM, Learn GH, Hahn BH, Alter HJ, Busch MP, Fierer DS, Ribeiro RM, Perelson AS, Bhattacharya T, Shaw GM. Single-genome sequencing of hepatitis C virus in donor-recipient pairs distinguishes modes and models of virus transmission and early diversification. J Virol. 2016;90:152–166. doi: 10.1128/JVI.02156-15. http://dx.doi.org/10.1128/JVI.02156-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell M, Esteban JI, Quer J, Genescà J, Weiner A, Esteban R, Guardia J, Gómez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman P, Libbrecht L, De Vos R, de Hemptinne B, Gevaert K, Vandekerckhove J, Roskams T, Leroux-Roels G. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology. 2005;41:847–856. doi: 10.1002/hep.20657. http://dx.doi.org/10.1002/hep.20657. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. http://dx.doi.org/10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Pawlotsky JM. Hepatitis C virus population dynamics during infection. Curr Top Microbiol Immunol. 2006;299:261–284. doi: 10.1007/3-540-26397-7_9. [DOI] [PubMed] [Google Scholar]
- Polenakovik H. New therapies for hepatitis C virus. Pril Makedon Akad na Nauk i Umet Oddelenie za Med Nauk. 2015;36:119–132. doi: 10.1515/prilozi-2015-0060. http://dx.doi.org/10.1515/prilozi-2015-0060. [DOI] [PubMed] [Google Scholar]
- Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BTM, Sharp PM, Shaw GM, Hahn BH. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. http://dx.doi.org/10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. Genetic diversity and evolution of hepatitis C virus–15 years on. J Gen Virol. 2004;85:3173–3188. doi: 10.1099/vir.0.80401-0. http://dx.doi.org/10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- TANOTI: a rapid BLAST-guided read mapper for small, divergent genomes, (manuscript communicated) [ http://www.bioinformatics.cvr.ac.uk/tanoti.php]
- Tarr AW, Khera T, Hueging K, Sheldon J, Steinmann E, Pietschmann T, Brown RJP. Genetic diversity underlying the envelope glycoproteins of hepatitis C virus: structural and functional consequences and the implications for vaccine design. Viruses. 2015;7:3995–4046. doi: 10.3390/v7072809. http://dx.doi.org/10.3390/v7072809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren K, de Jong YP, Meuleman P. HCV animal models and liver disease. J Hepatol. 2014;61:S26–S33. doi: 10.1016/j.jhep.2014.07.013. http://dx.doi.org/10.1016/j.jhep.2014.07.013. [DOI] [PubMed] [Google Scholar]
- Vercauteren K, Brown RJP, Mesalam AA, Doerrbecker J, Bhuju S, Geffers R, Van Den Eede N, McClure CP, Troise F, Verhoye L, Baumert T, Farhoudi A, Cortese R, Ball JK, Leroux-Roels G, Pietschmann T, Nicosia A, Meuleman P. Targeting a host-cell entry factor barricades antiviral-resistant HCV variants from on-therapy breakthrough in human-liver mice. Gut. 2015a doi: 10.1136/gutjnl-2014-309045. http://dx.doi.org/10.1136/gutjnl-2014-309045. [DOI] [PubMed]
- Vercauteren K, de Jong YP, Meuleman P. Animal models for the study of HCV. Curr Opin Virol. 2015b;13:67–74. doi: 10.1016/j.coviro.2015.04.009. http://dx.doi.org/10.1016/j.coviro.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Sherrill-Mix SA, Chang KM, Quince C, Bushman FD. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J Virol. 2010;84:6218–6228. doi: 10.1128/JVI.02271-09. http://dx.doi.org/10.1128/JVI.02271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ZJ, Andino R. Characterization of viral populations by using circular sequencing. J Virol. 2016;90:8950–8953. doi: 10.1128/JVI.00804-14. http://dx.doi.org/10.1128/JVI.00804-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeuzem S, Ghalib R, Reddy KR, Pockros PJ, Ben Ari Z, Zhao Y, Brown DD, Wan S, DiNubile MJ, Nguyen BY, Robertson MN, Wahl J, Barr E, Butterton JR. Grazoprevir-elbasvir combination therapy for treatment-naive cirrhotic and noncirrhotic patients with chronic hepatitis C virus genotype 1, 4, or 6 infection. A Randomized Trial Ann Intern Med. 2015;163:1–13. doi: 10.7326/M15-0785. http://dx.doi.org/10.7326/M15-0785. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nguyen D, Hu KQ. Chronic hepatitis C virus infection: a review of current direct-acting antiviral treatment strategies. N Am J Med Sci Boston. 2016;9:47–54. [PMC free article] [PubMed] [Google Scholar]