Abstract
Purpose
We sought to develop a whole-heart magnetic resonance angiography technique with 3D respiratory motion compensation and reduced scan time.
Methods
A novel respiratory motion compensation method was implemented that acquires a 1D navigator (NAV) and a low-resolution 3D-image of the heart (3D-LOC) just prior to the angiography data. The central 10% of SSFP k-space was fully acquired using NAV-gating, and then 10% of peripheral k-space was randomly undersampled to complete the scan. Spatial registration of the 3D-LOC information was used to correct the central and peripheral k-space lines for the bulk respiratory motion in 3 dimensions, and then the remaining k-space data was estimated using compressed sensing (CS). Ten volunteers each underwent 2 angiography acquisitions with 1 mm3 resolution: 1) conventional NAV with CS, and 2) the new 3D-LOC with CS.
Results
Compared to conventional NAV, the new 3D-LOC with CS technique had a shorter scan time (4.8 ± 1.1 vs. 6.3 ± 1.7 min, p <0.001), better objective vessel sharpness for all 3 coronary arteries (p<0.05), and no difference in subjective vessel sharpness for all 3 coronary arteries.
Conclusion
Compared to conventional NAV with CS, acceleration and respiratory motion correction using 3D-LOC with CS reduces scan time and improves objective vessel sharpness.
Keywords: whole-heart angiography, respiratory motion, navigator, motion compensation, compressed sensing
INTRODUCTION
Cardiac magnetic resonance has become a key imaging modality for procedural planning and ongoing surveillance in children and adults with congenital heart disease (1,2). One important cardiac magnetic resonance technique in this population is the electrocardiogram and respiratory-gated 3-dimensional steady-state free precession (3D-SSFP) sequence to acquire high-resolution anatomic datasets of the entire thorax, allowing for a comprehensive evaluation of intracardiac, coronary, and vascular abnormalities (3,4). An important limitation of 3D-SSFP, however, is its relatively long imaging time, typically lasting 5–15 minutes for 1.2 mm3 isotropic resolution (5). During a long acquisition, the patient’s heart rate, breathing pattern, and body position may change leading to reduced image quality or an incomplete scan (6).
The long scan time of a high-resolution 3D-SSFP sequence is mainly due to the need for motion compensation and its accompanying inefficient time utilization. To minimize cardiac motion, ECG-triggering is used to confine the 3D-SSFP data acquisition to only approximately 10% of the cardiac cycle, when cardiac motion is at its minimum (7). In the typical commercially available technique, respiratory motion is counteracted by limiting data acquisition to end-expiration. A respiratory navigator beam (NAV) is used to track the position of the right hemi-diaphragm and only accept 3D-SSFP data acquired within a narrow acceptance window during end-expiration (8). This respiratory gating procedure increases scan time 2–3 fold since patients usually spend 50–70% of the time outside the acceptance window (9,10). Moreover, right hemi-diaphragm motion does not accurately reflect respiratory-induced heart motion as it is only sensitive to displacement in the superior-inferior direction and is subject to hysteresis effects (11–13).
To reduce imaging time and improve respiratory motion compensation for 3D-SSFP, we previously developed a novel respiratory motion compensation algorithm, 3D-LOC (14). This technique used the startup pulses of the SSFP sequence to correct for the respiratory motion of the heart (15,16) by acquiring a low-resolution 3D localizing image (3D-LOC) immediately before the acquisition of the 3D-SSFP segmented k-space data. The 3D-LOCs are then registered offline to estimate and correct for the 3D bulk respiratory motion of the heart. In contrast to other respiratory compensation approaches (17–22), the 3D-LOC technique avoids the limitations of the NAV, corrects for bulk motion in all 3 directions, does not require binning, and is compatible with any k-space trajectory.
We now present an enhancement to the 3D-LOC 3D-SSFP technique aimed at improved respiratory-induced heart motion compensation and, importantly, a further reduction in imaging time by employing compressed sensing (CS) reconstruction (23,24). We hypothesize that 3D-LOC CS will achieve equivalent or better image quality compared to the conventional respiratory NAV with CS acquisition, but with a significant reduction in scan time since respiratory gating is used to acquire only a fraction of k-space.
METHODS
3D-LOC CS Technique
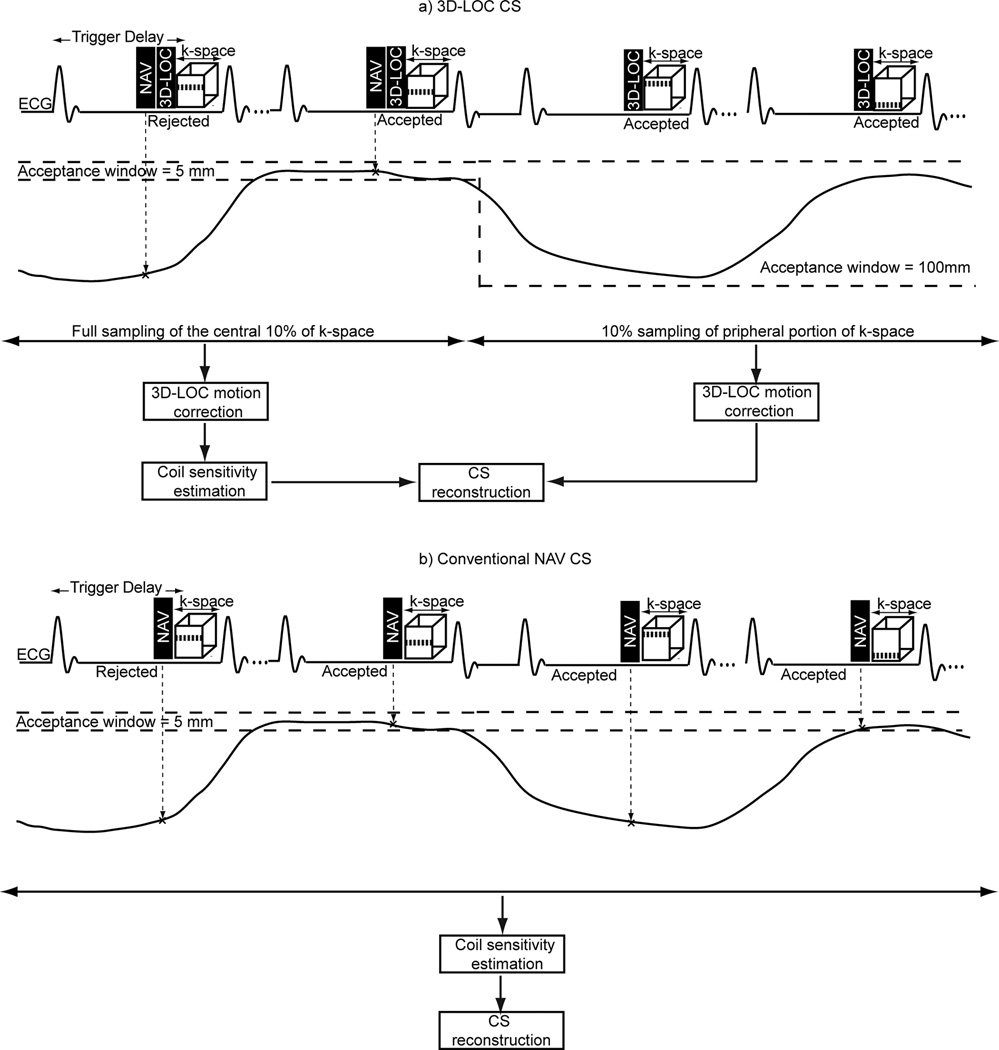
A schematic of the 3D-LOC CS technique is shown in Figure 1a. To minimize cardiac motion artifacts, the 3D-SSFP data are divided into multiple segments, each of which is acquired during the quiescent period of the cardiac cycle set by the trigger delay and shot duration. This pulse sequence is immediately preceded by a conventional 1-dimension NAV to monitor right hemi-diaphragm position and then the 3D-LOC acquisition of a single-shot low-resolution 3D volume. At the start of the scan, the central 10% of 3D-SSFP k-space is acquired using conventional respiratory NAV-gating with a 5 mm acceptance window. Then, the NAV window is opened widely to 100 mm resulting in 100% acceptance for the rest of the scan. During this time, 10% of the remaining peripheral 90% of 3D-SSFP k-space is randomly undersampled to complete the scan. The decision to acquire 10% of central k-space and 10% of peripheral k-space was based on a desire to achieve an approximately 5-fold acceleration suitable for clinical applications, and an assumption that acquiring 10% of central k-space would be necessary for acceptable image quality.
Figure 1.
Schematic diagrams of the proposed 3D-LOC CS (a) and conventional NAV CS (b). In the proposed 3D-LOC CS, data acquisition for the high-resolution 3D-SSFP image is immediately preceded by that of a NAV and 3D-LOC. Initially, the central 10% of 3D-SSFP k-space is acquired using NAV-gating, and then 10% of the peripheral 3D-SSFP k-space is randomly acquired without respiratory gating to expeditiously complete the acquisition. 3D-LOC images are used to correct the k-space lines for the bulk respiratory motion of the heart. The corrected k-space lines are then processed with a CS reconstruction algorithm to generate the 3D-LOC motion-corrected CS reconstructed image.
Spatial registration, motion compensation, and CS reconstruction are performed offline. The 3D-LOC image acquired with the first cardiac cycle is considered a reference. A rectangular region of interest is manually drawn around the heart in the reference 3D-LOC image to extract it from the surrounding region. The 3D-LOC images acquired at subsequent cardiac cycles, both with and without NAV-gating, are then registered to the segmented reference 3D-LOC image to measure the displacements of the heart in the superior-inferior, anterior-posterior, and right-left directions, as previously described (14). The estimated 3D translational displacement at each cardiac cycle is then used to retrospectively correct the 3D-SSFP k-space data acquired at that cycle (14). Finally, an iterative and nonlinear CS reconstruction algorithm (LOST) (29) along with coil sensitivity maps calculated from the central 10% of k-space are employed to estimate the unmeasured peripheral k-space data and generate the final 3D-LOC motion-corrected CS reconstructed 3D-SSFP image.
K-space Profile Order of 3D-LOC and 3D-SSFP
The 3D-LOC sequence used to acquire a low-resolution 3D volume also serves as the magnetization prepared startup pulses for 3D-SSFP sequence. Its k-space profile ordering consists of 13 startup pulses and has a reverse spiral-like pattern on Cartesian grids (Supporting Figure S1) to limit the signal oscillation of the net magnetization vector to the peripheral part of k-space and to sample the central point of k-space immediately before the 3D-SSFP sequence. The k-space profile ordering of the 3D-SSFP data is also modified to fully acquire the central 10% of k-space followed by 10% random undersampling of the remaining 90% of peripheral k-space (Supporting Figure S2). The central portion of k-space is filled in a manner that minimizes the phase jump within each shot (25). Assuming each shot consists of N k-space lines (i.e., N views), k-space is first sorted based on magnitude in the ky−kz plane (i.e., ) from low to high and divided equally into N groups. The k-space lines within each group are then sorted based on phase in the ky−kz plane (i.e., ). The first k-space lines from each group are concatenated to generate a shot to be acquired at each cardiac cycle.
Phantom Study
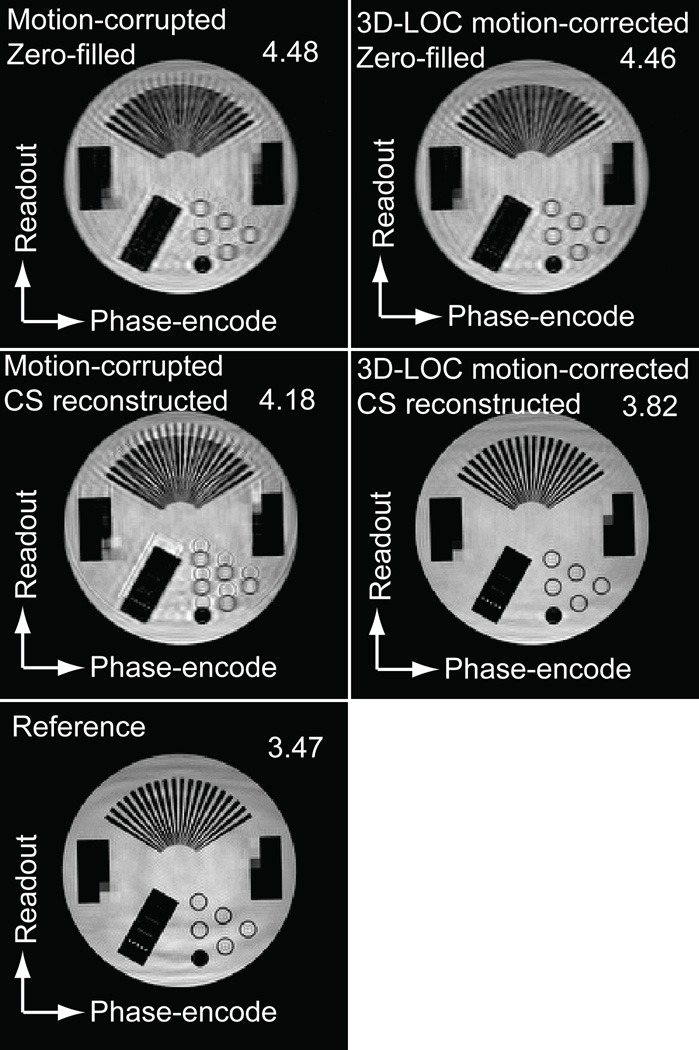
A phantom study was performed to assess the performance of 3D-LOC CS with respect to motion correction and estimation of unmeasured k-space lines. Using a Philips 1.5T Achieva scanner (Philips Medical Systems, Best, The Netherlands), a high-resolution phantom was imaged using the 3D-LOC CS sequence with the following imaging parameters: field of view 300 (SI) × 300 (RL) × 100 (AP) mm, voxel size 1.5 mm3 reconstructed to 0.65 × 0.65 × 0.75 mm3, flip angle 70°, echo time 1.9 ms, repetition time 3.8 ms, bandwidth 1.0 kHz, CS reduction factor of 3, and a 32-element receiver coil array. The scan was paused once in the middle of the acquisition to displace the phantom along the superior-inferior direction by 11 mm without moving the receiver coil arrays. For a reference image, the standard 3D-SSFP sequence with the same imaging parameters was used to acquire a fully sampled image of the static phantom.
Human Study
To investigate the performance of 3D-LOC CS, 10 healthy volunteers (7 males; age 39 ± 5 years) were examined on a Philips 1.5T Achieva scanner (Philips Medical Systems, Best, The Netherlands). The Boston Children’s Hospital Committee on Clinical Investigation approved the study, and written informed consent was obtained from each participant. In each subject, 2 whole-heart 3D-SSFP sequences were obtained. First, a conventional NAV 3D-SSFP sequence (Figure 1b) was acquired with the following imaging parameters: field of view 300 (SI) × 300 (RL) × 100 (AP) mm, voxel size 1 mm3 reconstructed to ≈0.5 mm3, flip angle 70°, echo time 1.95 ms, repetition time 3.9 ms, bandwidth 641 Hz, a respiratory NAV with a 5 mm acceptance window without tracking, CS reconstruction algorithm with a reduction factor of 5, and a 32-element receiver coil array. The second 3D-SSFP dataset was acquired using the 3D-LOC CS sequence with the same imaging parameters (Figure 1a).
For both the phantom and the human studies, 4 sets of 3D-LOC CS images were reconstructed: 1) motion-corrupted, zero-filled images: the acquired k-space lines were not corrected by 3D-LOC and the unmeasured peripheral k-space lines were filled with zeros; 2) motion-corrected, zero-filled images: the acquired k-space lines were corrected by 3D-LOC and the unmeasured peripheral k-space lines were filled with zeros; 3) motion-corrupted, CS reconstructed images: the acquired k-space lines were not corrected by 3D-LOC and unmeasured peripheral k-space lines were estimated using a CS reconstruction algorithm; and 4) motion-corrected, CS reconstructed images: the acquired k-space lines were corrected by 3D-LOC and the unmeasured peripheral k-space lines were estimated by a CS reconstruction algorithm. In the human study, the conventional NAV 3D-SSFP sequence was reconstructed in 2 ways: 1) with zero-filling, and 2) with the CS reconstruction algorithm.
To further assess the performance of 3D-LOC CS, a fully sampled, conventional NAV and 3D-LOC 3D-SSFP dataset was acquired in 3 additional healthy volunteers (3 females; age 33 ± 6 years). The imaging parameters were the same as above except a 7 mm acceptance window and 0.6 tracking factor were used to prospectively gate and track respiratory motion. From the raw k-space data, 4 image sets were retrospectively reconstructed: 1) fully sampled, conventional NAV: all of k-space was filled by data within the acceptance window; 2) fully sampled, 3D LOC motion-corrected: all of k-space was filled by consecutive data throughout the respiratory cycle and corrected by 3D-LOC; 3) Conventional NAV CS: the central 10% and a random 10% of peripheral k-space were filled by data within the acceptance window and CS reconstruction was performed; 4) 3D LOC CS: the central 10% of k-space was filled by data within the acceptance window, a random 10% of peripheral k-space was filled by consecutive data throughout the respiratory cycle and corrected by 3D-LOC, and CS reconstruction was performed.
Image Analysis
Image datasets were objectively and subjectively evaluated. For the phantom study, we calculated image sharpness using entropy as a focus criterion where lower entropy indicates better sharpness (26). For the human study, we used visualization of the coronary arteries as an indicator of overall image quality because their small size and mobility make them challenging targets to image clearly, and there are standardized metrics for analysis. Specifically, the vessel sharpness of the right coronary artery (RCA), left anterior descending coronary artery (LAD), and left circumflex coronary artery (LCX) was measured using a validated software tool (Soap-Bubble) (27). For the subjective assessment, images were independently evaluated by 2 experienced clinicians blinded to the imaging technique. Both graded each coronary artery using a 4-point scale (28): (1) poor or uninterpretable: coronary artery visible with markedly blurred borders, (2) fair: coronary artery visible with moderately blurred borders, (3) acceptable: coronary artery visible with mildly blurred borders, or (4) excellent: coronary artery visible with sharply defined borders. Scan times were recorded.
Statistical Analysis
Descriptive statistics are reported as mean ± standard deviation. A two-tailed paired Student t-test was used to compare vessel sharpness, and a non-parametric signed-rank test was used to compare the subjective scores. A p-value ≤ 0.05 was considered statistically significant. A correction for multiple comparisons was not performed because the study utilized a small number of planned statistical tests.
RESULTS
Phantom Study
The results of the phantom study are shown in Figure 2. As can be seen from the images and their entropy-based sharpness scores (a lower value is sharper), both 3D-LOC motion correction alone and CS reconstruction alone yielded significant image artifact. However, the combination of the 2 techniques accurately corrected for the motion and estimated the unmeasured k-space lines, achieving a result more similar to the reference image acquired without displacement or CS.
Figure 2.
Phantom study of 3D-LOC CS. A high-resolution phantom was imaged using the 3D-LOC CS 3D-SSFP sequence during which the scan was paused and the phantom moved 11 mm in the superior-inferior direction. Multiple reconstructions are shown along with their entropy-based sharpness score (a lower value is sharper). The reference image was acquired with no displacement and no CS.
Human Study
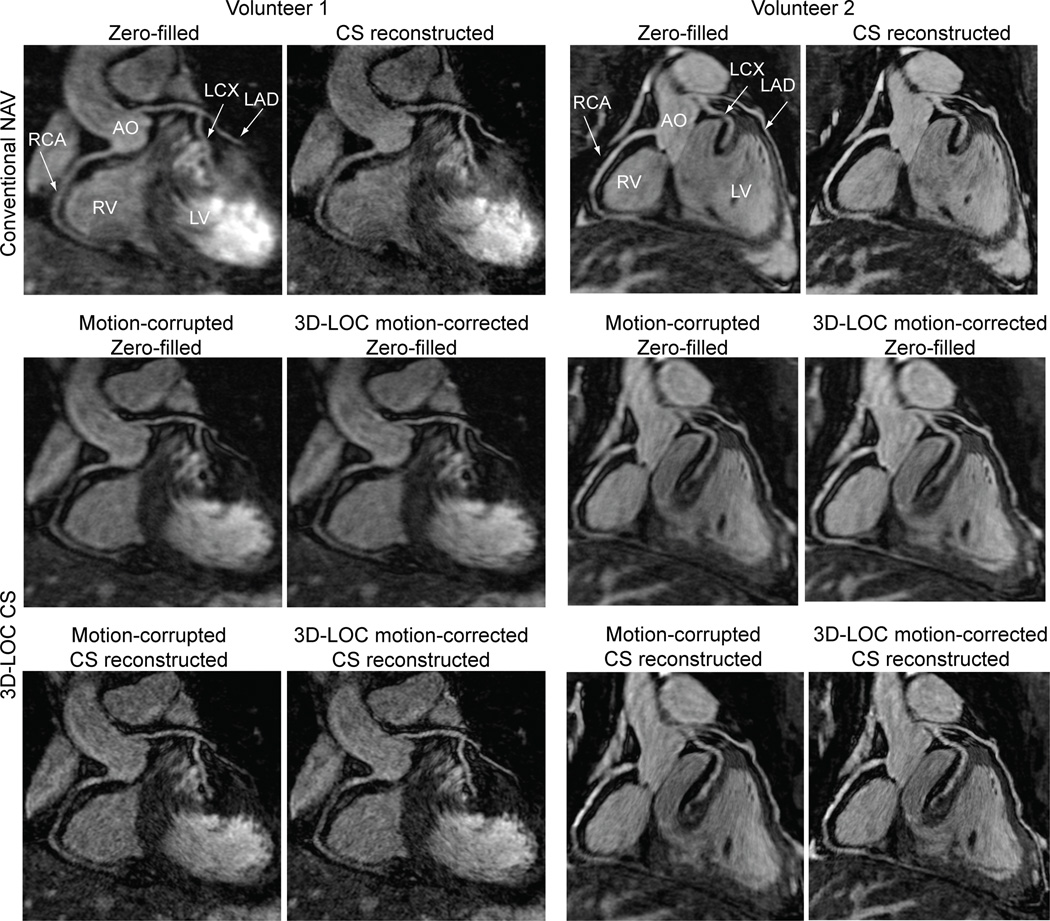
Ten healthy volunteers underwent whole-heart 3D-SSFP image acquisitions using both conventional NAV with CS and 3D-LOC CS with the same imaging parameters. All acquisitions were completed. Representative reformatted images with different reconstructions from 2 subjects are shown in Figure 3. The subjective visual scores and objective vessel sharpness measures for all subjects are shown in Table 1.
Figure 3.
Comparison of the conventional NAV with CS and 3D-LOC CS in 2 healthy volunteers. Coronal reformatted images depicting the proximal coronaries arteries with various reconstructions are shown.
Table 1.
Image Visual Score and Vessel Sharpness for 3D-SSFP Whole-heart 3D-SSFP (n=10).
| Conventional NAV with CS | 3D-LOC CS | |||||
|---|---|---|---|---|---|---|
| Zero-filled | CS reconstructed | Motion-corrupted Zero-filled |
Motion-corrected Zero-filled |
Motion-corrupted CS reconstructed |
Motion-corrected CS reconstructed |
|
| RCA visual score | 2.45±0.68 | 3.35±0.58 | 2.55±0.68 | 2.55±0.83 | 3.15±0.67 | 3.45±0.60 |
| RCA sharpness | 0.38±0.15 | 0.52±0.09 | 0.39±0.14 | 0.42±0.15 | 0.49±0.18 | 0.58±0.09 |
| LAD visual score | 2.55±0.50 | 3.25±0.72 | 2.65±0.47 | 2.65±0.67 | 3.05±0.76 | 3.35±0.75 |
| LAD sharpness | 0.43±0.09 | 0.52±0.12 | 0.43±0.06 | 0.49±0.07 | 0.53±0.09 | 0.57±0.08 |
| LCX visual score | 2.05±0.68 | 3.05±0.90 | 2.15±0.75 | 2.15±0.88 | 2.85±0.75 | 3.05±0.90 |
| LCX sharpness | 0.35±0.14 | 0.50±0.10 | 0.40±0.07 | 0.44±0.05 | 0.49±0.13 | 0.55±0.09 |
Values are mean ± standard deviation. Visual score: 1-poor to 4-excellent. Sharpness measure: 0-blurred to 1-sharp.
LAD, left anterior descending coronary artery; LCX, left circumflex coronary artery; RCA, right coronary artery.
For the conventional NAV sequence, objective vessel sharpness was significantly better with CS reconstruction than with zero-filled reconstruction for all 3 main coronary branches (RCA, p=0.038; LAD, p<0.001; LCX, p=0.013). The subjective visual scores were also significantly better with CS reconstruction for the all 3 coronary branches (RCA, p=0.016; LAD, p=0.031; LCX, p=0.008).
For the 3D-LOC CS images reconstructed with motion correction, vessel sharpness was significantly better with CS reconstruction than with zero-filled reconstruction for all 3 main coronary branches (RCA, p=0.039; LAD, p<0.001; LCX, p<0.001). Similarly, the visual scores were significantly better for all 3 coronary branches (RCA, p=0.016; LAD, p=0.031; LCX, p=0.016).
Compared to conventional NAV with CS reconstruction images, 3D-LOC with motion correction and CS reconstruction images had significantly better vessel sharpness for all 3 main coronary branches (RCA, p=0.015; LAD, p=0.012; LCX, p=0.008). Visual scores were not significantly different. The scan time for 3D-LOC CS was significantly shorter than conventional NAV with CS (4.8 ± 1.1 vs. 6.3 ± 1.7 min; p <0.001).
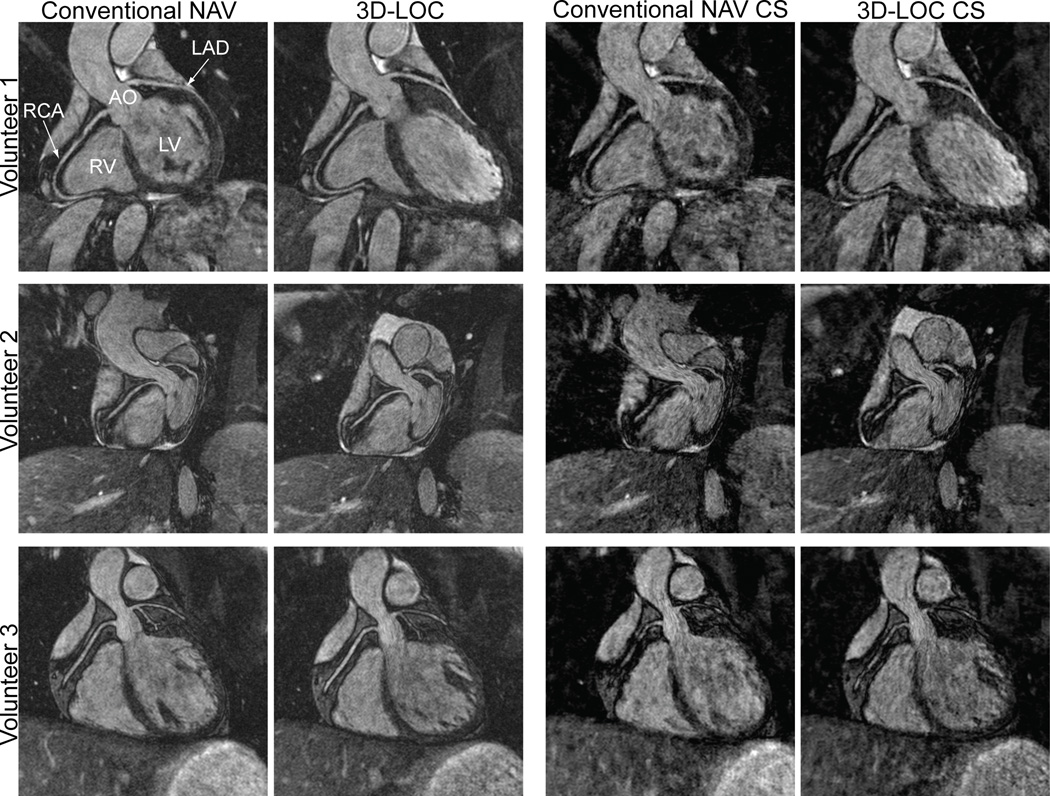
Three additional healthy volunteers underwent fully sampled, conventional NAV and 3D-LOC 3D-SSFP acquisitions. The 4 reconstructions described in detail in the methods section are shown in Figure 4. The small number of subjects precluded meaningful objective comparisons. Qualitatively, 3D-LOC respiratory motion compensation achieved similar image quality to conventional NAV. The undersampled k-space images had a lower signal-to-noise ratio compared to the fully sampled images resulting in some darkened regions throughout.
Figure 4.
Comparison of 4 reconstructions of a fully sampled 3D-SSFP dataset in 3 healthy volunteers. In the first 2 columns, k-space was fully sampled and respiratory motion compensation was performed with either a conventional NAV or 3D-LOC. In the last 2 columns, the central 10% and a random 10% of peripheral k-space were filled and the remainder of peripheral k-space was estimated using a CS reconstruction algorithm.
DISCUSSION
We have developed a novel enhancement to our previous 3D-LOC 3D-SSFP whole-heart magnetic resonance angiography technique aimed at improved respiratory-induced heart motion compensation and shorter imaging time by employing compressed sensing (CS) reconstruction. When applied to 10 healthy adult volunteers, our 3D-LOC CS angiography approach achieved encouraging results. Specifically, 3D-SSFP whole-heart imaging with 1 mm3 acquired resolution was obtained with a mean scan time of 4.8 minutes, mean subjective coronary image visual scores of 3.0–3.5, and mean coronary sharpness of 0.55–0.58.
Compared to conventional NAV with CS reconstruction images, 3D-LOC with CS reconstruction images had significantly better vessel sharpness and similar visual scores for all 3 coronary branches despite having nearly a 25% shorter mean scan time. These findings indicate that the 3D-LOC respiratory motion compensation approach is superior to NAV-gating with respect to both image quality and, as expected, scan efficiency. These benefits are similar to those found in our prior report on 3D-LOC (14) but are now extended to this new CS approach.
As with our approach, other techniques have recently been presented to acquire 3D-SSFP whole-heart angiography without respiratory gating and then retrospectively correct for respiratory-induced heart motion. Typically, these acknowledge that there is significant non-rigid heart deformation over the respiratory cycle, and therefore apply an affine transformation to compensate for this (18). Aitken et al. acquired a low resolution 2D image immediately before each 3D-SSFP shot and then used them to perform an offline, 2D affine transformation of the angiography data (29). Pang et al. (30) and Prieto et al. (31) provided a further improvement by correcting for affine motion in all 3 directions. They used either a 3D radial or Cartesian acquisition with a golden-angle spiral profile order to acquire the angiography data. Offline, low-resolution 3D images were reconstructed using an iterative sensitivity encoding algorithm for several bins (i.e., positions) within the respiratory cycle to estimate the affine transformation between each bin. This information was then used to generate the high-resolution, 3D motion-corrected data. Similar to all of these approaches, our 3D-LOC CS technique also aims to minimize non-rigid respiratory-induced heart motion; however, rather than using a complex post-processing algorithm for image registration, it addresses this issue during scanning by confining central k-space data acquisition to end-expiration, albeit at the expense of scan time. Moreover, it does not rely on a binning approach, which may suffer from an uneven division of data among the bins and inconsistent image registration (10).
Though our initial results with 3D-LOC CS are encouraging, there are several ways in which this strategy might be improved. First, the optimal percentage of the central part of k-space that should be NAV-gated requires further study. Higher percentages reduce non-rigid respiratory motion, coil sensitivity variations, and field inhomogeneity effects; however, these advantages must be balanced against the cost of a longer scan time. Second, during the NAV-gated portion of the scan, data goes unused when it is collected outside the NAV window. Improved scan efficiency could be achieved by smartly selecting whether the central or peripheral part of k-space is filled based on the NAV position. Third, 3D-LOC may reduce the impact of fat-saturation pre-pulses since it creates ≈60 ms delay between the fat-saturation pre-pulse and 3D-SSFP data acquisition; however, 3D-LOC could be either combined with the Dixon technique to appropriately suppress the fat (32) or performed before fat-suppression pre-pulses using interleaved acquisitions (33,34). Both the 3D-LOC CS and conventional NAV CS images have a reduced signal-to-noise ratio due to high spatial resolution and k-space undersampling (Figure 4). Therefore, it is worth exploring techniques to boost the signal-to-noise ratio of the magnetic resonance angiography data as this will increase image sparsity and improve the effectiveness of the CS reconstruction algorithm. One method to achieve this is to administer an intravascular contrast agent and null the myocardial signal with an inversion recovery pulse (35). Finally, our 3D-LOC CS technique was evaluated in only a small number of healthy subjects. Additional assessment is thus planned in a larger group of patients.
CONCLUSION
We developed a 3D-SSFP whole-heart magnetic resonance angiography technique with respiratory-induced heart motion correction in all 3 directions and CS acceleration. This technique yielded good image quality at 1 mm3 resolution with a scan time of approximately 5 minutes, and was superior to conventional NAV-gating. This approach will facilitate the acquisition of 3D whole-heart angiography in children and adults with congenital heart disease in a busy clinical environment. Moreover, it is amendable to a number of improvements to further enhance image quality and reduce scan time.
Supplementary Material
Acknowledgments
Dr. Moghari was supported by the Translational Research Program Fund and Office for Faculty Development at Boston Children’s Hospital. The study was supported in part by the Higgins Family Noninvasive Imaging Research Fund.
References
- 1.Kilner PJ, Geva T, Kaemmerer H, Trindade PT, Schwitter J, Webb GD. Recommendations for cardiovascular magnetic resonance in adults with congenital heart disease from the respective working groups of the European Society of Cardiology. Eur Heart J. 2010;31(7):794–805. doi: 10.1093/eurheartj/ehp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ACCF/ACR/SCCT/SCMR/ASNC/NASCI/SCAI/SIR 2006 appropriateness criteria for cardiac computed tomography and cardiac magnetic resonance imaging. A report of the American College of Cardiology Foundation Quality Strategic Directions Committee Appropriateness Criteria Working Group. Journal of the American College of Radiology : JACR. 2006;3(10):751–771. doi: 10.1016/j.jacr.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Sorensen TS, Korperich H, Greil GF, Eichhorn J, Barth P, Meyer H, Pedersen EM, Beerbaum P. Operator-independent isotropic three-dimensional magnetic resonance imaging for morphology in congenital heart disease: a validation study. Circulation. 2004;110(2):163–169. doi: 10.1161/01.CIR.0000134282.35183.AD. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen TS, Beerbaum P, Mosegaard J, Rasmusson A, Schaeffter T, Austin C, Razavi R, Greil GF. Virtual cardiotomy based on 3-D MRI for preoperative planning in congenital heart disease. Pediatr Radiol. 2008;38(12):1314–1322. doi: 10.1007/s00247-008-1032-5. [DOI] [PubMed] [Google Scholar]
- 5.Fenchel M, Greil GF, Martirosian P, Kramer U, Schick F, Claussen CD, Sieverding L, Miller S. Three-dimensional morphological magnetic resonance imaging in infants and children with congenital heart disease. Pediatr Radiol. 2006;36(12):1265–1272. doi: 10.1007/s00247-006-0314-z. [DOI] [PubMed] [Google Scholar]
- 6.McLeish K, Hill DL, Atkinson D, Blackall JM, Razavi R. A study of the motion and deformation of the heart due to respiration. IEEE T Med Imaging. 2002;21(9):1142–1150. doi: 10.1109/TMI.2002.804427. [DOI] [PubMed] [Google Scholar]
- 7.Felblinger J, Lehmann C, Boesch C. Electrocardiogram acquisition during MR examinations for patient monitoring and sequence triggering. Magn Reson Med. 1994;32(4):523–529. doi: 10.1002/mrm.1910320416. [DOI] [PubMed] [Google Scholar]
- 8.Ehman RL, Felmlee JP. Adaptive technique for high-definition MR imaging of moving structures. Radiology. 1989;173(1):255–263. doi: 10.1148/radiology.173.1.2781017. [DOI] [PubMed] [Google Scholar]
- 9.Lai P, Larson AC, Bi X, Jerecic R, Li D. A dual-projection respiratory self-gating technique for whole-heart coronary MRA. J Magn Reson Imaging. 2008;28(3):612–620. doi: 10.1002/jmri.21479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhat H, Ge L, Nielles-Vallespin S, Zuehlsdorff S, Li D. 3D radial sampling and 3D affine transform-based respiratory motion correction technique for free-breathing whole-heart coronary MRA with 100% imaging efficiency. Magn Reson Med. 2011;65(5):1269–1277. doi: 10.1002/mrm.22717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Riederer SJ, Ehman RL. Respiratory motion of the heart: kinematics and the implications for the spatial resolution in coronary imaging. Magn Reson Med. 1995;33(5):713–719. doi: 10.1002/mrm.1910330517. [DOI] [PubMed] [Google Scholar]
- 12.Spuentrup E, Manning WJ, Botnar RM, Kissinger KV, Stuber M. Impact of navigator timing on free-breathing submillimeter 3D coronary magnetic resonance angiography. Magn Reson Med. 2002;47(1):196–201. doi: 10.1002/mrm.10026. [DOI] [PubMed] [Google Scholar]
- 13.Nehrke K, Bornert P, Manke D, Bock JC. Free-breathing cardiac MR imaging: study of implications of respiratory motion--initial results. Radiology. 2001;220(3):810–815. doi: 10.1148/radiol.2203010132. [DOI] [PubMed] [Google Scholar]
- 14.Moghari MH, Roujol S, Henningsson M, Kissinger KV, Annese D, Nezafat R, Manning WJ, Geva T, Powell AJ. Three-dimensional heart locator (3D-LOC) for whole-heart coronary magnetic resonance angiography. Magn Reson Med. 2014;71(6):2118–2126. doi: 10.1002/mrm.24881. [DOI] [PubMed] [Google Scholar]
- 15.Yui M, Kassai Y, Kuhara S. Motion measurement using echoes during SSFP dummy cycles for whole-heart MR coronary artery. Proc 13th Int Soc Magn Reson Med-ISMRM. 2005:2158. [Google Scholar]
- 16.Henningsson M, Koken P, Stehning C, Razavi R, Prieto C, Botnar RM. Whole-heart coronary MR angiography with 2D self-navigated image reconstruction. Magn Reson Med. 2012;67(2):437–445. doi: 10.1002/mrm.23027. [DOI] [PubMed] [Google Scholar]
- 17.Keegan J, Gatehouse PD, Yang GZ, Firmin DN. Non-model-based correction of respiratory motion using beat-to-beat 3D spiral fat-selective imaging. J Magn Reson Imaging. 2007;26(3):624–629. doi: 10.1002/jmri.20941. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt JF, Buehrer M, Boesiger P, Kozerke S. Nonrigid retrospective respiratory motion correction in whole-heart coronary MRA. Magn Reson Med. 2011;66(6):1541–1549. doi: 10.1002/mrm.22939. [DOI] [PubMed] [Google Scholar]
- 19.Kolbitsch C, Prieto C, Buerger C, Harrison J, Razavi R, Smink J, Schaeffter T. Prospective high-resolution respiratory-resolved whole-heart MRI for image-guided cardiovascular interventions. Magn Reson Med. 2012;68(1):205–213. doi: 10.1002/mrm.23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang J, Bhat H, Sharif B, Fan Z, Thomson LE, LaBounty T, Friedman JD, Min J, Berman DS, Li D. Whole-heart coronary MRA with 100% respiratory gating efficiency: self-navigated three-dimensional retrospective image-based motion correction (TRIM) Magn Reson Med. 2014;71(1):67–74. doi: 10.1002/mrm.24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henningsson M, Prieto C, Chiribiri A, Vaillant G, Razavi R, Botnar RM. Whole-heart coronary MRA with 3D affine motion correction using 3D image-based navigation. Magn Reson Med. 2014;71(1):173–181. doi: 10.1002/mrm.24652. [DOI] [PubMed] [Google Scholar]
- 22.Ingle RR, Wu HH, Addy NO, Cheng JY, Yang PC, Hu BS, Nishimura DG. Nonrigid autofocus motion correction for coronary MR angiography with a 3D cones trajectory. Magn Reson Med. 2014;72(2):347–361. doi: 10.1002/mrm.24924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 24.Akcakaya M, Basha TA, Goddu B, Goepfert LA, Kissinger KV, Tarokh V, Manning WJ, Nezafat R. Low-dimensional-structure self-learning and thresholding: regularization beyond compressed sensing for MRI reconstruction. Magn Reson Med. 2011;66(3):756–767. doi: 10.1002/mrm.22841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akcakaya M, Basha TA, Chan RH, Rayatzadeh H, Kissinger KV, Goddu B, Goepfert LA, Manning WJ, Nezafat R. Accelerated contrast-enhanced whole-heart coronary MRI using low-dimensional-structure self-learning and thresholding. Magn Reson Med. 2012;67(5):1434–1443. doi: 10.1002/mrm.24242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atkinson D, Hill DL, Stoyle PN, Summers PE, Keevil SF. Automatic correction of motion artifacts in magnetic resonance images using an entropy focus criterion. IEEE Trans Med Imaging. 1997;16(6):903–910. doi: 10.1109/42.650886. [DOI] [PubMed] [Google Scholar]
- 27.Etienne A, Botnar RM, Van Muiswinkel AM, Boesiger P, Manning WJ, Stuber M. "Soap-Bubble" visualization and quantitative analysis of 3D coronary magnetic resonance angiograms. Magn Reson Med. 2002;48(4):658–666. doi: 10.1002/mrm.10253. [DOI] [PubMed] [Google Scholar]
- 28.Kim WY, Danias PG, Stuber M, Flamm SD, Plein S, Nagel E, Langerak SE, Weber OM, Pedersen EM, Schmidt M, Botnar RM, Manning WJ. Coronary magnetic resonance angiography for the detection of coronary stenoses. N Engl J Med. 2001;345(26):1863–1869. doi: 10.1056/NEJMoa010866. [DOI] [PubMed] [Google Scholar]
- 29.Aitken AP, Henningsson M, Botnar RM, Schaeffter T, Prieto C. 100% Efficient three-dimensional coronary MR angiography with two-dimensional beat-to-beat translational and bin-to-bin affine motion correction. Magn Reson Med. 2014 doi: 10.1002/mrm.25460. (in press) [DOI] [PubMed] [Google Scholar]
- 30.Pang J, Sharif B, Arsanjani R, Bi X, Fan Z, Yang Q, Li K, Berman DS, Li D. Accelerated whole-heart coronary MRA using motion-corrected sensitivity encoding with three-dimensional projection reconstruction. Magn Reson Med. 2014 doi: 10.1002/mrm.25097. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prieto C, Doneva M, Usman M, Henningsson M, Greil G, Schaeffter T, Botnar RM. Highly efficient respiratory motion compensated free-breathing coronary mra using golden-step Cartesian acquisition. J Magn Reson Imaging. 2014 doi: 10.1002/jmri.24602. (in press) [DOI] [PubMed] [Google Scholar]
- 32.Bornert P, Koken P, Nehrke K, Eggers H, Ostendorf P. Water/fat-resolved whole-heart Dixon coronary MRA: An initial comparison. Magn Reson Med. 2013 doi: 10.1002/mrm.24648. (in press) [DOI] [PubMed] [Google Scholar]
- 33.Powell J, Prieto C, Henningsson M, Koken P, Botnar R. CMRA with 100% navigator efficiency with 3D CMRA with 100% navigator efficiency with 3D. J Cardiovasc Magn Reson. 2014;16(Suppl 1):O8. [Google Scholar]
- 34.Henningsson M, Mens G, Koken P, Smink J, Botnar RM. A new framework for interleaved scanning in cardiovascular MR: Application to image-based respiratory motion correction in coronary MR angiography. Magn Reson Med. 2014 doi: 10.1002/mrm.25149. (in press) [DOI] [PubMed] [Google Scholar]
- 35.Makowski MR, Wiethoff AJ, Uribe S, Parish V, Botnar RM, Bell A, Kiesewetter C, Beerbaum P, Jansen CH, Razavi R, Schaeffter T, Greil GF. Congenital heart disease: cardiovascular MR imaging by using an intravascular blood pool contrast agent. Radiology. 2011;260(3):680–688. doi: 10.1148/radiol.11102327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.