Significance
All cellular life forms use a ring-shaped hexameric helicase during DNA replication. CMG (Cdc45, Mcm2–7, GINS) is the eukaryotic replicative helicase. CMG contains the ring-shaped hexameric Mcm2–7 that harbors the helicase motors. CMG is known to bind many other proteins, including a leading and lagging polymerase and primase. Thus, the threading of DNA through the CMG helicase at a replication fork determines the orientation of the associated polymerases at the replication fork, an important structural feature with many consequences that may direct future experimentation. This report uses cryo-EM single-particle reconstruction to image CMG that motored to a block site at a forked junction, enabling direct visualization of DNA threading through CMG.
Keywords: CMG helicase, DNA replication, DNA polymerase, origin initiation, replisome
Abstract
The eukaryotic CMG (Cdc45, Mcm2–7, GINS) helicase consists of the Mcm2–7 hexameric ring along with five accessory factors. The Mcm2–7 heterohexamer, like other hexameric helicases, is shaped like a ring with two tiers, an N-tier ring composed of the N-terminal domains, and a C-tier of C-terminal domains; the C-tier contains the motor. In principle, either tier could translocate ahead of the other during movement on DNA. We have used cryo-EM single-particle 3D reconstruction to solve the structure of CMG in complex with a DNA fork. The duplex stem penetrates into the central channel of the N-tier and the unwound leading single-strand DNA traverses the channel through the N-tier into the C-tier motor, 5′-3′ through CMG. Therefore, the N-tier ring is pushed ahead by the C-tier ring during CMG translocation, opposite the currently accepted polarity. The polarity of the N-tier ahead of the C-tier places the leading Pol ε below CMG and Pol α-primase at the top of CMG at the replication fork. Surprisingly, the new N-tier to C-tier polarity of translocation reveals an unforeseen quality-control mechanism at the origin. Thus, upon assembly of head-to-head CMGs that encircle double-stranded DNA at the origin, the two CMGs must pass one another to leave the origin and both must remodel onto opposite strands of single-stranded DNA to do so. We propose that head-to-head motors may generate energy that underlies initial melting at the origin.
Replicative helicases are hexameric rings in all domains of life (1–3). In bacteria and archaea, the replicative helicase is a homohexamer and encircles single-strand (ss) DNA at a replication fork. Some viral and phage replicative helicases are also ring-shaped hexamers, including bovine papilloma virus (BPV) E1, simian virus 40 (SV40) large T-antigen (T-Ag), and the T4 and T7 phage helicases. Unlike other replicative helicases, the eukaryotic replicative Mcm2–7 helicase is composed of six nonidentical but homologous Mcm subunits that become activated upon assembly with five accessory factors (Cdc45 and GINS tetramer) to form the 11-subunit CMG (Cdc45, Mcm2–7, GINS) (4–6). Numerous studies have outlined the process that forms CMG at origins in which the Mcm2–7 heterohexamer is loaded onto DNA as an inactive double hexamer in G1 phase, and becomes activated in S phase by several initiation proteins and cell-cycle kinases that assemble Cdc45 and GINS onto Mcm2–7 to form the active CMG helicases (7–9).
Helicases assort into six superfamilies (SF1–SF6) based on sequence alignments (10). The SF1 and SF2 helicases are generally monomeric and the SF3–SF6 helicases are hexameric rings used in DNA replication and other processes. The bacterial SF4 and SF5 helicases contain RecA-based motors and translocate 5′-3′, whereas the eukarytic SF3 and SF6 helicases contain AAA+ (ATPases associated with diverse cellular activities)-based motors and translocate 3′-5′ (3, 10). Examples of well-studied hexameric helicases include the viral BPV E1 and SV40 T-Ag (SF3), Escherichia coli DnaB, phage T4, and phage T7 replicative helicases (SF4), the bacterial Rho transcriptional terminator (SF5), and archaeal minichromosome maintenance (MCM) and eukaryotic Mcm2–7 (SF6). The current report examines CMG helicase from Saccharomyces cerevisiae.
Hexameric helicases are sculpted from dumbbell-shaped subunits with an N-terminal domain (NTD) and a C-terminal domain (CTD) that give the hexamer the appearance of two stacked rings that we refer to here as the N-tier ring and C-tier ring (1, 3). The CTD contains the ATP sites and this domain is the motor that drives translocation and DNA unwinding. It is generally accepted that hexameric helicases unwind DNA by the strand-exclusion model in which the helicase encircles and translocates along one strand while sterically excluding the other strand (1, 3, 11).
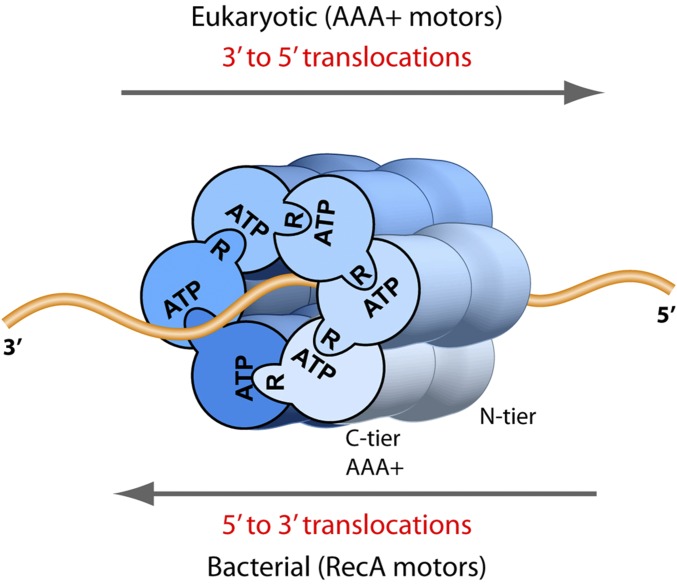
High-resolution crystal structures of helicases are essential to understand how translocation occurs, yet none have been crystallized with a replication fork, and only three have been crystalized with a ssDNA substrate. These structures include BPV E1 (12), Rho (3), and DnaB (13), representatives of SF3, SF5, and SF4 helicases, respectively. The polarity of DNA relative to the ATP sites is informative. In all hexameric helicases studied to date, the ATP sites are located at subunit interfaces and use residues from both subunits (e.g., P-loop ATP binding site in one, and arginine finger in the neighbor) (1, 3, 14). Interestingly, the ssDNA binds the C-tier motors of BPV E1, E. coli Rho, and E. coli DnaB in the same direction, such that when viewed 3′-5′ along DNA, the P-loops are clockwise to the neighboring arginine finger (Fig. 1), yet they translocate on DNA in opposite directions (3). Hence, in BPV E1 helicase the N-tier is pushed from behind by the C-tier motor, whereas in Rho and DnaB the C-tier motor is in front and pulls the N-tier ring behind during translocation (3, 12, 13). The object of this report is to determine whether the DNA orientation through the N-tier and C-tier rings of CMG is different or similar to the hexameric helicases of other superfamilies.
Fig. 1.
DNA translocation polarity in the RecA-based and AAA+-based superfamilies of bacterial and eukaryotic hexameric helicases. ATP sites are at subunit interfaces in both the bacterial RecA-based motors and eukaryotic AAA+ motors. ATP signifies the Walker A site and R signifies the arginine finger. Crystal structure analysis of BPV E1 (12), E. coli DnaB (13), and E. coli Rho (3), members of the SF3, -4, and -5 superfamilies, respectively, show that DNA binds the motors in similar fashion, illustrated here by looking down the DNA axis from the 3′ terminus, and into the motors from the C-surface view; the P loops lie clockwise to the arginine finger.
The archaeal MCM and eukaryotic Mcm2–7 of CMG are in the SF6 class and translocate 3′-5′ along the leading-strand template. High-resolution studies of MCM and CMG bound to their DNA substrates are lacking, but a negative-stain EM study of a streptavidin (SA)-tipped partial duplex bound to Drosophila melanogaster CMG (15), and FRET studies of an archaeal MCM (16), indicate that the C-tier is ahead and pulls the N-tier ring behind during translocation. This DNA orientation requires the DNA to pass through the helicase in the opposite direction compared with the SF3, SF4, and SF5 hexameric helicases. The Mcm SF6 class might bind DNA opposite to the other superfamilies, but we reexamined this herein because CMG is a central scaffold for replisome factors and the way DNA threads through CMG has important consequences for the orientation of proteins at a replication fork. In this report, we solved the structure of S. cerevisiae CMG bound to forked DNA by cryo-EM single-particle reconstruction. To our knowledge, this report is unique in visualizing a replicative helicase bound to a forked junction using a high-resolution technique. The results reveal that DNA traverses CMG 5′-3′ along the axis from the N-tier to the C-tier, and therefore DNA polarity through CMG is the same as determined for the other hexameric helicase superfamilies. Consistent with the 3′-5′ direction of translocation of CMG, the N-tier is first to reach the forked junction, followed by the C-tier. Our earlier work demonstrated that Pol ε binds the C-tier of CMG and Pol α-primase binds the N-tier (17). The new DNA orientation reverses our previous proposal of replisome architecture (17) that was based on the opposite DNA polarity through the D. melanogaster CMG and the archaeal MCM (15, 16). The N-tier ahead of the C-tier during CMG translocation has profound implications, not only for the replisome architecture but also for origin initiation. These implications are presented in Discussion.
Results
The CMG–ssDNA Complex.
To analyze whether the C-tier precedes the NTD during translocation of CMG, we initially assembled CMG onto a synthetic DNA fork containing a 25-mer double-stranded DNA (dsDNA) stem, a leading ssDNA 3′ dT40 tail, and a 16-mer 5′ (CCGA)4 ssDNA tail. The (CCGA)4 sequence was used for the lagging strand because CMG does not bind this sequence, whereas CMG binding to dT40 is readily apparent (18). CMG was mixed with a 4.2-fold molar excess of DNA fork and 0.2 mM Adenylylimidodiphosphate (AMPPNP) followed by gel filtration to remove excess DNA (SI Appendix, Fig. S1). Purified CMG–DNA–AMPPNP was analyzed by cryo-EM single-particle 3D reconstruction which yielded a 4.9-Å structure of CMG–DNA containing 14 nucleotides of ssDNA and 3 molecules of AMPPNP, referred to here as CMG–ssDNA (Fig. 2 and SI Appendix, Figs. S2–S4 and Table S1). CMG is known to act on the leading strand, but the forked junction was not visible. We presume that AMPPNP does not support CMG translocation to the forked junction. ATP would enable CMG to reach the fork, but then CMG would destroy the duplex by unwinding it. We devise a strategy later in this report that enables use of ATP without unwinding DNA and directly visualize CMG bound to a forked junction.
Fig. 2.
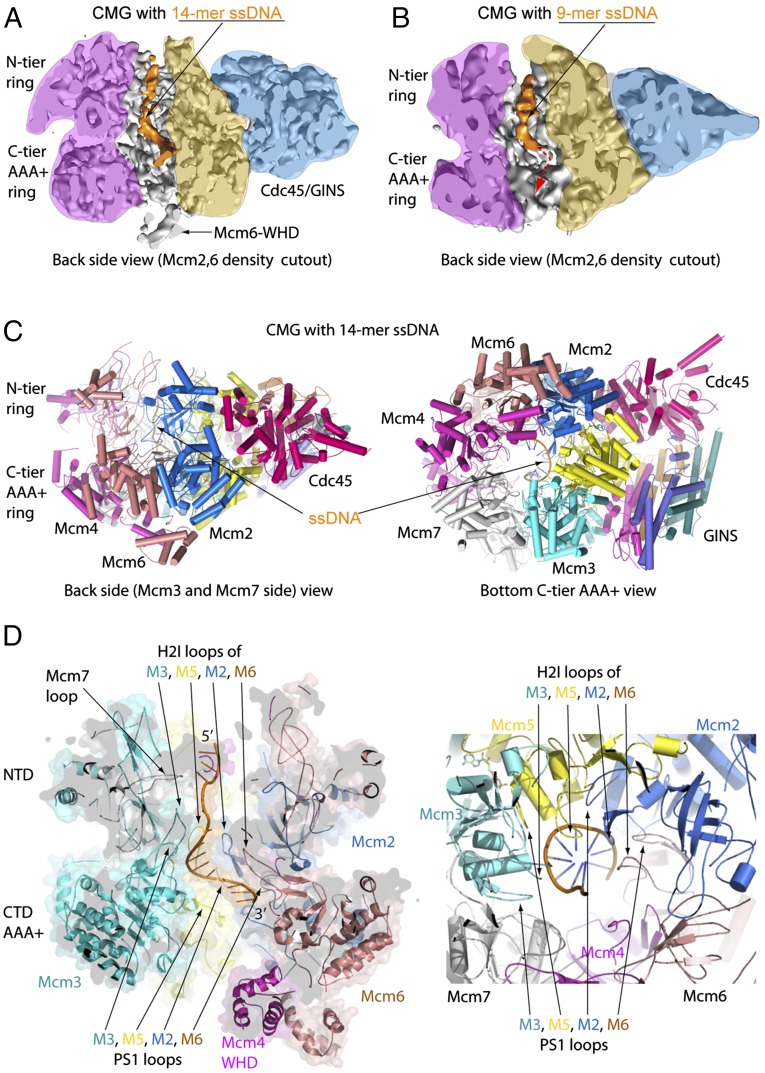
DNA densities and interactions with CMG in the CMG–ssDNA structure. CMG was mixed with a small forked DNA, then the CMG–DNA complex was purified by gel filtration (see also SI Appendix, Fig. S1). Two 3D reconstructions were obtained having different lengths of ssDNA. (A) Cut-open side view of cryo-EM map of CMG bound to 14 nucleotides at 4.9-Å resolution, and (B) with 9 nucleotides of ssDNA at 6.2-Å resolution. The dashed red arrow suggests the ssDNA threading down toward the C-tier ring. (C) Atomic model of CMG–ssDNA (14 bases) in a side (Left) and a bottom C-tier view (Right). The 14-base ssDNA is shown as stick in orange. (D) A cut-open side view (Left) and a bottom C-tier view (Right) of the CMG–ssDNA structure. The last eight bases follow a B-form helical path, surrounded by the H2I and PS1 loops of Mcm3, Mcm5, Mcm2, and Mcm6.
Six of the 14 nucleotides of ssDNA in CMG extend through the central channel of the N-tier ring in an approximately linear fashion and interact with loops of the (oligonucleotide/oligosaccharide-binding) OB folds in Mcm4 and Mcm7 (Fig. 2). The linear ssDNA passes through and binds to a narrow gap between the two upper OB loops (loop #2, between β-strands 3 and 4 of the OB domain) of Mcm7 and Mcm4 (SI Appendix, Fig. S4A). These loops are distinct from the lower OB-fold loops (loop #1, between β-strands 1 and 2 of the OB domain) that bind ssDNA perpendicular to the channel in an archaeal MCM N-tier (19). Importantly, the upper OB loops of Mcm4 and Mcm7 are well ordered even in the apo CMG structure, and they are the only two main projections into the central channel (see figure 1 C and E of ref. 20). The remaining eight nucleotides proceed into the C-tier motor ring and assume a right-handed B-form spiral, through interaction with the DNA-binding presensor-1 (PS1) and Helix-2 insertion (H2I) β-hairpin loops of Mcm subunits 3, 5, 2, and 6 (Fig. 2 and SI Appendix, Fig. S4B). A cryo-EM study of D. melanogaster CMG–DNA–ATPγS at 7–9.8 Å observed no nucletides in the N-tier but six nucleotides in the C-tier proximal to Mcm 7, 4, and 6, which are positioned on the opposite side of the AAA+ central channel (21). Superimposition of the S. cerevisiae CMG–AMPPNP–ssDNA and D. melanogaster CMG–ATPγS–ssDNA models, made by aligning the CTDs of Mcm6 and Mcm4, reveals that the ssDNA in these two models appear to belong to the same (leading) strand and are both in the B-form–like spiral (SI Appendix, Fig. S5). This observation indicates that the CTDs of all six Mcm proteins bind the leading strand during translocation, but DNA binding to particular subunits may be at different stages of the nucleotide cycle.
We also obtained a 6.2-Å structure of CMG–ssDNA containing only nine nucleotides of ssDNA, located mostly inside the N-tier ring (Fig. 2B). The 9-mer has the same conformation as the initial nine bases in the 14-mer CMG–ssDNA structure, revealing that ssDNA is held in a specific linear orientation along the axis of the central channel in the N-tier independent of DNA contacts to the C-tier. Assuming CMG threads onto the 3′ end of the leading-strand template (i.e., the known translocation strand of CMG), the 9-mer and 14-mer structures hint that the 3′ terminus first enters the N-tier ring (i.e., CMG–9-mer DNA) and then wiggles its way into the C-tier ring to produce the CMG–14-mer DNA structure. This is, in fact, the direction DNA threads into CMG, as is described later in this report.
ATP Site Occupancy.
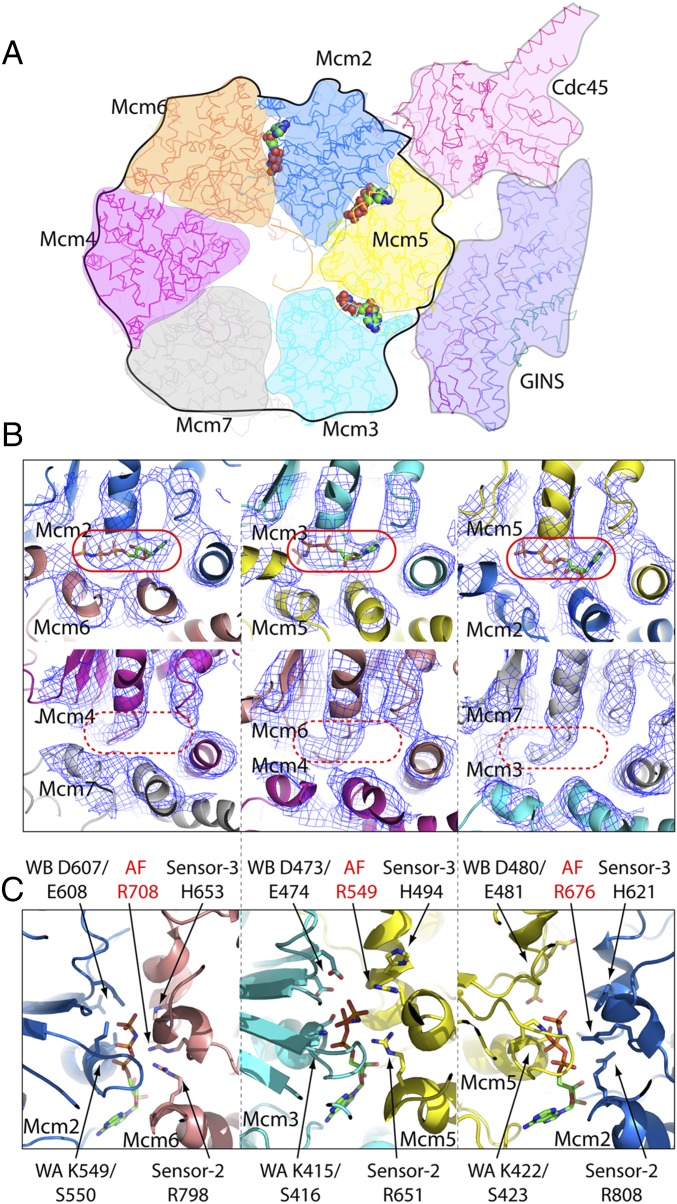
The 4.9-Å CMG–ssDNA structure provided sufficient resolution to determine ATP site occupancy (Fig. 3). Interestingly, only three of the six ATP sites in the CMG–DNA structure contain a bound nucleotide (the Mcm2/3, Mcm2/5, and Mcm5/6 sites), indicating that these consecutive ATP sites have the highest affinity for nucleotide. Interestingly, the subunits that comprise these sites are the same subunits that also bind the ssDNA. Hence, binding of a nucleotide to a subunit correlates with subunit binding to ssDNA. Two of these nucleotide-bound sites (Mcm2/3 and Mcm2/5) correspond to the two sites that are most required for helicase activity determined by mutagenesis of D. melanogaster CMG (5).
Fig. 3.
ATP site occupancy in the CMG–AMPPNP–ssDNA structure. (A) A cut-away overview of ATP sites viewed from the bottom C-tier side. The three modeled AMPPNP molecules are shown in spheres. (B) Electron density maps at the six nucleotide-binding subunit interfaces. The Upper row shows the three occupied sites (solid red ovals), and the Lower row shows the three unoccupied sites (dashed red ovals). (C) The three nucleotides and their interactions with the Mcm proteins. The subunit with the Walker A site is to the left, and the subunit with the Arg finger to the right. Walker A, Walker B motifs, sensor-2, sensor-3, and the arginine finger residues are labeled.
Our earlier cryo-EM study of apo CMG revealed two conformers, a compact conformer in which the C-tier ring is parallel to the N-tier ring, and an extended conformer in which the C-tier ring is tilted relative to the N-tier ring, thus extending the length of the central channel (20). It was proposed that the Mcm2/5 interface, known to open during Mcm2–7 loading onto the origin DNA, is closed and bound to nucleotide in the compact state. The CMG–ssDNA structure described herein is in the compact state (Figs. 2 and 3), consistent with nucleotide binding to the Mcm2/5 interface.
Conformation Changes of CMG Upon Binding ssDNA/AMPPNP.
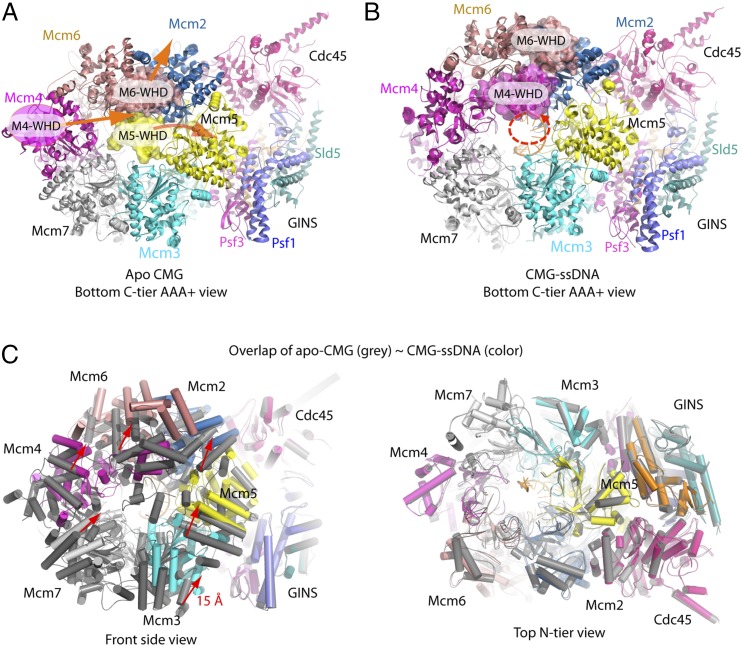
Our previous apoenzyme structure of CMG showed that the C-terminal winged helix domain (WHD) of Mcm5 occluded part of the central channel and was pinned in the channel by the WHD of Mcm6, restricting the central channel to a width that could only accommodate ssDNA (20). However, the CMG–ssDNA structure shows that the WHD domains no longer occlude the central channel (Fig. 4 A and B and SI Appendix, Fig. S6). The CMG–ssDNA structure shows a very different configuration of the WHD modules, and indicates that upon ssDNA binding, the Mcm5–WHD moves out of the central channel and becomes disordered, the Mcm6–WHD moves backward and away from the channel, and the Mcm4–WHD now partially occludes the channel opening. These changes make it likely that CMG can encircle dsDNA, consistent with reports demonstrating that Mcm2–7 matures into two fully assembled CMGs on dsDNA before origin DNA unwinding occurs (22–25).
Fig. 4.
DNA binding to the apo CMG causes a rearrangement of the WHDs that opens up the C-tier channel, and a major shift of the C-tier AAA+ ring toward the Cdc45 side, whereas the N-tier ring stays stationary. (A) The apo enzyme of CMG contained the WHD of Mcm5 inside the central channel, pinned there by the WHD of Mcm6. The Mcm4 WHD was not visualized in the apo structure (20); its location is marked by a magenta oval based on the double hexamer structure (57). (B) The CMG–ssDNA–AMPPNP structure. The orange arrows in A indicate that, upon ssDNA binding, the Mcm5–WHD moves out of the C-tier channel and becomes disordered, Mcm6–WHD moves back and away from the channel. The large void previously occupied by Mcm5–WHD and Mcm6–WHD is now partially occupied by Mcm4–WHD. The dashed red spiral in B indicates that the B-form ssDNA exits the C-tier ring unobstructed. (C) When ssDNA binds to the apo CMG helicase, the entire C-tier AAA+ ring of Mcm2–7 moves horizontally by about 15 Å toward the Cdc45 side (Left), but the N-tier ring does not move (Right).
Superpositioning the apo CMG with the CMG–ssDNA suggests that when ssDNA binds to CMG, the complete C-tier AAA+ ring of Mcm2–7 makes a horizontal movement toward the Cdc45 side of CMG by ∼15 Å, whereas the N-tier ring of Mcm2–7, and the GINS/Cdc45 do not move (Fig. 4C and Movies S1 and S2). We do not know the significance, if any, of this conformation change to CMG activity.
Comparison of ssDNA-Bound CMG to Other Hexameric Helicases.
BPV E1 contains AAA+ motors with conserved PS1 and H2I β-hairpin loops that bind ssDNA, and both Rho and DnaB also have loops in the C-tier that bind DNA (SI Appendix, Fig. S7) (3, 12, 13). In each case, the ssDNA is configured into a right-handed spiral. However, only in the CMG and the DnaB helicases is the ssDNA in the dsDNA-like B form; the central channels of Rho and E1 hexamers are too small for the ssDNA to assume the canonical B-form. A noteworthy difference between CMG and the E1 and Rho helicases is that the C-tier of CMG binds eight nucleotides using only four subunits, whereas all six subunits of E1 and Rho bind a total of six bases of ssDNA (3, 12). The six motor domains of DnaB bind 12 nucleotides (13). It is presumed that these lengths correspond to the step size, 2 bp/ATP for DnaB and 1 bp/ATP for E1 and Rho.
The DNA binding loops in E1 and Rho either point up or down relative to the axis of the central channel depending on the nucleotide bound state (3, 12). With ATP bound, the loops point up, against the direction of translocation, whereas in ADP and apo sites, the loops progressively position “downward.” The loop movements are due to a rotation of the domain to which the loop is attached and are proposed to move DNA through the central channel during ATP hydrolysis (12). The DNA binding loops of Mcm3, -5, -2, and -6 that interact with DNA in the CMG–ssDNA structure point upward relative to the central axis, and would move ssDNA in the 3′-5′ direction if they changed conformation to a “down” position. The DNA loops of Mcm subunits 4 and 7 are well ordered, and they do not bind DNA in our structure. However, they interact with the ssDNA in the cryo-EM structure of D. melanogaster CMG determined in the presence of ATPγS at 7.4-Å resolution (SI Appendix, Fig. S5) (21). Therefore, all six Mcm proteins are capable of binding the ssDNA in the C-tier motor ring, and together they bind 12 bases (i.e., one subunit binds two bases). This one-protein–two-nucleotide stoichiometry is the same as in the DnaB hexamer (13). However, a conformational change is likely required in order for Mcm4 and Mcm7 in the S. cerevisiae CMG to interact with DNA, perhaps in a different nucleotide state. We should note that our earlier cryo-EM analysis of CMG suggested that it functions as an inchworm ratchet during translocation, and this process may not require the loops to move; instead the C- and N-tiers move relative to one another during translocation (20). Clearly further studies are required to understand the translocation mechanism of CMG.
DNA Nanopointers Suggest the Polarity of ssDNA Binding to CMG.
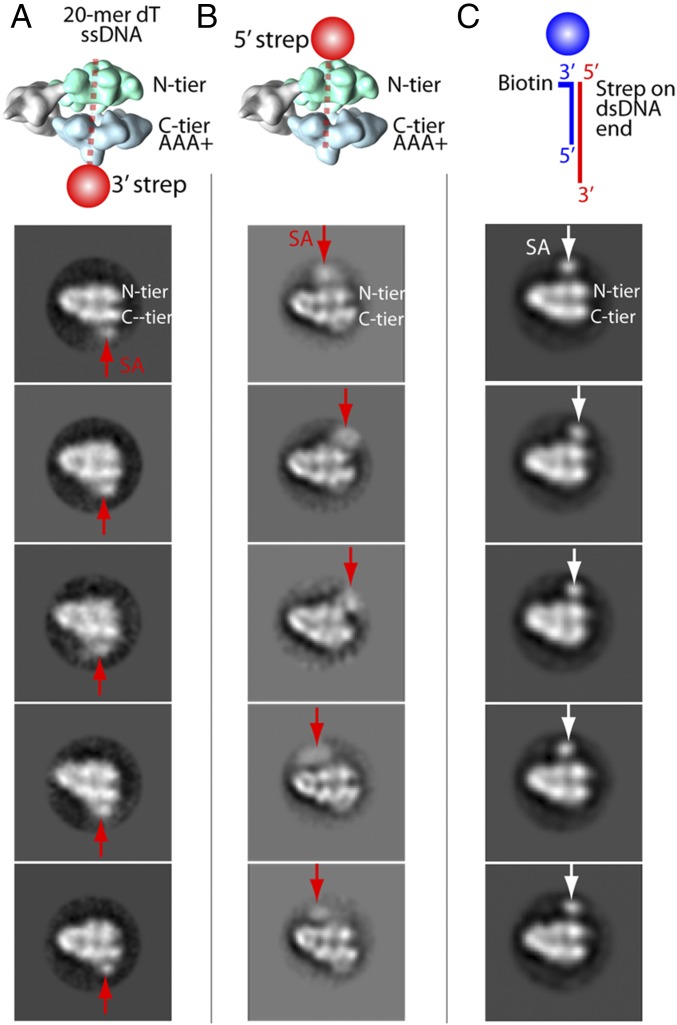
The resolution of CMG–ssDNA was insufficient to determine the polarity of the ssDNA chain within CMG, although this can be deduced from the CMG–forked DNA structure later in this report. As another means to assess the direction that DNA proceeds through CMG, we used nanopointer 20-mer dT oligonucleotides with SA bound to biotin at either the 3′ or 5′ end. This strategy was used in the study of D. melanogaster CMG, except the DNA was a partial duplex (15). In side view, the N-tier and C-tier of CMG are easily recognized. The SA bound to the 3′ biotinylated 20-mer nanopointer gave 2D averages of CMG side views having visible SA density only on the C-tier side of CMG (Fig. 5A). Because the 3′ biotin is covered by the SA, it cannot enter CMG, and therefore this result indicates that the 5′ terminus of DNA enters the C-tier ring. The SA bound to the 5′ biotinylated 20-mer nanopointer provided 2D averages of CMG side views with the SA density only on the N-tier side of CMG (Fig. 5B). The observation that the 3′ and 5′ biotinylated nanopointers bind opposite faces of CMG supports the expectation that DNA binds CMG with a preferred polarity. We also used a partial duplex having the same sequence as used in the D. melanogaster CMG study (15). This DNA has a 3′ 40-mer ssDNA of dT residues and a 20-mer duplex containing a biotin–SA conjugate. The side views showed the SA only on the N-tier side of CMG, and the extra distance of the SA from the CMG is likely due to the length of the rigid 20-mer duplex (Fig. 5C). Although this result is consistent with the 20-mer ssDNA nanopointers, we note that the nanopointer technique indicates that the DNA orientation through S. cerevisiae CMG is opposite that of D. melanogaster CMG. Therefore, we were unsure whether results using nanopointers can provide a faithful representation of DNA polarity through CMG, and we applied a different method to image CMG at a true replication fork.
Fig. 5.
Nanoscale pointers determine the direction of DNA through S. cerevisiae CMG. DNA 20-mer dT oligonucleotides biotinylated on either the 3′ (A) or 5′ (B) terminus were incubated with CMG and then with SA. (C) A tailed dsDNA from a previous work (15). Biotin was on the 3′ end of the shorter strand. SA labels the dsDNA end. The illustrations at the top of A and B denote the results observed in the selected 2D averages of CMG side views. For scale, the box size is 46.6 nm.
Cryo-EM of CMG on a DNA Forked Junction.
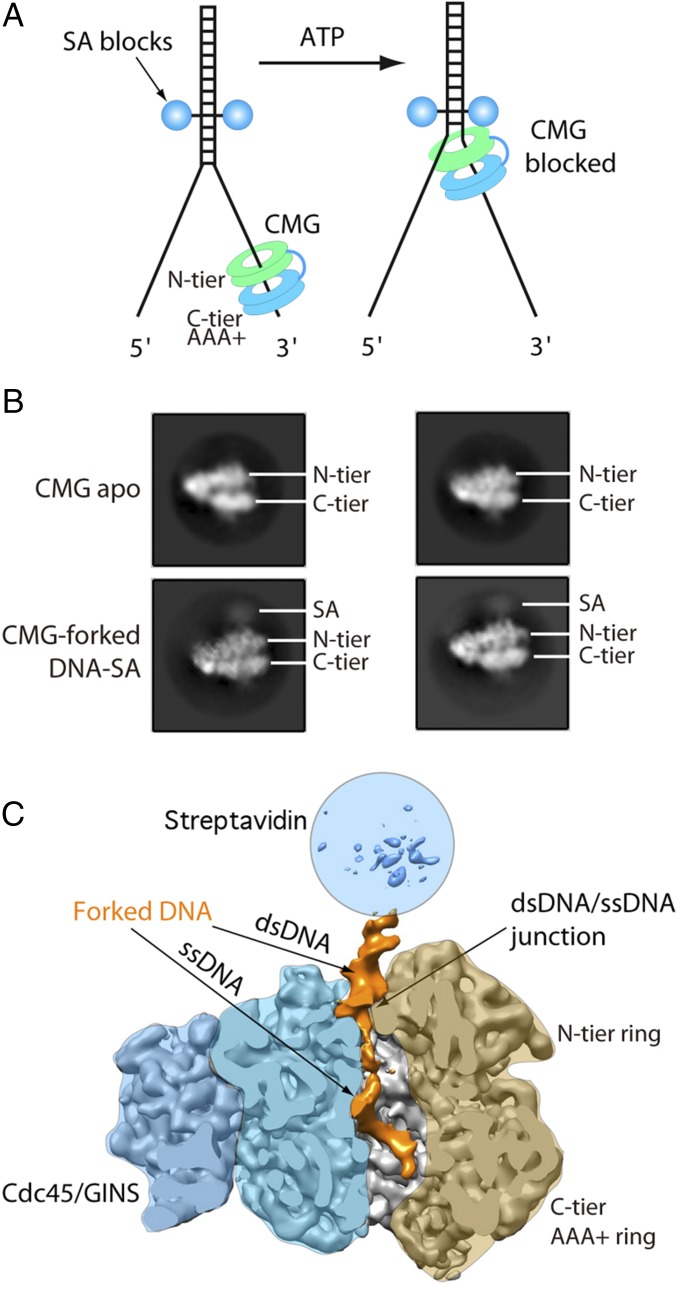
To directly visualize CMG at a forked DNA junction, we used ATP to allow CMG to translocate, and blocked CMG from unwinding the forked DNA by using two SA-biotin blocks on the duplex stem as illustrated in Fig. 6A. A dual SA block was sufficient to block forks in Xenopus extracts (11). SA was prebound to DNA, then CMG was added along with ATP for 6 min before cryo-grid preparation. The 2D class averages revealed the SA blocks as a diffuse intensity just above the N-tier of CMG (Fig. 6B). The 3D reconstruction produced a 6.2-Å structure, revealing CMG bound to the forked junction (Fig. 6C and SI Appendix, Figs. S8–S10). As expected from the experiments of Fig. 5 using 20-mer nanopointers, the N-tier ring faces the forked junction and the C-tier ring is behind the N-tier ring. dsDNA is tilted by 28° from the Mcm2–7 channel axis. Interestingly, dsDNA slightly enters the N-tier ring, followed by unwound ssDNA that traverses the central channel of the N-tier and C-tier rings (Figs. 6C and 7). The major and minor grooves of the duplex are clearly visible (Figs. 6 and 7 and SI Appendix, Fig. S8), thereby identifying the 3′ and 5′ ends, and that the ssDNA that proceeds down the central channel of CMG has a 5′-3′ polarity, consistent with the known translocation directionality of CMG. Superposition of the CMG–fork DNA structure with the CMG–ssDNA structure shows very little change in conformation (SI Appendix, Figs. S11 and S12), and the ssDNA 14-mer aligns nicely to the forked DNA (SI Appendix, Fig. S13). This polarity is also consistent with the known translocation direction of CMG. The same three ATP sites are filled as in the CMG–ssDNA structure, although the resolution cannot distinguish whether bound nucleotide is ATP, ADP, or a mixture (SI Appendix, Fig. S14). Given that the forked junction was not visualized in the CMG–ssDNA structure using AMPPNP, we presume the ATP was required for CMG to translocate along the leading-strand template to reach the forked junction. The presence of SA is reported to stop CMG about 10 bp before the biotinylated nucleotide (11), and therefore CMG may have unwound about 5 bp before being blocked. However, dsDNA near the SA blocks is not clearly visualized and we cannot be sure how many base pairs, if any, have become unwound. The lagging-strand template is not visible in the 3D reconstruction and therefore we assume it is too mobile to visualize by these methods. Because the dsDNA is tilted and enters the N-tier ring of CMG at a glancing angle, it seems likely that the unwound lagging strand will emerge from the central channel, and might pass through a gap between the Zn finger (ZF) domains of Mcm3 and Mcm5 to reach the nearby Pol α-primase (17).
Fig. 6.
Cryo-EM of CMG on a replication fork. (A) In the absence of ATP hydrolysis by using the nonhydrolysable AMPPNP, CMG is able to load itself onto the leading strand, but is unable to translocate to the fork junction (Left). This reflects the structures of CMG bound to the 14-base and 9-base ssDNA as shown in Figs. 2–4. However, in the presence of ATP hydrolysis, CMG moves to the junction and unwinds the double strand until it hits the dual SA blocks (Right). (B) Comparison of cryo-EM 2D class averages of apo-CMG (Upper) with CMG on a blocked fork (Lower). The SA density (SA) is fuzzy, but clearly located to the N-tier side, not the C-tier side, of the CMG structure. For scale, the box size is 41.6 nm. (C) A cut-open 3D cryo-EM map of the CMG–forked DNA at 6.1-Å resolution, in a front side view, with the front Mcm3 and Mcm7 largely removed to reveal the DNA density inside the central channel (orange). At this display threshold, the streptavidin density is very scattered, as marked by a light blue circle on top.
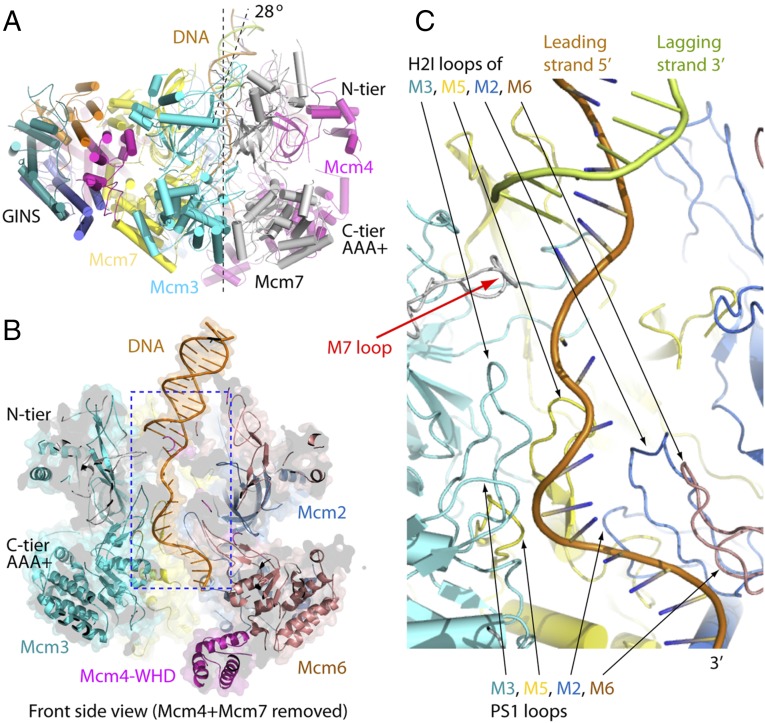
Fig. 7.
Interactions between CMG and DNA in the CMG–fork DNA structure. (A) A front side view of the atomic model of the CMG–fork DNA. Note that the dsDNA section is tilted to the right from the vertical central channel axis by about 28°. (B) A cut-open side view with most of the front Mcm4 being removed to show the forked DNA inside the central channel of the Mcm2–7. (C) An enlarged view of the region marked by a dashed blue rectangle in B. The H2I loops and PS1 loops of Mcm3 (M3, cyan), Mcm5 (M5, yellow), Mcm2 (M2, slate), and Mcm6 (M6, salmon) interact with the leading strand (in brown stick view). The lagging strand (in forest green stick view) stops shortly after the channel entrance by the protruding upper loop of the OB subdomain of Mcm7 (M7, gray).
The duplex stem of the forked junction is partially buried by the six ZF domains in the central channel of the N-tier and the upper loop of the OB-fold of Mcm7 appears to bind at the forked junction (Fig. 7C and SI Appendix, Fig. S13). It is tempting to speculate that this loop, and possibly together with the upper OB loop of Mcm4, may facilitate DNA unwinding, or may act to prevent both strands of a DNA fork from entering into the central channel. However, definitive conclusions about the exact role of the N-tier in helicase mechanism will require higher resolution to identify candidate-specific amino acid residues, followed by direct mutagenesis and biochemical characterization.
The CMG–forked DNA structure agrees with conclusions derived using 20-mer nanopointers, and both results demonstrate that DNA lies 5′-3′ along CMG when viewed from the N-tier to the C-tier. This DNA orientation is the same as observed in crystal structures of ssDNA bound to BPV E1 (SF3) (12), E. coli DnaB (SF4) (13), and E. coli Rho (SF5) (3). The result indicates that all four superfamilies of hexameric helicases bind DNA with the same polarity. Given the opposite directions of helicase with AAA+ and RecA motors, eukaryotic E1 and S. cerevisiae CMG AAA+ helicases translocate with the N-tier ring ahead and the C-tier motor behind, and bacterial RecA based helicases, E. coli Rho and DnaB, translocate 5′-3′, with the C-tier motor ahead of the N-tier ring.
Discussion
In principle, the 3′-5′ translocation of CMG could proceed with the C-tier motor ring ahead of the N-tier ring, or the other way around, with the N-tier ring ahead of the C-tier motor ring. This report determines that the C-tier ring follows the N-tier ring during translocation of S. cerevisiae CMG on DNA. The orientation of the N-tier and C-tier during translocation has important biological consequences, discussed below.
Implications of N-Tier Before C-Tier to Replisome Architecture.
We previously demonstrated that Pol ε and Pol α-primase bind opposite sides of S. cerevisiae CMG, with Pol ε on the C-tier side of CMG and Pol α-primase on the N-tier side of CMG (17). Earlier reports identified the C-tier ring of D. melanogaster CMG is ahead of the N-tier during translocation (15), consistent with FRET studies of an archaeal MCM (16). Given these earlier findings, we initially proposed Pol ε is on the leading face of S. cerevisiae CMG (i.e., because Pol ε binds the C-tier of CMG) and Pol α-primase below (17). The present study demonstrates that the DNA orientation in S. cerevisiae CMG is opposite that reported for D. melanogaster CMG. It is possible the two CMGs bind DNA in opposite orientations, or that DNA nanopointers do not faithfully represent DNA orientation for all CMGs and one must obtain structures of CMG at a DNA fork to determine the N-tier and C-tier orientation on DNA. The direction of DNA through S. cerevisiae CMG generalizes to the orientation of DNA through the hexameric helicase–ssDNA complexes of the SF3, -4, and -5 helicases solved by crystal structure analysis (i.e., Fig. 1) (3, 12, 13). The finding that the N-tier ring leads the C-tier ring during DNA translocation by S. cerevisiae CMG informs a revised architecture of the replisome at the fork (explained below).
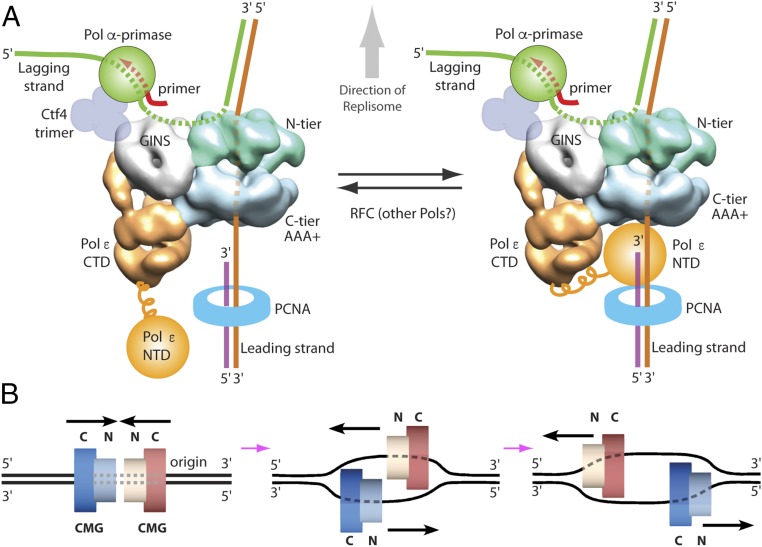
The new polarity of S. cerevisiae CMG on DNA, along with previously established structural information of S. cerevisiae CMG binding other replisome proteins, predicts a replisome architecture illustrated in Fig. 8A. The present report reveals that the 3′ end of DNA enters the N-tier of S. cerevisiae CMG, and this DNA threading places Pol ε at the “bottom” of CMG, where it can extend the leading strand immediately as the unwound DNA emerges from CMG. The new DNA threading places Pol α-primase at the top of the fork near the unwound lagging strand, a strategic position for immediate priming of the lagging-strand ssDNA template.
Fig. 8.
Implications of the N-tier ahead of C-tier direction of DNA translocation by CMG. (A) CMG in side view binds Pol α-primase and the Ctf4 trimer at the N-tier face, and the Pol ε is located on the C-tier AAA+ face. Given that the N-tier of CMG translocates ahead of the C-tier, the lagging strand is unwound from the leading strand at the N-tier, and excluded from the central channel, placing the lagging strand adjacent to Pol α-primase for immediate priming. Pol ε is positioned on the C-tier, below the helicase to immediately extend the leading-strand 3′ terminus upon exit from CMG. The Pol ε catalytic domain is in the NTD of Pol2 which has been proposed to be attached to the CTD of Pol ε by a flexible tether (see text for details) (17, 26, 27). This may allow the catalytic Pol ε NTD to transiently release the leading strand while remaining attached to CMG, exposing the leading 3′ terminus to RFC (or other proteins) for loading additional PCNA needed for Caf1-mediated nucleosome assembly and mismatch repair of the leading strand. (B) At the origin of initiation, two head-to-head CMGs are assembled around the duplex DNA (Top). Given the N-tier followed by the C-tier translocation polarity, the CMGs move toward one another, but are blocked when encircling dsDNA. Each CMG must transition to encircle ssDNA (Middle) to motor past one another and form bidirectional replication forks (Bottom).
A clear advantage of DNA threading 3′ into the N-tier of CMG, combined with the known Pol ε and Pol α-primase attachments to CMG, is that the amount of ssDNA exposed during replication is vastly minimized (i.e., discussed and compared in refs. 26 and 27). Additionally, threading 3′ DNA into the N-tier places the N-terminal Mcm2 histone binding motif in front of the fork, where it may interact with a parental H3–H4 tetramer (28, 29). This new arrangement, with the lagging machinery on top of the fork, may give an advantage to the lagging strand in inheriting parental nucleosomes and their epigenetic marks. It will be interesting to determine whether the spatial arrangement of the lagging-strand machinery relative to CMG plays a role in the inheritance of parental epigenetic marks on nucleosomes that favors the lagging-strand DNA compared with the leading strand.
Implications of N-Tier Ahead of C-Tier During CMG Translocation to Genome Integrity.
The POL2 gene of Pol ε consists of two halves: the N-half encodes the active polymerase and the C-half comprises an inactive polymerase (30). The C-half of POL2 is essential to cell viability and binds the accessory factors Dpb2, -3, and -4, whereas the active N-terminal polymerase encoded by POL2 is not essential, although cells are severely compromised in S-phase progression (31–33). Our earlier chemical cross-linking MS study showed that the N- and C-halves of Pol2 each have numerous internal cross-links, yet there is only one cross-link between the two halves of Pol2, suggesting the N- and C-halves of Pol2 are distinct folding units connected by a linker region that separates them spatially (SI Appendix, Fig. S15A) (17). Flexible domains within Pol ε were also observed in an early cryo-EM study of Pol ε (34) and is seen in 2D averages of Pol ε (SI Appendix, Fig. S15C). We noted earlier that the catalytic NTD of Pol2 may be mobile and averaged out in the CMG–Pol ε 3D reconstruction, thus explaining the noted loss of about 30% of the expected volume of Pol ε in CMG–Pol ε, as suggested previously (17, 26, 27).
The illustration in Fig. 8A suggests how a mobile catalytic domain of Pol ε may enable dynamic on/off action on the leading 3′ terminus. Specifically, the catalytic NTD of Pol2 may grip the leading 3′ terminus as it extrudes from CMG, and could periodically dissociate from the leading-strand 3′ terminus yet remain bound to the replisome through connection to the CTD of Pol ε that firmly binds CMG. This proposed dynamic action of Pol ε on/off DNA would provide access of other polymerases or replication factor C (RFC) to the leading 3′ terminus. Access of the RFC clamp loader to the leading 3′ terminus could be important to genomic integrity because it may enable loading of multiple proliferating cell nuclear antigen (PCNA) clamps, thereby explaining how the leading strand may become populated with PCNA. PCNA is required for mismatch repair (35, 36) and PCNA clamps are also required by Caf1 and Hir, nucleosome assembly factors that require PCNA for assembly of new nucleosomes on DNA (37, 38).
The CMG N-Tier/C-Tier Polarity on DNA Provides an Unrecognized Quality Control Mechanism at the Origin.
Translocation of CMG along DNA with the C-tier motors pushing the N-tier ring ahead has important implications for the mechanism of initiation at origins. The Mcm2–7 double hexamer is oriented head-to-head (N terminus to N terminus) in the PreRC during the G1 phase. In the S phase the Mcm2–7 double hexamer matures into head-to-head CMGs at the origin before ssDNA is generated (22–25). The two CMGs at the origin eventually separate because individual replication forks contain just one CMG (39, 40). If the C-tier moved ahead of the N-tier during CMG translocation, the head-to-head CMGs could simply motor away from one another in opposite directions, the current prevailing view. However, we demonstrate herein that the C-tier motor rings push the N-tier rings of CMG from behind, and therefore head-to-head CMGs push inward toward one another (Fig. 8B). Therefore, the two CMGs must pass one another to migrate out of the origin. For the two CMGs to pass they must first transition from dsDNA to encircle opposite strands of the parental DNA (Fig. 8B). Interestingly, a similar orientation of the two E. coli DnaB helicases has been noted that requires them to pass one another during initiation at the E. coli origin (41). Importantly, neither CMG can pass the other if one of the two CMGs encircles dsDNA. Thus, the head-to-head geometry, and N-tier to C-tier orientation during translocation, combine to provide a quality control mechanism that requires both CMGs to encircle ssDNA before either leaves the origin. Head-to-head hexameric helicases that encircle dsDNA have also been demonstrated to facilitate DNA melting (42), and thus the head-on orientation of two CMGs may provide energy for the initial unwinding of dsDNA at origins.
Materials and Methods
DNA Sequences.
DNA oligos were synthesized by IDT: (i) The replication fork (41/65-Tamn fork) was obtained upon annealing equimolar amounts of: a 41-mer lagging-strand oligo, (5′/5Biosg/GGCAGGCAGGCAGGCACACACTCTCCAATTCTCACTTCCTA -3′) and a 65-mer leading-strand oligo (5′/56TAMN/TAGGAAGTGAGAATTGGAGAGTGTGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT*T*T*T*T*T-3′). (ii) The SA-dual blocked fork was obtained upon annealing equimolar amounts of: a 45-mer lagging-strand oligo, (5′-GGCAGGCAGGCAGGCACACACTCTCCAATTA/iBiodT/CACTTCCTACTCTA-3′) and a 70-mer leading-strand oligo (5′/5Cy3/ TAGAGTAGGAAGTGA/iBiodT/AATTGGAGAGTGTGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT*T*T*T*T*T-3′).
Preparation of CMG–DNA–AMPPNP Samples for Cryo-EM.
Sixty-five microliters of 4.5 mg/mL CMG (380 pmols) freshly dialyzed in 300 mM NaCl, 10% (vol/vol) glycerol, 40 mM Tris-acetate pH7.5, 2 mM DTT, 2 mM Mg acetate, was mixed with 8 μL 41/65-Tamn fork (1,600 pmols); the CMG:DNA mix was further dialyzed in two steps: (i) 5% (vol/vol) glycerol/150 mM K-glutamate, then (ii) 2% (vol/vol) glycerol/50 mM K-glutamate in 25 mM Tris-acetate buffer pH 7.5, 2 mM Mg acetate, and 1 mM DTT. Both buffers contained 0.2 mM AMPPNP. After the second dialysis step, 70 μL of 3.2 mg/mL CMG:DNA was injected onto a Superose 6 Increase 3.2/300 (GE) gel-filtration column equilibrated in the second dialysis buffer without the glycerol. The complex separation was monitored at 260 nm, 280 nm, and 555 nm. Approximately 40-μL fractions were collected, analyzed on an SDS/PAGE gel [6% (wt/vol) polyacrylamide], and the CMG–DNA complex peak fractions were used for the preparation of cryo-grids.
Preparation of CMG-Blocked Fork DNA-ATP Samples for Cryo-EM.
Proteins were freshly dialyzed to remove the glycerol to less than 0.5% (verified with an Abbé refractometer). Thirteen microliters of 1.05 mg/mL CMG in 20 mM Tris-acetate pH 7.5, 40 mM K-glutamate, 40 mM KCl, 1 mM DTT, and 2 mM Mg acetate was mixed with 2 μL of 10 μM 45-Bio/70-Bio,Cy3 previously blocked with monovalent SA. The sample was incubated for 20 min at room temperature. Three microliters of the CMG:blocked fork was then incubated with 0.5 μL 1 mM ATP for 5 min at room temperature and immediately applied to grids.
Nanopointer Detection by Negative-Stain EM.
First, 2 µL of 1.65 µM of CMG was combined with 2 µL of 2.2 µM 20-mer ssDNA-biotin and incubated 5 min at room temperature. Then, 2 µL of 2.2 µM of monovalent SA was added and incubated an additional 5 min. We diluted the solution to 0.09 mg/mL and applied a 4-µL sample to a glow-discharged, carbon-coated copper grid and waited for 30 s. We blotted the excess sample with a piece of filter paper, added 2 µL of 2% (wt/vol) uranyl acetate aqueous solution, waited 30 s, blotted the excess stain, and repeated the stain application, leaving a very thin layer of stain solution to air dry. For the tailed dsDNA, we PAGE-purified the 20:60-mer hybrid using the same sequences as described previously (15); the staining procedure was the same as for the 20-mer ssDNAs. We recorded images either in a JEOL 2010F transmission electron microscope (200 kV) equipped with a Gatan 4k-by-4k CCD camera or in an FEI Tecnai Spirit (120 kV) equipped with an FEI Eagle 4k-by-4k CCD camera. Each dataset had between 200 and 300 micrographs, with at least 170,000 particles per dataset. Particle picking used either Swarm or Gaussian autopicking in EMAN2.1 (43), and exported the coordinate files to RELION 1.4 or RELION 2.0 (44). CTF calculations were performed using Gctf 1.06 (45), and the particles were binned 2× or 4× to speed up 2D classification. Finally, side views in which the SA label was clearly seen were selected for further 2D classification. Of a total of 29,423 particles in the side views of the dataset containing the 3′ biotin-labeled 20-mer dT ssDNA, 1,856 particles had clearly labeled SA (6.3%). The 20-mer dT ssDNA-5′SA dataset had 4,091 labeled particles of a total of 19,281 side view particles (21.2%). In the tailed dsDNA-containing dataset, 9,449 particles were clearly labeled by SA of a total of 53,527 side views (17.7%).
Sample Preparation and Cryo-EM.
S. cerevisiae CMG and Pol ε were purified as previously described (46, 47). To prepare EM grids, we first diluted each sample with 20 mM Tris-acetate, pH 7.5, 40 mM K-glutamate, 2 mM DTT, and 0.1 mM EDTA. Sample homogeneity was first verified by negative-stain EM. Negative stain of Pol ε was performed as described above for CMG with nanopointers. For cryo-grid preparation, we applied 3 μL of CMG–DNA complex at a final concentration of 0.6 mg/mL to glow-discharged C-flat 1.2/1.3 holey carbon grids, which were then incubated for 10 s at 10 °C and 95% humidity, blotted for 3 s, and plunged into liquid ethane using an FEI Vitrobot IV. We loaded the grids into a 300-keV FEI Titan Krios electron microscope and collected images automatically in low-dose mode at a magnification of 22,500× and a pixel size of 1.3 Å per pixel. A Gatan K2 Summit direct electron detector was used for image recording with an underfocus range from −1.2 to −3.4 μm under superresolution mode (0.5× binning). The dose rate was 10 e− per pixel-frame per second, and the total exposure time was 8 s. The total dose was divided into a 40-frame movie, and each frame was exposed for 0.2 s.
Image Processing and 3D Reconstruction.
We collected about 2,000 raw movie micrographs for CMG–DNA and 3,000 raw movie micrographs for CMG–bio–DNA–SA. The movie frames were first aligned and superimposed by the motioncorr program (48). Contrast transfer function parameters of each aligned micrograph were calculated with CTFFIND4 (49). We manually picked about 5,000 particles from different views to generate several good 2D averages that were used as templates for the subsequent automatic particle picking. Automatic particle picking was then performed for the whole dataset. Approximately 660,000 and half a million particles were initially picked for CMG–ssDNA and CMG–forked DNA, respectively. We then sorted them according to similarity to the 2D references. The bottom 10% of particles that had very low z-scores were deleted from the particle pools. The 2D classification of all of the remaining particles was performed and particles in bad classes were removed. Next, 395,443 good particles of the CMG–DNA sample and 243,796 good particles of the CMG–bio–DNA–SA sample were kept for the following 3D classification. For CMG–ssDNA, we derived six 3D models from the dataset: one model was identified having 14-mer ssDNA and their associated particles were combined for further refinement; one model was identified having 9-mer ssDNA and their associated particles were combined for further refinement; two models showed no DNA inside and closely resembled the closed and extended forms of apo CMG reported previously (20); two remaining models were distorted and those particles were discarded. A total of 83,043 and 83,002 particles were used for the final refinement of the CMG–9-mer and CMG–14-mer ssDNA, leading to the 6.2-Å map and 4.9-Å map, respectively (SI Appendix, Table S1). For the CMG–forked DNA dataset, five 3D models were derived: one model showed clear DNA and SA densities and their associated particles were combined for further refinement (58,511 particles); the other models either did not have DNA density or were distorted, and those particles were discarded, resulting in only one 3D map at 6.1 Å (SI Appendix, Table S1). All particle numbers for each of the three reported cryo-EM maps are detailed in SI Appendix, Figs. S3 and S9. Finally, all of the resolution estimations were based on gold-standard Fourier shell correlation calculations to avoid overfitting and reported resolutions were based on the Fourier shell correlation = 0.143 criterion. All of the density maps were corrected for modulation transfer function of the detector and by applying a negative B-factor sharping. All of the steps mentioned above, including particle autopicking, 2D classification, 3D classification, 3D refinement, and postprocess, were done in RELION 1.4 (44) and RELION 2.0 (50). Local resolution was estimated using ResMap (51). The negative-stain EM dataset of the full-length Pol ε contained 28,785 raw particles. These particles were sorted into 64 classes in EMAN2 (43). Six 2D class averages in views that showed a square-like feature were selected for comparison with the side views of the CMG–Pol ε.
Structural Modeling, Refinement, and Validation.
Models of all S. cerevisiae CMG subunits were directly extracted from the cryo-EM structure of the yeast CMG (20). Specifically, each Mcm protein was split into two parts, NTD and CTD, for subsequent docking. These models were rigid body-fitted into the 3D density map with COOT (52) and Chimera (53). The single-stranded or forked DNA was then manually built into the long density that ran through the Mcm2–7 hexamic structure in the program COOT (52). The entire CMG–DNA models were first refined by rigid body refinement of individual chains in the PHENIX program (54), and subsequently adjusted manually in COOT (52). The model was then refined in real space by phenix.real_space_refine (55) and in reciprocal space by PHENIX (54), with the application of secondary structure and stereochemical constraints. The structure factors (including phases) were calculated by Fourier transform of the experimental density map with the program Phenix.map_to_structure_factors (54). The final models were validated using MolProbity (56). Structural figures were prepared in Chimera (53) and Pymol (https://www.pymol.org).
Supplementary Material
Acknowledgments
We thank Dr. Nina Yao (The Rockefeller University) for illustrating Figs. 1, 6A, and 8 and Mark Ebrahim at the Rockefeller Evelyn Gruss Liper cryo-EM Recourse Center for technical assistance. This work was funded by NIH Grants GM111472 (to H.L.) and GM115809 (to M.E.O.); the Van Andel Research Institute (H.L); and the Howard Hughes Medical Institute (M.E.O.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The cryo-EM 3D maps have been deposited in the EMDataBank, www.emdatabank.org (EMD-8518, CMG-forked DNA; EMD-8519, CMG with 14-mer ssDNA; EMD-8520, CMG with 9-mer ssDNA). Atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID 5U8S, CMG-forked DNA; 5U8T, CMG with 14-mer ssDNA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620500114/-/DCSupplemental.
References
- 1.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18(2):243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka S, Araki H. Helicase activation and establishment of replication forks at chromosomal origins of replication. Cold Spring Harb Perspect Biol. 2013;5(12):a010371. doi: 10.1101/cshperspect.a010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomsen ND, Berger JM. Running in reverse: The structural basis for translocation polarity in hexameric helicases. Cell. 2009;139(3):523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell SD, Botchan MR. The minichromosome maintenance replicative helicase. Cold Spring Harb Perspect Biol. 2013;5(11):a012807. doi: 10.1101/cshperspect.a012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37(2):247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103(27):10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell SP, Labib K. Chromosome duplication in Saccharomyces cerevisiae. Genetics. 2016;203(3):1027–1067. doi: 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deegan TD, Diffley JF. MCM: One ring to rule them all. Curr Opin Struct Biol. 2016;37:145–151. doi: 10.1016/j.sbi.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 9.O’Donnell M, Langston L, Stillman B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol. 2013;5(7):a010108. doi: 10.1101/cshperspect.a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 11.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146(6):931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442(7100):270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 13.Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151(2):267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey MJ, Indiani C, O’Donnell M. Reconstitution of the Mcm2-7p heterohexamer, subunit arrangement, and ATP site architecture. J Biol Chem. 2003;278(7):4491–4499. doi: 10.1074/jbc.M210511200. [DOI] [PubMed] [Google Scholar]
- 15.Costa A, et al. DNA binding polarity, dimerization, and ATPase ring remodeling in the CMG helicase of the eukaryotic replisome. eLife. 2014;3:e03273. doi: 10.7554/eLife.03273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothenberg E, Trakselis MA, Bell SD, Ha T. MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J Biol Chem. 2007;282(47):34229–34234. doi: 10.1074/jbc.M706300200. [DOI] [PubMed] [Google Scholar]
- 17.Sun J, et al. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22(12):976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petojevic T, et al. Cdc45 (cell division cycle protein 45) guards the gate of the eukaryote replisome helicase stabilizing leading strand engagement. Proc Natl Acad Sci USA. 2015;112(3):E249–E258. doi: 10.1073/pnas.1422003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Froelich CA, Kang S, Epling LB, Bell SP, Enemark EJ. A conserved MCM single-stranded DNA binding element is essential for replication initiation. eLife. 2014;3:e01993. doi: 10.7554/eLife.01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan Z, et al. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat Struct Mol Biol. 2016;23(3):217–224. doi: 10.1038/nsmb.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abid Ali F, et al. Cryo-EM structures of the eukaryotic replicative helicase bound to a translocation substrate. Nat Commun. 2016;7:10708. doi: 10.1038/ncomms10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heller RC, et al. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146(1):80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012;31(9):2182–2194. doi: 10.1038/emboj.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22(4):343–349. doi: 10.1016/j.cub.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 25.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature. 2015;519(7544):431–435. doi: 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J, Yuan Z, Georgescu R, Li H, O’Donnell M. The eukaryotic CMG helicase pumpjack and integration into the replisome. Nucleus. 2016;7(2):146–154. doi: 10.1080/19491034.2016.1174800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donnell M, Li H. The eukaryotic replisome goes under the microscope. Curr Biol. 2016;26(6):R247–R256. doi: 10.1016/j.cub.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H, et al. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. Nat Struct Mol Biol. 2015;22(8):618–626. doi: 10.1038/nsmb.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Wang M, Yang N, Xu RM. Structure of the quaternary complex of histone H3-H4 heterodimer with chaperone ASF1 and the replicative helicase subunit MCM2. Protein Cell. 2015;6(9):693–697. doi: 10.1007/s13238-015-0190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV. Evolution of DNA polymerases: An inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol Direct. 2009;4:11. doi: 10.1186/1745-6150-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274(32):22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 32.Kesti T, Flick K, Keränen S, Syväoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3(5):679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 33.Ohya T, et al. The DNA polymerase domain of pol(epsilon) is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J Biol Chem. 2002;277(31):28099–28108. doi: 10.1074/jbc.M111573200. [DOI] [PubMed] [Google Scholar]
- 34.Asturias FJ, et al. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat Struct Mol Biol. 2006;13(1):35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 35.Pluciennik A, et al. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc Natl Acad Sci USA. 2010;107(37):16066–16071. doi: 10.1073/pnas.1010662107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goellner EM, et al. PCNA and Msh2-Msh6 activate an Mlh1-Pms1 endonuclease pathway required for Exo1-independent mismatch repair. Mol Cell. 2014;55(2):291–304. doi: 10.1016/j.molcel.2014.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87(1):95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 38.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11(7):463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 39.Ticau S, Friedman LJ, Ivica NA, Gelles J, Bell SP. Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell. 2015;161(3):513–525. doi: 10.1016/j.cell.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yardimci H, Loveland AB, Habuchi S, van Oijen AM, Walter JC. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol Cell. 2010;40(5):834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fang L, Davey MJ, O’Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4(4):541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 42.Kaplan DL, O’Donnell M. Twin DNA pumps of a hexameric helicase provide power to simultaneously melt two duplexes. Mol Cell. 2004;15(3):453–465. doi: 10.1016/j.molcel.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 43.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180(3):519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193(1):1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langston LD, et al. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci USA. 2014;111(43):15390–15395. doi: 10.1073/pnas.1418334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Georgescu RE, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21(8):664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192(2):216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11(1):63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 54.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 4):352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li N, et al. Structure of the eukaryotic MCM complex at 3.8 Å. Nature. 2015;524(7564):186–191. doi: 10.1038/nature14685. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.