Significance
Mutational inactivation of the VHL tumor suppressor gene is the signature lesion in the most common form of kidney cancer and causes inappropriate accumulation of the HIF transcription factor, which activates genes that normally facilitate adaptation to hypoxia but, in the context of kidney cancer, also promote tumorigenesis. Additional mutational events are needed, in conjunction with VHL loss, to cause kidney cancer. The most common of these are inactivating mutations of the PBRM1 tumor suppressor gene, which encodes a component [BRG1-associated factor 180 (BAF180)] of a multiprotein complex [Polybromo- and BRG1-associated factors-containing complex (PBAF)] that regulates the positions of nucleosomes throughout the genome. We describe here kidney cancer cell-based models for monitoring BAF180 function and show that loss of BAF180 accentuates the transcriptional response to HIF.
Keywords: kidney cancer, chromatin, PBAF, BAF180, hypoxia
Abstract
Most clear cell renal carcinomas (ccRCCs) are initiated by somatic inactivation of the VHL tumor suppressor gene. The VHL gene product, pVHL, is the substrate recognition unit of an ubiquitin ligase that targets the HIF transcription factor for proteasomal degradation; inappropriate expression of HIF target genes drives renal carcinogenesis. Loss of pVHL is not sufficient, however, to cause ccRCC. Additional cooperating genetic events, including intragenic mutations and copy number alterations, are required. Common examples of the former are loss-of-function mutations of the PBRM1 and BAP1 tumor suppressor genes, which occur in a mutually exclusive manner in ccRCC and define biologically distinct subsets of ccRCC. PBRM1 encodes the Polybromo- and BRG1-associated factors-containing complex (PBAF) chromatin remodeling complex component BRG1-associated factor 180 (BAF180). Here we identified ccRCC lines whose ability to proliferate in vitro and in vivo is sensitive to wild-type BAF180, but not a tumor-associated BAF180 mutant. Biochemical and functional studies linked growth suppression by BAF180 to its ability to form a canonical PBAF complex containing BRG1 that dampens the HIF transcriptional signature.
Biallelic inactivation of the VHL tumor suppressor gene is the usual initiating or truncal event in clear cell renal cell carcinoma (ccRCC), which is the most common form of kidney cancer (1–6). VHL loss, however, is not sufficient to cause ccRCC (4, 7–9). Other cooperating genetic events in ccRCC include copy number gain of chromosome 5q, copy number loss of chromosome 14q, and intragenic mutations affecting chromatin regulatory genes such as PBRM1, BAP1, ARID1A, SETD2, KDM5C, and KDM6A; PI3K pathway genes such as PTEN, PIK3CA, TSC1, and TORC1; and redox stress genes such as KEAP1 and NFE2L2 (1, 10–14).
PBRM1 is the gene that, after VHL, is most frequently mutated in ccRCC. Interestingly, PBRM1 and VHL reside at chromosomes 3p21 and 3p25, respectively. Accordingly, three genetic hits (intragenic cis mutations affecting PBRM1 and VHL, followed by loss of chromosome 3p) can cause biallelic loss of both PBRM1 and VHL. A similar situation exists for BAP1 and SETD2, which are also located on chromosome 3p21. PBRM1 and BAP1 mutations are largely mutually exclusive in ccRCC and define biologically distinct ccRCC subtypes (12).
The PBRM1 gene product, BRG1-associated factor 180 (BAF180), is part of the multisubunit Polybromo- and BRG1-associated factors-containing complex (PBAF) switch/sucrose nonfermentable (SWI/SNF) chromatin remodeling complex (15–18). Mutations affecting SWI/SNF components have been linked to multiple forms of cancers (15–18). For reasons that are not clear, however, there is a strong bias to mutate specific SWI/SNF components in specific types of cancer. In this regard, inactivating PBRM1 mutations are most common in ccRCC, followed by cholangiocarcinoma (19, 20), but are otherwise relatively uncommon in cancer.
siRNA-mediated knockdown of wild-type PBRM1 was reported to increase the proliferation of multiple ccRCC cell lines in monolayer culture and in soft agar (10). These effects were not, however, proven to be on-target, and were not interrogated in vivo. As a step toward understanding the role of BAF180 in ccRCC, we asked whether BAF180 participates in the canonical PBAF complex in ccRCC cell lines and whether loss of BAF180 measurably alters ccRCC behavior in cell culture and in mice.
Results and Discussion
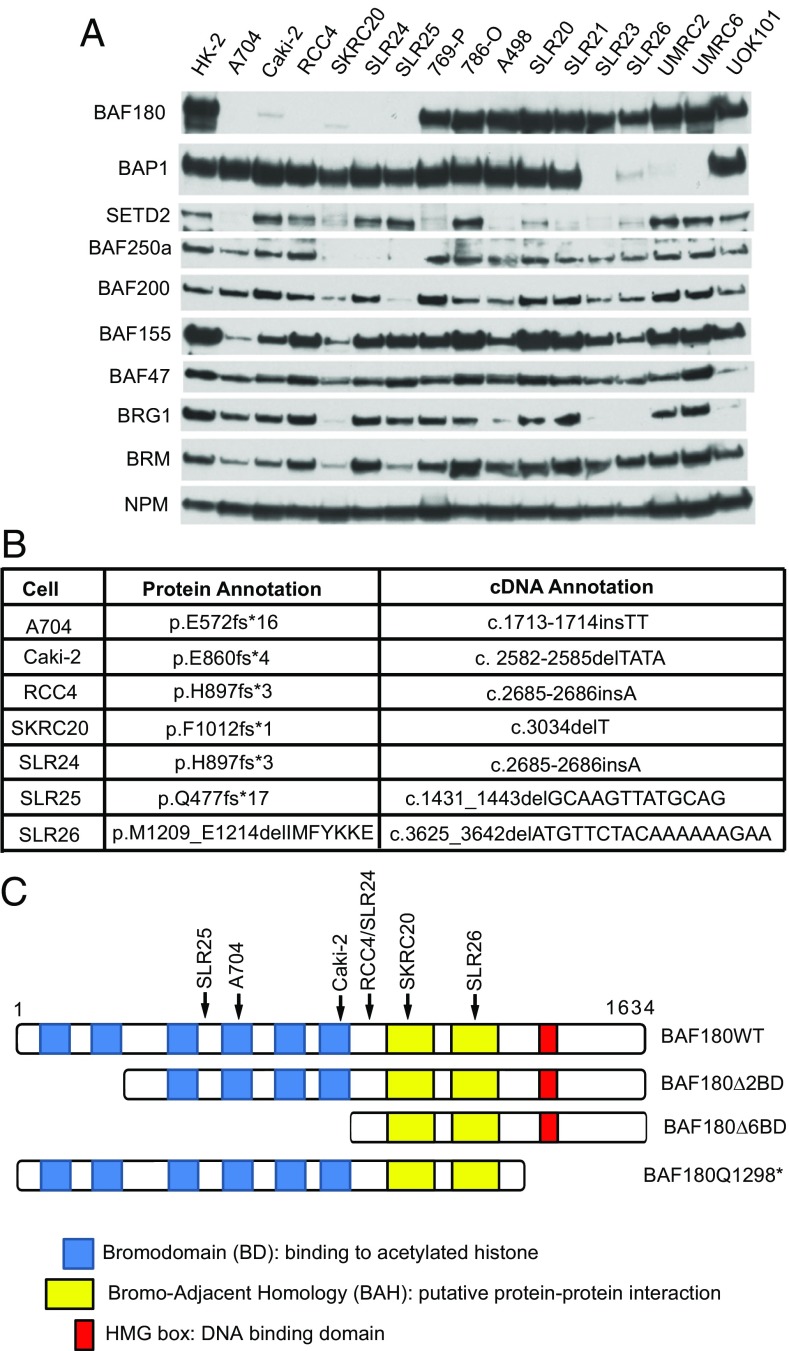
We examined the protein levels of BAF180, BAP1, SETD2, and various SWI/SNF components in 16 ccRCC cell lines, together with HK-2 immortalized renal epithelial cells, using immunoblot assays. All the 16 ccRCCs are pVHL-defective except for SLR20 and SLR21 (21). NPM was used as a loading control. BAF180 was undetectable in five cell lines (A704, RCC4, SKRC20, SLR24, and SLR25) and barely detectable in another (Caki-2; Fig. 1A and SI Appendix, Fig. S1). We also sequenced the PBRM1 cDNAs from these 16 ccRCC lines and identified frame-shift, presumably loss-of-function, mutations in the six cell lines with low or undetectable BAF180 (Fig. 1 B and C). In addition, we identified a small, in-frame, deletion in the PBRM1 cDNA in SLR26 cells (Fig. 1 B and C). The A704 and Caki-2 PBRM1 mutations have been reported previously (10, 22).
Fig. 1.
Identification of BAF180-defective clear cell renal carcinoma lines. (A) Immunoblot analyses of the indicated cell lines with rabbit polyclonal anti-BAF180 antibody. (B) Summary of PBRM1 mutations in the indicated cell lines. (C) BAF180 schematics. Arrows indicate locations of mutations listed in B.
Four lines had diminished or undetectable levels of the ccRCC suppressor protein BAP1 (SLR23, SLR26, UMRC2, and UMRC6), three had diminished or undetectable levels of the ccRCC suppressor BAF250a (SKRC20, SLR24, and SLR25), and eight had diminished or absent levels of the ccRCC suppressor SETD2 (A704, SKRC20, 769-P, A498, SLR20, SLR21, SLR23, and SRL26). In keeping with previous observations, loss of BAF180 and BAP1 appeared to be largely mutually exclusive (12). As noted earlier, however, the BAF180 present in SLR26 cells is possibly defective. In contrast, apparent loss of SETD2 was found both among cell lines lacking BAF180 and among cell lines lacking BAP1, consistent with genetic studies that have identified ccRCCs with coexisting SETD2 and PBRM1 mutations and ccRCCs with coexisting SETD2 and BAP1 mutations (2, 3, 6, 13, 20, 23).
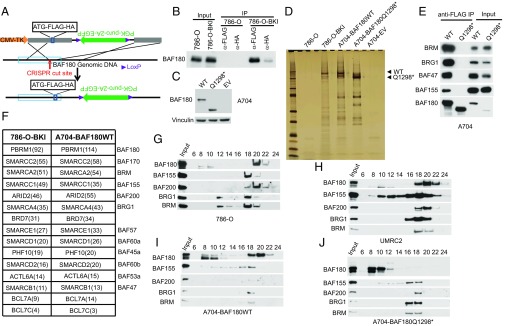
To begin to study the function of BAF180 in ccRCC, we first asked whether BAF180 forms PBAF complexes in this context and whether this biochemical property is disrupted by tumor-associated mutations. Toward this end, we used CRISPR-based gene editing to introduce a tandem FLAG-hemagglutinin (HA) epitope tag at the N terminus of the endogenous BAF180 ORF into 786-O cells (Fig. 2A). We confirmed that FLAG-HA-BAF180 could be immunoprecipitated with either an anti-FLAG antibody or an anti-HA antibody from these cells (786-O-BKI), but not from parental 786-O cells, although recovery of the tagged BAF180 was reproducibly higher with the anti-FLAG antibody than with the anti-HA antibody (Fig. 2B). In parallel, we infected A704 cells, which lack detectable endogenous BAF180 (Fig. 1A), with a lentivirus encoding FLAG-HA-tagged wild-type BAF180, a tumor-associated BAF180 mutant with a frame-shift mutation (Q1298*) (10), or the empty vector (EV) (Fig. 2C).
Fig. 2.
Detection of BAF180-associated proteins in clear cell renal carcinoma lines (A). Schematic for targeting vector used to introduce FLAG-HA epitope tags at N terminus of BAF180. (B) Immunoblot analysis of whole-cell extracts (Input) and immunoprecipitates (IP) from 786-O cells and 786-O cells after homologous recombination, using targeting vector shown in A (786-O-BKI). (C) Immunoblot analysis of A704 cells stably infected to produce FLAG-HA tagged versions of wild-type BAF180 (WT), BAF180 Q1298*, or with an EV. (D) Silver-stained gel of proteins captured from the indicated cell lines by sequential immunoprecipitation with anti-FLAG and anti-HA antibodies. After each round of immunoprecipitation, bound proteins were eluted with a 3×FLAG and HA peptide, respectively. (E) Immunoblot analysis of whole-cell extracts (Input) and anti-FLAG immunoprecipitates from A704 cells, as in C. (F) Proteins detected by mass spectroscopy after sequential immunoprecipitation, as in D. (G–J) Immunoblot analysis of cell extracts from the indicated cell lines after glycerol gradient centrifugation.
We next performed preparative anti-FLAG immunoprecipitations with these cell lines under stringent wash conditions. The bound proteins were then eluted with a FLAG-peptide, immunoprecipitated with an anti-HA antibody, eluted with an HA peptide, and either resolved by SDS/PAGE and detected by silver staining (Fig. 2D) or identified by mass spectrometry (Fig. 2F and SI Appendix, Table S1). As expected, we observed silver-stained protein bands with predicted molecular weights consistent with epitope-tagged BAF180 in immunoprecipitates prepared from the 786-O-BKI and A704-BAF180 cells and a slightly faster migrating band in the immunoprecipitates from A704-BAF180 (Q1298*) cells (Fig. 2D). No such bands were observed with the control samples prepared from parental 786-O cells and A704-EV cells.
Mass spectrometry analysis confirmed the recovery of BAF180 itself, as well as other members of the canonical PBAF complex, including BAF170, BAF155, BAF200, BAF57, BAF60a, BAF45a, BAF60b, BAF53a, and BAF47 and the SWI/SNF-associated proteins BCL7A and BCL7C. Surprisingly, we also recovered multiple peptides that are unique to the BRM DNA-dependent ATPase or unique to the BRG1 DNA-dependent ATPase, even though only BRG1 is believed to participate in PBAF complexes (15–18).
We next confirmed that exogenous wild-type BAF180, but not mutant (Q1298*) BAF180, coimmunoprecipitated with PBAF components in A704 cells (Fig. 2E). Consistent with this finding, the exogenous wild-type BAF180 in A704 cells, similar to the endogenous wild-type BAF180 in 786-O cells and UMRC2 cells, was detected in a high-molecular-weight complex containing other PBAF components after glycerol gradient centrifugation. In contrast, BAF180 (Q1298*) was not detected in this higher-order complex (Fig. 2 G–J).
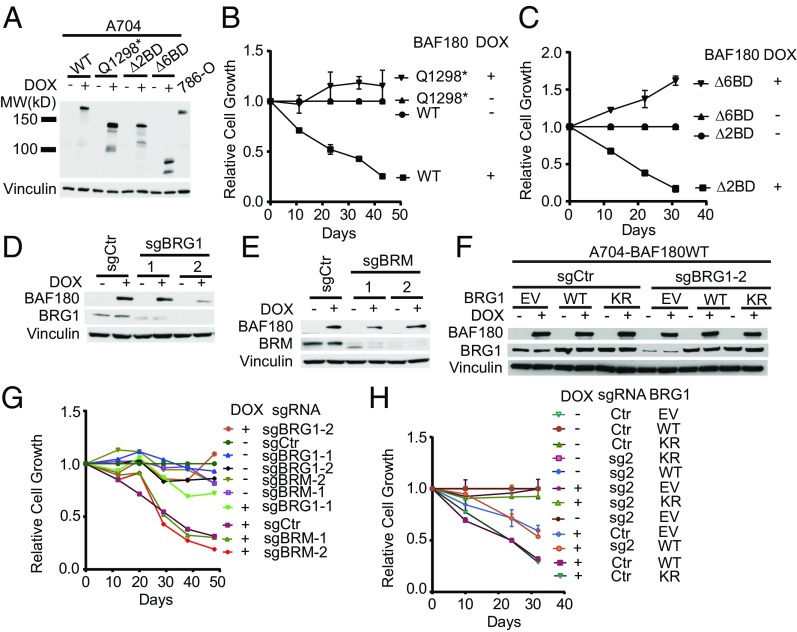
Structure function studies linking specific biochemical properties of BAF180 with ccRCC suppression will require cellular assays of BAF180-dependent phenotypes that are likely to translate into tumor suppression in humans. Toward this end, we infected A704 cells with lentiviral vectors expressing HA-tagged versions of wild-type BAF180, BAF180 (Q1298*), or BAF180 variants lacking two or six of its bromodomains (Δ2BD and Δ6BD, respectively) under control of a doxycycline (DOX)-inducible promoter (Figs. 1C and 3A). Treating these cells with DOX led to exogenous BAF180 protein levels that approximated the endogenous BAF180 levels in 786-O cells (Fig. 3A). Induction of wild-type BAF180, but not BAF180 (Q1298*), reproducibly inhibited A704 cell proliferation, but not the proliferation of BAF180-defective SKRC20, SLR24, or RCC4 cells (Fig. 3B and SI Appendix, Fig. S2). Our SLR25 cells were not tested because they were contaminated with mycoplasma. The differential sensitivity of A704 cells and SLR24 cells to reintroduction of wild-type BAF180 was also observed with an alternative DOX-inducible expression system (SI Appendix, Fig. S3A) that produced even higher levels of exogenous BAF180 (SI Appendix, Fig. S3 B–D).
Fig. 3.
BAF180 suppresses A704 cell proliferation in a bromodomain- and BRG1-dependent manner. (A) Rabbit polyclonal anti-BAF180 immunoblot analysis of A704 cells infected to produce the indicated BAF180 variants (Fig. 1C) in a DOX-inducible manner compared with 786-O cells. (B and C). Proliferation of A704 derivatives, as in A, grown in the presence or absence of DOX. For each line, values were normalized to the corresponding untreated (no DOX) sample at each point for that line. (D and E) Immunoblot analysis of A704 cells infected to produce wild-type BAF180 in a DOX-inducible manner and then superinfected to produce Cas9 and the indicated sgRNAs against BRG1 (D) or BRM (E). (F) Immunoblot analysis of A704 cells, as in D, that were superinfected with a virus expressing an sgRNA-resistant cDNA encoding wild-type (WT) or ATPase-defective (KR) BRG1. (G) Proliferation of A704 derivatives, as in D and E, grown in the presence or absence of DOX. For each line, values were normalized to sgCtr untreated samples at each point for that line. (H) Proliferation of A704 derivatives, as in F, grown in the presence or absence of DOX. For each line, values were normalized to the corresponding untreated samples at each point for that line.
The Δ2BD BAF180 variant, but not the Δ6BD variant, also suppressed A704 cell proliferation, suggesting either that BAF180 requires the presence of one or more of its C-terminal bromodomains to inhibit proliferation or that BAF180 must have four or more intact bromodomains to suppress proliferation (Fig. 3C). Why BAF180 has multiple bromodomains is currently not understood, and analyses of their histone binding preferences by others have produced conflicting results (24, 25).
Consistent with a recent report, inducible expression of wild-type BAF180 suppressed the proliferation of Caki-2 cells (SI Appendix, Fig. S3 E and F) (22). However, this effect was not specific to wild-type BAF180 because it was also observed with BAF180 (Q1298*; SI Appendix, Fig. S3F). We therefore focused on A704 cells in the experiments described here. It is possible that BAF180 would suppress the proliferation or fitness of the insensitive cell lines under conditions that more closely approximate in vivo conditions. Unfortunately, however, we were unable to grow the BAF180-defective cell lines in soft agar or in immunocompromised mice. It is also possible that these other cell lines sustained additional mutations in vivo or ex vivo that inured them to the antiproliferative effects of BAF180. In this regard, VHL−/− ccRCC lines also display variable sensitivity to restoration of pVHL function (26). Alternatively, BAF180 might serve primarily as a caretaker instead of a gatekeeper in these cell lines, with its loss promoting cancer by increasing DNA mutations and aneuploidy.
We next asked whether suppression of A704 cell proliferation by BAF180 was linked to restoration of PBAF function. To begin to address this, we used CRISPR to eliminate BRG1 or, according to our biochemical findings, BRM (Fig. 3 D and E). Eliminating the canonical PBAF ATPase BRG1, but not BRM, with two different guide RNAs (sgRNAs) rendered A704 cells insensitive to BAF180 (Fig. 3G). This effect was on-target because it was reversed by lentiviral expression of an sgRNA-resistant BRG1 mRNA, and specific because it required the ATPase function of BRG1 (Fig. 3 F and H).
The effects (or lack thereof) of inactivating a tumor suppressor gene in a cell line that is wild-type for that tumor suppressor gene can be difficult to interpret. For example, such cell lines could have evolved from a cell type in which loss of the tumor suppressor gene in question cannot promote transformation. Alternatively, such cell lines might have sustained extragenic mutations that are epistatic to the tumor suppressor gene in question, or otherwise render that tumor suppressor gene irrelevant. Nonetheless, we also asked whether eliminating BAF180 in BAF180-proficient cells would enhance their proliferation or fitness, motivated in part by Varela et al., who reported that decreasing BAF180 in PBRM1+/+ ccRCC lines such as 786-O, ACHN, TK10, and SN12C cells with siRNA technology increased cellular proliferation, migration, and soft agar growth (10). We designed 10 shRNAs against PBRM1, including three corresponding to the PBRM1 siRNAs used by Varela et al. (10), and identified five, including two of the three used by Varela et al., that reproducibly down-regulated BAF180 protein levels (SI Appendix, Fig. S4). None of these effective shRNAs, however, consistently enhanced the proliferation of 786-O compared with controls (SI Appendix, Fig. S4). The three PBRM1 shRNAs used by Varela et al. also did not enhance the proliferation of SN12C and UMRC2 cells in our hands (SI Appendix, Figs. S5 and S6). Similar findings have been observed by others (22). We do not fully understand the discrepancy between our findings and those reported by Varela et al.. We note that Varela et al. used acute transfection of an siRNA pool containing three independent PBRM1 siRNAs compared with acute transfection with a single, commercially obtainable control siRNA (10). We suspect their results might have been confounded by off-target effects and because each PBRM1 siRNA they used was present at one-third the concentration of the control siRNA.
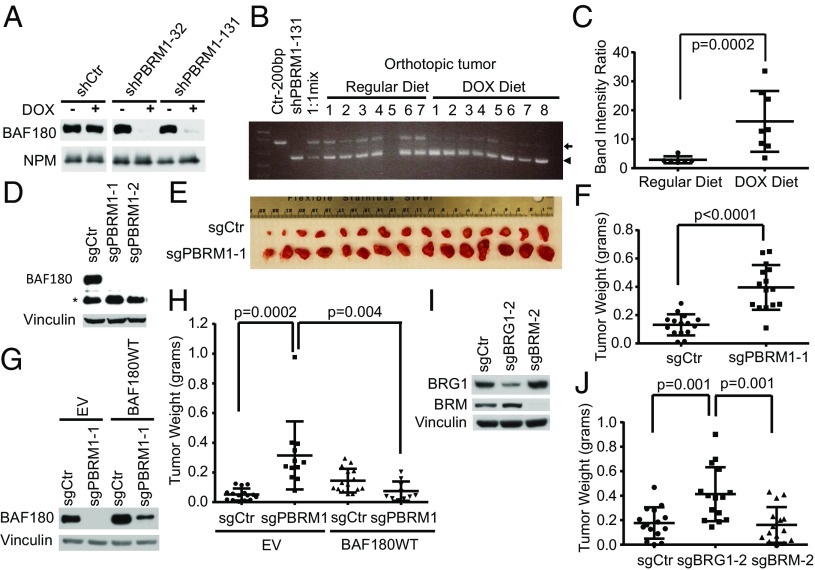
Nonetheless, we proceeded to ask whether eliminating BAF180 would enhance the growth of PBRM1+/+ ccRCC cells in vivo. In one set of experiments, we created 1:1 mixtures of UMRC2 cells expressing a DOX-inducible PBRM1 shRNA and UMRC2 cells expressing a DOX-inducible control shRNA (Fig. 4A), implanted them orthotopically in the kidneys of nonobese diabetic-SCID mice, and monitored the relative abundance of the two populations by PCR. In mice fed DOX-containing chow, the PBRM1 shRNA cells were enriched relative to control shRNA cells, indicating these cells had a fitness advantage in vivo (Fig. 4 B and C), despite the fact that these PBRM1 shRNAs caused a proliferative disadvantage ex vivo (SI Appendix, Fig. S6).
Fig. 4.
Down-regulation of BAF180 promotes clear cell renal carcinoma growth in vivo. (A) Immunoblot analysis of UMRC2 cells infected to produce the indicated shRNAs in a DOX-inducible manner. (B) PCR analysis of tumors formed by a 1:1 mixture of UMRC2 cells expressing a DOX-inducible BAF180 shRNA (sequence 131, arrowhead) and UMRC2 cells expressing a DOX-inducible control shRNA (arrow) in mice fed regular chow (Regular Diet; n = 7) or DOX-containing chow (DOX Diet; n = 8). The first three lanes contain PCR products generated by control shRNA vector, the PBRM1 shRNA vector, and a 1:1 mix of the vectors as controls. (C) Quantification of band intensity ratios (lower band/upper band) from B. (D) Immunoblot analysis of 786-O cells after CRISPR-based gene editing with PBRM1 sgRNAs or a control sgRNA. (E and F). Photographs (E) and weights (F) of tumors formed by 786-O cells, as in D (sgCtr and sgPBRM1-1). (G) Immunoblot analysis of 786-O cells from D (sgPBRM1-1), after infection with a virus expressing an sgRNA-resistant BAF180 mRNA or EV. (H) Weights of tumors formed by cells, as in G, after 8 wk. (I) Immunoblot analysis of 786-O cells after CRISPR-based gene editing with BRG1 and BRM sgRNAs. (J) Weights of tumors formed by cells, as in I.
In a complimentary set of experiments, we eliminated BAF180 in 786-O cells, using CRISPR-based gene editing with two different sgRNAs (Fig. 4D). Consistent with our shRNA-based results in ccRCC lines, presented earlier, eliminating BAF180 in 786-O did not affect their proliferation under standard cell culture conditions (SI Appendix, Fig. S7), but did enhance their ability to form s.c. tumors in immunocompromised mice (Fig. 4 E and F). This effect was specific because it was reversed by an sgRNA-resistant PBRM1 cDNA (Fig. 4 G and H). Similarly, CRISPR-based elimination of BRG1, but not BRM, enhanced s.c. tumor formation by 786-O cells (Fig. 4 I and J). Collectively, these results suggest that the canonical PBAF complex suppresses ccRCC in a BAF180- and BRG1-dependent manner. Interestingly, four of five ccRCC lines in which we could detect BAF180 had low or undetectable BRG1 levels, suggesting that in such cells, BRG1 loss provided an alternative mechanism for compromising BAF180 tumor suppressor function.
BAF180 regulates p53 and its downstream target p21 in some systems (27). We did not, however, detect consistent effects of BAF180 on p21 in A704 or 786-O cells (SI Appendix, Fig. S8). We therefore proceeded to examine the effects of BAF180 on transcription in the context of ccRCC cells.
The PBAF complex plays roles in nucleosome remodeling. Micrococcal nuclease sequencing (MNase-Seq) did not, however, detect gross changes in nucleosome positioning in isogenic A704 and UMRC2 cells that did or did not produce BAF180 (SI Appendix, Fig. S9). Similar results have been obtained after restoring the function of other SWI/SNF components in cancer cells (28).
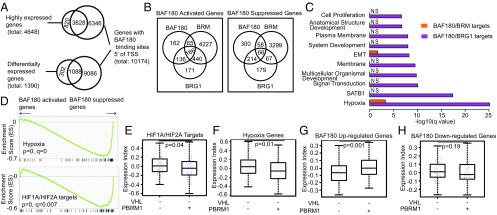
Next we performed anti-HA ChIP-Seq on A704 cells induced to express HA-BAF180 with DOX compared with DOX-treated A704 EV cells. In parallel, we measured changes in gene expression by RNA-Seq after DOX-induction of BAF180 in A704 cells in which either BRG1 or BRM was eliminated using CRISPR/Cas9. Specific BAF180-binding sites were detected throughout the genome, associated with more than 10,000 promoters (binding sites detected ≤5 kB 5′ putative transcription start sites; Fig. 5A). More than 80% of the top 25% highest-expressed genes were associated with BAF180-binding sites (Fig. 5A). Similarly, almost 80% of BAF180-responsive genes (false discovery rate < 0.01) had associated BAF180-binding sites (Fig. 5A).
Fig. 5.
BAF180 loss amplifies the HIF transcription signature in pVHL-defective ccRCC. (A) Venn diagrams showing overlap of genes with BAF180 binding detected 5′ of their putative transcription start sites (within 5 kB) with highly expressed (top 25% based on ∼18,000 genes) genes in A704 cells grown in the presence of DOX that were infected with a DOX-inducible BAF180 expression vector, as in Fig. 3A (Upper), and genes differentially expressed between these cells and DOX-treated EV cells (Lower). P values of both overlaps are <2.2e-16, using Fisher's exact test. (B) Venn diagrams showing mRNAs that are activated (or suppressed) by BAF180, BRG1, and/or BRM according to RNA-Seq of A704 cells, as in A (Lower) together with A704 cells grown in the presence of DOX that were infected with a DOX-inducible BAF180 expression vector and subsequently subjected to CRISPR/Cas9-mediated with sgBRG1-2 or sgBRM-2, as in Fig. 3D. For BAF180 up, BRG1 up, and BRM up, the overlap between any of the two is significant (P < 2.2e-16). BAF180 down and BRG1 down: P < 2.2e-16. BAF180 down and BRM down: P = 0.112. BRG1 down and BRM down: P = 2.89e-5. All these P values were obtained using Fisher's exact test. (C) Enriched gene ontology (GO) terms of genes regulated by BAF180 and BRM compared with BAF180 and BRG1. (D) GSEA of differentially expressed mRNAs identified in A (Lower). (E–H) mRNA expression indices for the indicated gene sets in ccRCC with the inferred genotypes based on the kidney cancer TCGA dataset.
Consistent with our biochemical results, we identified BAF180-responsive mRNAs that were BRG1-dependent, BRM-dependent, or both (Fig. 5B and SI Appendix, Fig. S10). For these and subsequent analyses, we included BAF180-responsive mRNAs, irrespective of whether they were transcribed from genes with BAF180-binding sites. As the antiproliferative effects of BAF180 in A704 cells required BRG1, but not BRM, we focused on BAF180-responsive mRNAs that were BRG1-dependent, but not BRM-dependent. These included 136 mRNAs that were induced by BAF180 and 214 mRNAs that were repressed by BAF180 (Fig. 5B and SI Appendix, Table S2).
Gene ontology analysis and gene set enrichment analysis (GSEA) revealed that BAF180-responsive, BRG1-dependent mRNAs were statistically significantly enriched for mRNAs linked to hypoxia, SATB1 target genes, and signal transduction (Fig. 5 C and D). Most notably, GSEA indicated that BAF180 suppresses HIF-inducible/hypoxia-inducible mRNAs in A704 cells (Fig. 5D and SI Appendix, Fig. S11). This was not peculiar to A704 cells because GSEA revealed that hypoxia-inducible mRNAs were also induced in 786-O cells after CRISPR-mediated elimination of BAF180 (SI Appendix, Fig. S11). In short, loss of BAF180 amplifies the HIF signature observed on VHL inactivation. Although SWI/SNF complexes regulate HIF1α expression in some models (29), the effects of BAF180 on the hypoxic response in A704 and 786-O cells were not linked to changes in HIF1α or HIF2α protein levels (SI Appendix, Fig. S11).
Interrogation of the ccRCC TCGA database revealed that the BAF180-responsive mRNAs we identified using the A704 cell system are similarly regulated by BAF180 in ccRCC tumors, with mRNAs that were up-regulated by BAF180 in vitro being higher in BAF180-proficient tumors, and BAF180-repressed mRNAs trending lower in the BAF180-proficent tumors. In particular, mRNAs induced by hypoxia or HIF are higher in ccRCC in which both VHL and PBRM1 are mutated compared with ccRCC in which VHL is mutated and PRBM1 is not (Fig. 5 E–H and SI Appendix, Fig. S12).
Most ccRCC cell lines do not require HIF under standard culture conditions, which could begin to explain why manipulating BAF180 function does not, with the exception of A704 cells, affect renal proliferation ex vivo, but does alter their growth in vivo (26). In this regard, we found that A704 cell proliferation was, in contrast to the other BAF180-defective ccRCC lines tested, inhibited by a pharmacological HIF2 antagonist (26) under standard monolayer conditions (SI Appendix, Fig. S13). Moreover, enhancement of HIF-dependent transcription could explain why both VHL and PBRM1 mutations are most commonly observed in ccRCC, which has been linked to deregulated hypoxic signaling. This model does not exclude other roles for BAF180 in renal carcinogenesis, including roles related to DNA damage repair and maintenance of mitotic fidelity.
A recent study suggested that BAF180 augments the hypoxic response, in contrast to our findings (22). Although we do not understand this discrepancy, the authors of that study used Caki-2 cells engineered to produce wild-type BAF180. We did not rely on this system because we found that both wild-type BAF180 and BAF180 Q1298* suppressed the proliferation of these cells.
A number of questions remain, including why BAF180 is the dominant PBAF complex member linked to clear cell carcinogenesis. Perhaps, for example, loss of the other PBAF components is antithetical to renal transformation because of functions they do not share with BAF180. It is also possible that these other components do not deregulate HIF to the same extent as BAF180 loss. Alternatively, the selection pressure to inactivate BAF180 in renal carcinogenesis might reflect inactivation of the PBAF complex and an as-yet-unappreciated noncanonical function of BAF180. Finally, given its role in the regulation of the HIF response, it will be interesting to see whether BAF180 status can serve as a predictive biomarker for therapeutics directed against HIF or its downstream targets. In this regard, one small study identified PBRM1 mutations in seven of 13 of patients who had partial or complete responses to VEGF inhibition lasting 3 or more years, and in only one 14 patients who progressed within 3 mo of therapy (30).
Supplementary Material
Acknowledgments
We thank members of the W.G.K. laboratory for useful discussions. X.S.L. is supported by Grant GM99409. W.G.K. is a Howard Hughes Medical Institute investigator and is supported by grants from the NIH.
Footnotes
Conflict of interest statement: W.G.K. receives consulting income and equity from Peloton Therapeutics, which is developing HIF2 inhibitors for treatment of kidney cancer.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619726114/-/DCSupplemental.
References
- 1.Kaelin WG. 2015. Molecular biology of kidney cancer. Kidney Cancer: Principles and Practice (Springer International Publishing, New York), eds Lara PN, Jonasch E. 2nd Ed, pp 31–57.
- 2.Gerlinger M, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46(3):225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mandriota SJ, et al. HIF activation identifies early lesions in VHL kidneys: Evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1(5):459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sankin A, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med. 2014;3(6):1485–1492. doi: 10.1002/cam4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schietke RE, et al. Renal tubular HIF-2α expression requires VHL inactivation and causes fibrosis and cysts. PLoS One. 2012;7(1):e31034. doi: 10.1371/journal.pone.0031034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montani M, et al. VHL-gene deletion in single renal tubular epithelial cells and renal tubular cysts: Further evidence for a cyst-dependent progression pathway of clear cell renal carcinoma in von Hippel-Lindau disease. Am J Surg Pathol. 2010;34(6):806–815. doi: 10.1097/PAS.0b013e3181ddf54d. [DOI] [PubMed] [Google Scholar]
- 9.Rankin EB, Tomaszewski JE, Haase VH. Renal cyst development in mice with conditional inactivation of the von Hippel-Lindau tumor suppressor. Cancer Res. 2006;66(5):2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo G, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2011;44(1):17–19. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 12.Peña-Llopis S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet. 2012;44(7):751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duns G, et al. Targeted exome sequencing in clear cell renal cell carcinoma tumors suggests aberrant chromatin regulation as a crucial step in ccRCC development. Hum Mutat. 2012;33(7):1059–1062. doi: 10.1002/humu.22090. [DOI] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499(456):43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45(6):592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadoch C, Crabtree GR. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci Adv. 2015;1(5):e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer. 2011;11(7):481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 18.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28(14):1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 19.Jiao Y, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45(12):1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, et al. SQSTM1 is a pathogenic target of 5q copy number gains in kidney cancer. Cancer Cell. 2013;24(6):738–750. doi: 10.1016/j.ccr.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowdhury B, et al. PBRM1 regulates the expression of genes involved in metabolism and cell adhesion in renal clear cell carcinoma. PLoS One. 2016;11(4):e0153718. doi: 10.1371/journal.pone.0153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charlop-Powers Z, Zeng L, Zhang Q, Zhou MM. Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Cell Res. 2010;20(5):529–538. doi: 10.1038/cr.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kupitz C, Chandrasekaran R, Thompson M. Kinetic analysis of acetylation-dependent Pb1 bromodomain-histone interactions. Biophys Chem. 2008;136(1):7–12. doi: 10.1016/j.bpc.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho H, et al. On-target efficacy of a HIF2alpha antagonist in preclinical kidney cancer models. Nature. 2016;539(7627):107–111. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci USA. 2010;107(32):14280–14285. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolstorukov MY, et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc Natl Acad Sci USA. 2013;110(25):10165–10170. doi: 10.1073/pnas.1302209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenneth NS, Mudie S, van Uden P, Rocha S. SWI/SNF regulates the cellular response to hypoxia. J Biol Chem. 2009;284(7):4123–4131. doi: 10.1074/jbc.M808491200. [DOI] [PubMed] [Google Scholar]
- 30.Fay AP, et al. Whole-exome sequencing in two extreme phenotypes of response to VEGF-targeted therapies in patients with metastatic clear-cell renal cell carcinoma. J Natl Compr Canc Netw. 2016;14(7):820–824. doi: 10.6004/jnccn.2016.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.